Abstract
Calreticulin and calnexin are key components in maintaining the quality control of glycoprotein folding within the endoplasmic reticulum. Although their lectin function of binding monoglucosylated sugar moieties of glycoproteins is well documented, their chaperone activity in suppressing protein aggregation is less well understood. Here, we use a series of deletion mutants of calreticulin to demonstrate that its aggregation suppression function resides primarily within its lectin domain. Using hydrophobic peptides as substrate mimetics, we show that aggregation suppression is mediated through a single polypeptide binding site that exhibits a Kd for peptides of 0.5–1 μm. This site is distinct from the oligosaccharide binding site and differs from previously identified sites of binding to thrombospondin and GABARAP (4-aminobutyrate type A receptor-associated protein). Although the arm domain of calreticulin was incapable of suppressing aggregation or binding hydrophobic peptides on its own, it did contribute to aggregation suppression in the context of the whole molecule. The high resolution x-ray crystal structure of calreticulin with a partially truncated arm domain reveals a marked difference in the relative orientations of the arm and lectin domains when compared with calnexin. Furthermore, a hydrophobic patch was detected on the arm domain that mediates crystal packing and may contribute to calreticulin chaperone function.
Keywords: Endoplasmic Reticulum (ER), Glycoprotein Biosynthesis, Lectin, Peptide Interactions, X-ray Crystallography, Calreticulin, Molecular Chaperone, Protein Folding
Introduction
Soluble calreticulin (Crt)7 and membrane-bound calnexin (Cnx) are glycoprotein-specific chaperones of the endoplasmic reticulum (ER). They are components of the quality control machinery that prevents the premature release of folding intermediates from the ER, and they promote proper folding by preventing aggregation and by providing a suitable environment in which the associated ERp57 enzyme catalyzes thiol oxidation and isomerization. If proper folding cannot be achieved, the glycoproteins are marked for ER-associated degradation (1–4).
Crt and the ER luminal portion of Cnx share a similar structure (5–7). They consist of a globular lectin domain and an elongated hairpin-like arm domain, the tip of which comprises the binding site for ERp57 (8, 9). The arm domain is also known as the P domain because it consists of two proline-rich sequence motifs that are repeated in tandem three or four times in Crt and Cnx, respectively. Whereas the structure of the ER luminal portion of Cnx has been solved (7), only the individual lectin and arm domain structures have been determined for Crt by x-ray (6) and NMR (5) methods, respectively. These studies revealed the presence of a bound Ca2+ ion within the lectin domain that is crucial for chaperone stability (10, 11) as well as the location of the oligosaccharide binding site. The lectin domain of both chaperones binds monoglucosylated N-linked oligosaccharides on newly synthesized glycoproteins (12–14), and the removal and re-addition of the terminal glucose residue on these oligosaccharides regulates lectin-based cycles of chaperone interaction (15). Cnx and Crt also interact with many folding glycoproteins through polypeptide-based interactions (for review, see Ref. (4). This has been demonstrated in in vitro experiments where both Cnx and Crt can suppress the aggregation of non-glycosylated substrates (16, 17) as well as in cells where lectin-deficient mutants of the chaperones retain function in the biogenesis of class I histocompatibility molecules (18, 19).
The lectin sites of Cnx and Crt have been well characterized through structural and mutagenesis studies, including the recent determination of the structure of the Crt lectin domain in complex with Glc1Man3 tetrasaccharide (6, 7, 9, 20, 21). In contrast, the polypeptide binding sites are less well characterized. Early experiments in this regard used in vitro aggregation suppression assays that were performed either in the absence of Ca2+ or at temperatures ranging from 42 to 50 °C (9, 20, 22–25). The fact that Ca2+ plays a major role in stabilizing these chaperones as well as their relatively low melting temperatures (46–49 °C) suggest that the observed polypeptide-based interactions may have been influenced by partial protein unfolding. However, more recent studies have focused on characterizing polypeptide-based interactions under physiological conditions of the ER lumen. In the case of Cnx, it potently suppresses the in vitro aggregation of non-glycosylated firefly luciferase at 37 °C and 0.4 mm Ca2+ (10). Furthermore, deletion mutagenesis mapped a single polypeptide binding site to its lectin domain at a location distinct from the oligosaccharide binding site. Detailed substrate specificity studies were not undertaken, but two hydrophobic peptides were shown to bind to this site with micromolar affinity (10). Crt also suppresses the in vitro aggregation of non-glycosylated proteins under physiological conditions (17, 26). The specificity of its polypeptide binding site has been extensively characterized in vitro using a competitive enzyme-linked immunosorbent assay. Crt binds to denatured but not native proteins, and analysis of a broad range of peptide substrates revealed that binding required a length of at least five residues that was hydrophobic in character (27, 28). Several binding sites for peptides or proteins have been mapped onto Crt (29–31), but the relationship between these sites and the site that mediates suppression of protein aggregation remains unknown. Furthermore, the relative roles of the arm and lectin domains in either aggregation suppression or direct peptide binding are unclear.
In this study we investigate structural and functional relationships of the arm and lectin domains of Crt. We use various arm truncation mutants to show that the arm domain contributes to the aggregation suppression function of Crt, but the primary polypeptide binding site is located within the lectin domain. This site binds hydrophobic peptides with micromolar dissociation constants and is distinct from Crt sites that interact with partners such as GABARAP and thrombospondin. We also provide the first crystal structure of Crt with a partial arm domain and identify a hydrophobic patch on the arm that may contribute to Crt chaperone function.
EXPERIMENTAL PROCEDURES
Materials
The hep1 peptide (ELTGAARKGSGRRLVKGPD), 6KSGG peptide (KKKKKKSGGSGGSGGSC) with an amidated C terminus and fCLV peptide (FITC-Ahx-CLVLFVAMWSD, where Ahx is NH2-(CH2)5-COOH) were synthesized by GenScript Corp. (Piscataway, NJ). The 6KAAF peptide (KKKKKKAAFAAFAAFAA) and the KHP peptide (KHPYAYLAAAIAAEVAGTTALKLSK) were provided by Dr. Charles Deber, SickKids, Toronto. Human GABARAP (TrEMBL MM46, accession number O95166) was obtained from Dr. Dieter Willbold, Forschungszentrum Jülich, Germany. All peptides were purified by reverse-phase HPLC, dissolved in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2, separated into aliquots, and stored at −70 °C. The G1M3 oligosaccharide (Glcα1–3Manα1–2Manα1–2Man-OH) was purchased from the Alberta Research Council (Edmonton, Alberta, Canada).
Mutagenesis of Calreticulin cDNA, Protein Expression, and Purification
Amino acid numbering refers to the mouse Crt sequence, with residue 1 corresponding to the first residue of the signal sequence. All primers for PCR-based amplification or mutagenesis are listed in Table 1. Full-length Crt (residues 18–417) and its arm domain (arm, residues 207–300) were amplified in standard PCR reactions using as template mouse Crt cDNA in pcDNA3.1 (19). Both PCR-generated fragments were digested with NdeI and BamHI and ligated into the expression vector pET15b-Tev, a modified pET15b vector (Novagen) that encodes an N-terminal His6 tag followed by a tobacco etch virus (Tev) protease cleavage site. The unpaired Cys at residue 163 in Crt was subsequently mutated to Ser using QuikChange mutagenesis with pET15b-Tev-Crt as template. The resulting pET15b-Tev-CrtCS was then used as the template to generate two variants of Crt with a truncated arm domain, Δarm1 (Δaa239–272) and Δarm2 (Δaa223–286). The mutagenic primers for these constructs introduced a GGSG and a GSG linker, respectively, at the site of truncation. The lectin domain (residues 18–206 and 301–417) was obtained by PCR amplification using pET15b-Tev-CrtCS as template, and the primers incorporated a GSGSG linker between residues 206 and 301. Each construct was verified by DNA sequencing. Crt and its various deletion mutants were expressed in Escherichia coli BL21-CodonPlus cells (Stratagene) and purified as previously described for calnexin (10). Typical yields from 2 liters of bacterial culture were 23 mg (Crt), 20 mg (Δarm1), 16 mg (Δarm2), 7 mg (lectin), and 32 mg (arm).
TABLE 1.
