Abstract
RIG-I is a major innate immune sensor for viral infection, triggering an interferon (IFN)-mediated antiviral response upon cytosolic detection of viral RNA. Double-strandedness and 5′-terminal triphosphates were identified as motifs required to elicit optimal immunological signaling. However, very little is known about the response dynamics of the RIG-I pathway, which is crucial for the ability of the cell to react to diverse classes of viral RNA while maintaining self-tolerance. In the present study, we addressed the molecular mechanism of RIG-I signal detection and its translation into pathway activation. By employing highly quantitative methods, we could establish the length of the double-stranded RNA (dsRNA) to be the most critical determinant of response strength. Size exclusion chromatography and direct visualization in scanning force microscopy suggested that this was due to cooperative oligomerization of RIG-I along dsRNA. The initiation efficiency of this oligomerization process critically depended on the presence of high affinity motifs, like a 5′-triphosphate. It is noteworthy that for dsRNA longer than 200 bp, internal initiation could effectively compensate for a lack of terminal triphosphates. In summary, our data demonstrate a very flexible response behavior of the RIG-I pathway, in which sensing and integration of at least two distinct signals, initiation efficiency and double strand length, allow the host cell to mount an antiviral response that is tightly adjusted to the type of the detected signal, such as viral genomes, replication intermediates, or small by-products.
Keywords: Double-stranded RNA, Innate Immunity, Pathogen-associated Molecular Pattern (PAMP), Pattern Recognition Receptor, RNA Viruses, RNA-Protein Interaction, Viral Immunology
Introduction
Crucial events upon viral infection of a host organism are the efficient recognition of the pathogen and the mounting of a set of antiviral defense mechanisms, most importantly the activation of the type I IFN system. The promptness as well as the strength of this initial innate immune response is key in setting the overall course of the disease and, in many instances, determines whether the virus will eventually be cleared. RIG-I,3 a member of the cytosolic RIG-I like receptor family, has received increasing attention over the past years (1, 2). It detects viral RNA products and upon recognition associates with the adapter molecule MAVS (formerly also known as Cardif, IPS-1, or VISA) (3–6) via its two N-terminal caspase activation and recruitment domains. MAVS then relays the danger signal, ultimately leading to phosphorylation and activation of the latent transcription factor IFN regulatory factor 3 (IRF-3), which promotes transcription of the IFN-β gene as well as a panel of IFN-stimulated genes (ISGs), most prominently ISG56 (gene symbol IFIT1) (7).
RIG-I has been investigated intensively, and its vital role in the host defense against a variety of viruses has been established (8–11). Although the molecular identity of the respective RNA ligands during viral infection has not been determined unequivocally, the triphosphorylated 5′-terminus (5′-ppp) of viral or in vitro transcribed RNA has been recognized as a primary motif for recognition by RIG-I (12–14), particularly when present in the context of a double-stranded blunt end (15, 16). In fact, negative strand RNA viruses detected by RIG-I either possess complementary sequences at the termini of their genomes, leading to the formation of blunt end panhandle structures, or they are prone to produce large amounts of defective interfering particles containing panhandle forming subgenomes. It has recently been demonstrated that RIG-I associates almost exclusively with such 5′-ppp, blunt end RNAs in influenza- or Sendai virus-infected cells (17, 18). However, there have also been reports on RIG-I signaling in response to dsRNA lacking a 5′-ppp, like RNaseL cleavage products (19) or poly(I·C) (10, 20–22).
Although many studies have addressed ligand specificity, also because of increasing interest in defining optimal stimulatory molecules for immunotherapy (23), comparatively little is known about the molecular mechanisms of RNA detection by RIG-I and its translation into an antiviral signal. Individual mechanistic aspects, however, have been studied recently. Importantly, crystallographic analyses have shown that the C-terminal domain of RIG-I can bind the terminus of dsRNA, both triphosphorylated and nonphosphorylated (24–27), which is in line with reported ligand specificities. Major shortcomings of all structural studies so far are, that it has not been possible to crystallize the full RIG-I protein, or at least a fragment including the helicase domain, and that the dsRNA used for co-crystallization has been extremely short (8–14 bp). Longer substrates, like poly(I·C), were shown to also induce RIG-I activation but putatively involving the helicase domain rather then the RIG-I C-terminal domain (20, 25). Nevertheless, it was reported that the stimulatory potential is higher for short dsRNA (21), although the underlying mechanism remains enigmatic, and helicase/translocase activity has been shown to increase on longer substrates (28).
In our present study, we have now combined quantitative signaling assays, cell-free biochemical analyses, and single molecule microscopy. Our findings will help to understand how RIG-I simultaneously senses the various described signals and how it integrates these signals into a tightly adjusted response. The presented model provides an example of a complex adaptive achievement of the immune system to detect viral invaders with maximum sensitivity while maintaining self-tolerance.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
Huh7.5/RIG-I cells were described previously (22). Mouse embryonic fibroblasts were kindly provided by Osamu Takeuchi (Osaka, Japan) and Gunther Hartmann (Bonn, Germany). All murine and human cells were maintained in Dulbecco modified Eagle's medium supplemented with 2 mm l-glutamine, nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum. Medium for Huh7.5/RIG-I cells was additionally supplemented with 1 mg/ml G418 (Invitrogen).
RNA Methods
For generation of differently long dsRNA, PCR products were amplified from a pCDNA3.1-based plasmid containing the human Toll-like receptor 3 coding sequence (cDNA sequence accession NM_003265.2). PCR primers contained the T7 promoter sequence allowing for transcription of either the forward (F) or reverse (R) strand as shown in supplemental Fig. S1. Sequences of the PCR primers are given below (underlined, T7 promoter; italic, six nucleotide extension to ensure blunt ends; normal type, sequence complementary to templateDNA): common-F fwd, TAATACGACTCACTATAGGGAGATGATGCTTTCTCTTGGTTGGGC; common-R fwd, GGGAGATGATGCTTTCTCTTGGTTGGGC; ds100-F rev, GGGAGATCTCCATTCCTGGCCTGTGA; ds100-R rev, TAATACGACTCACTATAGGGAGATCTCCATTCCTGGCCTGTGA; ds200-F rev, GGGAGAGCATCAGTCGTTGAAGGCTTG; ds200-R rev, TAATACGACTCACTATAGGGAGAGCATCAGTCGTTGAAGGCTTG; ds400-F rev, GGGAGAACCACCAGGGTTTGCGTGTTT; 400-R rev, TAATACGACTCACTATAGGGAGAACCACCAGGGTTTGCGTGTTT;ds1600-F rev, GGGAGAGAACTTCAGGGTCAGCTTGC; and 1600-R rev, TAATACGACTCACTATAGGGAGAGAACTTCAGGGTCAGCTTGC. For in vitro transcription of the 40-bp dsRNA (ds40), two dsDNA oligonucleotides (40-F and 40-R), analogous to PCR products, were purchased (MWG Operon) and used as a template: 40-F, TAATACGACTCACTATAGGGAGATGATGCTTTCTCTTGGTTGGGCCACCTATCTCCC; and 40-R, TAATACGACTCACTATAGGGAGATAGGTGGCCCAACCAAGAGAAAGCATCATCTCCC. As a nonphosphorylated control (5′-OH), the two strands of ds40 were commercially synthesized chemically and HPLC-purified by MWG Operon.
