Abstract
IL-27 induces stronger proliferation of naive than memory human B cells and CD4+ T cells. In B cells, this differential response is associated with similar levels of IL-27 receptor chains, IL-27Rα and gp130, in both subsets and stronger STAT1 and STAT3 activation by IL-27 in naive B cells. Here, we show that the stronger proliferative response of CD3-stimulated naive CD4+ T cells to IL-27 is associated with lower levels of IL-27Rα but higher levels of gp130 compared with memory CD4+ T cells. IL-27 signaling differs between naive and memory CD4+ T cells, as shown by more sustained STAT1, -3, and -5 activation and weaker activation of SHP-2 in naive CD4+ T cells. In the latter, IL-27 increases G0/G1 to S phase transition, cell division and, in some cases, cell survival. IL-27 proliferative effect on naive CD4+ T cells is independent of MAPK, but is dependent on c-Myc and Pim-1 induction by IL-27 and is associated with induction of cyclin D2, cyclin D3, and CDK4 by IL-27 in a c-Myc and Pim-1-dependent manner. In BCR-stimulated naive B cells, IL-27 only increases entry in the S phase and induces the expression of Pim-1 and of cyclins A, D2, and D3. In these cells, inhibition of Pim-1 inhibits IL-27 effect on proliferation and cyclin induction. Altogether, these data indicate that IL-27 mediates proliferation of naive CD4+ T cells and B cells through induction of both common and distinct sets of cell cycle regulators.
Keywords: Cell Cycle, Cell Division, Cellular Immune Response, Cytokine, Cytokine Action, Immunology, Lymphocyte, Signal Transduction, IL-27, Human
Introduction
IL-27 is a heterodimeric cytokine of the IL-12 family composed of two subunits, EBV-induced gene 3 (EBI3) and p28 (1, 2). In humans, IL-27 is expressed at high levels by activated antigen-presenting cells and placental trophoblast cells (2–4). Its receptor comprises two chains, IL-27Rα (also called TCCR or WSX-1) and the shared cytokine receptor signaling subunit gp130 (5). This receptor is expressed by a large variety of immune and non-immune cells resulting in pleiotropic fonctions for IL-27 (6–13).
The first identified biological function for IL-27 was its ability to increase the proliferation of naive, but not memory, murine, and human CD4+ T cells (2). Subsequently, many studies have focused on the role of IL-27 on naive CD4+ T cells and have shown that IL-27 is a key regulator of naive CD4+ T cell differentiation. Depending on the context, IL-27 can promote the initiation of Th1 response via T-bet and IL-12 receptor induction, or inhibit Th2 or Th17 responses by down-regulating GATA3 or RORγt expression, respectively (reviewed in Refs. 14–16). Fewer studies have investigated the role of IL-27 on differentiated CD4+ T cells. Studies performed in mice showed that several biological effects of IL-27 besides proliferation, such as induction of RORγt expression or modulation of IL-17 or IFN-γ production, were differentially regulated in naive and effector/memory CD4+ T cells (17–19). These data suggested that IL-27 signaling varies depending on the stage of murine CD4+ T cell differentiation.
Knowledge on the signaling pathways induced by IL-27 mostly arises from studies performed using murine naive CD4+ T cells. In these cells, IL-27 activates the Jak/STAT pathway, in particular STAT1, STAT3, and STAT5 (20–22). In the IL-27 receptor, IL-27Rα has been shown to constitutively interact with Jak1 and contribute to STAT1 activation, while gp130 that constitutively interacts with Jak1, Jak2, and Tyk2, contributes to STAT3 activation (20, 23, 24). By using murine T cells specifically deficient for either STAT1 or STAT3, STAT3 was shown to be indispensable for IL-27 proliferative effect on murine naive CD4+ T cells, whereas STAT1 was found to be dispensable (22, 24). In addition to Jak/STAT pathway, p38 and erk1/2 MAPKs have been shown to be activated by IL-27 in murine naive CD4+ T cells and involved in IL-27-induced Th1 differentiation (25).
Previously, we have shown that human B cells responded differently to IL-27 depending on their stage of differentiation (8). Thus, although naive and memory tonsillar B cells expressed comparable levels of IL-27 receptor at the cell surface, either constitutively or upon CD40 or Ig stimulation, naive B cells exhibited a stronger response than memory B cells to IL-27. STAT1 and -3 activation by IL-27 was stronger in naive than memory B cells, and several IL-27-mediated biological effects, such as T-bet or cell surface molecule induction, were more robust in activated naive than memory B cells. One striking difference between both B cell subsets was their proliferative response to IL-27, in that IL-27 significantly increased proliferation of naive, but not memory, B cells (8).