Oligonucleotides used in this study
Restriction sites are underlined, stop codons are shown in italics, and introduced codons are depicted in bold.
| Primer | Sequence 5′ → 3′ | Modifications |
|---|---|---|
| wt-Crt forward | atatatatatcatatggaccctgccatctatttcaaagag | NdeI |
| wt-Crt reverse | atatatggatccttagagctcatccttggcttggccag | BamHI, Stop |
| Arm forward | atatatatatcatatgaagataaaggaccctgatgctg | NdeI |
| Arm reverse | atatatggatccttaggagtattcagggttgtcaatttc | BamHI, Stop |
| Δarm1 forward | ccaagcctgaggactgggacaagggcggtagtggtggcgagtggaaaccacgtcaaattgacaacccag | GGSG linker |
| Δarm1 reverse | gacgtggtttccactcgccaccactaccgcccttgtcccagtcctcaggcttggaatctgtggggtcatcg | GGSG linker |
| Δarm2 forward | gactgggatgaacgaggcagtggcggtacctggatacacccagaaattgacaaccctgaatactcccccgatgc | GSG linker |
| Δarm2 reverse | cagggttgtcaatttctgggtgtatccaggtaccgccactgcctcgttcatcccagtcttccggcttggcagcatc | GSG linker |
| Lectin forward | gactttctgccacccaagggcagcgggagcggccccgatgcaaatatctatgcctatgatagttttgc | GSGSG linker |
| Lectin reverse | ggcatagatatttgcatcggggccgctcccgctgcccttgggtggcagaaagtcccaatcatcctcc | GSGSG linker |
| C163S forward | gtgctgatcaacaaggatatccggtctaaggatgatg | C163S |
| C163S reverse | gtgtgtgaattcatcatccttagaccggatatccttg | C163S |
For crystallization, a variant of the Crt Δarm1 construct was created. This variant, termed CrtΔarm1ΔC368, featured the same arm domain truncation as Δarm1 as well as the C163S mutation (see above) but also included a shorter C terminus ending at residue 368. This variant was inserted into the pET29a vector (Amersham Biosciences) and expressed in E. coli BL21(DE3) in LB medium as an N-terminal and C-terminal His-tag fusion. Cells were harvested and broken in 50 mm Tris, 300 mm NaCl, 3 mm CaCl2, pH 8.0. The fusion protein was purified by affinity chromatography on Ni2+-charged Sepharose resin, and the N-terminal tag was removed by cleavage with thrombin, leaving a Gly-Ser-Met N-terminal extension and a Leu-Glu-His-His-His-His-His-His C-terminal extension. The cleaved protein was additionally purified using size-exclusion chromatography with 20 mm Tris, 100 mm NaCl, 3 mm CaCl2, pH 7.5 buffer.
Crystallization
Initial crystallization conditions were identified utilizing hanging drop vapor diffusion using the PACT screen (Qiagen). The best crystals were obtained at 22 °C by mixing a 0.6-μl drop of CrtΔarm1ΔC368 (10 mg/ml in 20 mm Tris, 100 mm NaCl, 3 mm CaCl2, pH 7.5) with 0.6 μl of reservoir solution containing 25% (w/v) polyethylene glycol 1500 and 25 mm sodium malonate, 37.5 mm imidazole, 37.5 mm boric acid, pH 9.0, and suspending over 0.6 ml of reservoir solution. No additional cryoprotectant was necessary. For data collection, crystals were picked up in a nylon loop and flash-cooled in a N2 cold stream (Oxford Cryosystem). The crystals contain one molecule in the asymmetric unit (Z = 4) corresponding to Vm = 2.2 Å3 Da−1 and a solvent content of 44%.
Structure Solution and Refinement
The native dataset from a crystal of CrtΔarm1ΔC368 was collected using a single wavelength (0.9779 Å) regime on an ADSC Quantum-210 CCD detector (Area Detector Systems Corp.) at beamline A1 at the Cornell High Energy Synchrotron Source (Table 2). Data processing and scaling were performed with HKL2000 (32). The starting phases were obtained using molecular replacement with the Crt lectin domain structure (PDB code 3o0v) using PHASER (33). The resulting model was extended manually with the help of the program Coot (34) and was improved by several cycles of refinement using the program REFMAC (35) followed by the translation-libration-screw (TLS) refinement (36). Of 332 residues in the construct, the final model does not include the N-terminal GSM residues remaining after thrombin cleavage, residues Asp-226—Lys-238, the GGSG linker that replaces the outermost repeat motifs of the arm domain, Gly-273–Pro-283, and the C-terminal residues 368KLEHHHHHH. In addition, 1 calcium ion and 26 water molecules were included in the model. The final model has good stereochemistry with no outliers in the Ramachandran plot computed using PROCHECK (37).
TABLE 2.
Data collection and refinement statistics
| CrtΔarm1C368 | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions, a, b, c (Å) | 43.06, 71.70, 108.39 |
| Resolution (Å) | 50-2.55 (2.59-2.55)a |
| Rsym | 0.053 (0.408) |
| I/σI | 23.6 (2.2) |
| Completeness (%) | 96.9 (77.9) |
| Redundancy | 3.7 (2.3) |
| Refinement | |
| Resolution (Å) | 59.8-2.57 |
| No. reflections | 10,321 |
| Rwork/Rfree | 0.234/0.285 |
| No. atoms | |
| Protein | 2332 |
| Calcium ions | 1 |
| Water | 26 |
| B-Factors | |
| Protein | 44.9 |
| Calcium ions | 54.0 |
| Water | 54.3 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.06 |
| Ramachandran statistics (%) | |
| Most favored regions | 87.8 |
| Additional allowed regions | 12.2 |
a The highest resolution shell is shown in parentheses.
Aggregation Assay
Firefly luciferase (FL, Promega) was dissolved at a concentration of 300 μm in 20 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm CaCl2, 0.5% glycerol, and stored in aliquots at −70 °C. They were thawed only once before use. Crt constructs were equilibrated in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 in the presence or absence of peptides (20 μm) for 10 min at 37 °C. FL was subsequently added to 3 μm final concentration in a total volume of 150 μl. Aggregation was monitored continuously over 1 h at 37 °C by measuring light scattering at 360 nm using a Shimadzu 1601 spectrophotometer equipped with a temperature-controlled cuvette holder. Data were recorded every 6 s.
Fluorescence Measurements
Crt constructs (1 μm) were equilibrated for a period of 1 h at 25 °C in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 in the presence or absence of various peptides (20 μm). Tryptophan fluorescence emission spectra were then recorded at 25 °C from 290 to 390 nm using an excitation wavelength of 280 nm. A Fluorolog 3 Horiba Jobin Yvon fluorescence spectrofluorometer was used with excitation and emission slit widths set to 2 and 5 nm, respectively. When titrating with various peptides, samples were equilibrated for 10 min between the titration steps. Fluorescence measurements were corrected by subtracting the peptide contribution to fluorescence intensity.
Circular Dichroism Experiments
Solutions of Crt or deletion mutants (3.5 μm) were prepared in 8 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1.4 mm CaCl2 and equilibrated for 10 min at 37 °C (room temperature for lectin and arm constructs) before CD spectra were recorded between 200 and 260 nm. In thermal denaturation experiments, Crt or its deletion mutants (3.5 μm) were equilibrated in 20 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm EDTA containing either 1.0 or 1.4 mm CaCl2 for 10 min at room temperature. Melting curves were recorded in a 1-mm path length cuvette from 20 to 70 °C at 228 nm with a scan rate of 2 °C/min. SigmaPlot 2004 Version 9.0 was used to fit the thermal denaturation data to a standard equation by nonlinear least-squares regression assuming a two-state transition process. Tm values represent the midpoint temperature of the thermal unfolding transition. A Jasco J-810 spectropolarimeter equipped with a PTC-423S temperature controlling unit was used for all CD measurements. The results are shown as mean residue molar ellipticity [ϴ] with units converted to degrees cm2 dmol−1.