In vitro transcription was performed according to common procedures using T7 polymerase (Promega). Standard reaction time was 4 h, which was extended by 2 h for transcripts shorter than 200 nucleotides. After in vitro transcription, template DNA was DNase I-digested (Promega). Two methods were applied to clear annealed in vitro transcripts from side products and unincorporated nucleotides: 1) single-stranded or annealed transcripts were subjected to standard gel electrophoresis using low melting agarose; RNA molecules of the expected size were excised, extracted from the gel by agarase treatment and repeated organic extraction, and subsequently precipitated with 2.5 m (NH)4-acetate and 2 volumes ethanol; and 2) 500 μl of annealed transcripts were applied to an ÄKTA chromatography system (GE Life Sciences) equipped with a Superose 6 10/300 GL column and 400-μl fractions containing only dsRNA of the expected size were collected and concentrated by standard sodium acetate/ethanol precipitation. In either case, dsRNA was solved in water and quality-validated by capillary gel electrophoresis (Qiaxcel; Qiagen) or analytical size exclusion chromatography on a Superose 6 PC 3.2/30 column (GE Life Sciences). RNA was quantified using photospectrometry or the QuantIt RNA kit and Qubit fluorometer (Invitrogen).
Shrimp alkaline phosphatase (Fermentas) was used to remove the terminal phosphates of in vitro transcribed RNA. RNA was incubated 30 min at 37 °C with 0.5 units of enzyme/pmol RNA. The reaction was stopped by heating to 65 °C for 10 min.
Hepatitis C virus RNA was generated by T7 (positive strands) or T3 (negative strands) polymerase (Promega) mediated in vitro transcription of a genotype 2a (JFH-1) subgenomic replicon using the construct pFKi389neoNS3–3′_∂g_JFH-1 wild type or with the ΔGDD deletion in NS5B (nonreplicating). Vesicular stomatitis virus and encaphalomyocarditis virus viral RNA was prepared by pelleting virus from cell-free infected cell culture supernatants by ultracentrifugation and subsequent RNA extraction using the NucleoSpin RNA II kit (Macherey-Nagel). Rabies virus leader RNA (in vitro transcribed and chemically synthesized) was a gift from Karl-Klaus Conzelmann (Munich, Germany).
ISG56 Promoter Reporter Assay
RIG-I signaling in Huh7.5/RIG-I and murine embryonic fibroblast (MEF) cells was assessed as described previously (22). Briefly, the cells were co-transfected with the ISG56 promoter firefly luciferase construct pGL3B/561 (gift from Ganes Sen) and Renilla luciferase pRL-SV40 (Promega) in a ratio of 3:1 using Effectene transfection reagent (Qiagen) for Huh7.5/RIG-I or Lipofectamine 2000 reagent (Invitrogen) for MEFs. Six hours after reporter transfection, the cells were stimulated by transfection of defined RNA using Lipofectamine 2000 (Invitrogen). Particular care was taken to always use an identical ratio of nucleic acid: Lipofectamine 2000 reagent to keep liposome-RNA complex composition (charge per lipoplex) constant. Therefore, when transfecting less than 1 μg of RNA/well (24-well format), the transfection mix was supplemented with poly(C) as a carrier to a final concentration of 1 μg/well. Transfection with 1 μg of poly(C) only was therefore used as a negative control (see Fig. 1). Firefly and Renilla luciferase were measured 16 h post-stimulation. Firefly luciferase was normalized to Renilla luciferase activity. dsRNA concentrations are given as pmol/well; conservatively assuming 2 × 105 cells/well and 100% transfection efficiency, 1 pmol/well would translate into 3 × 106 dsRNA molecules/cell. All of the experiments were performed using duplicate measurements in duplicate wells, and the data are given as averages ± S.D. One representative experiment of at least two independent repetitions is shown if not stated otherwise.
FIGURE 1.
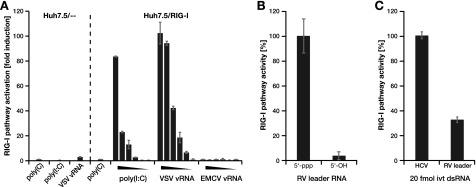
Huh7.5/RIG-I cells are a sensitive RIG-I indicator system. A–C, Huh7.5/- - (vector control) or Huh7.5/RIG-I cells were stimulated by transfection with different viral RNAs, and RIG-I pathway activation was assessed using ISG56 promoter reporter activity. A, cells were transfected with no RNA (mock), 1 μg/well homopolymeric ssRNA poly(C), the dsRNA analog poly(I·C), or RNA prepared from particles of vesicular stomatitis virus (VSV) or encephalomyocarditis virus (EMCV). The triangles indicate titrations from 1 to 10−5 μg/well. B, cells were stimulated with 0.1 μg/well 58 nucleotides rabies virus (RV) leader RNA generated by in vitro transcription (5′-ppp) or chemically (5′-OH). C, cells were stimulated with 0.1 μg/well (20 fmol) of double-stranded in vitro transcripts of replication incompetent hepatitis C virus (HCV) RNA or with an equal number (20 fmol) of rabies virus leader in vitro transcribed dsRNA molecules.