The molecular basis underlying the differential response of naive and memory CD4+ T cells to IL-27 has not been previously analyzed. Therefore, in this study, we analyzed the possible cellular events and signaling pathways that could be differentially regulated between naive and memory CD4+ T cells and result in the stronger proliferative response of naive CD4+ T cells to IL-27. Next, we investigated in both naive CD4+ T cells and naive B cells, the mechanisms by which IL-27 regulates their proliferation. We delineated the precise effects of IL-27 on cell survival, cell division, and cell cycle progression, and investigated the effects of IL-27 on the expression of various molecules known to regulate cell proliferation, such as cyclins, cyclin-dependent kinases (CDKs),2 and CDK inhibitors.
EXPERIMENTAL PROCEDURES
Isolation of Human CD4+ T Cell Subsets and Naive B Cells
Naive and memory CD4+ T cell subsets were isolated from peripheral blood mononuclear cells of adult healthy donors (Etablissement Français du Sang, Centre Necker-Cabanel, Paris), as described (26). Total CD4+ T cells were isolated by negative selection by using the CD4+ T cell isolation kit II according to the manufacturer's instructions (Miltenyi Biotech). These cells were further separated into naive or memory CD4+ T cells by incubation with CD45RO microbeads and separation by using a LS column (Miltenyi Biotech). The negative fraction (naive CD4+ T cells) was collected and the positive fraction was passed again over a MS column to enrich in CD45RO+ cells. Naive CD4+ T cells were also purified in a single-step procedure using the naive CD4+ T cell isolation kit II (Miltenyi Biotech). Naive and memory CD4+ T cell fractions contained less than 3% non-CD4+ T cells and showed ≥95% purity based on CD45RA and CD45RO markers. For analysis of IL-27 expression by naive and memory CD4+ T cells, cells were also purified by electronic cell sorting on a FACSAria cytometer (IRNEM cell sorting facility).
Human naive B cells were isolated from pediatric tonsils obtained at Necker Hospital, as previously described (8). Briefly, IgD+ naive B cells were purified by incubation of tonsillar mononuclear cells with anti-IgD mAb (IA6–2, BD Biosciences) followed by goat anti-mouse IgG microbeads (Miltenyi Biotech) and magnetic separation by using a LS column (purity: 95–99% IgD+). In all cases, human samples were obtained with informed consent of the patient and processed in accordance with French ethical guidelines.
T and B Cell Culture
In most cases, naive and memory CD4+ T cells were stimulated with plate (Costar)-coated CD3ϵ mAb (UCHT1, R&D Systems, 3 μg/ml unless otherwise specified) in the absence or presence of soluble CD28 mAb (CD28.2, BD Biosciences, 0.5 μg/ml) in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS, l-glutamine and antibiotics (complete RPMI medium). Alternatively, CD4+ T cell subsets were stimulated with beads (ratio of 1 bead per 2 cells) coated with CD3 mAb (10 μg/ml, T cell activation/expansion kit, Miltenyi Biotec). In some experiments, neutralizing mAbs for IL-2 (MQ1–17H12, BD Biosciences) and IL-2R (B-B10 CD25 mAb, Diaclone) were used at 10 μg/ml each. Recombinant sIL-27Rα-Fc, comprising the extracellular domain of IL-27Rα fused to the Fc region of human IgG1 (TCCR/WSX-1/Fc chimera, R&D Systems), or a control irrelevant human IgG1, were used at 2 μg/ml. Purified B cells (2 × 106 per ml) were cultured in complete RPMI medium in the presence of F(ab′)2 fragment of goat polyclonal anti-human μ Ab (Jackson ImmunoResearch Laboratories, 5 μg/ml unless otherwise specified).
Recombinant IL-27 corresponding to single-chain EBI3p28 fusion protein (Schering-Plough Biopharma) was used at 50 ng/ml. The following chemical inhibitors were used: U0126, SB203580, and SP600125 (Calbiochem), Pim-1 inhibitor 2 (Santa Cruz Biotechnology) and c-Myc inhibitor (10058-F4, Sigma-Aldrich). Because chemical inhibitors were diluted in DMSO, DMSO was added at a similar dilution in the controls. Inhibitors were added 1 h before the addition of IL-27. At the concentrations used, no toxic effect of the inhibitors was observed.
Cell Surface Staining
For cell surface staining, purified naive and memory CD4+ T cells were saturated in PBS containing 20% normal human serum before incubation with specific Abs. All staining and washes were performed in FACS buffer (PBS containing 2% FBS and 0.01% sodium azide). The following PE-conjugated mouse mAbs (all from BD Biosciences) were used: anti-gp130 mAb (AM64), anti-human HLA-A, -B, -C mAb (G46–2.6), CD95 mAb (DX2), and isotype control mAb (MOPC-21). IL-27Rα was detected with anti-TCCR/WSX-1 mAb (191106, R&D Systems) that had been biotinylated with the biotin protein labeling kit from Roche, and PE-conjugated streptavidin (BD Biosciences).
