Abstract
The RING finger E3 ubiquitin ligase Siah2 is implicated in control of diverse cellular biological events, including MAPK signaling and hypoxia. Here we demonstrate that Siah2 is subject to regulation by the deubiquitinating enzyme USP13. Overexpression of USP13 increases Siah2 stability by attenuating its autodegradation. Consequently, the ability of Siah2 to target its substrates prolyl hydroxylase 3 and Spry2 (Sprouty2) for ubiquitin-mediated proteasomal degradation is attenuated. Conversely, inhibition of USP13 expression with corresponding shRNA decreases the stability of both Siah2 and its substrate Spry2. Thus, USP13 limits Siah2 autodegradation and its ubiquitin ligase activity against its target substrates. Strikingly, the effect of USP13 on Siah2 is not mediated by its isopeptidase activity: mutations in its ubiquitin-binding sequences positioned within the ubiquitin-specific processing protease and ubiquitin-binding domains, but not within putative catalytic sites, abolish USP13 binding to and effect on Siah2 autodegradation and targeted ubiquitination. Notably, USP13 expression is attenuated in melanoma cells maintained under hypoxia, thereby relieving Siah2 inhibition and increasing its activity under low oxygen levels. Significantly, on melanoma tissue microarray, high nuclear expression of USP13 coincided with high nuclear expression of Siah2. Overall, this study identifies a new layer of Siah2 regulation mediated by USP13 binding to ubiquitinated Siah2 protein with a concomitant inhibitory effect on its activity under normoxia.
Keywords: Deubiquitination, Hypoxia, Protein Degradation, Protein-Protein Interactions, Ubiquitin, Ubiquitination, Siah2, Ubc13
Introduction
Siah proteins are RING finger E3 ubiquitin ligases implicated in the ubiquitination and proteasome-dependent degradation of substrate molecules, which also limit their own availability through self-ubiquitination and degradation (1–3). Siah was first identified in Drosophila melanogaster as seven in absentia (sina), which regulates formation of the R7 photoreceptor through control of tramtrack stability (4, 5). Three murine Siah genes (Siah1a, Siah1b, and Siah2) share significant homology with the two human (SIAH1 and SIAH2) orthologues (6, 7).
Siah2 is an important regulator of pathways activated under stress and hypoxia. Among substrates reportedly regulated by Siah under these conditions are TRAF2, α-ketoglutarate dehydrogenase, Spry2 (Sprouty2), and two of the three prolyl hydroxylases (8–10). Given the role of PHD proteins as oxygen sensors and regulators of HIF1α, the master transcription factor responding to hypoxia, Siah is implicated in the control of hypoxia, particularly within the range of 2–6% oxygen at which PHD proteins retain sufficient activity (11, 12). The role of Siah2 in tumor development has been extensively studied. Inhibition of Siah reportedly attenuates breast, pancreatic, lung, prostate, and melanoma tumors (13–16). Mechanistically, Siah2 contributes to tumor development and metastasis via its control of diverse substrates, as shown in a melanoma model (14).
A search for novel Siah2 interacting factors led to identification of the isopeptidase USP13 (17). This protease shares a 54.8% identity with its isoform USP5 (18, 19). Both proteins contain four ubiquitin-associated (UBA)2 domains and a ubiquitin-specific processing protease (UBP) domain. The UBP domain of USP13 harbors a catalytic site, a zinc finger domain, and two UBA domains. Sequence alignment of USP13 and other UBA-containing proteins revealed significant homology with the DNA repair protein Rad23 and the E3 ligase Cbl-b. Interestingly, overexpression of the UBA domain of Cbl-b stabilized ubiquitinated Siah1 (20).
Similar to other thiol proteases, USP13 contains consensus cysteines and histidines, which likely function in catalysis. Whereas USP5 is implicated as a specialized deubiquitinating enzyme responsible for disassembly of unanchored polyubiquitin chains in vivo (19), the function of USP13 is unknown. Although USP13 contains all four putative ubiquitin-binding domains present in USP5, it has not been shown to hydrolyze polyubiquitin (21) but rather serves as a protease for ISG15, whereby it reduces ISGylation (21). Little is known about the substrate specificity of USP13.
We show here that USP13 binds to Siah2 primarily through its UBA domains, thereby increasing Siah2 stability while attenuating its activity. Notably, we observed that reduced USP13 expression, as seen in melanoma cell lines under hypoxia, contributes to increased Siah2 activity.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
HeLa and 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with calf serum (10%) and antibiotics. Melanoma cell lines Mel501, UACC-903, and Lu1205 were maintained in Dulbecco's modified Eagle's medium supplemented with fetal bovine serum (10%) and antibiotics. Cell cultures were maintained at 37 °C in 5% CO2. Antibodies against the FLAG epitope, β-actin, and USP13 were purchased from Sigma. Anti-HA, anti-Myc, anti-Sprouty, anti-GFP, and anti-Siah2 were purchased from Santa Cruz. Polyclonal HIF-1α antibody was a generous gift from Dr. Gary Chiang. Cycloheximide and MG132 were purchased from Sigma. All of the USP13 point mutants were prepared using a QuikChange site-directed mutagenesis kit (Stratagene). The jetPRIME DNA transfection reagent was purchased from Genesee Scientific. Human USP13 in pcmv6-xl4 vector was purchased from Origene.
Hypoxia Treatment
Cells were exposed to hypoxia (2% O2) in a hypoxia workstation (In Vivo 400; Ruskin Corp.) and then processed immediately on ice.
Plasmids and Transfection
The following mutants were generated with the indicated primers in site-directed mutagenesis of USP13 plasmid template: SalUSP13F, GTCGACATGCAGCGCCGGGGCGCCCTGTTC; NotUSP208R, GCGGCCGCACCACTTGGAGGAATCCTGACTCC;, SalUSP209F, GTCGACTGGAAGTGTGCCAGATGCGAC; NotUSP640R, GCGGCCGCTTTTGAGTCATCAGGAATGACTATGGGGG; SalUSP641F, GTCGACGATCGCCTGATGAACCAATTG; and NotUSP863R, GCGGCCGCGCTTGGTATCCTGCGGTAAAAGTAC. The mutations were confirmed by DNA sequencing. Human USP13 in pcmv6-xl4 vector was subcloned into pcDNA3 vector with a FLAG or Myc tag and into pET-28a. Truncated forms of USP13 were generated by PCR and cloned into pcDNA3 vector with a Myc tag. FLAG-tagged, Myc-tagged, and HA-tagged mouse Siah2 and FLAG-tagged Siah2 RING mutant in pcDNA3.1 vector have been described previously (10). GST-Siah2 and GST-Siah2 RING mutant were expressed in pGEX-4T1 vector. The cells were transfected using the jetPRIME DNA transfection reagent kit according to the manufacturer's protocol.