Protein Purification
The GST gene was cloned in front of human RIG-I, separated by the consensus cleavage site for the PreScission protease (GE Life Sciences). Recombinant GST-RIG-I expressing baculovirus was generated using the Bac-to-Bac expression system (Invitrogen). For virus production as well as protein expression, Sf9 cells were used. For protein purification, cell pellets were resuspended in binding buffer 150 mm NaCl, 30 mm Tris (pH 7.4), 10 μm ZnCl2, 10 mm DTT, 1 mm EDTA, supplemented with complete protease inhibitor cocktail (Roche) and lysed by three or four cycles of freeze/thawing (liquid nitrogen). RIG-I was affinity-purified using glutathione-Sepharose 4B (GE Life Sciences) according to the manufacturer's recommendations. Beads were washed thoroughly in binding buffer, and RIG-I was cleaved off the GST tag by PreScission protease at 4 °C. Purity was checked in SDS-PAGE (supplemental Fig. S3), and monomeric RIG-I was further purified by preparative size exclusion chromatography on an ÄKTA chromatography system, equipped with a Superose 6 10/300 GL column (GE Life Sciences). Protein was quantified using the QuantIt protein kit and Qubit fluorometer (Invitrogen).
ATPase Activity Assay
RIG-I protein concentration was 1.5 pmol/well, and dsRNA concentrations are indicated for the respective experiments. The reactions were performed in 20 mm Tris (pH 7.5), 40 mm NaCl, 4 mm MgAc2, 0.5 mm EDTA in the presence of 1 mm ATP and incubated for 30 min at 37 °C. Generated free phosphate was quantified using the QuantiChrom ATPase/GTPase assay kit (BioAssay Systems) according to the manufacturer's protocol. All of the samples were measured in duplicate or triplicate wells, and data are given as averages ± S.D.; one representative experiment of at least two independent repetitions is shown.
Analytical Size Exclusion Chromatography (SEC)
Analytical SEC was performed on a Superose 6 PC 3.2/30 column, and absorbance was monitored at 215 nm (protein) and 254 nm (RNA). Baseline calculations and peak quantifications were done with Unicorn 5.11 software (GE Life Sciences). For binding experiments, 0.1–1 μg of dsRNA were incubated with 3 μg of RIG-I in 50 μl of binding buffer (see above) for 30 min at room temperature before applying to SEC. 100-μl fractions were collected for further quantitative analysis.
Quantification of collected SEC fractions was performed by vacuum blotting 30 μl of each fraction to nitrocellulose or Hybond-N+ membrane (GE Life Sciences) for immunolabeling and Northern hybridization, respectively. RIG-I was detected by monoclonal RIG-I antibody (Alme-1; Alexis), followed by quantification on the ODYSSEY infrared imaging system (Licor). Northern hybridization was performed using a radioactively labeled probe corresponding to the first 40 bp of the dsRNA (oligonucleotide 40-F, see above) following a standard protocol. Quantification was done by phosphorimaging and QuantityOne software (Bio-Rad).
Scanning Force Microscopy (SFM)
10–40 nm dsRNA, 10 nm RIG-I, or a mixture of both were briefly incubated in 20 μl of buffer (10 mm HEPES, pH 8.0, 10 mm NaCl, 2 mm MgCl2) and adsorbed to a freshly cleaved mica surface for 5 min. 20 μl of buffer supplemented with 4 mm NiSO4 was added to fix the samples on the surface. The surface was washed with water and then dried. SFM images were obtained with a Nanoscope III (Digital Instruments) operating in tapping mode. Images of a 2 × 2-μm area were acquired using etched silicon tips (type NHC; Nanosensors) at a scan rate of 2–3 Hz and a resolution of 256 pixels/μm. The images were flattened with the Nanoscope IIIa software, and three-dimensional surface plots were generated using ImageJ software (National Institutes of Health).
Statistical Analyses and Nonlinear Regression
Significance was tested by one-way analysis of variance (see Fig. 7) or the extra sum-of-squares F-test using the Prism 5 software (GraphPad) for statistical comparison of dose-response or length-response regressions. A difference stated to be “highly significant” in the main text generally corresponds to a p value <0.01. Hill regression of dose-response data, including calculation of parameters and their confidence intervals, was done in Prism 5 or Matlab (The Mathworks), using the optimization toolbox.
RESULTS
Viral 5′-Triphosphorylated RNAs Are Differentially Recognized by RIG-I
The human hepatoma cell line Huh7.5 is deficient for RIG-I signaling because of a dominant negative mutation (29). Moreover, because of a lack of Mda54 and Toll-like receptor 3 (30) expression, the cells exhibit no IRF-3 activation upon transfection of immunostimulatory RNAs like poly(I·C) or vesicular stomatitis virus viral RNA (Fig. 1A, left panel). We have previously shown that Huh7.5 stably transduced with wild-type RIG-I (Huh7.5/RIG-I) exhibit a strong, RIG-I-dependent IRF-3 activation upon stimulation (22), which was not due to up-regulation of any other factors (like Mda5 or Toll-like receptor 3) as evidenced by genome-wide transcriptional profiling.4 The validity of this cell line as a highly sensitive and specific RIG-I indicator system was further established by its efficient and dose-dependent IRF-3 activation upon transfection of poly(I·C) and vesicular stomatitis virus viral RNA, while still being unresponsive against single-stranded poly(C) and encaphalomyocarditis virus viral RNA (Fig. 1A, right panel). Further, we tested this system for its 5′-ppp dependence by transfecting 58-nucleotide-long in vitro transcripts of rabies virus leader RNA or synthetic, 5′-hydroxylated (5′-OH) RNA of the same sequence (12, 24) (kind gifts from Karl-Klaus Conzelmann, Munich, Germany). As expected, only the 5′-ppp variant stimulated the RIG-I pathway (Fig. 1B). We then wanted to compare RIG-I stimulation by RNAs of different viral origins. To this end, we used in vitro transcribed rabies virus leader RNA and full genomic (but replication-incompetent) hepatitis C virus RNA. In both cases, we annealed positive and negative strand transcripts to warrant a fully double-stranded conformation, which has been shown to be required for 5′-ppp recognition. Strikingly, we observed significantly stronger pathway activation upon transfection of hepatitis C virus as compared with rabies virus leader dsRNA (Fig. 1C), even though we employed equal amounts of 5′-ppp termini (20 fmol of dsRNA in both cases). We therefore hypothesized that the main distinction between rabies virus leader (58 nucleotides) and hepatitis C virus (∼9600 nucleotides) RNA is their length, and thus, RIG-I is able to sense the length of the double-stranded region of an RNA, as has been suggested before (21, 28).