FITC Labeling of Crt Substrates and Anisotropy Measurements
Various peptide and protein substrates of Crt were dissolved in 1 ml of 50 mm HEPES, pH 7.5, at a concentration of 200 μm. Tris(2-carboxyethyl)phosphine (1 mm) was added, and the reaction mixtures were incubated for 15 min at room temperature. Fluorescein-5-maleimide dissolved in DMF was added at 5:1 molar excess to the Crt substrate, and the samples were incubated for 2 h in the dark. The reactions were quenched by adding a 20-fold molar excess of β-mercaptoethanol over labeling reagent and incubated for 30 min in the dark. The samples were then centrifuged at 16,000 × g for 10 min. The supernatants were loaded on a preparative C-18 reverse phase column, and the peptides/proteins were eluted using a gradient of acetonitrile and water, both with 0.1% trifluoroacetic acid. Initially, a small amount of the reaction mixture (50 μl) was run to identify the elution time of the labeled peptides/proteins (at 215 nm). Subsequently, the entire reaction mixture was run in the dark without using the UV detector, and the eluate corresponding to the labeled peptides/proteins was collected. Peptide and protein concentrations were determined using the bicinchoninic acid assay.
Fluorescence Anisotropy Measurements
Various FITC-labeled Crt substrates were mixed with individual Crt constructs in 150 μl of 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 at the indicated concentrations and incubation times as described in the figure legends corresponding to each specific experiment. Using a 10-mm quartz cuvette, fluorescence anisotropy was measured at 25 °C on a Fluorolog 3 Horiba Jobin Yvon fluorescence spectrofluorometer equipped with polarizers with excitation and emission slit widths set to 3.75 nm. The excitation and emission wavelengths were 491 and 518 nm, respectively. The increment was set to 0.5 nm, and the integration time was set to 2000 ms. A total of five scans were recorded and averaged.
Reduction and Carboxymethylation of α-Lactalbumin
Bovine α-lactalbumin (500 μm, Sigma) was reduced and carboxymethylated (RCMLA) as previously described (38). RCMLA was dialyzed, separated into aliquots, and stored at −70 °C in 20 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm CaCl2.
RESULTS
Conformational Assessment of Calreticulin Deletion Mutants
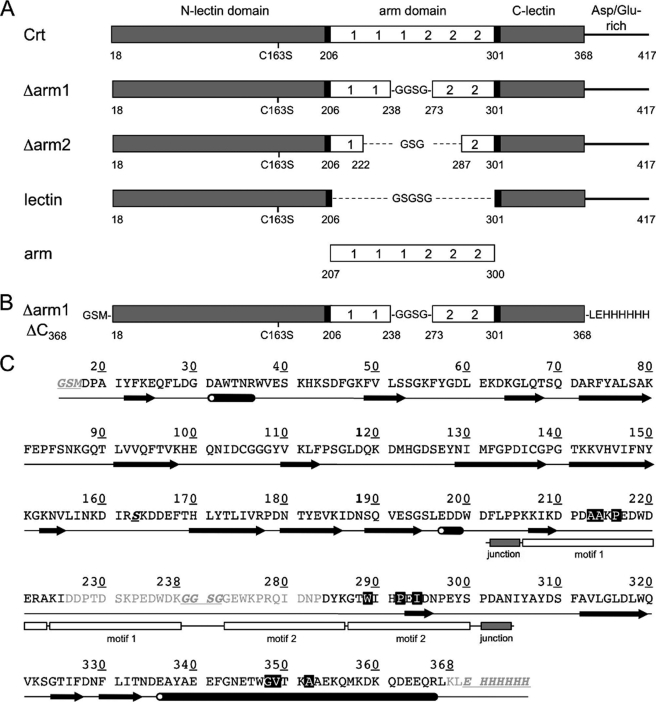
To localize the polypeptide binding site that mediates the chaperone function of Crt, several deletion mutants were constructed that included the individual arm and lectin domains as well as progressive truncations of the arm domain (Fig. 1A). The structural integrity of these constructs was assessed using far-UV CD spectroscopy. The CD spectrum of wild type Crt shown in Fig. 2A had a negative band at ∼226 nm and closely resembled that described previously for Crt in the presence of Ca2+ (11). Likewise, the CD spectra of the isolated lectin domain and the arm domain, the latter with its strong negative maximum at 230 nm, were similar to published spectra (39). The Δarm1 and Δarm2 constructs exhibited spectra intermediate between that of wild type Crt and its lectin domain, consistent with stepwise truncations of the arm (Fig. 2A).
FIGURE 1.
Crt constructs used in this study. A, the linear sequences of the Crt constructs are shown with regions corresponding to the arm domain and globular lectin domain depicted as white and gray rectangles, respectively. Black rectangles denote a short junction region between the lectin and arm domains. Numbers 1 and 2 within the arm domain represent two proline-rich, tandem-repeated sequence motifs. Each motif 1 is paired with the corresponding motif 2 in the hairpin configuration of the arm domain, resulting in three modular units (5). Δarm1 represents Crt with an arm domain lacking amino acids 239–272, corresponding to the most distal repeat motifs. Δarm2 is shortened further by the removal of the next motif pair, corresponding to amino acids 223–286. For the lectin mutant, the complete arm domain was removed, i.e. residues 207–300. Deleted amino acids are shown as dashed lines that were replaced by the indicated Gly-Ser linkers. All constructs contain the indicated C163S mutation to minimize intermolecular disulfide formation during purification. B, calreticulin construct used in crystallization is shown. The Δarm1ΔC368 construct is identical to Δarm1 except for removal of the Asp/Glu-rich C-terminal segment after residue 368 and the presence of the non-Crt sequences GSM and LEHHHHHH at the N and C termini, respectively. C, shown is the amino acid sequence of the fragment crystallized. Non-Crt residues are italicized and underlined. Residues not seen in the crystal structure are shown in gray. The positions of secondary structure elements and the arm domain repeat motifs are shown below the sequence. Residues making hydrophobic contacts in the crystal are boxed in black.
FIGURE 2.
Characterization of Crt domains. A, shown are the far-UV CD spectra of Crt constructs. The CD spectra of 3.5 μm Crt or deletion constructs in 8 mm Hepes, pH 7.4, 150 mm NaCl, and a free Ca2+ concentration of 0.4 mm were monitored at 37 °C (room temperature for lectin and arm). B, shown are thermal stabilities of Crt and its deletion mutants. The constructs (3.5 μm) were equilibrated in 20 mm Hepes, pH 7.4, 150 mm NaCl and a free Ca2+ concentration of 0.4 mm, and thermal denaturation curves (20–70 °C) were recorded by monitoring the change in CD signal at 228 nm.
To assess the stabilities of the Crt constructs, their thermal unfolding curves were measured from 20 to 70 °C. In the presence of Ca2+, a midpoint temperature of unfolding (Tm) of 46.3 °C was calculated for wild type Crt, in agreement with previous findings (11, 26) (Fig. 2B and Table 3). Partial truncation of the arm domain by roughly 33 and 66% in the Δarm1 and Δarm2 constructs, respectively, did not lead to significant destabilization of Crt, whereas complete removal of the arm domain reduced the Tm to 38.5 °C. The isolated arm domain also exhibited a markedly lower Tm of 30 °C. The reduced stabilities of the isolated lectin and arm domain have been noted previously (39) and contrast with Cnx where the individual domains exhibit only modestly reduced stabilities (10). Despite the reduced stabilities of the lectin and arm domains of Crt, we demonstrated previously that they retain the ability to bind monoglucosylated oligosaccharide and ERp57, their respective ligands (6, 9). In the absence of Ca2+, the thermal stabilities of the Crt constructs were significantly decreased except for the arm domain (Table 3). This indicates that all constructs containing the lectin domain bind Ca2+, consistent with the bound Ca2+ ion detected in the crystal structure of the Crt lectin domain (6). Collectively, these findings indicate that the truncation mutants used in this study retain native structure.
TABLE 3.
