Abstract
Cellular biosynthesis of macromolecules often involves highly branched enzyme pathways, thus cellular regulation of such pathways could be rather difficult. To understand the regulatory mechanism, a systematic approach could be useful. We genetically analyzed a branched biosynthetic pathway for glycosphingolipid (GSL) GM1 using correlation index-based responsible enzyme gene screening (CIRES), a novel quantitative phenotype-genotype correlation analysis. CIRES utilizes transcriptomic profiles obtained from multiple cells. Among a panel of B cell lines, expression of GM1 was negatively correlated with and suppressed by gene expression of CD77 synthase (CD77Syn), whereas no significant positive correlation was found for enzymes actually biosynthesizing GM1. Unexpectedly, a GM1-suppressive phenotype was also observed in the expression of catalytically inactive CD77Syn, ruling out catalytic consumption of lactosylceramide (LacCer) as the main cause for such negative regulation. Rather, CD77Syn seemed to limit other branching reaction(s) by targeting LacCer synthase (LacCerSyn), a proximal enzyme in the pathway, because they were closely localized in the Golgi apparatus and formed a complex. Moreover, turnover of LacCerSyn was accelerated upon CD77Syn expression to globally change the GSL species expressed. Collectively, these data suggest that transcriptomic assessment of macromolecule biosynthetic pathways can disclose a global regulatory mechanism(s) even when unexpected.
Keywords: Gene Expression, Glycolipids, Glycosylation, Golgi, Microarray, Biosynthetic Pathway Branching, Genotype-Phenotype Correlation, Glycosyltransferase
Introduction
Cellular macromolecules are biosynthesized by multistep enzymatic biosynthetic pathways that often show extensive branching. Many glycans are biosynthesized by ordered multistep reactions that involve multiple glycosyltransferases (1–4). At the same time, glycan biosynthetic pathways branch at the substrates utilized by multiple enzymes (5–8). Therefore, a given branched biosynthetic pathway (both the biosynthetic pathway and its branches) contains numerous candidate enzymes that can potentially regulate the expression of a glycan product. Moreover, the regulatory mechanism of these enzymes may vary. Hence, a systematic approach is most suitable for understanding highly branched pathways. We examined how currently unclear branching points on glycan biosynthesis are regulated by using a phenotype-genotype analysis.
Glycosphingolipid (GSL)2 is composed of hydrophobic ceramide and hydrophilic glycan(s). Expression of GSL is a regulated event, as only a limited number of GSLs occur in a particular cell type in a given state (3, 9). The first step of GSL biosynthesis is controlled by the cytosol-facing enzyme ceramide glucosyltransferase, which biosynthesizes glucosylceramide (GlcCer) from ceramide (10, 11) (Fig. 1A). FAPP2 or MDR1 transports GlcCer to the Golgi apparatus (12, 13), where GlcCer is converted into lactosylceramide (LacCer) by Golgi-luminal LacCer synthase (LacCerSyn; encoded by B4GALT6) (10, 14). The commitment for various series of GSL biosynthesis takes place at the usage of LacCer, a common precursor for complex GSLs. Globo series GSLs are biosynthesized by CD77/Gb3 synthase (CD77Syn; encoded by A4GALT) (15, 16) and lacto series GSLs are biosynthesized by Lc3 synthase (Lc3Syn; encoded by B3GNT5) (17), whereas ganglio series are biosynthesized by GM3 synthase (GM3Syn; encoded by ST3GAL5) (18). Due to the complexity of branching at LacCer, the identification of the regulatory mechanisms of GSL expression has been a major challenge, even after identification of the enzymes involved (Fig. 1A) (19). Ganglio series biosynthesis may be an efficient enzyme step because LacCerSyn and GM3Syn participate in multienzyme complexes, by which channeling of LacCer to the GM3Syn reaction may occur (20, 21). This complex, however, may change its sub-Golgi compartment localization in response to other expressed proteins (22, 23) and the regulatory mechanism has not yet been fully clarified. Nevertheless, GSL expression is regulated by various cellular events, such as the activation of human B cells in the germinal center (24). Different GSL species appear to function in cellular signaling events, although individual functional differences have not been fully explored. Moreover, the expressions of particular GSL species are targeted by various bacterial toxins, and thus have pathological significance. For example, the shiga toxin, cholera toxin, and heat-labile enterotoxin targets CD77, GM1, and GD1a, respectively (Fig. 1A) (25–27).
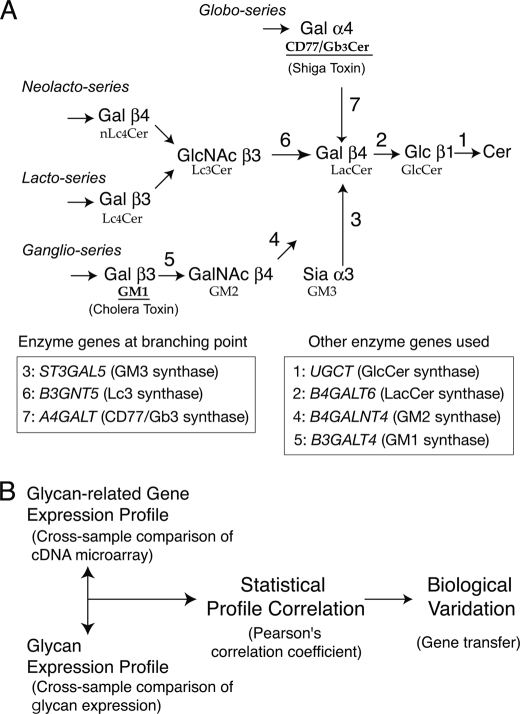
FIGURE 1.
Biosynthetic pathway of GSL and the CIRES procedure. A, biosynthetic pathway for GSLs. Biosynthetic pathways for GSLs comprising various series are illustrated. Arrows indicate specific glycosyltransferase reactions for biosynthesis. Biosynthetic pathways of globo-series (top), lacto series (middle), and ganglio series (bottom) incorporating GSL branching at the glycosyltransferase reaction for GM3Syn/ST3GAL5 (depicted as 3), Lc3Syn/B3GNT5 (6), and CD77Syn/A4GALT (7), respectively. The biosynthesis of these three series of glycans share the same substrate, LacCer. The names of the GSLs that terminate with the indicated sugars are shown below. GM1 and CD77 are epitopes for the B subunit of the cholera toxin and shiga toxin, respectively. B, the general concept of CIRES is illustrated. The main methodology and factors involved in CIRES were previously reported (29). Briefly, the glycan expression profile was statistically tested against the expression profiles of glycan-related genes that were obtained by cross-sample comparison of cDNA microarray experiments. The presence of glycan-related genes, which exhibit similar (positively correlated) or dissimilar (negatively correlated) profiles to glycan expression, are listed as candidate(s) responsible genes. Genes exhibiting statistical significance by the Pearson correlation index analyses were then experimentally tested for their ability to change the expression of glycans on the cell surface via gene-transfer experiments.
The expressions of many cellular phenotypes are mainly controlled by the regulation of gene expression (28). Correlation index-based responsible enzyme gene screening (CIRES) is a novel systematic strategy to quantitatively assess the phenotype-genotype correlation. This method was developed to identify genetically dominant enzyme gene(s) that may quantitatively regulate the expression of cell surface glycans (29). CIRES involves the statistical comparison of multisample profiles between genotypes (glycan-related gene expression profiles obtained by DNA microarray) and cell surface phenotypes (glycan expression). Correlations found between these profiles are used to build hypotheses, and subsequent genetic manipulation of cells provides experimental verification (Fig. 1B). CIRES has been successfully used to screen glycosyltransferase genes responsible for the level of glycan expression on the cell surface (29, 30). Thus, we used CIRES to analyze other complicated GSL biosynthetic pathways, with a focus on the regulation of branching.