RIG-I Pathway Activation Systematically Increases with dsRNA Length
The majority of RIG-I ligand studies (14) employed very short dsRNAs (typically 10–40 nucleotides) at very high concentrations. Consequently, the influence of the terminal structure likely has been overrepresented relative to the impact of other parameters like ligand concentration or dsRNA length. We therefore characterized the dependence of RIG-I signaling on dsRNA length in the context of fixed molecule (i.e. 5′-ppp) numbers, effectively blinding out any effect mediated by the termini. We generated in vitro transcribed RNA from PCR products of an arbitrary human cDNA sequence. PCR primers were designed such that there was one common forward primer, resulting in homologous products of increasing length depending on the position of the reverse primer (40, 100, 200, 400, and 1600 bp). Single-stranded RNA products were annealed to form blunt-ended dsRNA (supplemental Fig. S1 for details). Because ubiquitous side products of shorter and longer size generated during in vitro transcription can lead to false conclusions about the nature of the real stimulatory RNA species (15, 16), we purified the desired products by preparative gel electrophoresis and/or SEC and employed stringent quality control of the final RNAs using capillary electrophoresis (Fig. 2A and supplemental Fig. S1) or analytical SEC (see below).
FIGURE 2.
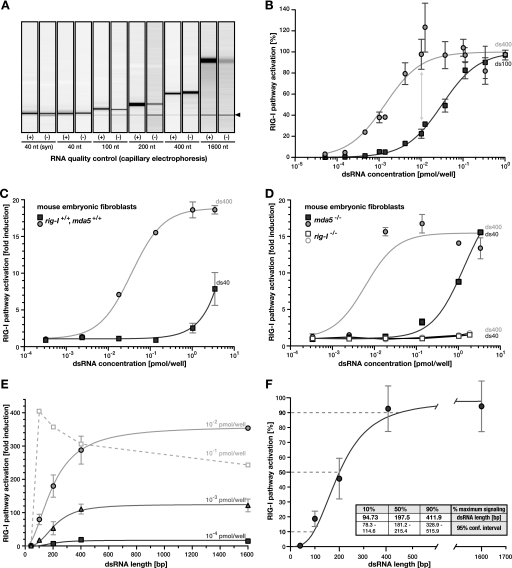
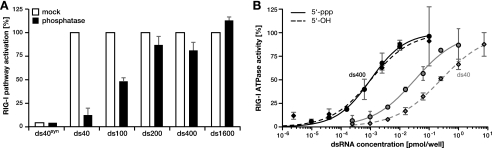
RIG-I signaling systematically increases with dsRNA length. A, RNA used for transfection was extensively purified and quality controlled. A graphical representation of capillary electrophoresis of purified positive (+) and negative (−) strand in vitro transcripts is shown. The band marked with the black arrowhead corresponds to the internal calibrator of the system. B, Huh7.5/RIG-I cells were stimulated with dsRNA of 100 bp (ds100, squares) or 400 bp (ds400, circles) length, and ISG56 promoter reporter activity was determined. Dose-response was standardized and fitted using Hill regression. Data points were combined from two independent repetitions of the experiment. C and D, MEFs were transfected with ds100 or ds400 and ISG56 promoter reporter activity was assessed. MEFs were derived from mice without (C) or with (D) a homozygous deletion of the rig-I or mda5 gene, respectively (as indicated). E, ISG56 promoter-reporter activity was assessed in Huh7.5/RIG-I cells upon stimulation with equal numbers of dsRNA molecules of increasing length. Independent response curves for three different concentrations are given (as indicated). Paradoxical behavior for unphysiological molecule numbers is shown as a dashed line. F, the three absolute response curves from E were standardized to the respective maximum signal (100%), and data points were averaged (±S.D.). A Hill model was fitted to the data, which was significantly more accurate than standard mass action kinetics (p < 0.0001 in extra sum-of-square F-test). dsRNA lengths required to elicit 10%, 50%, and 90% of the maximum signal were calculated from the regression and are given with the respective 95% confidence intervals in the inset.
These highly purified dsRNAs allowed us to directly examine the effect of dsRNA length on recognition by RIG-I. We transfected Huh7.5/RIG-I cells with dsRNA of 100 and of 400 bp in length, respectively, and measured IRF-3 activation by ISG56 promoter reporter assays. As we titrated molar concentrations of the RNAs over a range of several orders of magnitude, we paid particular attention to keeping RNA-liposome complex formation and transfection conditions constant by using nonstimulatory poly(C) as a carrier. Transfection of the dsRNAs resulted in dose-dependent IRF-3 activation, which could be well described by nonlinear regression using a standard Hill model (Fig. 2B). Remarkably, we observed up to 5-fold higher activity for the 400-bp dsRNA as compared with the 100-bp dsRNA at identical numbers of molecules, i.e. 5′-ppp moieties (Fig. 2B, gray arrow). To exclude a cell type- or species-specific phenomenon, we validated this length dependence of RIG-I RNA recognition also in MEF (Fig. 2C). Experiments in MEFs derived from mice with a homozygous deletion of the rig-I or mda5 gene, respectively, demonstrated that the presence of RIG-I was necessary and sufficient to elicit this signaling, ruling out any dependence of the effect on Mda5 (Fig. 2D).
To further characterize this RNA length sensing by RIG-I, we performed systematic stimulation assays in Huh7.5/RIG-I cells using fixed numbers of increasingly long dsRNA molecules. Already for extremely low concentrations of 0.1 fmol/well, corresponding to less than 300 molecules of dsRNA/cell by conservative calculation, RIG-I pathway activation increased steadily with increasing length of the transfected dsRNA (Fig. 2E and logarithmic plot in supplemental Fig. S2B). The same length dependence was also observed for 10- and 100-fold higher molecule numbers (Fig. 2E). Only at unphysiologically high concentrations, we observed a paradoxical effect with a tendency to inverse this relation, likely because of the consumption and sequestration of the whole cellular RIG-I pool on long dsRNA (Fig. 2E, dashed line). This behavior has been described before upon stimulation with high amounts of enzymatically shortened poly(I·C) (21). However, within the dynamic range of the system of at least 3 orders of magnitude of ligand concentrations, the normalized response behavior was virtually identical, highlighting the robustness of this signal-to-length relation even against substantial differences in the amount of stimulatory molecules (Fig. 2F). Half-maximal RIG-I stimulation required dsRNA of ∼198 bp in length and 90% of the maximal stimulation was reached with dsRNA of ∼411 bp. Signaling was less than 10% for dsRNA below ∼95 bp (Fig. 2F), suggesting that the RIG-I sensor system is optimized for recognition of dsRNA of several hundreds of base pairs in length, rather than short RNA oligonucleotides such as the ones were widely used for previous RIG-I/ligand interaction studies. Of note, length dependence of RIG-I signaling exhibited sigmoidal characteristics (p < 0.0001 when tested against standard mass action kinetics using the extra sum-of-square F-test), indicating the involvement of cooperative effects.