Immunoprecipitation and Western Blotting
The cells were harvested in lysis buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA, 1 mm sodium orthovanadate, 1 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A. To immunoprecipitate FLAG-tagged proteins, lysates were incubated with M2 beads (Sigma) overnight, beads were washed three times, and precipitated proteins were eluted with 1 mg/ml of FLAG peptide. To extract whole cell lysates, the cells were harvested using assay buffer (50 mm Tris-HCl pH7.5, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mm EDTA, 1 mm sodium orthovanadate, 1 mm PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Cell lysates were subjected to SDS-PAGE, and proteins were transferred onto a nitrocellulose membrane (Osmonics Inc.). The membrane was probed with primary antibodies followed by a secondary antibody conjugated with fluorescent dye and detected by the Odyssey detecting system (Amersham Biosciences).
In Vitro Ubiquitination and Deubiquitination Assay
GST-Siah2 was purified from the bacteria using glutathione-Sepharose (Amersham Biosciences). USP13 protein was obtained from 293T cells transfected with FLAG-USP13. After 24 h, FLAG-USP13 was immunoprecipitated and eluted with 1 mg/ml of FLAG peptide. Purified GST-Siah2 attached on glutathione beads was subjected to an in vitro ubiquitination assay in ubiquitination buffer (50 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 0.5 mm dithiothreitol, 2 mm NaF) supplemented (or not) with purified ubiquitin (2 μg), 2 mm ATP, E1 (50 ng) (Boston Biochem, Cambridge, MA), purified E2 (UbcH5b) (250 ng) for 45 min at 37 °C. The reactions were washed four times with buffer containing (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA). GST-Siah2 bound to beads was then incubated in deubiquitination reaction buffer (25 mm Tris, pH 7.5, 150 mm NaCl, 1 mm DTT) for deubiquitination assay with or without the eluted USP13 for 1 h at 37 °C. After washing four times, GST-Siah2 was eluted with SDS loading buffer and subjected to Western blot analysis with anti-ubiquitin antibody. The same membrane was reprobed with anti-GST antibody.
In Vivo Ubiquitination Assay
293T cells were transfected with 3 μg of FLAG-Siah2 and/or USP13. 24 h later, the cell pellets were lysed, and FLAG-Siah2 was immunoprecipitated as described previously (22). Bead-bound proteins were then eluted in SDS sample buffer and immunoblotted with anti-ubiquitin antibody. The same membrane was reprobed with anti-FLAG antibody.
For analysis of Siah2 and Skp2 deubiquitination by USP13, 293T cells were transfected with 3 μg of FLAG-USP13, with ubiquitin and Myc-Siah2, or with ubiquitin and Myc-Skp2. 24 h after transfection, FLAG-USP13 was immunoprecipitated and eluted with 1 mg/ml of FLAG peptide. Cells transfected with Myc-Siah2 or Myc-Skp2 were treated with MG132 (20 μm) for 3 h prior to protein preparation. Myc-Siah2 or Myc-Skp2 were immunoprecipitated by incubating the cells lysates with anti-Myc antibody (3 μg) for 3 h, followed by incubation with protein A/G beads for 1 h. The immunoprecipitated Myc-Siah2 or Myc-Skp2 bound to the beads were incubated in a reaction buffer (25 mm Tris, pH 7.5, 150 mm NaCl, 1 mm DTT) with or without the eluted USP13 for 1 h at 30 °C. After washing four times, the precipitated Myc-Siah2 and Myc-Skp2 were eluted with SDS loading buffer and subjected to Western blot analysis with anti-ubiquitin antibody. The same membrane was reprobed with anti-Myc antibody.
In Vitro Ubiquitination Assay and GST Pulldown
GST-Siah2 and GST-Siah2 RING mutant were purified from the bacteria using glutathione-Sepharose (Amersham Biosciences). His-USP13 was expressed and purified from the bacteria using Ni2+-nitrilotriacetic acid-agarose as described previously (23). GST-Siah2 and GST-Siah2 RING mutant attached on glutathione beads were subjected to an in vitro ubiquitination assay in ubiquitination buffer (50 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 0.5 mm dithiothreitol, 2 mm NaF) supplemented with ubiquitin (2 μg), 2 mm ATP, E1 (50 ng) (BostonBiochem, Cambridge, MA), and purified E2 (UbcH5b) (250 ng) for 45 min at 37 °C. GST-fused proteins attached on glutathione beads were washed four times with washing buffer (50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 1% Triton X-100, 1 mm EDTA). At this point HIS-USP13 purified proteins were mixed with the indicated GST fusion proteins and incubated overnight at 4 °C with shaking. The beads were washed extensively in washing buffer and eluted with sample buffer for loading on to the gel.
Reverse Transcription PCR and Real Time PCR
Total RNA was extracted using a total RNA miniprep kit (Sigma) and digested with DNase I. cDNA was synthesized using oligo(dT) and random hexamer primers for SYBR Green quantitative PCR analysis. H3.3A served as an internal control. Triplicate samples were used for quantitative PCR analysis. PCR primers were designed using Primer3, and their specificity was verified by BLAST searches. PCR products were limited to 100–200 bp. The primers used were: hH3.3AR, AAGCAGACTGCCCGCAAAT; hH3.3AR, GGCCTGTAACGATGAGGTTTC; HUSP13F, CCTTCTCCTACGACTCTCCCA; and HUSP13R, CAGACGCCCCTCTTACCTTCT.
shRNA Transfection
USP13 shRNAs were purchased from Sigma (Sigma MISSION pLKO.1-Puro3 shRNA bacterial glycerol stock). shRNAs and negative controls were introduced into HeLa or Lu1205 cells using jetPRIME as described previously (25).