We analyzed the branching point of LacCer in GSL biosynthesis using CIRES to understand the genetic characteristics of the branching regulatory mechanism, which is a frequent subject of biochemical studies. Our genetic analyses of B cell lines revealed that the globo series dominated the ganglio series in this biosynthetic pathway at the branching point of the common precursor, LacCer. This genetic dominance is based on LacCerSyn regulation by CD77Syn. In contrast, actual consumption of LacCer to biosynthesize Gb3/CD77 is not required for the suppression of ganglio series biosynthesis. Thus, this study proposes a novel mechanism for LacCerSyn regulation at the branching point of the biosynthetic pathway.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmid DNA, and Antibiotics
The Namalwa and Ramos B cell lines were obtained from Health Science Research Resources Bank and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (JRH Bioscience, Lenexa, KS), sodium pyruvate, non-essential amino acids, and 2-mercaptoethanol. COS-7 and CHO-K1 cells were cultured in α-minimal essential medium supplemented with 10% fetal bovine serum. Retrovirus was produced by transient transfection of modified pMSCV vectors (Clontech, Mountain View, CA) to Plat-A amphotropic packaging cells (31) using the calcium-phosphate method as previously reported (29). The various plasmid DNAs used in this study are summarized under supplemental Table S2. Aspartic acid residues in the DXD motif of CD77Syn (residues 192 and 194) were point mutated to threonine residues by PCR-based mutagenesis to give rise to the CD77Syn-TXT mutant. CHO-K1 cells were successively transfected with three vectors harboring resistance genes to antibiotics obtained from Nacalai Tesque (Kyoto, Japan) or Invivogen (San Diego, CA): G418 (1000 μg/ml), blasticidin (10 μg/ml), and zeocin (125 μg/ml).
Antibodies and Other Probes
Biotin-xx-conjugated cholera toxin B subunit was obtained from Invitrogen. Anti-CD77 monoclonal antibody (mAb) (clone 38-13) was obtained from IMMUNOTECH (Marseille, France). Anti-giantin rabbit polyclonal antibodies (pAb) (RPB-114C) were kindly provided by Dr. H-M. Shin (Kyoto University). Anti-GM130 (clone 35) and calnexin (clone 37) were from BD Transduction Laboratories (Franklin Lakes, NJ). Anti-HA mAb (HA.11) was from Covance (Berkeley, CA) and anti-HA PAb (y-11) was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-FLAG mAb (M2) and anti-PDI pAb (P7496) were from Sigma. The following labeled probes were used to detect signals: anti-rat IgM (phycoerythrin, Rockland), streptavidin (phycoerythrin, Caltag), anti-mouse IgG1 (Alexa 488, Invitrogen), anti-rabbit IgG (Alexa 568, Invitrogen, or HRP, DAKO), and anti-mouse IgG (HRP, Zymed Laboratories Inc.).
Flow Cytometry (FCM)
Approximately 5 × 105 cells in 100 μl of FACS buffer (1% BSA and 0.1% NaN3 in PBS(−)) were incubated with anti-glycan probes at room temperature for 1 h. Secondary staining was carried out for 30 min. Data were obtained using FACScan (BD Bioscience) and analyzed using FlowJo software (Tristar, San Carlos, CA). For cross-comparison of the staining signal among cell lines with different autofluorescence, the mean florescence intensity (MFI) of background staining was roughly adjusted to MFI = 10 and the relative staining signal was expressed as a ratio of staining MFI divided by control MFI as previously reported (29).
Statistical Analyses of Glycan Expression Profiles
Other than the use of different glycan-binding probes, the correlation index analyses involved in the CIRES procedure, which included a systematic comparison of the relative profiles of gene expression obtained by cDNA microarray and glycan expression obtained by FCM in six cell lines, were essentially the same as those previously reported (29, 30). A full list of the gene expression profiles used to estimate Pearson correlation coefficients is available as a supplemental table in previous reports (29, 30).
Retrovirus-mediated Gene Transfer
MSCVs carrying the intended glycosyltransferases were prepared by transient transfection of the modified MSCV vector, pMSCV-IRES-EGFP, which bicistronically encodes an enhanced green fluorescent protein (GFP) open reading frame under control of an internal ribosomal entry site. Vectors were transiently transfected into a Plat-A packaging cell line (31) and culture supernatant was filtered and used for spin infection. Infected cells were cultured for 2 weeks to stabilize expression and stained with anti-glycan probe, whereby a GFP-positive population was regarded as containing gene-transferred cells and GFP-negative cells were considered controls. Thus, a mixture of both populations was stained in a single tube for two-color FCM. The resultant MFI for phycoerythrin was used as an indication of the cell-surface expression of the glycan epitope. Staining of the GFP-negative population was monitored to confirm that it was similar to that of non-infected/vector-infected cells. This method ensured reliable staining with anti-glycan probes to evaluate quantitative differences in glycan expression.
Thin Layer Chromatographic Analysis of GSLs
The GFP-positive population of retrovirus-infected cells was sorted by FACSAriaII (BD Biosciences) to enrich glycosyltransferase-expressing cells. Cellular GSLs were isolated as previously reported (32). In short, neutral GSLs contained in the lower phase of the Folch partition were per-acetylated and purified with a Florisil column, whereas the acidic GSL fraction was prepared by repeated partitioning with the salt-containing upper phase of the Folch partition. GSL fractions equivalent to 1 mg of cellular protein were applied to a silica-based TLC plate and separated with a solvent (chloroform, methanol, 0.2% CaCl2, 60:35:8). Orcinol was used for the visualization of sugar moieties in GSLs and charring was carried out to gain sensitivity for the detection after orcinol staining (33). For detection of GM1, 670 ng/ml of biotinylated CTxB was overlaid and the signal was detected with Konica Immunostain HRP-1000 (Seikagaku). This system could detect roughly 30 ng of GM1 on TLC plate.
Immunofluorescent Detection of Glycosyltransferase Localization
COS-7 cells were grown on glass coverslips and transiently transfected with C-terminal HA-tagged glycosyltransferases harboring the CMV promoter for expression with Lipofectamine reagent (Invitrogen). Cells were harvested 30 h after transfection to avoid overexpression of glycosyltransferases. Cells were fixed with methanol at −20 °C for 10 min. Slips were blocked with 5% BSA, 0.05% Tween 20 in TBS and stained with anti-HA tag antibody (HA.11) and organelle markers. Fluorescent images were obtained using an inverted microscope (IX70; Olympus, ×100 objective lens) equipped with a CCD camera (Cool SNAP HQ/OL; Photometrics) and Methamolph software (Olympus, Tokyo, Japan). The obtained images were subsequently processed with Photoshop software (Adobe). To avoid overexpression-induced mislocalization of glycosyltransferase, cells with moderate HA.11 signals were chosen to obtain images, although strongly stained cells also showed essentially the same staining pattern 30 h post-transfection. The use of COS-7 cells resulted in a more defined intracellular localization of the Golgi-localized enzyme when compared with that of CHO-K1, HeLa, or HT1080 cells. Thus, the images of transfected COS-7 cells were analyzed in detail.
Transfection and Co-immunoprecipitation of Glycosyltransferases
LacCerSyn-FLAG was transiently co-transfected with CD77Syn-HA or GM3Syn into COS-7 cells using Lipofectamine (Invitrogen). Cells were harvested 48 h after transfection by trypsinization. Cell pellets were washed with PBS and lysed with sonication in TDE lysis buffer (50 mm Tris-HCl, pH 7.6, 1 mm DTT, 1 mm EDTA) containing protease inhibitor mixture (Nacalai Tesque). Post-nuclear supernatants were ultracentrifuged (55,000 rpm for 30 min) in MLA-130 rotor (Beckman) and the resultant pellets were sonicated in TL buffer (1% Triton X-100, 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1 mm EDTA) containing a protease inhibitor mixture to extract the membrane fraction of the cells. The ultracentrifuge supernatants of these membrane extracts were used as membrane fractions. These were immunoprecipitated with either HA.11 (anti HA antibody) or M2 (anti-FLAG antibody) and Protein G-Sepharose (Amersham Biosciences, Uppsala, Sweden), and co-immunoprecipitates were detected by immunoblotting with M2 or y-11, respectively. To assess competition between GM3Syn-HA and non-tagged CD77Syn/CD77Syn-TXT in complex formation with LacCerSyn-FLAG, the respective genes were successively introduced to CHO-K1 cells. CHO-K1 cells were employed because this cell line tends to stably hold introduced transgenes. To ensure polyclonality of the cells, a double transfectant clone (expressing LacCerSyn-FLAG and GM3Syn-HA) of CHO-K1 cells (selected with G418 and blasticidin) was transfected with the third constructs and polyclonally selected with zeocin.