In Vitro Enzymatic Activity Correlates with Signaling
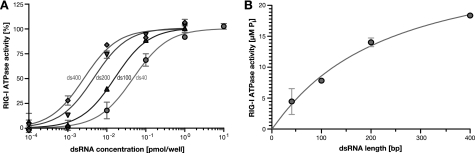
To rule out an epiphenomenon caused by unrelated cellular processes or by technical issues like varying transfection efficiencies, we next performed cell-free biochemical experiments. Therefore, we expressed human RIG-I fused to GST in Sf9 insect cells via a baculovirus-based system. Following glutathione affinity chromatography, the GST tag was cleaved off from the protein, and native, monomeric RIG-I was isolated by preparative SEC. The purity of the protein preparation and its monomeric state were validated in SDS-PAGE and analytical SEC (supplemental Fig. S3). We then assessed its functionality in enzymatic assays exploiting the described ATPase activity residing in the DExD/H-box helicase like domain. As described previously, ATPase activity was virtually undetectable in the absence of RNA and increased with classical dose-response behavior in titrations of dsRNA ligand (Fig. 3A). Remarkably, in ATPase assays with equimolar concentrations of increasingly long dsRNA, RIG-I activity again strictly correlated with the length of the ligand (Fig. 3B). From a standard mass action regression, we calculated half-maximal activity to be reached for dsRNA of 253 bp (S.E. ± 60 bp). Fitting a Hill-model yielded a similar result (221 bp) with a slightly better goodness of fit; however, the difference was statistically not significant. Accordingly, the dose-response relation for longer dsRNA shifted significantly left toward lower molar concentrations as compared with short (40 bp) dsRNA (Fig. 3A). These observations further substantiate the notion of RIG-I being most sensitive for dsRNAs of several hundred base pairs in length. Moreover, they demonstrated that length sensing occurs at the stage of RIG-I binding or its catalytic activity and is independent of additional cellular processes.
FIGURE 3.
RIG-I enzymatic activity increases for longer dsRNA. A, purified recombinant RIG-I was incubated with increasing amounts of purified, differently long dsRNA (400 bp, diamonds; 200 bp, inverted triangles; 100 bp, triangles; 40 bp, circles), and generation of inorganic phosphate by ATP hydrolysis was assessed. Dose-response was standardized and fitted using Hill regression. B, RIG-I was incubated with 0.01 pmol of dsRNA of increasing length, and ATPase activity was measured as in A. The values were regressed using standard mass action kinetics, because a Hill model was not significantly more accurate.
RIG-I Cooperatively Multimerizes on dsRNA
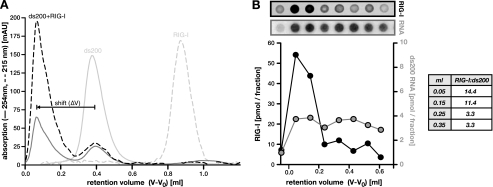
The observed increase in RIG-I signaling and ATPase activity with increasing length of the dsRNA ligand could in principle be due to either 1) higher or more sustained activity of individual RIG-I molecules when bound to longer dsRNA or 2) the accumulation of higher numbers of RIG-I molecules binding to one molecule of dsRNA. To differentiate between these two possibilities, we performed binding experiments and analyzed complex formation in SEC. Recombinant, monomeric RIG-I and purified dsRNAs were individually subjected to analytical SEC to determine their chromatographic behavior (supplemental Fig. S4A). We then preincubated RIG-I together with dsRNA of a given length to allow for complex formation before analyzing the mixture in SEC. For all cases tested (40-, 100-, and 200-bp dsRNA could be resolved on the employed SEC matrix), we observed a significant reduction of free monomeric RIG-I and dsRNA, accompanied by the appearance of a new peak with a lower retention volume, i.e. a larger size (hydrodynamic radius), representing RIG-I-dsRNA complexes (Fig. 4A and supplemental Fig. S4, B–D). We determined the shifting distance between plain dsRNA and the respective complex (ΔV; Fig. 4A), which corresponds to the contribution of RIG-I to the total size of the complex. Although ΔV does not directly allow the determination of the number of bound RIG-I molecules, we found it to be very small for complexes of 40- and 100-bp dsRNA but substantial in the case of 200-bp complexes (supplemental Fig. S4E). This indicates that 200-bp dsRNA is bound by significantly more RIG-I molecules than short dsRNA. To quantitatively assess the stoichiometry of such a complex, we isolated the SEC fractions containing the 200-bp dsRNA-RIG-I complex and performed dot-blot analyses. We quantified the RNA and protein content by Northern hybridization or immunodetection, respectively, and found 10–15 RIG-I molecules/200-bp dsRNA (Fig. 4B). This was also backed by approximate calculations based on the absorbance (215 and 254 nm) of the respective peaks, leading to an estimate of ∼20 molecules/200-bp dsRNA (not shown). Taking into account the relative molecule sizes, this stoichiometry arguably corresponds to dsRNA that is fully covered with RIG-I. However, considering the observed complete consumption of monomeric RIG-I but only partial depletion of free dsRNA (Fig. 4A), this cannot be explained by simple stochastic binding. Stochastic binding would rather yield a gaussian distribution of complexes, ranging from “naked” to fully occupied dsRNA. Our experiments, on the contrary, revealed the formation of a clearly defined complex that was well separable from residual free dsRNA by SEC (Fig. 4A). This argues for strong cooperative effects, facilitating multimerization of RIG-I along dsRNA once initial binding has occurred.
FIGURE 4.
Oligomeric binding of RIG-I to long dsRNA. A, purified recombinant RIG-I was preincubated with 200-bp dsRNA and subsequently analyzed in SEC. Absorption was assessed at 215 nm (protein, dashed line) and 254 nm (RNA, solid line). Elution profiles of the individual components are shown for reference (light color). Shifting distance between plain dsRNA and RIG-I-dsRNA complexes is indicated (ΔV). B, contiguous fractions of the eluate were dot-blotted onto membranes and subjected to immunodetection of RIG-I (black) or Northern hybridization of the RNA (gray), respectively. Signals were quantified using a serially diluted standard. Calculated molar ratios are given in the inset. Note that fractionation was technically limited, with each dot representing the average RNA or protein content of one 100-μl fraction. This leads to a markedly reduced resolution as compared with the absorption profile in A.