Tissue Microarray Analysis
Melanoma tissue microarrays have been described previously (24) and were processed as in Ref. 25 with minor modifications for immunofluorescent staining. Melanoma tissue microarrays were rehydrated and processed for immunohistochemistry. Antigen retrieval was performed using Dako target retrieval solution, followed by peroxidase block for 30 min with 3% hydrogen peroxide. Slides were incubated with mouse Siah2 (1:300 dilution) or rabbit USP13 (1:100 dilution) antibody diluted in Dako antibody diluent overnight at 4 °C. After incubation, the slides were washed three times with PBS/Tween-20 and incubated with Dako Labeled Polymer-HRP (anti-mouse and anti-rabbit) for 1 h at RT. The slides were then washed four times with PBS/Tween-20, developed with SG, and counterstained with nuclear Fast Red. Immunostaining performed in the absence of primary antibodies was used as negative controls. Scoring of TMA staining was performed by a board certified dermatopathologist. Nuclear and cytoplasmic compartments were scored separately. Staining intensity was scored on a scale of 0 to 3 with the following designations: 0, no staining; 1, weak intensity; 2, moderate intensity; and 3, intense staining.
RESULTS
USP13 Binds to and Stabilizes Siah2
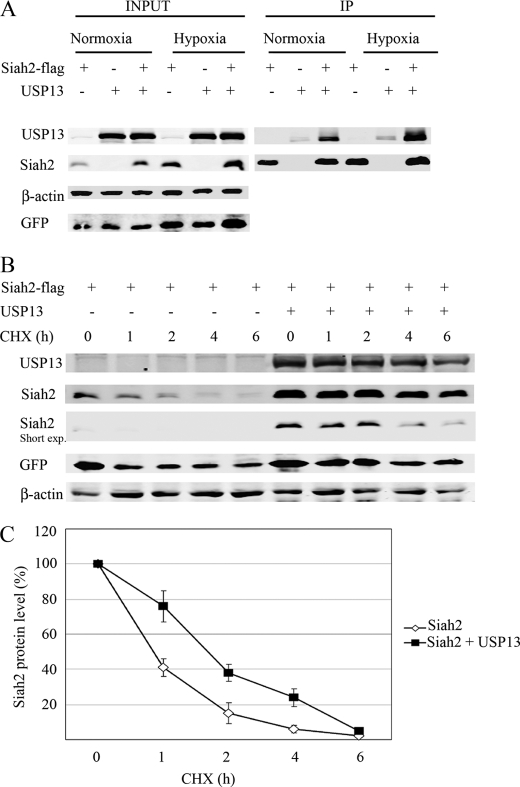
In studying the ubiquitin ligase Siah2, we have assessed possible post-translational modifications that would affect its E3 ligase activity. Phosphorylation of Siah2 was shown to affect its subcellular localization and activity (26, 27). Here we asked whether Siah might also be regulated by a deubiquitinating enzyme, a mechanism reported for several RING finger E3 ligases (28, 29). To do so, we evaluated whether Siah2 protein expression was altered following ectopic expression of different deubiquitinating enzymes. Among deubiquitinating enzymes found to alter Siah2 expression was USP13. To determine whether Siah2 and USP13 interacted, we made lysates of 293T cells co-expressing FLAG-tagged wild-type Siah2 and USP13 and immunoprecipitated FLAG-tagged Siah2 followed by immunoblotting with an antibody against USP13. This analysis revealed an interaction between the two proteins both in normoxic conditions and in cells that were maintained in hypoxia (2% O2) (Fig. 1A). To determine whether USP13 altered Siah2 stability, we monitored Siah2 half-life in the presence of the protein synthesis inhibitor cycloheximide and found that USP13 expression in 293T cells prolonged the half-life of Siah2 from 50 to 90 min (Fig. 1, B and C). Like other RING finger E3 ubiquitin ligases, Siah2 limits its own expression by autodegradation; therefore, the ability of USP13 to increase Siah2 stability implies that it may modulate Siah2 degradation and targeted ubiquitination.
FIGURE 1.
USP13 binds to and stabilizes Siah2. 293T cells were transfected with FLAG-Siah2, USP13, and a GFP expression vector. Total cell lysates were analyzed for GFP expression to assess transfection efficiency. A, cells were kept 6 h in normoxia or in hypoxia (2% O2) before cell lysis. FLAG-Siah2 was immunoprecipitated (IP) with FLAG antibody, and immunocomplexes were subjected to immunoblotting with the indicated antibodies. B, cycloheximide (CHX) chase of Siah2 in the absence and presence of USP13. 293T cells transfected with FLAG-Siah2 and USP13 were treated with the protein synthesis inhibitor cycloheximide (40 μg/ml) for 1, 2, 4, and 6 h, and cell lysates were subjected to Western blot to detect FLAG-Siah2. β-Actin served as the loading control. C, degradation curves of Siah2 by cycloheximide chase in the presence or absence of USP13. The graph represents the mean value of band density in 3 experiments.
USP13 Decreases Siah2 Ubiquitin Ligase Activity
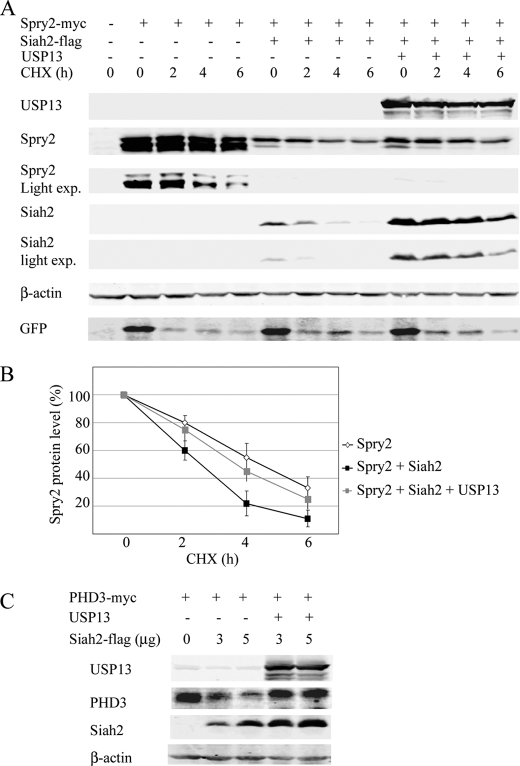
To determine whether USP13 altered Siah2 function as an E3 ubiquitin ligase, we monitored the effect of Siah2 on degradation of known substrates in the presence of USP13. To this end, we monitored changes in Spry2 (Fig. 2A), which binds directly to Siah2, and in PHD3 (Fig. 2C), a substrate that requires an adaptor protein for interaction with Siah2 (10). Whereas Siah2 expression effectively reduced Spry2 half-life from 4 h to about 2 h (Fig. 2B), co-expression of USP13 prolonged Spry2 half-life to 3 h (Fig. 2B). Similarly, steady state levels of PHD3 protein were higher when USP13 was co-expressed with Siah2 (Fig. 2C). These results suggest that USP13 attenuates the E3 ligase activity of Siah2.