Metabolic Labeling of LacCerSyn-FLAG
The polyclonal triple stable cells described above were metabolically pulse labeled with [35S]cysteine and methionine (EXPRE35S35S Protein labeling mix, PerkinElmer Life Sciences) for 30 min and chased in the medium containing enriched cold cysteine and methionine for the indicated times. Cells were recovered with trypsinization and frozen in a deep freezer as cell packs. Cells were lysed with Nonidet P-40/Triton X-100 (1% each) containing lysis buffer and immunoprecipitated with M2 anti-FLAG antibody. The amounts of cell lysate used for immunoprecipitation were adjusted according to the size of the cell pack. Immunoprecipitated samples were subjected to 10% SDS-PAGE and the signals of the visualized bands were quantified with BAS2500 (Fujifilm, Tokyo) after the gels were dried.
RESULTS
Dominant Effect of CD77Syn on GSL Expression in B Cells
Within the biosynthetic pathway of GSLs, LacCer can be used by a variety of enzymes to give rise to various series of GSLs. Therefore, this branching point in the pathway could be interpreted as the point of lineage commitment to specific biosynthetic pathways (Fig. 1A). CIRES may be useful for understanding the regulation of glycan biosynthesis at pathway branching positions given its ability to identify regulatory enzyme genes even when the enzyme reaction is not directly involved in the formation of the glycan in question (29). The B subunits of two bacterial toxins, the shiga toxin and cholera toxin, were employed to probe CD77 and GM1, respectively, in FCM. When the relative expression of a panel of six B cell lines was compared, the staining profiles of CD77 and GM1 were distinct. CD77 was strongly expressed in the germinal center-like B lymphomas Daudi, Ramos, and Raji (Fig. 2A), whereas prominent expression of GM1 was found only in Namalwa cells (Fig. 2A) that lacked expression of CD77. Given that the shiga toxin B subunit and the anti-CD77 antibody showed identical staining patterns for these cell lines, we used anti-CD77 for the remainder of the study. The GSL expression profiles were compared with the gene expression profiles for glycan-related genes in identical sets of cell lines. The expression profile of the CD77Syn (A4GALT) gene positively correlated (r = 0.83) with that of CD77 and negatively correlated with that of GM1 (r = −0.75; Fig. 2B and supplemental Table S1). In contrast, none of the gene expression profiles for GM3Syn (ST3GAL5), GM2 synthase (B4GALNT4), or GM1 synthase (B3GALT4) exhibited significant positive correlations, although these are genes whose products are involved in actual GM1 biosynthesis. A full list of the correlated glycan-related genes is provided under supplemental Table S1. Here, we focus on the presence of the negative correlation because such a relationship may be attributable to unexpected regulatory mechanisms.
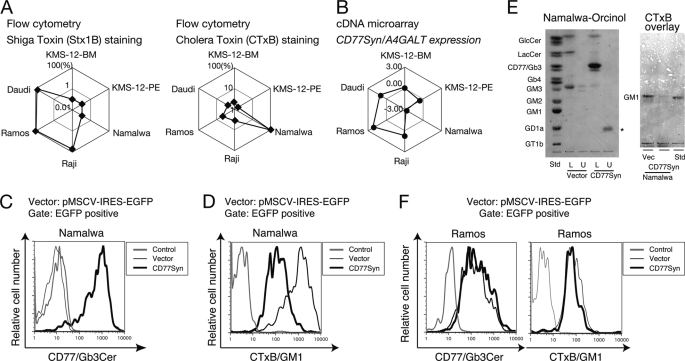
FIGURE 2.
Dominant effect of CD77Syn on GSL expression. A, relative expression profile obtained by FCM staining using shiga toxin B subunit (left) or cholera toxin B subunit (right) plotted in web-graph format. The relative strengths of the toxin epitope expressions in a set of six cells are expressed on a log-scale on diagonal lines of a hexagon. Thus, plots located at the edge of the hexagon indicate stronger expression. Cells with the strongest expression were set to 100% for each staining. B, relative gene expression profile of CD77Syn/A4GALT among a set of six cell lines. The intensity of gene expression is indicated as the logarithmic value of the relative CD77Syn expression signal, which was normalized with the value of the control RNA in the cDNA microarray experiments (29). C and D, anti-CD77- (C) or CTxB- (D) stained FCM results of cells infected with vector or CD77Syn are overlaid. The bold line indicates expression of CD77 (C) or CTxB epitope (D) in CD77Syn-expressed cells, and the thin line indicates the results for vector-infected cells. The gray line corresponds to the negative control. E, GSL analyses on TLC. A GFP-positive population was sorted using the cell sorter for control vector- and CD77Syn-infected Namalwa cells. GSL fractions were prepared from Folch partition. U indicates the upper layer enriched with the acidic GSL fraction, whereas L indicates the lower layer enriched with the neutral GSL fraction. The standard GSL (Std) used is indicated. Ceramide monohexoside and ceramide dihexoside are indicated as GlcCer and LacCer, respectively. GSL was visualized by orcinol staining, and the single band indicated with an asterisk was judged as orcinol-negative due to the difference in color. Each lane containing purified GSLs corresponds to 1 mg of protein. GM1 was visualized with a CTxB overlay. F, CD77Syn infection in Ramos cells. Ramos cells were infected with CD77Syn or control vector virus and the GFP-positive fraction was assessed for GSL expression (left, CD77; right, CTxB) by FCM.
CD77Syn Expression Determines the Expression of GSL Species
First, we hypothesized that the CD77Syn reaction would dominate at the branching point using LacCer. Consistent with this hypothesis, retrovirus-mediated ectopic expression of CD77Syn changed the expression profile of GSL in Namalwa cells, where induction of CD77 (Fig. 2C) and concomitant reduction of GM1 was found in FCM (Fig. 2D). In this assay, we utilized a vector in which EGFP is conjugated with an internal ribosomal entry site to monitor glycosyltransferase gene expression, which provided an internal staining control in the same tube. Moreover, when GFP-positive populations were pooled by a cell sorter and their GSL fractions were analyzed by thin layer chromatography (TLC), prominent induction of CD77 and reduction of GM3 were observed upon CD77Syn expression (Fig. 2E). When CTxB was overlaid, a reduction in the GM1 band was also observed (Fig. 2E). These results indicate that CD77Syn controls a global change in the GSL lineage. In contrast to the Namalwa cells, the introduction of CD77Syn to Ramos cells had a minimal effect on GSL biosynthesis (Fig. 2F). We reasoned that strong endogenous expression of CD77Syn (Fig. 2B) could be responsible for the lack of effect in Ramos cells because the total flow of GSL biosynthesis is less likely to be changed by further introduction of CD77Syn.