RIG-I Covers the Entire dsRNA Molecule as a Linear Multimer
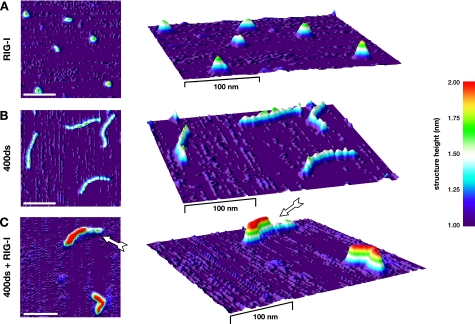
To confirm the above findings from SEC by a complementary method, we directly visualized RIG-I-dsRNA complexes by SFM. Samples were incubated in identical buffer conditions as in the SEC experiments and scanned at high resolution. As expected for a monomeric preparation, RIG-I alone was a homogeneous solution of single globular particles with similar sizes (Fig. 5A). Similar to DNA (31), dsRNA appeared as “threads” with an average length of 0.29 (±0.02) nm/base pair and an apparent structure height of 1.5–1.7 nm (Fig. 5B and supplemental Fig. S5). In contrast, RIG-I-dsRNA complexes were still of linear, “thread”-like shape, but appeared slightly shorter and thicker than unbound dsRNA and had a significantly higher profile (≥2.0 nm; Fig. 5C and supplemental Fig. S5). Most frequently, the dsRNA was decorated with RIG-I along its entire length. In >90% of those instances, in which part of the dsRNA remained uncovered, we observed exactly one continuous stretch of RIG-I (Fig. 5C, arrow). This further substantiated the notion of RIG-I binding occurring as a cooperative, linear multimerization along its dsRNA ligand.
FIGURE 5.
RIG-I covers dsRNA as linear multimers. RIG-I (A) or 400-bp dsRNA (B) alone or RIG-I-dsRNA complexes (C) were applied to a mica surface and subjected to SFM. Images were transformed into three-dimensional surface plots, and height information was mapped to false-color shading. x/y and z dimensions are not to scale (x/y, scale bar is 100 nm; z, see color scheme). The arrow in C indicates a contiguous stretch of bound RIG-I on partially uncovered dsRNA, suggesting incomplete cooperative multimerization.
Affinity of the Primary Binding Event Modulates RIG-I Activation
In such a scenario, a combination of the initial binding affinity on one hand, and the degree of multimerization on the other hand, would determine the eventual strength of pathway activation. While on one hand the above results provided evidence for a direct relation between length of the dsRNA and the final amount of multimerized RIG-I molecules, binding initiation on the other hand, is likely to depend on the presence of characteristic structural motifs. Based on literature reports (12, 13, 26–28), we speculated that RIG-I would have a significantly higher affinity to 5′-ppp than to nonphosphorylated termini or internal portions of the dsRNA. To test our hypothesis, we enzymatically dephosphorylated the dsRNA by alkaline phosphatase treatment. Dephosphorylation efficiency was very high, as shown by a >95% reduction of radioactive γ-phosphate in a test sample without affecting RNA integrity (supplemental Fig. S6). As a control, we used chemically synthesized 40-bp RNA (ds40syn), which is fully devoid of terminal phosphates. We then transfected Huh7.5/RIG-I cells with mock-treated or dephosphorylated dsRNA of different length, using the respective concentrations for half-maximal signaling to exploit the full dynamic range of the system (compare supplemental Fig. S2A). For short dsRNA (40 bp), offering very limited space for internal binding, enzymatic removal of the 5′-ppp virtually abrogated RIG-I-mediated signaling (Fig. 6A). In contrast, dephosphorylated 100-bp dsRNA was impaired to only ∼50%, whereas dsRNA ≥200 bp provided sufficient internal binding sites to effectively compensate for the removal of the phosphates (Fig. 6A). Similar results were obtained in cell-free ATPase activity assays with recombinant RIG-I. Also in this setting, we observed a significant (p < 0.001) decrease in RIG-I activity for nonphosphorylated as compared with 5′-ppp containing 40-bp dsRNA, whereas no difference was detectable for dsRNA of 400 bp (Fig. 6B). These findings confirm our hypothesis that the level of RIG-I activation is determined by both the length of the dsRNA as well as the presence of certain, stimulatory motifs, like 5′-terminal triphosphosphates. It is noteworthy that the absence of such highly efficient motifs can be compensated by low efficiency binding initiation on sufficiently long double-stranded regions.
FIGURE 6.
Long dsRNA can compensate for the lack of a high affinity motif in RIG-I activation. A and B, purified in vitro transcribed dsRNA of different length was dephosphorylated enzymatically with shrimp alkaline phosphatase. A, Huh7.5/RIG-I cells were transfected with mock (open bars) and phosphatase-treated (solid bars) dsRNA using the respective concentrations for half-maximal signaling (previously determined, see supplemental Fig. S2A). ISG56 promoter-reporter activity was determined and normalized to the respective mock-treated sample. B, triphosphorylated (5′-ppp, solid lines) and unphosphorylated (5′-OH, dashed lines) 400-bp (black) and 40-bp (gray) dsRNA was analyzed in RIG-I ATPase assays. The values were standardized and fitted using Hill regression.
DISCUSSION
RIG-I was demonstrated to recognize the triphosphorylated 5′-terminus of RNA in the context of a short double-stranded blunt end, like the panhandles found in full-length or defective interfering genomes of many negative strand RNA viruses (12, 13, 15–18). The structural basis of the recognition of such short, 5′-ppp dsRNAs by the C-terminal domain of RIG-I has been studied extensively (24, 25, 27, 32). However, it has been shown that RIG-I also mediates signaling in response to nontriphosphorylated dsRNA (10, 19–22). Mechanistically, little has been known on how RIG-I detects properties such as double-strandedness or triphosphorylation and how it integrates these distinct cues into downstream signaling strength. In the present study, we now explicitly investigated RIG-I-mediated responses to defined RNAs and linked them to the underlying biochemical and structural events.