FIGURE 2.
USP13 decreases Siah2 ubiquitin ligase activity. A, 293T cells were transfected with an expression vector encoding GFP and Myc-Spry2 alone or with FLAG-Siah2 and or USP13. Total cell lysates were analyzed for GFP expression to assess transfection efficiency. The cells were treated with the protein synthesis inhibitor cycloheximide (CHX, 40 μg/ml) for 2, 4, and 6 h, and the cell lysates were subjected to Western blotting for detection of Myc-Spry2, FLAG-Siah2, and USP13. exp., exposure. B, degradation curves of Spry2 by cycloheximide chase in the presence of Siah2 alone or with USP13. The graph represents the mean value of band density in 3 experiments. C, 293T cells were transfected with Myc-PHD3 and increasing amounts of FLAG-Siah2 in the presence or absence of USP13. Total cell lysates were probed for the presence of USP13, Myc-PHD3, FLAG-Siah2, and β-actin.
USP13 Knockdown Decreases Siah2 Protein Level and Increases Siah2 Activity
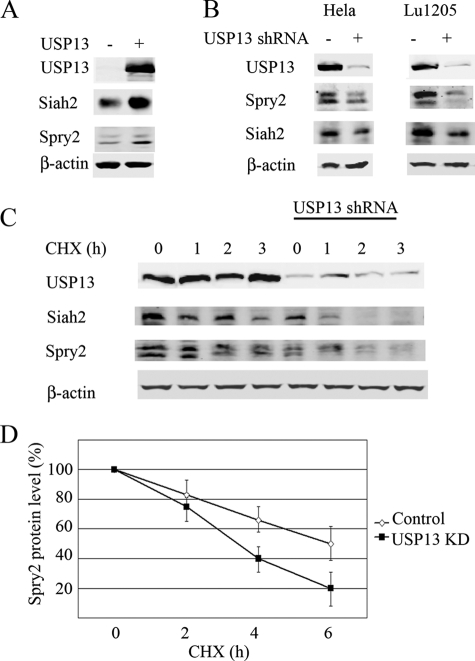
We next assessed the effect of USP13 on endogenous Siah2 protein. The specificity of Siah2 antibodies used in this study was confirmed using MEFs from Siah2 wild-type and Siah2 knock-out mice (30) (supplemental Fig. S1). Exogenous expression of USP13 resulted in increased expression of both endogenous Siah2 and Spry2 (Fig. 3A). To analyze this effect in loss-of-function studies, we previously constructed several shRNAs targeting USP13 and selected one that effectively inhibited USP13 expression at the RNA and protein levels (supplemental Fig. S2). The efficiency of USP13 knockdown was previously validated by quantitative RT-PCR and immunoblotting (25). Analysis of different USP13-KD by different USP13 shRNAs confirmed the major biochemical phenotypes (supplemental Fig. S2). Cells transfected with empty pLKO.1-Puro3 vector served as control. We then assessed changes in Siah2 and its substrates in HeLa and melanoma (Lu1205) cell lines in which USP13 expression was inhibited. In both lines we observed a decrease in steady state levels of Siah2 protein level (Fig. 3B). Interestingly, shUSP13 expression also decreased Spry2 levels (Fig. 3B). To confirm that this decrease is due to changes in its stability, we determined Spry2 half-life in shUSP13-expressing cells and found that it was reduced from 6 h in untreated cells to 3 h in USP13 knockdown cells (Fig. 3, C and D). This observation implies that Siah2 is more active in cells in which USP13 expression is inhibited and is consistent with the notion that active Siah2 limits its own availability by autodegradation and hence is less stable. These observations also agree with earlier reports demonstrating that Siah2 self-degradation requires its ubiquitin ligase activity (1, 3). We next assessed Siah2 half-life in 293T cells that were transfected with WT or RING mutant forms of Siah2, in the presence or absence of USP13. The half-life of Siah2 WT but not RING mutant was prolonged by USP13 (supplemental Fig. S6). These findings support the short half-life of Siah2, dependent of its intact RING domain, consistent with its self-degradation capacity (10).
FIGURE 3.
USP13 knockdown decreases Siah2 protein levels and increases Siah2 activity. A, HeLa cells were transfected with Myc-USP13 and endogenous levels of Siah2 and Sprouty2 from total cell lysates were detected by Western blot analysis. B, HeLa and melanoma Lu1205 cells were transfected with control shRNA or shRNA targeting USP13. Endogenous Siah2 and Spry2 from total cell lysates were detected by Western blot analysis. C, cycloheximide (CHX) chase of Siah2 in cells with USP13 knocked down. Following the addition of cycloheximide cells were harvested after 1, 2, and 3 h, and cell lysates were subjected to Western blot to detect Spry2 and Siah2. β-Actin served as the loading control in A–C. D, degradation curves of Spry2 by cycloheximide chase in control or USP13-KD. The graph represents the mean value of band density in 3 experiments..