Thermal stabilities of various Crt constructs
Tm values were calculated by fitting the melting curves to an equation describing a two-state denaturation process using SigmaPlot software.
|
Tm |
||
|---|---|---|
| 0.4 mm Ca2+ | 0 mm Ca2+ | |
| °C | ||
| wt-Crt | 46.3 | 38 |
| Δarm1 | 46.2 | 37.5 |
| Δarm2 | 45.5 | 37.5 |
| Lectin | 38.5 | 30 |
| Arm | 30 | 28 |
Participation of Crt Domains in Suppressing Firefly Luciferase Aggregation
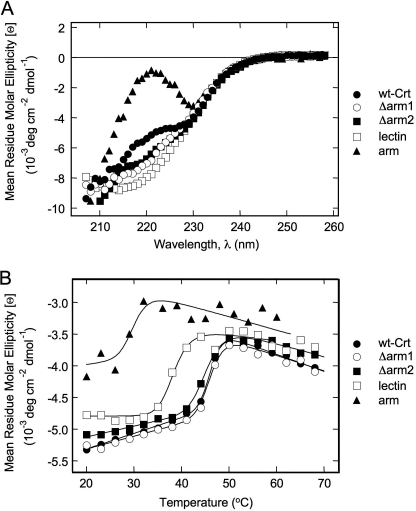
To localize the domain responsible for the aggregation suppression (chaperone) function of Crt, the various truncation mutants were tested for their abilities to prevent the aggregation of FL. FL is a non-glycosylated protein that has been used in the past as a substrate in aggregation suppression assays to characterize polypeptide-based interactions with Cnx under physiological conditions of the ER, i.e. 37 °C and 0.4 mm Ca2+ (10, 16). Initially, we compared the abilities of Crt and its truncation mutants to suppress the aggregation of FL at a 3-fold molar excess of chaperone over substrate. As shown in Fig. 3A, wild type Crt completely suppressed FL aggregation under these conditions, whereas the Δarm1 construct only partially suppressed aggregation; the Δarm2 construct was even less potent than Δarm1. To express their relative potencies in a more quantitative manner, we compared the concentrations of Crt and its truncation mutants required to suppress the aggregation of 3 μm FL to 40–45% of the level observed with FL alone (supplemental Fig. S1), which is a more sensitive assay than the complete suppression of FL aggregation. For wild type Crt, a 5 μm concentration suppressed FL aggregation to 40–45%. The concentrations of Δarm1 and Δarm2 required to suppress FL aggregation to the same extent were 11 and 14 μm, respectively. In other words, Δarm1 and Δarm2 were 2.2- and 2.8-fold less potent than wild type Crt, respectively, indicative of a role for the arm domain in aggregation suppression. Unfortunately, due to their reduced thermal stabilities, neither the individual arm nor lectin domains could be tested in this 37 °C assay. Consequently, we explored whether small peptides could be used as substrate mimetics to further characterize the polypeptide binding site of Crt at temperatures where all of the deletion constructs remained stable.
FIGURE 3.
Aggregation suppression function of Crt and its deletion mutants in the absence or presence of hydrophobic peptides. A, shown is the aggregation suppression capacity of Crt mutants. Crt and its deletion mutants were incubated at the indicated concentrations in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 for 1 h at 37 °C. After the addition of 3 μm FL, aggregation was recorded at 37 °C every 6 s by monitoring light scattering at 360 nm. Data points are shown every 5 min. B, hydrophobic peptides compete with Crt in the suppression of FL aggregation. Before each aggregation measurement, the indicated peptides (20 μm) were equilibrated with 5 μm Crt in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 for 1 h at 37 °C. FL (3 μm) was added, and light scattering was monitored continuously at 360 nm for 1 h at 37 °C. Data points are plotted every 5 min.
Hydrophobic Peptides Block the Ability of Crt to Suppress FL Aggregation
In an effort to identify substrate mimetics, we tested whether several hydrophobic peptides could compete with Crt in suppressing FL aggregation. A Crt:FL ratio was again chosen to provide only ∼40% aggregation suppression as a sensitive readout of the effects of different peptides. None of the four peptides tested affected the aggregation of FL on their own (data not shown). In contrast, when Crt was preincubated for 1 h with peptide 6KAAF (KKKKKKAAFAAFAAFAA), its ability to subsequently suppress FL aggregation was almost fully blocked (Fig. 3B). To rule out the possibility that the positively charged hexalysine segment was responsible for the inhibitory effect of the 6KAAF peptide, we tested the more hydrophilic peptide 6KSGG (KKKKKKSGGSGGSGGS) and observed that it had no significant effect on the aggregation suppression function of Crt (Fig. 3B). Two other largely hydrophobic peptides were tested as well, fCLV (FITC-CLVLFVAMWSD) and peptide KHP (KHPYAYLAAAIAAEVAGTTALKLSK), corresponding to the first transmembrane segment of the Hsmr protein from Halobacterium salinarum. The fCLV peptide blocked the ability of Crt to suppress FL aggregation to a similar extent as the 6KAAF peptide, whereas the KHP peptide was somewhat less potent (Fig. 3B). The experiments were repeated with the 6KAAF and KHP peptides and the Δarm1 and Δarm2 truncation mutants with similar results, except that the two peptides exhibited similar potencies in blocking the ability of these mutants to suppress FL aggregation (supplemental Fig. S2). Given that the 6KAAF, fCLV, and KHP peptides were effective competitors in the aggregation assay, they are good candidates for substrate mimetics whose binding can report on the location and characteristics of the polypeptide binding site on Crt responsible for its aggregation suppression function.
Polypeptide Binding Occurs at a Single Site in the Lectin Domain of Crt
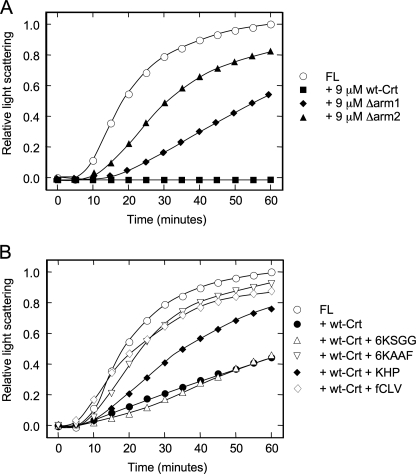
The affinity and stoichiometry of peptide binding to Crt were investigated using changes in intrinsic tryptophan fluorescence of Crt upon incubation with the 6KAAF and 6KSGG peptides. The 6KSGG peptide served as a negative control because it did not compete with Crt in the aggregation suppression assay. As expected, the 6KSGG peptide did not alter the fluorescent emission spectrum of Crt (Fig. 4A). However, Crt fluorescence decreased significantly in the presence of the 6KAAF peptide, suggesting a conformational change associated with complex formation. The change in fluorescent emission at the peak wavelength of 340 nm was then monitored to produce titration profiles with increasing peptide concentration (Fig. 4B). We observed saturable binding of the 6KAAF peptide to Crt, and the peptide binding isotherm could be fit to an equation describing single site binding with a Kd of 0.84 μm. The equivalence point was obtained by drawing lines of best fit through the initial linear and saturation portions of the binding isotherm corresponding to an approximate stoichiometry of 1.15 μmol of 6KAAF peptide bound/μmol of Crt (40, 41).
FIGURE 4.
Binding of peptides to Crt constructs as assessed by Trp fluorescence. A, shown are Trp fluorescence emission spectra of Crt. Crt (1 μm) was incubated in the absence or presence of peptides (20 μm) in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 for 1h at 25 °C. With an excitation wavelength at 280 nm, the Trp fluorescence was measured from 290 to 390 nm, and emission values were plotted every 5 nm. B, shown are titration of Crt and deletion mutants with peptides. Crt constructs (1 μm) were equilibrated in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 for 1 h at 25 °C before the addition of peptide. Intrinsic fluorescence was recorded at 340 nm with an excitation wavelength of 280 nm with 10-min equilibration periods between each titration step. Results are plotted as the change in fluorescence emission (ΔF) divided by the maximum change in fluorescence emission between 20 and 0 μm 6KAAF peptide (ΔFmax). SigmaPlot was used to fit the data to an equation describing single site binding. Dashed lines are lines of best fit for determination of 6KAAF peptide binding to Crt (equivalence point = 1.15 μm 6KAAF peptide; stoichiometry = 1.15:1).
To localize the polypeptide binding site to an individual domain, the binding experiments with the 6KAAF peptide were repeated with the Δarm2 truncation construct as well as with the individual arm and lectin domains. This was possible because the binding experiments could be performed at 25 °C, a temperature where all constructs were stable. As shown in Fig. 4B and Table 4, the removal of two-thirds of the arm domain (Δarm2) or its complete removal (lectin) had little impact on peptide binding with Kd values of 1.1 and 1.2 μm, respectively. Consistent with this finding, the arm domain alone exhibited a negligible capacity to bind to the 6KAAF peptide. Therefore, the polypeptide binding site resides within the globular lectin domain of Crt.