Although it is unambiguous that the 5′-ppp is a potent recognition motif, the structural characteristic common to all stimulatory RIG-I ligand RNAs tested so far has been double-strandedness; even in cases where single-stranded RNA had been described as having a stimulatory potential, recent studies strongly suggest this to be caused by inadvertently generated dsRNA in the course of in vitro transcription (14–16). We therefore devised an experimental strategy that allowed us to isolate the effect of the double strand from that of the 5′-ppp. By using fixed molecule numbers (fixed numbers of 5′-termini), the impact of the 5′-ppp was effectively canceled out, and thus, the observed differences in signaling or biochemical activity could be solely ascribed to the varying length of the double-strand. In these experiments, we found a very strong positive correlation between ligand length and RIG-I activation, both in promoter-reporter based signaling assays and in enzymatic activity assessments (Figs. 2 and 3). In fact, dsRNAs of several hundred bp in length were more efficient by orders of magnitudes at triggering IRF-3 activation than 40-bp dsRNA (supplemental Fig. S2). Experiments in MEF cells showed that this effect was not specific for one cell line and, importantly, that it was independent of Mda5, a sister molecule of RIG-I that has been described to recognize higher order networks of very long RNA (33). Concordant with this notion, only poly(I·C) or dsRNA several thousand bp in length elicited measurable Mda5 signaling in our hands.5 Previous studies on RIG-I, on the other hand, mostly employed high concentrations of extremely short dsRNA (<50 bp), thereby overriding direct recognition of the double strand by terminus-mediated effects. Strikingly, the sole previous study explicitly dealing with RNA length (21) reported an inverse correlation to what we describe here. Kato et al. (21) could show higher RIG-I activity in response to enzymatically shortened poly(I·C). However, throughout their experiments, Kato et al. used a high (1 μg/ml) and fixed amount (by weight) of RNA for stimulation, irrespective of its length, whereas we used titrations with controlled molarities. In fact, whereas the phenotype that we describe was very robust across orders of magnitude of dsRNA concentrations (Fig. 2, E and F), we too observed a paradoxical effect when applying saturating amounts of RNA for stimulation (Fig. 2E, dashed line). This effect could actually be even more pronounced if we had used constant amounts of RNA by weight instead of molecule numbers.
To explain the observed stronger activation of RIG-I with long dsRNA, in principle, two hypotheses were conceivable: one dsRNA is always bound by the same number of RIG-I molecules (e.g. by a dimer (24)), but these molecules exhibit a higher activity on longer dsRNA, or longer dsRNA is bound by a higher number of RIG-I molecules. In support of the first hypothesis, an elegant study in fact demonstrated periodic translocation of RIG-I along its dsRNA ligand, with the average duration of translocation increasing with the length of the double strand (28). However, also in that study, dsRNA never exceeded 50 bp in length, and additionally, multiple RIG-I binding was not explicitly addressed. We therefore directly investigated the stoichiometry and structural constitution of RIG-I-dsRNA complexes in gel filtration (SEC) and by SFM. In SEC, we found higher numbers of RIG-I molecules binding to longer dsRNA. For 200-bp dsRNA, we determined 10–15 bound RIG-I molecules, which corresponds to a binding footprint of one to two helical turns (∼11 bp), arguing for complete coverage of the RNA by RIG-I (Fig. 4). This was also confirmed by SFM analysis, where we primarily found fully decorated RNA double-strands (Fig. 5 and supplemental Fig. S5). These findings clearly demonstrate that RIG-I multimerizes along dsRNA. They do not rule out, however, that there still might be a certain degree of translocation along the dsRNA (28), possibly as a concerted periodic movement of the whole RIG-I assembly. In fact, binding assays (SEC and SFM) were conducted in the absence of ATP, arguing for a separate, ATP-dependent activity of RIG-I distinct from plain binding. Translocation on or unwinding of dsRNA are two such energy-dependent functions, both of which have been described for RIG-I (25, 28). However, how they may be linked to signaling has not yet been fully understood (25, 28, 34).
Another striking aspect was the predominance of fully assembled complexes in SEC, consuming virtually the whole pool of RIG-I but leaving a considerable amount of dsRNA entirely uncovered (Fig. 4A). This cannot be easily explained by an independent association of each individual RIG-I molecule with dsRNA. In such a case, one would expect a broad gaussian distribution of complex stoichiometries with a range from completely uncovered to fully covered dsRNA. Rather, our observation hinted toward a cooperative mode of RIG-I multimerization, in which the initial binding event is of low affinity and, therefore, rate-limiting, whereas subsequent condensation of further RIG-I molecules is strongly facilitated. Consequently, completion of multimerization on a partly covered dsRNA would be strictly favored over binding to a new, uncovered dsRNA region. Such a model is also in full agreement with our SFM studies, where we predominantly observed fully assembled RIG-I-dsRNA complexes or dsRNA, of which exactly one part was decorated with a stretch of RIG-I, leaving the remainder uncovered (Fig. 5, arrow, and supplemental Fig. S5). Complexes with only one or a few interspersed RIG-I molecules occurred very rarely. Although all of our experiments support the notion of cooperative multimerization of RIG-I, further studies will be required to unambiguously address and characterize this cooperativity, e.g. by surface plasmon resonance using terminally immobilized dsRNA molecules.
Because multimerization is not rate-limiting, the affinity of the primary binding event is crucial in determining RIG-I substrate selectivity. Numerous studies have previously suggested that RIG-I would have a significantly higher affinity for 5′-triphosphorylated termini than for nonphosphorylated RNA or internal double-stranded regions (14, 24, 28, 32). This was confirmed by our experiments, which showed a detrimental effect of dephosphorylation for short (40 bp) dsRNA (Fig. 6). Interestingly, the impact of the 5′-ppp vanished as dsRNA length increased, indicating that availability of a huge amount of less efficient and nonspecific internal initiation sites can in effect compensate for the lack of a highly efficient initiation motif like the 5′-ppp (Fig. 6). Although beyond the scope of this study, it is conceivable that many more factors can modulate the efficiency of RIG-I binding initiation. For example, two studies have reported sequence specificity of RIG-I binding and activation in the context of hepatitis C virus recognition (35, 36). Moreover, RNA modifications, like pseudo-uridylation or 2′-O-methylation, were shown to affect recognition of dsRNA by RIG-I (12, 27, 35). In effect, the presence (e.g. 5′-ppp or sequence) or absence (e.g. RNA modifications) of such motifs would consequently determine the affinity of the primary RIG-I binding. As a second cue, RIG-I then reads out the double-strandedness of the RNA by cooperative multimerization, amplifying the signal overproportionally with increasing length of the dsRNA (Fig. 2F). By this integration of (at least) two distinct signals, the RIG-I system ensures highest possible sensitivity while avoiding detection of host RNA, which in general does not carry 5′-ppp, is heavily modified, and never exhibits consecutive duplex regions of several hundred base pairs.