USP13 UBA Domains Bind Siah2
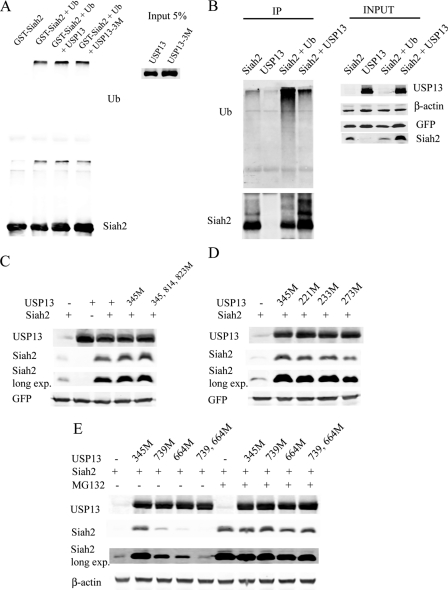
We next undertook experiments designed to determine how USP13 regulates Siah2 self-degradation and targeted ubiquitin ligase activity. Given that USP13 is an isopeptidase, we asked whether it exhibited deubiquitinating activity, thus altering ubiquitin chains formed on Siah2. To this end, we monitored degree of ubiquitin chains coupled to Siah2 in the presence and absence of USP13, both in vivo and in vitro. In vitro ubiquitination of Siah2 was performed using GST-Siah2 protein bound to glutathione beads followed by incubation with immunopurified FLAG-USP13 (see “Experimental Procedures”). Based on homology of the USP13 catalytic site with that of USP5 and other USP (19, 31), we mutated putative catalytic sites C345A, H814A, and H823A in USP13 (supplemental Fig. S3). The length of ubiquitin chains conjugated to Siah2 was not altered in the presence of wt or triple mutant (C345A,H814A,H823A) USP13 in vitro (Fig. 4A). We next undertook in vivo assessment in 293T cells transfected with FLAG-Siah2 alone or with Myc-USP13. Siah2 co-transfected with HA-ubiquitin served as a positive control for ubiquitination. Although USP13 expression increased Siah2 levels, it did not alter the level of ubiquitin chains bound to Siah2 (Fig. 4B), consistent with observations made in vitro. To further investigate USP13 catalytic function, we compared the level of Siah2 ubiquitination with that of Skp2, which was found, in our independent studies, to be affected by USP13 catalytic activity (25). Although USP13 decreased Skp2 ubiquitination, it did not alter the degree of Siah2 ubiquitination (supplemental Fig. S5). These data suggest that USP13 modulation of Siah2 stability is independent of its isopeptidase activity.
FIGURE 4.
USP13 regulates Siah2 protein stability via noncatalytic UBA domains. A, in vitro ubiquitination reactions for GST-Siah2 followed by deubiquitination assay in presence of USP13. The ubiquitination was monitored by Western blot analysis using anti-ubiquitin antibody. The amount of GST-Siah2 used in these reactions is shown in the lower panel using anti-Siah2 antibody. Ub, ubiquitin. B, 293T cells were transfected with FLAG-Siah2/Myc-USP13 and, where indicated, with a ubiquitin expression construct and lysed by a heat-denaturing method. Protein extracts were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with anti-ubiquitin antibody. The amount of immunoprecipitated Siah2 was determined using anti-FLAG antibody. C and D, cells were co-transfected with a GFP-expressing vector, FLAG-Siah2, and USP13 wild-type or mutant constructs (M). After 24 h, the steady levels of FLAG-Siah2 and USP13 were detected with corresponding antibodies. Total cell lysates were analyzed for GFP expression to assess transfection efficiency. exp., exposure. E, cells were co-transfected with FLAG-Siah2 and USP13 wild-type or mutant constructs (M) for 24 h. Where indicated, the cells were incubated with MG132 (20 μm) for an additional 4 h. Cell lysates were prepared, and USP13 and FLAG-Siah2 levels were monitored by immunoblotting with anti-FLAG and anti-USP13 antibodies. β-Actin served as the loading control.
We then assessed the effects of a single mutant (C345A), a triple mutant (C345A,H814A,H823A) USP13 and wild-type USP13 on Siah2 steady state expression levels. Notably, both single (C345A) and triple (C345A,H814A,H823A) mutant USP13 constructs affected Siah2 expression similar to that seen upon expression of wild-type USP13 (Fig. 4C). These observations further suggests that USP13 effect on Siah2 stability is elicited independent of its catalytic activity.
We then asked whether USP13 alters Siah2 activity by binding to its ubiquitin chains through its UBP or UBA domains. To do so we mutated residues 221, 233, and 273 within the USP13 UBP (supplemental Fig. S3), which correspond to USP5 ubiquitin-binding residues. Expression of the catalytic inactive (C345M) mutant or two of the UBP single mutants (W221A or K233A) did not markedly change Siah2 expression levels (Fig. 4D). However expression of USP13 F273A mutant promoted a 2-fold decrease in Siah2 protein level compared with wild-type USP13 (Fig. 4D), suggesting a partial involvement of the USP13-UBP domain in Siah2 stability.
Strikingly, mutations within the UBA domains (supplemental Fig. S3), which were previously shown to bind ubiquitin in USP5, abolished the USP13 effect on Siah2. Expression of USP13 single mutants (M664E or M739E) partially attenuated the ability of USP13 to stabilize Siah2, whereas the M664E,M739E double mutant no longer affected Siah2 stability (Fig. 4E). These data suggest that the two UBA domains of USP13 bind to ubiquitinated Siah2 and may block its proteasomal degradation.
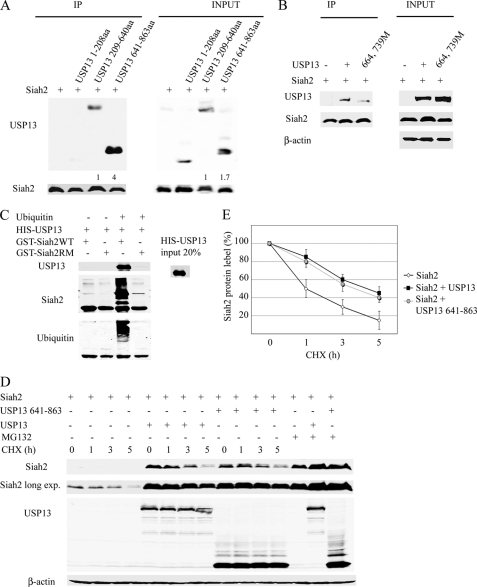
To confirm that Siah2 binds to USP13 UBA and UBP domains, we generated three USP13 deletion mutants: the N terminus (amino acids 1–208), the central part containing the UBP domain (amino acids 209–640), and C-terminal region containing both UBA domains (amino acids 641–863). Co-expression of each with Siah2 in 293T cells revealed strong interaction between Siah2 and the C-terminal USP13 domain containing the UBA domains and a weaker interaction between Siah2 and the central part, which can also bind ubiquitin (Fig. 5A). Only after a longer exposure, we could detect a very faint interaction between Siah2 and the amino terminal part (data not shown).