TABLE 4.
Dissociation constants (μm) for binding of Crt constructs to different substrates
| fCLVa |
6KAAFb | fRCMLaa | ||
|---|---|---|---|---|
| −OS | +OS | |||
| wt-Crt | 0.56 | 0.66 | 0.84 | 2.3 |
| Δarm1 | 0.56 | 0.6 | 10 | |
| Δarm2 | 0.9 | 0.8 | 1.1 | 20 |
| Lectin | 1.2 | 33 | ||
| Arm | NDc | NDc | ||
a Kd values were obtained from fluorescence anisotropy experiments.
b Kd values were obtained from intrinsic fluorescence experiments.
c ND, no detectable binding.
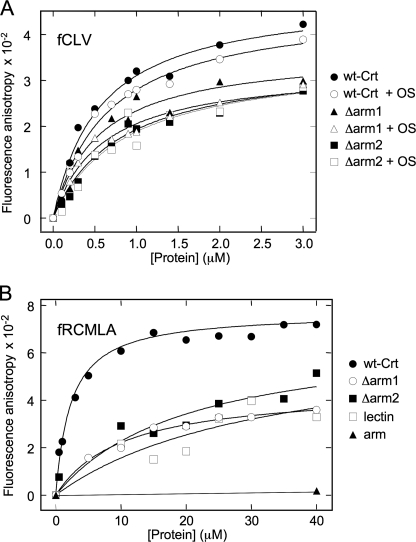
Given that the lectin site of Crt has significant hydrophobic character (6), we sought to determine whether the polypeptide binding site is distinct from the oligosaccharide binding site. We used Glc1Man3 tetrasaccharide, which binds to Crt with a Kd similar to that of the native Glc1Man9GlcNAc2 oligosaccharide (1–1.5 μm (42, 43)) and tested whether it could compete with peptide binding to Crt. As a fluorescent peptide binding reporter, we used the FITC-labeled fCLV peptide. This peptide was as effective as the 6KAAF peptide in blocking the ability of Crt to suppress FL aggregation (Fig. 3B). Using fluorescence anisotropy, 1 μm fCLV peptide was titrated with increasing concentrations of Crt either in the absence or presence of Glc1Man3. As shown in Fig. 5A, saturable binding was observed in the absence of tetrasaccharide, and the data could be fit to an equation describing single site binding with a Kd of 0.56 μm, similar to that observed with the 6KAAF peptide. This was only slightly affected by the presence of 100 μm Glc1Man3, with a Kd of 0.66 μm for fCLV binding to Crt (Fig. 5A, Table 4). Similarly, the presence of excess tetrasaccharide had minimal impact on the binding of fCLV to either the Δarm1 or Δarm2 arm truncation constructs (Fig. 5A, Table 4). Given that both tetrasaccharide and peptide bind with micromolar affinities to Crt and that the addition of a 100-fold excess of Glc1Man3 over peptide had minimal impact on peptide binding, these studies indicate distinct binding sites for the two ligands.
FIGURE 5.
Effects of G1M3 tetrasaccharide and substrate size on polypeptide binding by Crt. A, shown is the effect of G1M3 tetrasaccharide on the binding of Crt constructs to fluorescein-labeled CLV peptide. Fluorescein-labeled CLV peptide (1 μm) was incubated with Crt constructs at indicated concentrations in the presence (100 μm) or absence of oligosaccharide (OS) in 20 mm Hepes, pH 7.4, 150 mm NaCl, 0.4 mm CaCl2 for 1 h at 25 °C before measurement of fluorescence anisotropy. The anisotropy value of fCLV peptide alone was subtracted from each sample to obtain the anisotropy change. Changes in fluorescence anisotropy were recorded at 518 nm with an excitation wavelength of 491 nm. The binding curves were fit to an equation describing single site binding using SigmaPlot. B, Binding of fluorescein-labeled RCMLA to Crt and its deletion mutants. Fluorescein labeled RCMLA (10 nm) was incubated with Crt constructs at the indicated concentrations in 20 mm Hepes, pH 7.4, 150 mm NaCl, 0.4 mm CaCl2 for 1 h at 25 °C before measurement of fluorescence anisotropy changes as described in panel A.
The Arm Domain of Crt Influences the Binding of Large Substrates
The finding that the polypeptide binding site of Crt resides within its lectin domain appeared inconsistent with our observation that arm domain truncations reduce the effectiveness of this chaperone in suppressing the aggregation of FL (Fig. 3A). Because the peptide and FL substrates used in the two types of experiments differ substantially in size, we tested the idea that the arm domain might selectively enhance the binding of large substrates. RCMLA is a 14-kDa protein that remains soluble despite a non-native conformation with little secondary structure (44). It competes with Crt in the FL aggregation assay (data not shown) and, hence, should be a useful large substrate in binding experiments with the various Crt truncation mutants. Changes in fluorescence anisotropy were used to monitor binding of fluorescein-labeled RCMLA with wild type Crt as well as the Δarm1, Δarm2, lectin, and arm constructs (Fig. 5B). For all constructs possessing the lectin domain, the titration data could be fitted to a single-site binding equation, whereas no binding could be detected for the arm domain of Crt. The calculated dissociation constants for wild Crt, Δarm1, Δarm2, and lectin were 2.3, 10, 20, and 33 μm, respectively (Table 4). Therefore, the progressive shortening of the arm domain resulted in progressively less effective binding to this large substrate, mirroring the trend observed in the aggregation suppression assay with the 61-kDa FL substrate. This is in marked contrast to the binding by Crt of the small 6KAAF and fCLV peptide substrates, which was essentially unaffected by arm domain truncation (Table 4).
Binding of Additional Known Peptide Ligands to Crt
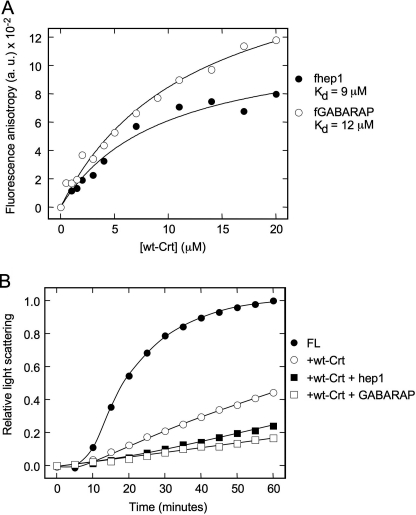
We wanted to explore the relationship between the Crt polypeptide binding site defined in this study with other sites that had been described previously with defined peptide or protein ligands. Crt has been shown to bind to thrombospondin to mediate the anti-cell adhesive activity of the latter protein (30). This interaction has been mapped to residues 36–53 of Crt and to a segment of thrombospondin defined by the hep1 peptide (ELTGAARKGSGRRLVKGPD). In addition, the ubiquitin-like molecule GABARAP (14 kDa), which interacts with the GABAA receptor, has been shown to bind to Crt both in a two-hybrid screen and in vitro (31). Its site of interaction with Crt has been mapped to Crt residues 195–205. Both hep1 and GABARAP were fluorescein-labeled, and their binding to Crt was monitored by fluorescence anisotropy. The binding isotherms could be fit to an equation describing single site binding with a Kd of 9.5 and 12.5 μm, respectively (Fig. 6A). We also assessed the capacity of these Crt ligands to act as competitors in the aggregation suppression assay with FL. As shown in Fig. 6B, neither the hep1 peptide nor GABARAP blocked the ability of Crt to suppress FL aggregation, suggesting that they bind to sites on Crt that are distinct from that which confers its aggregation suppression capability. Interestingly, the combination of Crt with either the hep1 or GABARAP peptide resulted in increased aggregation suppression compared with Crt alone. This was not due to an ability of the peptides to prevent FL aggregation directly as control experiments containing only peptides and FL displayed aggregation profiles indistinguishable from FL on its own (data not shown). It is conceivable that the binding of hep1 or GABARAP peptides to Crt improves the aggregation suppression activity of the chaperone, possibly through allosteric effects on the binding site responsible for the aggregation suppression function.
FIGURE 6.