Minor Contribution of Non-correlated Enzyme Genes in GSL Expression
To determine the regulatory competence of other glycosyltransferases on the GSL biosynthetic pathway (Fig. 1A), we also introduced genes encoding synthases for GlcCer, LacCer, GM3, GM2, GM1, and Lc3 to Namalwa and Ramos cells (Fig. 3, A–D). GM3Syn may occur in alternatively spliced forms (34). Because RT-PCR analysis showed that both Namalwa and Ramos cells expressed the longer isoform of GM3Syn, we used the cDNA of this isoform for further examinations. Consistent with the correlation results, introduction of the CD77Syn gene had the most prominent effect on the expression of both GM1 and CD77 in Namalwa cells (Fig. 3, A and B). In Ramos cells, in which CD77Syn is endogenously expressed, overexpression of GlcCer synthase had the most prominent effect; i.e. a greater than 6-fold increase in CD77 expression was detected without altering the GM1 expression, indicating that GlcCer synthase determines the total flow of GSL biosynthesis in Ramos cells (Fig. 3C). The GSL fraction of the GlcCer synthase-infected GFP-positive pool also exhibited clear induction of CD77 by TLC analysis, whereby a subtle increase in GlcCer but not LacCer or ganglio series GSL was noted (Fig. 3E). This was consistent with the hypothesis that LacCer utilization can be determined by the expression of CD77Syn. In Ramos cells, GM3Syn increased the expression of GM1 up to 4-fold (Fig. 3D). Thus, the abundant expression of GM3Syn was somewhat able to overcome dominance to use LacCer for ganglio series GSLs. These results suggest the presence of simple competition among enzymes to utilize LacCer at the biosynthetic branch. However, previous reports have indicated that branching enzymes CD77Syn, Lc3Syn, and GM3Syn exhibit the same order of Km values toward LacCer, although the reports varied in their values, i.e. 54.5 μm for CD77Syn (15), 80 μm for GM3Syn (35), 3.2 μm for GM3Syn, and 8.0 μm for Lc3Syn (36). Moreover, these data may not take into account molecular crowding, another factor to be considered within the Golgi apparatus (37). Therefore, we asked whether enzymatic conversion of LacCer to CD77 is required for CD77Syn to exhibit dominant “activity” to suppress ganglio series expression.
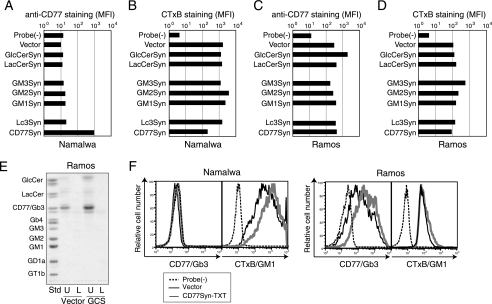
FIGURE 3.
Effect of overexpression of relevant enzymes in the pathway on the cell surface expression of CD77 or GM1. A–D, expression of CD77 (A and C) or CTxB epitope (B and D) on Namalwa (A and B) or Ramos (C and D) cells infected with various glycosyltransferase genes. The log-scaled signal strength (MFI) in staining was plotted for the indicated glycosyltransferases. Probe(−) indicates the background level of fluorescence detected for the fluorochrome-conjugated secondary probe alone. E, GSL profile analysis of GlcCerSyn-infected Ramos cells on TLC. A GFP-positive population was sorted for vector control (Vector) and GlcCerSyn (GCS)-infected Ramos cells. TLC was carried out as described in the legend to Fig. 2E and GSLs were visualized with orcinol staining. U and L indicate the upper and lower fractions from Folch partitioning, respectively. The standard GSL (Std) used are indicated on the left. Each lane contains a GSL fraction from cells corresponding to 0.65 mg of protein. F, effect of the CD77Syn mutant on GSL expression. A retrovirus vector for CD77Syn-TXT was prepared and used to infect Namalwa or Ramos cells as described in the legend to Fig. 2C. Expression of GSL was monitored with FCM using anti-CD77 or CTxB. Dashed lines indicate the negative control of the staining and bold gray lines indicate probe staining with control virus infection. Thin lines indicate probe staining of infected cells with retrovirus encoding CD77Syn-TXT.
Enzyme Activity-independent Dominance of CD77Syn over Other Enzyme Genes in the Pathway
CD77Syn is a member of the conventional glycosyltransferase family, whose luminal catalytic domain contains a DXD motif for enzyme activity (38–40). We site directly point-mutagenized the DXD motif into TXT to create a catalytically inactive mutant. When the TXT mutant was introduced to Namalwa cells, no induction of CD77 was observed (Fig. 3F), as expected. However, a reduction in GM1 was still observed (Fig. 3F). To measure the reduction rate between CD77Syn and its TXT mutant, sorted GFP-positive cells were compared via side by side staining. The rate of GM1 reduction was greater in CD77Syn-introduced cells than in TXT mutants (supplemental Fig. S1A). This difference could be due to some degree of catalytic consumption of the LacCer. Alternatively, because the GFP signal, which infers the degree of transgene expression, was greater in CD77Syn cells, the level of protein expression between the two cell types could account for the difference. The expression level of the mutant appeared to be important because the CTxB signal tended to be weaker in the cells exhibiting stronger GFP signals. CD77Syn-expressing cells exhibited stronger GFP signaling with weaker CTxB signaling when superimposed with that of TXT mutants (supplemental Fig. S1B). In any case, the observed reduction in GM1 with the TXT mutant provides evidence against the possibility that enzymatic conversion of LacCer to CD77 by CD77Syn overexpression is the primary cause of the dominant effect of CD77Syn in suppressing GM1 expression.
When the TXT mutant was introduced into Ramos cells, which express endogenous CD77Syn, CD77 expression was reduced, indicating that CD77Syn-TXT competes with the endogenous CD77Syn for the intracellular niche (Fig. 3F). Consistent with wild-type CD77Syn expression, the TXT mutant exhibited only subtle inhibition of GM1 in Ramos cells, indicating that the inhibitory effect of wild-type and TXT mutants on GM3 biosynthesis is similar, resulting in no additive effect in CD77Syn-sufficient Ramos cells. Taken together, these results indicate that genetic dominance of CD77Syn was caused by the occupation of a putative intracellular niche; therefore, catalytically inactive TXT mutants are able to exhibit a dominant-negative phenotype. Moreover, these results clearly rule out the notion that the substrate in the Golgi apparatus is distributed evenly, and is thus accessible to downstream enzymes in the pathway. Rather, depending on the CD77Syn, the accessibility of LacCer to GM3Syn appeared to be regulated in the B cells that endogenously regulate GSL expression. To determine whether or not CD77Syn exhibit a dominant function at branching of GSL in different cell types that is not known for GSL alteration, we expressed CD77Syn and CD77Syn-TXT in COS-7 cells expressing GD1b and GM1 as major and minor gangliosides, respectively (41). Non-conjugated GFP was co-transfected to monitor transfected cells in transient expression experiments. GFP positive cells in CD77Syn-expressed cells did not increase CD77 staining, indicating that dominance in the CD77Syn expression could be cell-type specific. CD77Syn-TXT cells showed suppression of GM1 (supplemental Fig. S2), thus CD77Syn-TXT could alter glycosylation even in the cell type that does not regulate GSL species.