FIGURE 5.
USP13 UBA domains and to a lesser extent the UBP domain bind to Siah2. A, 293T cells were co-transfected with Siah2 and three deletions USP13 encoding its N-terminal (amino acids (aa) 1–208), central part (amino acids 208–640), and C-terminal (amino acids 641–863) for 24 h and incubated with MG132 (20 μm) for an additional 4 h. The cell lysates were prepared, Siah2 was immunoprecipitated (IP) with anti-FLAG antibody, and the levels of FLAG-Siah2 and USP13 were monitored by immunoblotting with anti-FLAG and anti-USP13 antibodies. Relative amounts of USP13 were quantified by ImageJ software. B, cells were co-transfected with FLAG-Siah2 and USP13 wild-type or double mutant (at positions 664 and 739) forms for 24 h and then incubated with MG132 (20 μm) for an additional 4 h. Siah2 proteins were immunoprecipitated with anti-FLAG antibody, and immunocomplexes were subjected to immunoblotting with anti-USP13 antibody. C, GST-Siah2 and GST-Siah2 RING mutant (GST-Siah2RM) attached to the glutathione beads were subjected or not to in vitro ubiquitination, followed by GST immunoprecipitation in the presence of HIS-USP13. Western blot analysis was performed using anti-USP13 and anti-ubiquitin antibodies. Western blot with Siah2 antibodies revealed the amount of ubiquitin ligases used in these reactions. D, cells were co-transfected with FLAG-Siah2 and either full-length Myc-USP13 or the C-terminal part of Myc-USP13 (amino acids 641–863). Following addition of cycloheximide (CHX) for 1, 3, and 5 h, the cells were harvested, and the cell lysates were subjected to Western blot to detect USP13 and Siah2. In A–C, β-actin serves as the loading control. E, Siah2 degradation curves by cycloheximide chase alone or in the presence of full-length USP13 or the C-terminal deletion mutant. The graph represents the mean value of band density in 3 experiments.
To assess whether mutations in the USP13 UBA domains impaired USP13 binding to Siah2, we transiently expressed Siah2 with either wild-type or the M664E-M739E double mutant form of USP13. Notably, association of Siah2 with the double mutant was markedly reduced (Fig. 5B), suggesting that these residues are required for association with Siah2 and that USP13 binds to Siah2 ubiquitin chains. To verify that only ubiquitinated Siah2 binds to USP13, GST-Siah2 and GST-Siah2RM were subjected or not to ubiquitination in vitro and used in a GST pulldown assay with HIS-USP13 protein. Only ubiquitinated Siah2 could bind to USP13 (Fig. 5C).
Notably, expression of the USP13 C-terminal containing the UBA domains was sufficient to stabilize Siah2 protein, albeit less efficiently that wild-type USP13 (Fig. 5D). Co-expression of USP13 C-terminal or USP13 full-length prolonged Siah2 half-life from 2 to 5 h (Fig. 5E).
Hypoxia-reduced Expression of USP13 Correlates with Increased Siah2 Activity
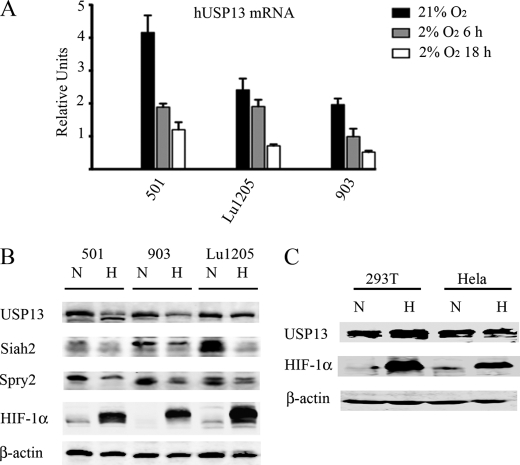
The finding that USP13 regulates Siah2 stability and activity led us to explore possible physiological conditions in which USP13 affects Siah2. Given our earlier observations that Siah2 is more active under hypoxia (26), we assessed possible changes in Siah2 regulation by USP13 under low oxygen concentrations. Notably, quantitative PCR analysis revealed a decrease in USP13 transcript levels in different melanoma cells maintained under hypoxia for 6–24 h (Fig. 6A), which was confirmed at the protein level (Fig. 6B). Decreased USP13 expression seen in cells maintained for 24 h under hypoxia coincided with reduced expression of Siah2 as well as reduced levels of its substrate Spry2 (Fig. 6B). However, not all cell lines tested presented decreased USP13 under hypoxia (Fig. 6C and supplemental Fig. S4), suggesting that USP13 down-regulation in hypoxia may be cell type-dependent. These findings indicate that USP13 down-regulation in hypoxic conditions contributes to increased Siah2 activity. These data also points to a novel mechanism of Siah2 regulation under hypoxic conditions.
FIGURE 6.
Reduced USP13 expression and increased Siah2 activity in melanoma cells under hypoxia. A, quantitative real time PCR detection of relative transcription levels of USP13 mRNA in different melanoma cell lines in normoxia and after 6 and 18 h in hypoxia (2% O2). All of the values were calculated using the ΔΔCt method as relative quantity value, using histone 3.3A as internal control. The results are shown as the means ± S.E. of three independent experiments. B, melanoma cell lines 501, UACC-903, and Lu1205 were kept in normoxic or subjected to hypoxic (2% O2) conditions for 24 h. The cells were harvested, and cell lysates were subjected to Western blot analysis to detect USP13, Siah2, Spry2, and HIF-1α. β-Actin served as the loading control. C, 293T and HeLa cells were kept in normoxic or subjected to hypoxic (2% O2) conditions for 24 h. The cells were harvested, and cell lysates were subjected to Western blot analysis to detect USP13 and HIF1α. β-Actin served as the loading control.
USP13 Expression Coincides with Siah2 Expression in Melanoma TMA
To determine the significance of UPS13 effect on Siah2 expression under hypoxia, we have subjected a large cohort TMA consisting of >500 melanoma samples for analysis of Siah2 and USP13 expression.