Interaction of Crt with other peptide and protein ligands. A, shown is binding of Crt to fluorescein-labeled GABARAP and hep1 peptide. Crt at the indicated concentrations in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 was incubated with the fluorescein-labeled substrates (10 nm) at 37 °C for 1 h before measurement of fluorescence anisotropy changes as in Fig. 5. B, GABARAP and hep1 peptide do not compete with Crt in the suppression of FL aggregation. Before aggregation measurements, GABARAP and hep1 peptide (20 μm) were equilibrated with Crt (5 μm) in 20 mm Hepes, pH 7.4, 150 mm NaCl, and 0.4 mm CaCl2 for 1 h at 37 °C. FL was then added at a final concentration of 3 μm, and light scattering was monitored at 360 nm for 1 h at 37 °C. Light scattering was measured every 6 s and plotted every 5 min.
Crystallization of CrtΔarm1ΔC368 and Structure Determination
In an effort to gain further insight into how the arm and lectin domains of Crt participate in its chaperone functions, we crystallized a variant of the Δarm1 mutant. This variant, termed CrtΔarm1ΔC368 (Fig. 1B and C), lacks the acidic C terminus that we previously showed interferes with crystal formation (6). It also contains the non-Crt sequences Gly-Ser-Met and Leu-Glu-His6 at its N and C termini, respectively. The crystals diffracted to 2.6 Å using synchrotron radiation with one molecule of CrtΔarm1ΔC368 in the asymmetric unit.
The structure was determined by molecular replacement using the structure of the Crt lectin domain (6). The initial electron density map showed clear electron density for the first arm domain module proximal to the lectin domain. This was followed by weak patches of electron density for the second module of the arm domain. No electron density was observed for Asp-226—Lys-238, Gly-273—Pro-283 or the GGSG linker (Table 2).
Structure of Crt with a Partially Truncated Arm Domain
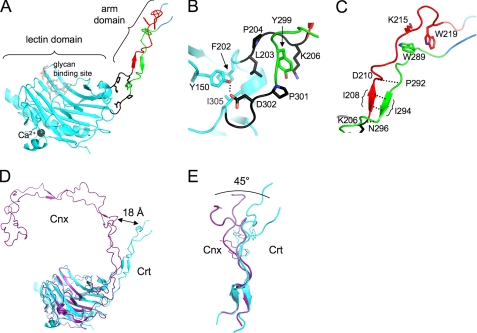
The structure of the lectin domain of CrtΔarm1ΔC368 is nearly identical (root mean square deviation of 0.22 Å over 230 Cα atoms) to that previously determined for Crt with the arm domain deleted (6). The lectin domain adopts a fold formed by two large β-sheets, a seven-stranded concave β-sheet and a six-stranded convex β-sheet, and three α-helices including a long C-terminal α-helix (Fig. 7A). A small β-sheet is positioned on the side of the β-sandwich. The carbohydrate binding site is located on the concave β-sheet, whereas the arm domain is inserted into the lectin domain and separates its N- and C-terminal parts. There is a single calcium ion bound on the opposite side of the lectin domain.
FIGURE 7.
Structure of mouse Crt with a partially truncated arm domain. A, shown is a schematic representation of the lectin domain (cyan), arm domain (red/green), and junction (black). For orientation, the position of Glc1Man3 tetrasaccharide previously observed in the structure of the lectin domain is included (6). B, shown is an enlarged view of the junction region where the arm domain leaves the lectin domain. The junction is positioned by lectin domain residues Tyr-150, Phe-202, Ile-305 and the hydrogen bond (dotted line) between Tyr-150 and Asp-302. Residue Tyr-299 of the arm domain is sandwiched between Pro-301 and Lys-206 to contribute to the rigidity of the junction region. C, shown is an enlarged view of the first module of the arm domain. The arm is stabilized by a small β-sheet with hydrogen bonds between residues in motif 1 (red) and motif 2 (green) and a small hydrophobic cluster formed by the aromatic rings of Trp-219 and Trp-289 and the aliphatic part of the side chain of Lys-215. D, shown is an overlay of the lectin domains of the Crt (cyan) and Cnx (PDB entry 1jhn; purple) structures. The Cα atoms of corresponding residues near the end of the first modules are separated by ∼18 Å. E, shown is an overlay of the first modules of the Crt and Cnx arm domains. Changes in the packing of the tryptophan and lysine residues that form the characteristic small hydrophobic core lead to a roughly 45° angular change in the shape of the arm repeat.
The CrtΔarm1ΔC368 structure reveals a small structured region, which we term the junction region, that links the arm domain to the Crt lectin domain. In the Cnx structure, this region was disordered and incompletely modeled (7). The junction region starts at Leu-203 and ends at Lys-206 on one strand and residues Pro-301 to Asn-304 on the other. The region is structured by the aromatic ring of Tyr-299 from the arm domain, which is surrounded by the side chains of Leu-203, Lys-206, and Pro-301, forming a small hydrophobic core (Fig. 7B). The reason for the difference between the Crt and Cnx structures is unclear; it could result from crystal packing, or the linker region could be intrinsically more flexible in Cnx. Sequence alignment between Crt and Cnx reveals low sequence conservation that may be responsible for the differences in structure (supplemental Fig. S3).
The first arm domain module starts from a short two-stranded β-sheet formed by Lys-207–Lys-209 and Glu-293–Glu-295. Its conformation is similar to that observed in the NMR structure of the Crt arm domain (5). The small hydrophobic core is formed by the two tryptophan residues (Trp-219 and Trp-289) that stack against each other and the side chain of a conserved lysine residue (Lys-215) (Fig. 7C). The second module of the arm domain could not be modeled due to disorder in the crystals.
The Arm Domains of Crt and Cnx Adopt Different Orientations
Comparison of the Crt and Cnx structures shows that the relative orientations of the arm domains are markedly different (Fig. 7D). In Crt, the arm domain is less curved, and the angle between the arm domain and the lectin domain is larger by 30°. By the end of the first module, the distance between analogous residues in Crt and Cnx is ∼18 Å. Sequence comparison shows that the type 1 repeats of the Crt arm domain are two residues shorter than those in Cnx, which likely affects overall curvature of the arm domains (7). Interestingly, the overlay of the first modules of the Crt and Cnx arm domains also reveals differences in packing of aromatic residues, which may contribute to the differences in curvature (Fig. 7E). Furthermore, this reveals a significant and previously unappreciated conformational plasticity within an individual module.
In contrast to the static x-ray structures, NMR studies of the Crt arm domain suggested the existence of hinge-like motions between the individual modules (5). In our crystal, this mobility is likely to be responsible for the lack of electron density for the second module. The B-factors for the Crt arm domain (∼60 Å2) are markedly higher than for the globular lectin domain (∼40 Å2) (supplemental Fig. S4). Given the limited number of structures, the extent to which crystal contacts affect the overall orientation of the arm domain is difficult to assess. In the Cnx structure, the arm domain wraps around the globular domain of another Cnx molecule. In our structure, the orientation of the Crt arm domain may also be affected by intermolecular contacts (see below and Fig. 8A).
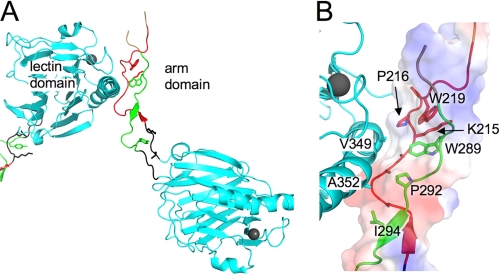
FIGURE 8.
Crystal contacts identify a putative hydrophobic binding site on the arm domain. A, the arm domain makes contacts with the C-terminal helix of the neighboring Crt molecule in the crystal. B, shown is an enlarged view of the interaction site. Residues involved in the contacts are shown as sticks and labeled. The surface is colored according to charge distribution: positive charge is in blue, negative charge is in red, and non-polar regions are in gray.