Subcellular Localization of Branching Enzymes
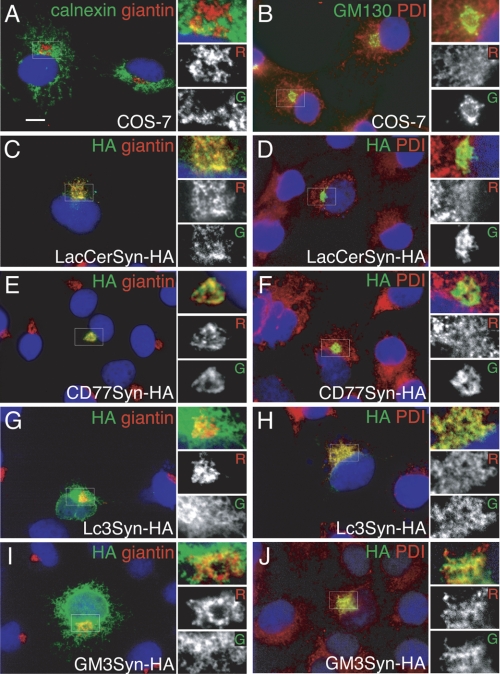
To visualize the putative intracellular niche that CD77Syn may be occupying, we compared the intracellular localization of CD77Syn with other GSL synthases related to LacCer in B cells. The localization of these branching glycosyltransferases was not directly compared in the same cells. Because B cell lines have limited cytoplasmic space, we used COS cells, which have been extensively used for examining Golgi retention signaling of glycosyltransferases (42), thus could be useful in this study. The short N-terminal tail may be involved in the determination of intracellular localization of glycosyltransferase, thus C-terminal HA-tagged glycosyltransferase constructs were transiently expressed in COS-7 cells. Attempts to conjugate a larger tag, such as GFP, resulted in the failed expression of glycosyltransferases, probably due to folding problems. To visualize intracellular localization, we compared the localities of tagged enzymes for LacCerSyn, CD77Syn, Lc3Syn, and GM3Syn to conventional markers for the Golgi apparatus, giantin, or GM130, and the endoplasmic reticulum, calnexin, or peptidyl disulfide isomerase (PDI) in co-staining experiments (Fig. 4, A and B). In B lymphoma cells, among β1–4-galactosyltransferases reported to biosynthesize LacCer, B4GALT6 rather than B4GALT5 (43) was prominently expressed by the cDNA microarray (29), and shRNA-mediated knockdown of B4GALT5 did not alter GSL expression. These results are similar to those previously reported in knock-out mouse liver cells (44). Thus, B4GALT6 was used for subsequent experiments. This assay showed that LacCerSyn-HA and CD77Syn-HA were primarily localized to giantin-positive compartments, likely the Golgi apparatus (Fig. 4, C–F). In contrast, Lc3Syn-HA were primarily localized to the endoplasmic reticulum, as judged from co-localization with PDI (Fig. 4, G and H). GM3Syn-HA showed widespread localization, with partial co-localization with both PDI and giantin (Fig. 4, I and J). Golgi localization could be key for biosynthetic branching, as the degree of Golgi localization was in good correlation with the effect of GSL biosynthetic dominance in the retrovirus-mediated gene transfer experiment; i.e. the efficiency was similar to that of CD77Syn (exhibited 88% reduction in GM1 in Namalwa cells), GM3Syn (exhibited 42% reduction in CD77 in Ramos cells), and Lc3Syn (exhibited no reduction; Fig. 3, B and C). In any case, in the putative intracellular niche, CD77Syn seemed to localize closer to LacCerSyn. However, this was not sufficiently conclusive evidence for the dominance of CD77Syn given that GM3Syn could also co-localize with LacCerSyn in the Golgi apparatus (Fig. 4I), indicating that compartmentalization is not completely rigid.
FIGURE 4.
Intracellular localization of glycosyltransferases related to LacCer. COS-7 cells were transiently transfected with HA-tagged glycosyltransferases and their intracellular localization was detected with indirect immunofluorescent studies (anti-HA, green channel, Alexa 488) with Golgi-localized giantin (red channel, Alexa 568; A, C, E, G, and I) or endoplasmic reticulum-localized PDI (red channel; B, D, F, H, and J). Control staining of non-transfected COS-7 cells was also carried out (A and B) with endoplasmic reticulum-localized calnexin (green; A) or Golgi-localized GM130 (green; B). HA-tagged LacCerSyn (C and D), CD77Syn (E and F), Lc3Syn (G and H), and GM3Syn (I and J) were transfected. Small boxes at the right of the images are magnified and shown on the left for each channel (R, red; G, green) to better examine overlap in localization. DAPI was used to counterstain the nucleus (blue). Bar = 10 μm.
Complex Formation of CD77Syn with LacCerSyn
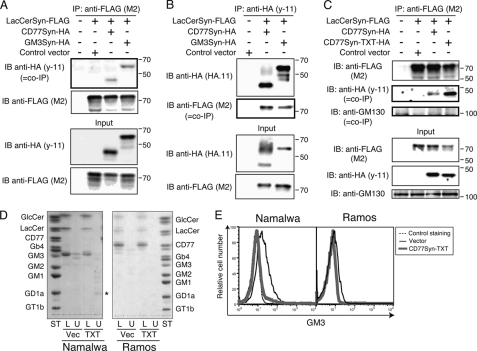
It was previously reported that LacCerSyn and GM3Syn are co-complexed, and this complex formation is offered as evidence for the efficient conversion of LacCer to GM3, as this physical kin association could ease the process of channeling the substrate from one to the other (20). However, it was not clear how CD77Syn fit into this process, because, at least genetically, the efficiency of CD77Syn in the pathway appeared greater than that of GM3Syn in B cells (Figs. 2 and 3). Therefore, we examined whether CD77Syn could form a complex with LacCerSyn. As previously reported, when LacCerSyn was co-expressed with GM3Syn, the two enzymes formed a co-immunoprecipitable complex (Figs. 5, A and B). CD77Syn also co-immunoprecipitated with LacCerSyn (Fig. 5, A and B) indicating that such complex formation is not limited to LacCerSyn with GM3Syn. This complex did not contain microscopically co-localized giantin, which is a Golgi resident tether protein. The co-immunoprecipitation was not detectable when the lysates of independently transfected cells were mixed at the step of immunoprecipitation (supplemental Fig. S3). Thus, complex formation requires their co-expression in the same Golgi apparatus, similar to the case of GM1 and GM2 synthases (20).
FIGURE 5.
Complex formation of LacCerSyn with CD77Syn. A, LacCerSyn-FLAG was transiently co-transfected with CD77Syn-HA or GM3Syn-HA in COS-7 cells. The membrane fraction (Input) was prepared from transfectant and immunoprecipitated with M2 anti-FLAG mAb and Protein G-Sepharose. Immunoprecipitate and co-immunoprecipitate (bold box, co-IP) were monitored by immunoblotting with M2 (FLAG) mAb and y-11 (HA) pAb, respectively. B, similar to the experiments in A, except that HA-tagged proteins were immunoprecipitated, and co-immunoprecipitation of LacCerSyn-FLAG was examined by immunoblotting. C, complex formation of a CD77Syn mutant with LacCerSyn. COS-7 cells were transiently transfected with LacCerSyn-FLAG and either CD77Syn-HA or CD77Syn-TXT-HA as indicated at the top. Immunoprecipitation and immunoblotting were carried out as in A. GM130 was used as a control. D, GSL expression profiling visualized by TLC. Sorted GFP-positive cell fractions were assessed for GSL expression as described in the legend to Fig. 2F for Namalwa and Ramos cells infected with MSCV-IRES-GFP-CD77Syn-TXT (TXT) or a control virus (Vec). GSLs were visualized by orcinol staining to ensure that they possessed a sugar moiety. Purified GSLs spotted on each lane correspond to 1 or 0.65 mg of protein for Namalwa or Ramos cells, respectively. E, reduction in GM3 expression due to the CD77Syn-TXT mutant visualized by FCM. Sorted GFP-positive populations were assessed by anti-GM3 (GMR6, Seikagaku Corp., Tokyo) mouse mAb and phycoerythrin-conjugated anti-mouse IgM. Only vector-infected Namalwa cells exceeded the detection limit in the experiment. This could be due to the preferential expression of GM3 within intracellular organelles or a weak reactivity of the antibody. In any case, CD77Syn-TXT infection diminished the expression of detectable GM3.