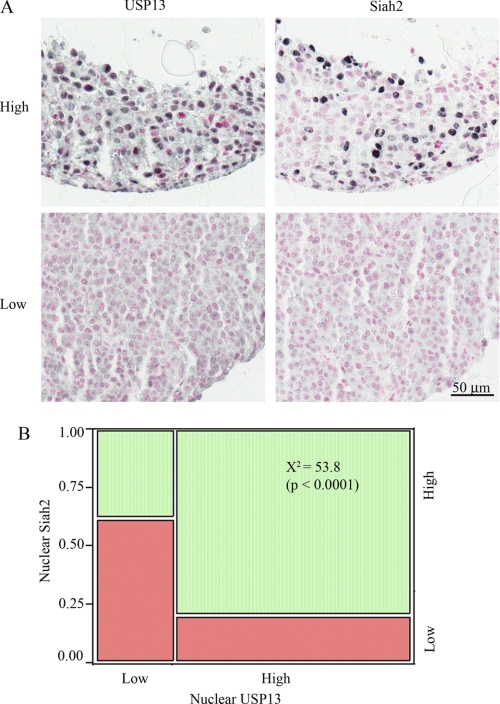
USP13 expression was either cytoplasmic or nuclear, whereas Siah2 expression was primarily nuclear. Examples of strong nuclear staining are shown in Fig. 7A. High nuclear expression of USP13 was found to correlate with higher expression of Siah2 (p < 0.0001), as shown in Fig. 7B. However, USP13 and nuclear Siah2 staining were not associated with survival in either the primary or metastatic cohorts.
FIGURE 7.
High nuclear expression of USP13 correlates with higher expression of Siah2 in melanoma tumors. A, melanoma tissue microarrays were stained with a mouse Siah2 antibody or a rabbit USP13 antibody, followed by HRP-conjugated anti-mouse or anti-rabbit antibody, respectively. The immunostaining was visualized with SG (black/blue), and the nuclei were counterstained with nuclear fast red (pink). B, chi-squared analysis of low (red) and high (green) nuclear USP13 (x axis) and nuclear Siah2 expression (y axis) in 371 melanoma histospots. USP13 and Siah2 are strongly co-expressed, χ2 = 53.8 (p < 0.0001).
DISCUSSION
RING finger E3 ubiquitin ligases are known for their capacity to autoubiquitinate, which limits their availability to ubiquitinate target substrates. Evidence for post-translational modification of E3 ligases and the effect of deubiquitinating enzymes has been demonstrated for the Mdm2-MdmX-Hausp-p53 complex (32). Here we demonstrate the effect of the deubiquinating factor USP13 on Siah2 stability and E3 ligase activity our finding reveals that USP13 binding to ubiquitinated Siah2 attenuates Siah2 self-degradation and increases Siah2 stability. Consequently, Siah2 ability to ubiquitinate substrate proteins is reduced, as demonstrated for Spry2 and PHD3, which represent substrates that bind to Siah2 directly or through an adaptor protein, respectively. Notably, increased Siah2 expression seen following USP13 expression coincides with increased expression of its substrates; conversely, USP13 inhibition reduces expression of Siah2 and its substrates. Overall, these findings point to an inverse correlation between Siah2 expression and activity, with the more active form of Siah2 equally potent against its substrates and itself. One would expect that additional factors govern whether Siah2 targets itself or other substrates. For example, MdmX and Daxx are implicated in control of Mdm2/Hausp regulation of p53 or Mdm2 autodegradation (32).
Significantly, USP13 expression is reduced in cells subjected to hypoxia, thereby relieving Siah2 inhibition and increasing its activity. Notably, this change was identified in several melanoma cell lines but not in 293 or HeLa cells, suggesting that control of USP13 and concomitant effects on Siah2 under hypoxia are cell type and tissue-specific. One would expect that physiological or pathological conditions attenuating USP13 expression coincide with elevated Siah2 activity. Indeed, analysis of TMA consisting of over 500 melanoma tumors identified that tumors with high nuclear expression of USP13 also exhibited high expression of Siah2. These findings identify a physiological role for USP13 in the regulation of Siah2 expression. Co-expression of Siah2 and USP13 in nuclei is likely to result in a less active form of Siah, given our finding that USP13 attenuates Siah2 ubiquitin ligase activity while increasing it stability. At the same time, Siah2 expression in cytosol may not be detected given its higher activity, which also limits its stability.
Notably, Siah2 regulation by USP13 does not require USP13 isopeptidase activity: mutation of three putative USP13 catalytic sites had no effect on Siah2. Of note, we have observed that the same mutations attenuate the effect of USP13 on Skp2 (25). Instead, mutation of residues contacting ubiquitin in the zinc finger UBP domain and in the USP13 UBA domains (based on homology with USP5) effectively attenuated the ability of USP13 to affect Siah2. Proteins containing a UBA domain reportedly interact with ubiquitin. Interestingly, the UBA domain of the E3 ubiquitin ligase Cbl-b shows high homology with the USP13 UBA domain, can bind Siah1, inhibiting its degradation (20). Consistent with this finding, the UbL-UBA protein hHR23 reportedly binds to the E3 ligase mdm2 through its UBA domain and blocks p53 degradation (33).
How binding of USP13 to ubiquitin chains conjugated to Siah2 affect its autodegradation and targeted ubiquitination requires further study. Such associations likely impede Siah2 recognition by the proteasome, preventing its degradation. Binding of USP13 to ubiquitinated Siah may also cause structural changes that attenuate ubiquitin transfer to substrates. Notably, USP13 association with ubiquitinated Siah2 does not interfere with Siah2 binding to its substrates, suggesting that targeted ubiquitination might be affected because of altered conformation of this complex. The possibility that ubiquitinated Siah2 may need to be removed before it can elicit ubiquitination of its substrates cannot be excluded.
Overall, the present study identifies a physiologically relevant mechanism for control of the ubiquitin ligase Siah2 by USP13. Reduced USP13 expression under hypoxia alleviates its inhibition of Siah2, enhancing Siah2 autodegradation and targeted ubiquitination. The USP13 effect on Siah2 is mediated via USP13 ubiquitin-binding domains, which recognize ubiquitinated Siah2. USP13 control of Siah2 also reveals an inverse correlation between Siah2 expression and activity.