The Arm Domain Contains a Putative Substrate Binding Site
Analysis of the crystal contacts shows that the first module of the arm domain interacts with the long C-terminal helix of another Crt molecule (Fig. 8A). The contacts are largely hydrophobic, as the arm domain curls around the helix, forming a shallow groove by the side chains of Ala-213, Ala-214, Pro-216, Trp-289, Pro-292, and Ile-294 (Fig. 8B). The interacting surface of the α-helix is centered on Gly-348, Val-349, and Ala-352. This observation may be relevant to our finding that the arm domain, when combined with the lectin domain, contributes to the suppression of FL aggregation and to the binding of large substrates (Figs. 3A and 5) but on its own is unable to bind hydrophobic substrates (Fig. 4 and Table 4). The identified contacts result in a relatively small buried surface area of 795 Å2, suggesting a low binding affinity. Thus, in the context of full-length Crt, the hydrophobic site on the arm domain could play a complementary role in binding large non-native proteins but is unlikely to bind hydrophobic substrates on its own.
DISCUSSION
We took a deletion mutagenesis approach to examine the contribution of the lectin and arm domains to the aggregation suppression (chaperone) activity of Crt and to localize the polypeptide binding site responsible for this function. Biophysical characterization of the mutants revealed that the individual lectin and arm domains were substantially less stable than the intact protein with melting temperatures 8 and 16 °C, respectively, below that of wild type Crt. This is in good agreement with previous studies on the isolated domains by Bouvier and co-workers (39) but contrasts with the findings of Del Cid et al. (26), who noted no loss of stability with the isolated lectin domain. The reason for this discrepancy is unclear but may reflect differences in the assays used (CD versus fluorescent dye binding) or slight differences in truncation boundaries and the size of the bridging linker used to construct the lectin domain. Despite the stabilizing influence of the domains on one another, little difference was noted between the structure of the isolated lectin domain (6) and the lectin domain solved in this study in the context of a partial arm domain. This indicates that the arm domain does not substantially affect the conformation of the lectin domain and is consistent with the fact that the isolated arm and lectin domains retain the ability to bind ERp57 and monoglucosylated oligosaccharide, respectively (9). We also confirmed by thermal denaturation studies a previous report showing that the stabilizing influence of Ca2+ on Crt occurs within the lectin domain and not the arm domain (39), a finding supported by the detection in the crystal structure of a Ca2+ ion within the lectin domain.
Using the various truncation mutants, we demonstrate that Crt suppresses the aggregation of non-native proteins mainly through its lectin domain. To characterize the site(s) responsible, several hydrophobic peptides were used as substrate mimetics in direct binding assays, which revealed that the chaperone activity is conferred through a single polypeptide binding site that is distinct from the lectin site. These findings are strikingly similar to our previous studies on the soluble ER luminal segment of Cnx, S-Cnx (10). The lectin domains of both proteins bind identical hydrophobic peptides with dissociation constants in the micromolar range, and the binding is unaffected either by the addition of a 100-fold molar excess of monoglucosylated oligosaccharide or, in the case of Cnx, by ablating lectin function through mutagenesis (Table 4 and Ref. 10).
Numerous studies over the years have implicated Crt in a bewildering array of processes that occur not only within the ER but in locations as diverse as the cytosol, nucleus, cell surface, and extracellular environment. These include autoimmune disease, complement activation, tumor immunogenicity, cytolytic T cell activity, apoptosis, angiogenesis, wound healing, cardiogenesis, and adipogenesis among others (45). In many of these processes, interactions between Crt and diverse proteins have been documented that are unlikely to involve lectin-oligosaccharide interactions either due to cellular location or to the fact that the protein or interacting segment is not glycosylated or bears complex glycans. Such interactions include Crt binding within the cytosol to the tail of α-integrins, to the ubiquitin-like protein GABARAP (31), and to glucocorticoid and androgen receptors as well as at the cell surface to complement component C1q, thrombospondin, CD91, and laminin (for review, see Ref. 45). In two cases, thrombospondin and GABARAP, the binding site on Crt has been identified as residues 35–53 and 195–205, respectively (30, 31). Consequently, it was of interest to explore whether these interaction sites were similar or distinct from the site that we identified as being responsible for Crt aggregation suppression function.
We were able to reproduce the binding between Crt and either GABARAP or the thrombospondin-derived peptide hep1. However, unlike peptides such as K6AAF or fCLV that bind to the site on Crt responsible for its aggregation suppression activity, neither GABARAP nor hep1 competed with Crt in the FL aggregation suppression assay. Thus, we conclude that neither the GABARAP nor thrombospondin interaction sites is responsible for the chaperone function of Crt. A recent x-ray structural study of the lectin domain of human Crt suggested the presence of an additional peptide binding site that overlapped with the lectin site (29). This was based on the observation that within the crystal lattice, an N-terminal peptide extension arising from the construction of the recombinant protein was found to bind to a portion of the lectin site in a neighboring molecule defined by residues Phe-46, Phe-74, Cys-105, Met-131, Asp-135, Cys-137, Asp-317, and Trp-319. It was proposed that this peptide binding site might be responsible for the chaperone function of Crt. However, we think this is unlikely for two reasons. First, the peptide extension mediating this interaction (KGSIEGR) is much more polar than the hydrophobic peptides shown in the present study to bind to the chaperone site of Crt. Furthermore, many of the residues in the proposed peptide binding site, including Asp-135, Met-131, Asp-317, and Trp-319, are known to mediate interactions with monoglucosylated oligosaccharide and thus would be unavailable when the lectin site is occupied. The present study clearly shows that occupancy of the lectin site with Glc1Man3 tetrasaccharide has no impact on the binding of hydrophobic peptides to the chaperone site of Crt.
Because none of the known peptide binding sites mediate the aggregation suppression function of Crt, the precise delineation of the chaperone site remains a high priority. Efforts to accomplish this through mutagenesis have been made in the past. For example, mutation of His-153 in Crt (numbered His-170 in the current study) impaired its ability to suppress the aggregation of malate dehydrogenase at 45 °C (46). However, this mutation was found to destabilize Crt, a situation that would be further exacerbated by the elevated temperature. Thus, it is unclear whether the impairment of chaperone function is due to the mutated residue or to some indirect conformational perturbation. In another study mutagenesis of Crt was undertaken that targeted hydrophobic residues predicted to be surface-exposed based on a homology model of the Crt structure (17). However, examination of the subsequent crystal structure of the lectin domain of Crt revealed that the side chains of about half of the residues selected were directed into the core of the protein or were buried by oligosaccharide. Neither these nor the other mutants impaired the aggregation suppression function of Crt under physiological conditions of the ER (17). Our efforts to delineate the chaperone site by co-crystallizing hydrophobic peptides with Crt have been unsuccessful to date. Consequently, mutagenesis experiments are ongoing to test candidate surface-exposed hydrophobic sites for this function.
Our current experiments also demonstrate a role for the arm domain of Crt in suppressing the aggregation of non-native protein substrates such as FL. A progressive reduction in chaperone potency was noted as the arm domain was increasingly truncated. Such truncations also progressively reduced the binding affinity of Crt for the large, non-native protein substrate RCMLA but had no effect on its binding to small hydrophobic peptides. These findings are remarkably similar to our previous studies on S-Cnx wherein we showed that the length of the arm domain correlates with binding affinity for large substrates such as FL and RCMLA but not with small peptides (10). Given that the isolated arm domains of either S-Cnx or Crt exhibit no detectable binding to these large substrates, we previously suggested that the arm domains contribute to binding affinity by sterically constraining large protein substrates that enter the cavity between the arm and lectin domains (10). Thus, protein aggregation would be suppressed by a combination of binding to exposed hydrophobic patches on the folding substrate (through the polypeptide binding site on Cnx and Crt) as well as by physically sequestering the substrate from the vicinity of other folding proteins. An additional possibility is suggested by crystal contacts observed in the structure we solved of Crt with a partially truncated arm domain. Hydrophobic contacts were observed between the innermost arm domain module and the C-terminal α-helix of another Crt molecule. Such contacts may also occur between this module and large substrates, thereby contributing to overall binding affinity. However, given the small surface area of the contacts, they would be unlikely to bind hydrophobic substrates on their own. Because small hydrophobic patches are also present in the remaining arm domain modules of Cnx (7) and possibly Crt as well, it is conceivable that they may also contribute to the binding of large substrates. Extensive mutagenesis studies will be required to evaluate the relative contributions of the “steric constraint” and “multi-point contact” models for the role of the arm domain in suppressing substrate aggregation. Regardless of mechanism, it is likely that the arm domain plays a significant role in substrate interaction in cells as removal of two-thirds of this domain (Crt Δarm2 mutant) results in a loss of Crt interaction with class I histocompatibility molecules (47).