We next examined the ability of CD77Syn-TXT to form a complex with LacCerSyn, because this mutant retained the ability to repress ganglio series biosynthesis without consuming LacCer. Consistent with its ability to inhibit ganglio series biosynthesis, CD77Syn-TXT also formed an enzyme complex with LacCerSyn (Fig. 5C). It was worth noting that the TXT mutant expressing COS-7 cells had less LacCerSyn expression in the lysate (Fig. 5, B and C) thus mutation may cause stronger interaction of the enzymes. This reduction of LacCerSyn may explain why we detected reduced GM1 expression in COS-7 cells only upon CD77Syn-TXT transfection (supplemental Fig. S2). This result prompted us to assess the level of GSLs in CD77Syn-TXT introduced into Namalwa and Ramos cells. Because retrovirus-mediated gene transfer to Namalwa cells did not reach 100%, we sorted a GFP-positive population of control or CD77Syn-TXT virus-infected cells. Consistent reduction in GM3 occurred in CD77Syn-TXT-infected cells in both TLC (Fig. 5D) and FCM (Fig. 5E), whereas GM1 was detectable only in FCM (Fig. 3F). Although it is expected that biosynthetic inhibition at the branching point utilizing LacCer could cause its accumulation, CD77Syn-TXT cells showed slightly reduced LacCer expression. In contrast, the observed expression of GlcCer was similar to controls. Thus, GlcCer incorporation into the GSL biosynthetic pathway appeared unchanged. These data indicate that LacCer usage for GSL, such as GM1 biosynthesis, is regulated by the presence of CD77Syn and that this regulatory dominance could be achieved via a biological process, such as the formation of an intra-Golgi complex with LacCerSyn.
Possible Compartmentalization of CD77Syn and GM3Syn to Utilize the LacCerSyn Complex
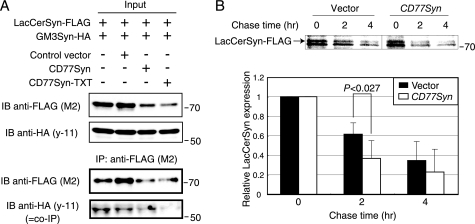
Because both CD77Syn and GM3Syn can localize to the Golgi apparatus and form complexes with LacCerSyn, an important question is whether LacCerSyn is freely accessible to these distal enzymes. Cargo proteins that traverse the Golgi apparatus can be differentially sorted into their own destinations in relationship to glycosylation (45). We hypothesized that ganglio series and globo series GSL biosynthesis could be distinctly functionally compartmented to explain the above findings. Alternatively, if CD77Syn and GM3Syn function primarily in the same compartment of the Golgi apparatus, LacCerSyn should be accessible to both of the distal enzymes. Therefore, one could expect competition between complexes for LacCerSyn. To examine these possibilities, we tested whether CD77Syn could dominantly form a complex with LacCerSyn to out-compete GM3Syn, resulting in dominance in product formation. We first developed a double transfectant cell line expressing both LacCerSyn-FLAG and GM3Syn-HA, which form an intra-Golgi complex (Fig. 6A). We then polyclonally introduced CD77Syn or CD77Syn-TXT and compared them to a control vector to assess changes in complex formation between LacCerSyn and GM3Syn. Expression of CD77Syn and CD77Syn-TXT both resulted in reduced LacCerSyn in the membrane fraction used for immunoprecipitation (Fig. 6A), which is somewhat consistent with the reduction in LacCer found in Ramos cells and CD77Syn-expressed Namalwa cells in TLC (Figs. 2E and 3E). Accordingly, GM3Syn-LacCerSyn co-immunoprecipitation was reduced. In contrast, expression of GM3Syn in the membrane fraction did not change, suggesting that the reduction was specific to LacCerSyn. These data indicate that LacCerSyn was not freely available to GM3Syn when CD77Syn was expressed; thus, CD77Syn and GM3Syn are less likely to compete for LacCerSyn in the same compartment of the Golgi apparatus. Rather, it is likely that CD77Syn acts more proximally to LacCerSyn in the pathway than GM3Syn, possibly in different subcompartments, thus favoring globo series GSL biosynthesis.
FIGURE 6.
CD77Syn expression alters LacCerSyn turnover. A, polyclonal triple stable cell lines expressing LacCerSyn-FLAG, GM3Syn-HA with vector, CD77Syn, or CD77Syn-TXT were subjected to a co-immunoprecipitation assay as described in the legend to Fig. 5A. Expressions of LacCerSyn and GM3Syn were examined in both membrane fraction (input) and immunoprecipitated (IP) fractions. B, polyclonal triple stable cell lines expressing LacCerSyn-FLAG, GM3Syn-HA with vector, or CD77Syn were pulse labeled for 30 min and chased for the indicated times to examine LacCerSyn turnover. The radioactivity of the immunoprecipitated LacCerSyn band in the SDS-PAGE gel was visualized by BAS2500. The bottom graph indicates the relative mean values of LacCerSyn signals in triplicate individual experiments, whereby a faster turnover of LacCerSyn in CD77Syn-expressing cells was detected. The bars at the top of each column represent the mean ± S.E., and the p value was calculated using a Student's t test. IB, immunoblot.
Change in the Turnover of LacCerSyn in the Presence of CD77Syn
The apparent loss of LacCerSyn upon CD77Syn expression (Fig. 6A) indicated possible functional compartmentalization between biosynthetic enzymes for the globo and ganglio series. Yet, when immunofluorescent signals were compared, neither GM3Syn nor CD77Syn seemed to localize in the specified sub-Golgi structure, given that they were both co-localized with the authentic Golgi marker giantin, indicating that such putative compartmentalization may not be seen at the microscopic level even if it exists. As an alternative approach, we focused on LacCerSyn turnover given the apparent loss of LacCerSyn, because a considerable number of reports have demonstrated that changes in the sorting of a protein may alter its turnover (46–49). Moreover, the sorting of the Golgi glycosyltransferase could be a regulated event (50). We carried out pulse-chase experiments on the above polyclonal triple stable cells to determine whether the turnover rate of LacCerSyn expression would increase. We pulsed these triple stable cells with [35S]cysteine and methionine, then chased the cells for the indicated amounts of time. The radioactivity of immunoprecipitated LacCerSyn did not differ between controls and CD77Syn-expressing cells immediately after the labeling. However, the reduction rate was greater in CD77Syn-expressing cells as a function of time (Fig. 6B). It was unlikely that CD77Syn expression caused overall acceleration of protein turnover, because the expression of GM3Syn appeared unaffected (Fig. 6A). Taken together, these data indicate that complex formation with CD77Syn may change the turnover rate of LacCerSyn within the cells. These data do not conflict with the notion that expression of CD77Syn may trigger alternate sorting of LacCerSyn to a functional compartment that is distinct from the putative GM3-biosynthesizing compartment. Collectively, we propose that a change in CD77Syn expression in activated B cells may cause the conversion of LacCer by separate mechanisms, namely, the catalytic consumption of LacCer (although it may not be a major cause, as anticipated), complex formation with LacCerSyn to ease substrate transfer from upstream enzymes, and alteration of LacCerSyn turnover to limit LacCerSyn accession to other branching enzymes. Supplemental Fig. S4 illustrates this using a flow model.
DISCUSSION
We used genetic techniques to analyze biosynthetic pathways by applying novel quantitative phenotype-genotype correlation analysis; specifically, we used cell surface GSL expression and glycan-related gene expression to examine phenotype and genotype characteristics, respectively. We focused on genes that showed negative correlations between phenotype and genotype, because such a relationship could represent a novel regulatory mechanism. Subsequent analysis revealed a mechanism explaining the genetic dominance of CD77Syn in GSL regulation at a biosynthetic branching point. The genetic analyses presented here provide novel insights into the architecture of biosynthetic pathway branching regulation. Our data support the possible glycosyltransferase interaction-mediated alteration of functional compartmentalization of enzymes involved in pathway branching.
The Impact of Pathway Branching Control at LacCer in the Regulation of GSL Biosynthesis
It is noteworthy that ectopic expression of CD77Syn and CD77Syn-TXT both resulted in a reduction in LacCerSyn by Western blotting (Fig. 6A). However, being similar to Ramos cells, CD77Syn-introduced Namalwa cells exhibited very strong CD77 expression, even when using charring detection, which mirrors the level of GSLs better than orcinol detection. Therefore, the question remains how CD77Syn could suppress LacCerSyn yet efficiently produce CD77.