Supplementary Material
Acknowledgments
We thank Keith Wilkinson for advice and Meera Shah, Meifan Chen, and members of the Ronai lab for discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant CA128814 (to Z. A. R.). This work was also supported by XXXXXX.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- UBA
- ubiquitin-associated
- UBP
- ubiquitin-specific processing protease
- PHD
- prolyl hydroxylase
- TMA
- tissue microarray analysis.
REFERENCES
- 1. Hu G., Fearon E. R. (1999) Mol. Cell. Biol. 19, 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tang A. H., Neufeld T. P., Kwan E., Rubin G. M. (1997) Cell 90, 459–467 [DOI] [PubMed] [Google Scholar]
- 3. Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li S., Li Y., Carthew R. W., Lai Z. C. (1997) Cell 90, 469–478 [DOI] [PubMed] [Google Scholar]
- 5. Boehm J., He Y., Greiner A., Staudt L., Wirth T. (2001) EMBO J. 20, 4153–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Della N. G., Senior P. V., Bowtell D. D. (1993) Development 117, 1333–1343 [DOI] [PubMed] [Google Scholar]
- 7. Hu G., Chung Y. L., Glover T., Valentine V., Look A. T., Fearon E. R. (1997) Genomics 46, 103–111 [DOI] [PubMed] [Google Scholar]
- 8. Habelhah H., Frew I. J., Laine A., Janes P. W., Relaix F., Sassoon D., Bowtell D. D., Ronai Z. (2002) EMBO J. 21, 5756–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nadeau R. J., Toher J. L., Yang X., Kovalenko D., Friesel R. (2007) J. Cell. Biochem. 100, 151–160 [DOI] [PubMed] [Google Scholar]
- 10. Nakayama K., Frew I. J., Hagensen M., Skals M., Habelhah H., Bhoumik A., Kadoya T., Erdjument-Bromage H., Tempst P., Frappell P. B., Bowtell D. D., Ronai Z. (2004) Cell 117, 941–952 [DOI] [PubMed] [Google Scholar]
- 11. Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 12. Stolze I. P., Tian Y. M., Appelhoff R. J., Turley H., Wykoff C. C., Gleadle J. M., Ratcliffe P. J. (2004) J. Biol. Chem. 279, 42719–42725 [DOI] [PubMed] [Google Scholar]
- 13. Qi J., Nakayama K., Cardiff R. D., Borowsky A. D., Kaul K., Williams R., Krajewski S., Mercola D., Carpenter P. M., Bowtell D., Ronai Z. A. (2010) Cancer Cell 18, 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qi J., Nakayama K., Gaitonde S., Goydos J. S., Krajewski S., Eroshkin A., Bar-Sagi D., Bowtell D., Ronai Z. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16713–16718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed A. U., Schmidt R. L., Park C. H., Reed N. R., Hesse S. E., Thomas C. F., Molina J. R., Deschamps C., Yang P., Aubry M. C., Tang A. H. (2008) J. Natl Cancer Inst. 100, 1606–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt R. L., Park C. H., Ahmed A. U., Gundelach J. H., Reed N. R., Cheng S., Knudsen B. E., Tang A. H. (2007) Cancer Res. 67, 11798–11810 [DOI] [PubMed] [Google Scholar]
- 17. Timms K. M., Ansari-Lari M. A., Morris W., Brown S. N., Gibbs R. A. (1998) Gene 217, 101–106 [DOI] [PubMed] [Google Scholar]
- 18. Lacombe T., Gabriel J. M. (2002) FEBS Lett. 531, 469–474 [DOI] [PubMed] [Google Scholar]
- 19. Reyes-Turcu F. E., Horton J. R., Mullally J. E., Heroux A., Cheng X., Wilkinson K. D. (2006) Cell 124, 1197–1208 [DOI] [PubMed] [Google Scholar]
- 20. Davies G. C., Ettenberg S. A., Coats A. O., Mussante M., Ravichandran S., Collins J., Nau M. M., Lipkowitz S. (2004) Oncogene. 23, 7104–7115 [DOI] [PubMed] [Google Scholar]
- 21. Catic A., Fiebiger E., Korbel G. A., Blom D., Galardy P. J., Ploegh H. L. (2007) PLoS One 2, e679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Habelhah H., Laine A., Erdjument-Bromage H., Tempst P., Gershwin M. E., Bowtell D. D., Ronai Z. (2004) J. Biol. Chem. 279, 53782–53788 [DOI] [PubMed] [Google Scholar]
- 23. Russell N. S., Wilkinson K. D. (2005) Methods Mol. Biol. 301, 207–219 [DOI] [PubMed] [Google Scholar]
- 24. McCarthy M. M., DiVito K. A., Sznol M., Kovacs D., Halaban R., Berger A. J., Flaherty K. T., Camp R. L., Lazova R., Rimm D. L., Kluger H. M. (2006) Clin. Cancer Res. 12, 3856–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen M., Gutierrez G. J., Ronai Z. A. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 9119–9124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khurana A., Nakayama K., Williams S., Davis R. J., Mustelin T., Ronai Z. (2006) J. Biol. Chem. 281, 35316–35326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Calzado M. A., de la Vega L., Möller A., Bowtell D. D., Schmitz M. L. (2009) Nat. Cell Biol. 11, 85–91 [DOI] [PubMed] [Google Scholar]
- 28. Brooks C. L., Li M., Hu M., Shi Y., Gu W. (2007) Oncogene 26, 7262–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. (2003) Nature 424, 801–805 [DOI] [PubMed] [Google Scholar]
- 30. Frew I. J., Dickins R. A., Cuddihy A. R., Del Rosario M., Reinhard C., O'Connell M. J., Bowtell D. D. (2002) Mol. Cell. Biol. 22, 8155–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu M., Li P., Li M., Li W., Yao T., Wu J. W., Gu W., Cohen R. E., Shi Y. (2002) Cell 111, 1041–1054 [DOI] [PubMed] [Google Scholar]
- 32. Brooks C. L., Gu W. (2006) Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brignone C., Bradley K. E., Kisselev A. F., Grossman S. R. (2004) Oncogene 23, 4121–4129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.