Taken together, our results suggest a differential response behavior of the RIG-I system. Short 5′-ppp dsRNA structures, like the panhandle in the genomes of many viruses and particularly their defective interfering particles, elicit only a weak response when present at low concentrations. This would be the case right after particle entry into the host cell and might suffice to put the cell in an alert, hypersensitive state. In contrast, detection of direct markers of intracellular virus replication, like a few inadvertently liberated long, double-stranded replicative intermediates or high concentrations of 5′-ppp panhandle-containing subgenomic byproducts, triggers mounting of the full-blown antiviral defense program. Our study provides a first insight into the molecular mechanisms, which allow RIG-I to detect different features of nonself RNA and to integrate these cues into a flexible antiviral response that is tightly adjusted to the specific danger situation.
Supplementary Material
Acknowledgments
We are grateful to Ulrike Herian for outstanding technical assistance. We further thank Alena Grebe and Stefan Winheim for technical support.
This work was supported by Deutsche Forschungsgemeinschaft Grant FOR1202 “Mechanisms of Persistence of Hepatotropic Viruses” (to R. B. and V. L.) and Bundesministerium für Bildung und Forschung Grant 01KI0786 (to R. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
M. Binder, unpublished observations.
M. Binder and F. Eberle, unpublished data.
- RIG-I
- retinoic acid-inducible gene-I
- 5′-ppp
- 5′-terminal triphosphate
- dsRNA
- double-stranded RNA
- IFN
- interferon
- IRF-3
- interferon regulatory factor 3
- ISG56
- interferon stimulated gene of 56 kDa
- Mda5
- melanoma differentiation associated gene 5
- SEC
- size exclusion chromatography
- SFM
- scanning force microscopy
- MEF
- murine embryonic fibroblast.
REFERENCES
- 1. Rehwinkel J., Reis e Sousa C. (2010) Science 327, 284–286 [DOI] [PubMed] [Google Scholar]
- 2. Yoneyama M., Fujita T. (2007) J. Biol. Chem. 282, 15315–15318 [DOI] [PubMed] [Google Scholar]
- 3. Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 4. Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 5. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 6. Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 7. Grandvaux N., Servant M. J., tenOever B., Sen G. C., Balachandran S., Barber G. N., Lin R., Hiscott J. (2002) J. Virol. 76, 5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 9. Loo Y. M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M. A., García-Sastre A., Katze M. G., Gale M., Jr. (2008) J. Virol. 82, 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. (2005) J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 11. Habjan M., Andersson I., Klingström J., Schümann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Mühlberger E., Mirazimi A., Weber F. (2008) PLoS One 3, e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 13. Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 14. Schlee M., Hartmann G. (2010) Mol. Ther. 18, 1254–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlee M., Roth A., Hornung V., Hagmann C. A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K. A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. (2009) Immunity 31, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt A., Schwerd T., Hamm W., Hellmuth J. C., Cui S., Wenzel M., Hoffmann F. S., Michallet M. C., Besch R., Hopfner K. P., Endres S., Rothenfusser S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baum A., Sachidanandam R., García-Sastre A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rehwinkel J., Tan C. P., Goubau D., Schulz O., Pichlmair A., Bier K., Robb N., Vreede F., Barclay W., Fodor E., Reis e Sousa C. (2010) Cell 140, 397–408 [DOI] [PubMed] [Google Scholar]
- 19. Loo Y. M., Owen D. M., Li K., Erickson A. K., Johnson C. L., Fish P. M., Carney D. S., Wang T., Ishida H., Yoneyama M., Fujita T., Saito T., Lee W. M., Hagedorn C. H., Lau D. T., Weinman S. A., Lemon S. M., Gale M., Jr. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hausmann S., Marq J. B., Tapparel C., Kolakofsky D., Garcin D. (2008) PLoS One 3, e3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T. S., Fujita T., Akira S. (2008) J. Exp. Med. 205, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Binder M., Kochs G., Bartenschlager R., Lohmann V. (2007) Hepatology 46, 1365–1374 [DOI] [PubMed] [Google Scholar]
- 23. Barchet W., Wimmenauer V., Schlee M., Hartmann G. (2008) Curr. Opin. Immunol. 20, 389–395 [DOI] [PubMed] [Google Scholar]
- 24. Cui S., Eisenächer K., Kirchhofer A., Brzózka K., Lammens A., Lammens K., Fujita T., Conzelmann K. K., Krug A., Hopfner K. P. (2008) Mol. Cell 29, 169–179 [DOI] [PubMed] [Google Scholar]
- 25. Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. (2008) Mol. Cell 29, 428–440 [DOI] [PubMed] [Google Scholar]
- 26. Lu C., Ranjith-Kumar C. T., Hao L., Kao C. C., Li P. (2010) Nucleic Acids Res. 39, 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., Hartmann G., Patel D. J. (2010) Nat. Struct. Mol. Biol. 17, 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Myong S., Cui S., Cornish P. V., Kirchhofer A., Gack M. U., Jung J. U., Hopfner K. P., Ha T. (2009) Science 323, 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sumpter R., Jr., Loo Y. M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S. M., Gale M., Jr. (2005) J. Virol. 79, 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li K., Chen Z., Kato N., Gale M., Jr., Lemon S. M. (2005) J. Biol. Chem. 280, 16739–16747 [DOI] [PubMed] [Google Scholar]
- 31. Rivetti C., Guthold M., Bustamante C. (1996) J. Mol. Biol. 264, 919–932 [DOI] [PubMed] [Google Scholar]
- 32. Lu C., Xu H., Ranjith-Kumar C. T., Brooks M. T., Hou T. Y., Hu F., Herr A. B., Strong R. K., Kao C. C., Li P. (2010) Structure 18, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pichlmair A., Schulz O., Tan C. P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., Reis e Sousa C. (2009) J. Virol. 83, 10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gee P., Chua P. K., Gevorkyan J., Klumpp K., Najera I., Swinney D. C., Deval J. (2008) J. Biol. Chem. 283, 9488–9496 [DOI] [PubMed] [Google Scholar]
- 35. Uzri D., Gehrke L. (2009) J. Virol. 83, 4174–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr. (2008) Nature 454, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.