We found that only CD77Syn-sufficient Ramos cells induced globe series-skewed expression by overexpression of GlcCerSyn (Fig. 3), thus the quantity of GlcCerSyn, not LacCerSyn is likely to control the total flow rate of the GSL biosynthesis in CD77Syn-sufficient cells. Nevertheless, regulation of the GSL species seemed to take place at the LacCer branching point. Therefore, we concentrated on branching regulation to address the above question. One clue was the lack of LacCer accumulation upon expression of CD77Syn-TXT (Fig. 5D). When CD77Syn forms a physical complex with LacCerSyn, CD77Syn complexing may limit the free GlcCer accessibility of LacCerSyn. Taken together, we propose a feasible possibility: induced CD77Syn may compartmentalize LacCerSyn so that ongoing biosynthesis of LacCer is efficiently coupled with subsequent branching to CD77 biosynthesis. In such a scenario, the CD77Syn compartment has a shorter turnover of contents, and thus accelerated LacCerSyn turnover is observed (Fig. 6B). Regardless of the mechanism, CD77Syn appears to play a key role in regulating LacCerSyn during GSL expression in B cells, where activation-dependent CD77 biosynthesis is potentiated (supplemental Fig. S4). The above mentioned effect of CD77Syn-TXT supports the idea that functional compartmentalization takes place in GSL biosynthesis branching. This may be achieved by the glycosyltransferase-resident protein interaction (23). More recently, Yamaji et al. (51) reported that induced expression of transmembrane BAX inhibitor motif-containing family proteins could suppress CD77Syn. Thus, CD77Syn activity at the branching point can be dependent on the microenvironment of the Golgi, which would include the expression of BAX inhibitor motif-containing family proteins or other unidentified protein(s). Expression profiles of these proteins may determine cell-type specificity in GSL expression. What is needed for a more precise understanding of this putative compartmentalization model is mutant B cells that lack the ganglio series suppressive activity of CD77Syn. Such synthetic biological analysis of a rescuing CD77Syn-TXT mutant would provide information with regards to the putative molecule responsible for compartmentalization. We are currently attempting to isolate such mutant cells for this purpose.
Global Change in GSL to Globo Series
The characterizations in this study indicated that CD77Syn is a good candidate enzyme to globally change the expression of the GSL species at LacCer branching (10, 52), limiting the expression of LacCerSyn available to other pathways. Thus, induction of globo series GSL and a concomitant reduction in ganglio series GSL could be regulated by transcriptional activation of CD77Syn alone. Expression of CD77 is specific to the germinal center B cells in secondary lymphoid organs, thus CD77 has been utilized to mark germinal center cells such as centroblasts. Indeed, induction of CD77Syn was reported in in vitro-activated human B cells in a microarray study (53). Thus, the above mechanism appears to be responsible for GSL expression in activated B cells during human germinal center reactions. Similarly, CD77-negative (and ganglio series GSL-expressing) M1 cells can be converted into a strongly CD77-positive state in 72 h in response to differentiation to macrophagic cells (54). Such a drastic change in the GSL profile could be explained by the function of CD77Syn to control LacCerSyn. The physiological/functional meaning of such global GSL conversion in each specific cell type is an important issue to be examined in the future, and which we are currently pursuing using manipulated cells only with the regulatory enzyme(s) identified.
Advantages of the CIRES Technique for Genetic Screening at Branching Points
CIRES is a novel method for phenotype-genotype analysis that utilizes the quantitative cellular phenotype to determine the gene(s) regulating the strength of the phenotype as modulator(s) of the system (29, 55). In the present study, we applied CIRES to quantitatively evaluate differences in GSL expression. From correlation analysis and overexpression experiments, we first hypothesized that the biosynthetic pathway branches for the utilization of LacCer two steps upstream of the GM1 synthase reaction, which could be a key step for GM1 biosynthesis, and that this branching is regulated by CD77Syn, although this enzyme is not in the biosynthetic pathway of GM1. This hypothesis was based on the observed negative correlation, and was a key step in identifying the presence of transferase activity-independent complex formation of LacCerSyn with CD77Syn and changes in LacCerSyn turnover by CD77Syn. To the best of our knowledge, CIRES is the only systematic approach for determining such negative relationships between enzymes, and thus could be an especially powerful tool for the identification of regulatory enzymes in highly branched glycan biosynthesis pathways, where a bona fide regulatory enzyme may not be directly mapped within the biosynthetic pathway of the glycan (e.g. the CD77Syn reaction for GM1 expression). Such regulatory enzyme step mapping should be combined with the introduction of “dominant-negative” glycosyltransferase, rather than knocking down the gene, to informatively analyze the regulation. We strongly recommend this strategy, especially when a negative correlation is found via profile analyses.
Supplementary Material
Acknowledgments
We thank Dr. Hisashi Narimatsu and Dr. Takashi Sato (National Institute of Advanced Industrial Science and Technology) for providing full-length cDNA clones of glycosyltransferases, Dr. Yasuyuki Imai (University of Shizuoka) for shiga-like toxin B subunit, and Dr. Toshio Kitamura (University of Tokyo) for Plat-A retrovirus packaging cells.
This work was supported by the Takeda Science Foundation, Mizutani Foundation of Glycoscience, and a Grant-in-aid for Scientific Research from the Ministry of Science and Culture, Sports and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S4.
- GSL
- glycosphingolipid
- CD77
- Gb3Cer/Galα1–4Galβ1–4Glcβ1–1-ceramide
- Cer
- ceramide
- FCM
- flow cytometry
- GM1
- Galβ1–3-GalNAcβ1–4-(Siaα2–3)-Galβ1–4-Glcβ1–1-ceramide)
- GM3
- Siaα2–3-Galβ1–4-Glcβ1–1-ceramide
- LacCer
- lactosylceramide (Galβ1–4-Glcβ1–1-ceramide)
- MFI
- mean fluorescent intensity
- MSCV
- murine stem cell virus
- PDI
- peptidyl disulfide isomerase
- EGFP
- epidermal growth factor protein
- CTxB
- cholera toxin B
- GM2
- GalNAcβ1–4-(Siaα2–3)-Galβ1–4Gloβ1–1 Cer
- GD1a
- Siaα2–3Galβ1–3GalNAcβ1–4(Siaα2–3)-Galβ1–4Glcβ1–1 Cer.