Supplementary Material
Acknowledgments
For providing various Crt substrates, we gratefully acknowledge Drs. Arianna Rath and Charles Deber (6KAAF and KHP) and Dr. Dieter Willbold (GABARAP). We also thank Dr. Ronnie Lum for the FITC-labeling protocol, initial RCMLA probes, and helpful discussions as well as Myrna Cohen-Doyle for excellent technical assistance. Data acquisition at the Macromolecular Diffraction (MacCHESS) facility at the Cornell High Energy Synchrotron Source was supported by the National Science Foundation Award DMR 0225180 and the National Institutes of Health Award RR-01646.
This work was supported by Canadian Institutes of Health Research Grants MOP-53310 and MOP-81277 (to D. B. W. and K. G.) and the Canadian Cancer Society (to D. B. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (code 3RG0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Crt
- calreticulin
- Cnx
- calnexin
- ER
- endoplasmic reticulum
- FL
- firefly luciferase
- GABARAP
- 4-aminobutyrate type A (GABAA) receptor-associated protein
- RCMLA
- reduced and carboxymethylated α-lactalbumin
- G1M3 oligosaccharide
- (Glcα1-3Manα1-2Manα1-2Man-OH
- Tev
- tobacco etch virus.
REFERENCES
- 1. Hebert D. N., Molinari M. (2007) Physiol. Rev. 87, 1377–1408 [DOI] [PubMed] [Google Scholar]
- 2. Rutkevich L. A., Williams D. B. (2011) Curr. Opin Cell Biol. 23, 157–166 [DOI] [PubMed] [Google Scholar]
- 3. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams D. B. (2006) J. Cell Sci. 119, 615–623 [DOI] [PubMed] [Google Scholar]
- 5. Ellgaard L., Riek R., Herrmann T., Güntert P., Braun D., Helenius A., Wüthrich K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3133–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kozlov G., Pocanschi C. L., Rosenauer A., Bastos-Aristizabal S., Gorelik A., Williams D. B., Gehring K. (2010) J. Biol. Chem. 285, 38612–38620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 8. Frickel E. M., Riek R., Jelesarov I., Helenius A., Wuthrich K., Ellgaard L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leach M. R., Cohen-Doyle M. F., Thomas D. Y., Williams D. B. (2002) J. Biol. Chem. 277, 29686–29697 [DOI] [PubMed] [Google Scholar]
- 10. Brockmeier A., Brockmeier U., Williams D. B. (2009) J. Biol. Chem. 284, 3433–3444 [DOI] [PubMed] [Google Scholar]
- 11. Li Z., Stafford W. F., Bouvier M. (2001) Biochemistry 40, 11193–11201 [DOI] [PubMed] [Google Scholar]
- 12. Ware F. E., Vassilakos A., Peterson P. A., Jackson M. R., Lehrman M. A., Williams D. B. (1995) J. Biol. Chem. 270, 4697–4704 [DOI] [PubMed] [Google Scholar]
- 13. Spiro R. G., Zhu Q., Bhoyroo V., Söling H. D. (1996) J. Biol. Chem. 271, 11588–11594 [DOI] [PubMed] [Google Scholar]
- 14. Hammond C., Braakman I., Helenius A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hebert D. N., Foellmer B., Helenius A. (1995) Cell 81, 425–433 [DOI] [PubMed] [Google Scholar]
- 16. Brockmeier A., Williams D. B. (2006) Biochemistry 45, 12906–12916 [DOI] [PubMed] [Google Scholar]
- 17. Jeffery E., Peters L. R., Raghavan M. (2011) J. Biol. Chem. 286, 2402–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leach M. R., Williams D. B. (2004) J. Biol. Chem. 279, 9072–9079 [DOI] [PubMed] [Google Scholar]
- 19. Ireland B. S., Brockmeier U., Howe C. M., Elliott T., Williams D. B. (2008) Mol. Biol. Cell 19, 2413–2423 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Culina S., Lauvau G., Gubler B., van Endert P. M. (2004) J. Biol. Chem. 279, 54210–54215 [DOI] [PubMed] [Google Scholar]
- 21. Thomson S. P., Williams D. B. (2005) Cell Stress Chaperones 10, 242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ihara Y., Cohen-Doyle M. F., Saito Y., Williams D. B. (1999) Mol. Cell 4, 331–341 [DOI] [PubMed] [Google Scholar]
- 23. Martin V., Groenendyk J., Steiner S. S., Guo L., Dabrowska M., Parker J. M., Müller-Esterl W., Opas M., Michalak M. (2006) J. Biol. Chem. 281, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 24. Rizvi S. M., Mancino L., Thammavongsa V., Cantley R. L., Raghavan M. (2004) Mol. Cell 15, 913–923 [DOI] [PubMed] [Google Scholar]
- 25. Thammavongsa V., Mancino L., Raghavan M. (2005) J. Biol. Chem. 280, 33497–33505 [DOI] [PubMed] [Google Scholar]
- 26. Del Cid N., Jeffery E., Rizvi S. M., Stamper E., Peters L. R., Brown W. C., Provoda C., Raghavan M. (2010) J. Biol. Chem. 285, 4520–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duus K., Sandhu N., Jørgensen C. S., Hansen P. R., Steinø A., Thaysen-Andersen M., Højrup P., Houen G. (2009) Protein Pept. Lett. 16, 1414–1423 [DOI] [PubMed] [Google Scholar]
- 28. Sandhu N., Duus K., Jørgensen C. S., Hansen P. R., Bruun S. W., Pedersen L. Ø., Højrup P., Houen G. (2007) Biochim. Biophys. Acta 1774, 701–713 [DOI] [PubMed] [Google Scholar]
- 29. Chouquet A., Païdassi H., Ling W. L., Frachet P., Houen G., Arlaud G. J., Gaboriaud C. (2011) PLoS One 6, e17886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goicoechea S., Pallero M. A., Eggleton P., Michalak M., Murphy-Ullrich J. E. (2002) J. Biol. Chem. 277, 37219–37228 [DOI] [PubMed] [Google Scholar]
- 31. Thielmann Y., Weiergräber O. H., Mohrlüder J., Willbold D. (2009) FEBS J. 276, 1140–1152 [DOI] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 36. Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 37. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 38. Bösl B., Grimminger V., Walter S. (2005) J. Biol. Chem. 280, 38170–38176 [DOI] [PubMed] [Google Scholar]
- 39. Tan Y., Chen M., Li Z., Mabuchi K., Bouvier M. (2006) Biochim. Biophys. Acta 1760, 745–753 [DOI] [PubMed] [Google Scholar]
- 40. Arnaud N., Georges J. (1997) Analyst 122, 143–146 [Google Scholar]
- 41. Urbanová N., Kádár M., Tóth K., Bogáti B., Andruch V., Bitter I. (2008) Anal Sci. 24, 727–733 [DOI] [PubMed] [Google Scholar]
- 42. Patil A. R., Thomas C. J., Surolia A. (2000) J. Biol. Chem. 275, 24348–24356 [DOI] [PubMed] [Google Scholar]
- 43. Kapoor M., Srinivas H., Kandiah E., Gemma E., Ellgaard L., Oscarson S., Helenius A., Surolia A. (2003) J. Biol. Chem. 278, 6194–6200 [DOI] [PubMed] [Google Scholar]
- 44. Okazaki A., Ikura T., Nikaido K., Kuwajima K. (1994) Nat. Struct. Biol. 1, 439–446 [DOI] [PubMed] [Google Scholar]
- 45. Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M. (2009) Biochem. J. 417, 651–666 [DOI] [PubMed] [Google Scholar]
- 46. Guo L., Groenendyk J., Papp S., Dabrowska M., Knoblach B., Kay C., Parker J. M., Opas M., Michalak M. (2003) J. Biol. Chem. 278, 50645–50653 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y., Kozlov G., Pocanschi C. L., Brockmeier U., Ireland B. S., Maattanen P., Howe C., Elliott T., Gehring K., Williams D. B. (2009) J. Biol. Chem. 284, 10160–10173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.