REFERENCES
- 1. Roseman S. (1970) Chem. Phys. Lipids 5, 270–297 [DOI] [PubMed] [Google Scholar]
- 2. Kornfeld R., Kornfeld S. (1985) Annu. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 3. van Echten G., Sandhoff K. (1993) J. Biol. Chem. 268, 5341–5344 [PubMed] [Google Scholar]
- 4. Young W. W., Jr. (2004) J. Membr. Biol. 198, 1–13 [DOI] [PubMed] [Google Scholar]
- 5. Schachter H. (2000) Glycoconj. J. 17, 465–483 [DOI] [PubMed] [Google Scholar]
- 6. Roseman S. (2001) J. Biol. Chem. 276, 41527–41542 [DOI] [PubMed] [Google Scholar]
- 7. Kolter T., Proia R. L., Sandhoff K. (2002) J. Biol. Chem. 277, 25859–25862 [DOI] [PubMed] [Google Scholar]
- 8. Varki A., Esko J. D., Colley K. J. (2009) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler E. E. eds) 2nd Ed., Chapter 3, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 9. Hakomori S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10231–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lannert H., Gorgas K., Meissner I., Wieland F. T., Jeckel D. (1998) J. Biol. Chem. 273, 2939–2946 [DOI] [PubMed] [Google Scholar]
- 11. Ichikawa S., Hirabayashi Y. (1998) Trends Cell Biol. 8, 198–202 [DOI] [PubMed] [Google Scholar]
- 12. D'Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C. C., van der Spoel A. C., Platt F. M., Hannun Y. A., Polishchuk R., Mattjus P., De Matteis M. A. (2007) Nature 449, 62–67 [DOI] [PubMed] [Google Scholar]
- 13. Lala P., Ito S., Lingwood C. A. (2000) J. Biol. Chem. 275, 6246–6251 [DOI] [PubMed] [Google Scholar]
- 14. Nomura T., Takizawa M., Aoki J., Arai H., Inoue K., Wakisaka E., Yoshizuka N., Imokawa G., Dohmae N., Takio K., Hattori M., Matsuo N. (1998) J. Biol. Chem. 273, 13570–13577 [DOI] [PubMed] [Google Scholar]
- 15. Kojima Y., Fukumoto S., Furukawa K., Okajima T., Wiels J., Yokoyama K., Suzuki Y., Urano T., Ohta M., Furukawa K. (2000) J. Biol. Chem. 275, 15152–15156 [DOI] [PubMed] [Google Scholar]
- 16. Keusch J. J., Manzella S. M., Nyame K. A., Cummings R. D., Baenziger J. U. (2000) J. Biol. Chem. 275, 25315–25321 [DOI] [PubMed] [Google Scholar]
- 17. Togayachi A., Akashima T., Ookubo R., Kudo T., Nishihara S., Iwasaki H., Natsume A., Mio H., Inokuchi J., Irimura T., Sasaki K., Narimatsu H. (2001) J. Biol. Chem. 276, 22032–22040 [DOI] [PubMed] [Google Scholar]
- 18. Ishii A., Ohta M., Watanabe Y., Matsuda K., Ishiyama K., Sakoe K., Nakamura M., Inokuchi J., Sanai Y., Saito M. (1998) J. Biol. Chem. 273, 31652–31655 [DOI] [PubMed] [Google Scholar]
- 19. Levine T. P. (2007) J. Cell Biol. 179, 11–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giraudo C. G., Daniotti J. L., Maccioni H. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giraudo C. G., Maccioni H. J. (2003) J. Biol. Chem. 278, 40262–40271 [DOI] [PubMed] [Google Scholar]
- 22. Uliana A. S., Crespo P. M., Martina J. A., Daniotti J. L., Maccioni H. J. (2006) J. Biol. Chem. 281, 32852–32860 [DOI] [PubMed] [Google Scholar]
- 23. Quintero C. A., Valdez-Taubas J., Ferrari M. L., Haedo S. D., Maccioni H. J. (2008) Biochem. J. 412, 19–26 [DOI] [PubMed] [Google Scholar]
- 24. Schwartz-Albiez R., Dörken B., Möller P., Brodin N. T., Monner D. A., Kniep B. (1990) Int. Immunol. 2, 929–936 [DOI] [PubMed] [Google Scholar]
- 25. Jacewicz M., Clausen H., Nudelman E., Donohue-Rolfe A., Keusch G. T. (1986) J. Exp. Med. 163, 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fishman P. H., Moss J., Osborne J. C., Jr. (1978) Biochemistry 17, 711–716 [DOI] [PubMed] [Google Scholar]
- 27. Connell T. D., Holmes R. K. (1995) Mol. Microbiol. 16, 21–31 [DOI] [PubMed] [Google Scholar]
- 28. Barton N. H., Keightley P. D. (2002) Nat. Rev. Genet. 3, 11–21 [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto H., Takematsu H., Fujinawa R., Naito Y., Okuno Y., Tsujimoto G., Suzuki A., Kozutsumi Y. (2007) PLoS One 2, e1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Naito Y., Takematsu H., Koyama S., Miyake S., Yamamoto H., Fujinawa R., Sugai M., Okuno Y., Tsujimoto G., Yamaji T., Hashimoto Y., Itohara S., Kawasaki T., Suzuki A., Kozutsumi Y. (2007) Mol. Cell. Biol. 27, 3008–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morita S., Kojima T., Kitamura T. (2000) Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 32. Saito T., Hakomori S. I. (1971) J. Lipid Res. 12, 257–259 [PubMed] [Google Scholar]
- 33. Kyogashima M., Tamiya-Koizumi K., Ehara T., Li G., Hu R., Hara A., Aoyama T., Kannagi R. (2006) Glycobiology 16, 719–728 [DOI] [PubMed] [Google Scholar]
- 34. Uemura S., Yoshida S., Shishido F., Inokuchi J. (2009) Mol. Biol. Cell 20, 3088–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Preuss U., Gu X., Gu T., Yu R. K. (1993) J. Biol. Chem. 268, 26273–26278 [PubMed] [Google Scholar]
- 36. Nakamura M., Tsunoda A., Sakoe K., Gu J., Nishikawa A., Taniguchi N., Saito M. (1992) J. Biol. Chem. 267, 23507–23514 [PubMed] [Google Scholar]
- 37. Weiss M., Nilsson T. (2003) Traffic 4, 65–73 [DOI] [PubMed] [Google Scholar]
- 38. Breton C., Bettler E., Joziasse D. H., Geremia R. A., Imberty A. (1998) J. Biochem. 123, 1000–1009 [DOI] [PubMed] [Google Scholar]
- 39. Munro S., Freeman M. (2000) Curr. Biol. 10, 813–820 [DOI] [PubMed] [Google Scholar]
- 40. Li J., Rancour D. M., Allende M. L., Worth C. A., Darling D. S., Gilbert J. B., Menon A. K., Young W. W., Jr. (2001) Glycobiology 11, 217–229 [DOI] [PubMed] [Google Scholar]
- 41. Anastasia L., Holguera J., Bianchi A., D'Avila F., Papini N., Tringali C., Monti E., Villar E., Venerando B., Muñoz-Barroso I., Tettamanti G. (2008) Biochim. Biophys. Acta 1780, 504–512 [DOI] [PubMed] [Google Scholar]
- 42. Colley K. J. (1997) Glycobiology 7, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kumagai T., Sato T., Natsuka S., Kobayashi Y., Zhou D., Shinkai T., Hayakawa S., Furukawa K. (2010) Glycoconj. J. 27, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kumagai T., Tanaka M., Yokoyama M., Sato T., Shinkai T., Furukawa K. (2009) Biochem. Biophys. Res. Commun. 379, 456–459 [DOI] [PubMed] [Google Scholar]
- 45. Scheiffele P., Peränen J., Simons K. (1995) Nature 378, 96–98 [DOI] [PubMed] [Google Scholar]
- 46. Stack J. H., Horazdovsky B., Emr S. D. (1995) Annu. Rev. Cell Dev. Biol. 11, 1–33 [DOI] [PubMed] [Google Scholar]
- 47. Schekman R., Orci L. (1996) Science 271, 1526–1533 [DOI] [PubMed] [Google Scholar]
- 48. Wendland B., Emr S. D., Riezman H. (1998) Curr. Opin. Cell Biol. 10, 513–522 [DOI] [PubMed] [Google Scholar]
- 49. Klionsky D. J., Emr S. D. (2000) Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weiss M., Nilsson T. (2000) FEBS Lett. 486, 2–9 [DOI] [PubMed] [Google Scholar]
- 51. Yamaji T., Nishikawa K., Hanada K. (2010) J. Biol. Chem. 285, 35505–35518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Allende M. L., Li J., Darling D. S., Worth C. A., Young W. W., Jr. (2000) Glycobiology 10, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 53. Shaffer A. L., Emre N. C., Lamy L., Ngo V. N., Wright G., Xiao W., Powell J., Dave S., Yu X., Zhao H., Zeng Y., Chen B., Epstein J., Staudt L. M. (2008) Nature 454, 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kannagi R., Levery S. B., Hakomori S. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 2844–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aderem A. (2005) Cell 121, 511–513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.