Thymidine Incorporation Assay
Proliferation was measured by assessing DNA synthesis by thymidine incorporation. Naive and memory CD4+ T cells (5 × 104 per well) or B cells (2 × 105 per well) were cultured in triplicate in 96-well plate for 4 or 3 days, respectively. [3H]Thymidine (0.5 μCi per well, Amersham Biosciences) was added for the last 8 h of the last day of incubation. After collecting cells on a filter, [3H]thymidine incorporation was measured in a β-scintillation counter.
Cell Division, Cell Cycle, and Cell Survival Analyses
For measure of cell division, purified B or T cell subsets were labeled at day 0 with Vybrant® CFDA S.E. cell tracer (Invitrogen). Cells (1 × 107 per ml) were incubated with 1 μm Vybrant® CFDA S.E. cell tracer in PBS containing 2% FBS for 5 min at room temperature and washed twice with PBS-10% FBS before culture. After 3 to 5 days of culture, CFSE-labeled cells were analyzed on Canto II (BD Biosciences). For cell cycle analysis, cells (1 × 106 per ml) were incubated at the term of the culture with 10 μm Vybrant® DyeCycle violet stain (Invitrogen) in complete RPMI medium for 30 min at 37 °C and immediately analyzed by flow cytometry on FACSCanto II. To quantify cell survival/apoptosis, cultured T or B cells were washed twice in PBS and double-stained with annexin V-FITC conjugate and propidium iodide (PI) following the manufacturer's instructions (apoptosis detection kit, BD Biosciences) and analyzed on FACSCanto II. Data were analyzed using FlowJo software.
Western Blot Analysis
Cells were washed in ice-cold PBS and lysed for 1 h on ice in lysis buffer (1% Nonidet P-40, 50 mm Tris, pH 7.4, 150 mm NaCl, 3% glycerol, 1.5 mm EDTA) supplemented with protease inhibitors (1 mm PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin) and phosphatase inhibitors (1 mm Na3VO4 and 10 mm NaF). Cell lysate was centrifuged for 15 min at 13,000 × g and the supernatant was assayed for protein concentration using the microBCATM protein assay (Pierce). Cell lysates (from 50 to 100 μg) were subjected to SDS-PAGE and transferred to nitrocellulose for immunoblotting. Blots were incubated with the following primary Abs:rabbit polyclonal Abs specific for phospho-STAT1, phospho-STAT3, phospho-STAT5, phospho-SHP-2, STAT1, STAT5, or SHP-2, all from Cell Signaling Technology; mouse anti-STAT3 mAb (F-2), mouse anti-Pim-1 mAb (12H18), rabbit polyclonal anti-c-Myc Ab (N-262), goat polyclonal anti-cyclin A Ab (C-19), rabbit polyclonal anti-cyclin D3 Ab (C-16), mouse anti-cyclin E mAb, mouse anti-p21 mAb (F-5), goat polyclonal anti-CDK2 Ab (M2), rabbit polyclonal CDK4 Ab (H-22), goat polyclonal CDK6 Ab (C-21) and goat polyclonal anti-actin Ab (I19), all from Santa Cruz Biotechnology; mouse anti-cyclin D2 mAb (G132–43) and mouse anti-p27 mAb (Kip1/p27), both from BD Biosciences. Binding of primary Abs was detected with HRP-conjugated anti-mouse (GE Healthcare) or anti-goat (Santa Cruz Biotechnology) Abs, or HRP-conjugated protein A (GE Healthcare). Peroxidase reaction was developed with chemiluminescence reagents (Pierce).
Real-time Quantitative PCR Analysis (RTqPCR)
Total RNA was isolated by TRIzol extraction followed by DNase I digestion (Invitrogen), or by using the RNeasy Plus Micro kit (Qiagen). RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase and oligo(dT) or random hexamer primers (Invitrogen). RTqPCR was performed using Taqman Universal PCR Master mix and TaqMan® gene expression assays (Applied Biosystems). For each sample, triplicate reactions were run for 40 cycles on 7900HT or StepOne Plus thermal cycler (Applied Biosystems). Levels of target mRNA were normalized relative to levels of β2-microglobulin or large ribosomal protein (RPLPO) mRNA, and relative mRNA expression was calculated using the comparative cycle threshold (CT) method (ΔΔCT method). Only CT values below 36 were considered.
Statistical Analysis
Statistical analysis was performed by paired Student's t test or Mann-Whitney test. A p value < 0.05 was considered to indicate statistical significance.
RESULTS
Analysis of Naive and Memory CD4+ T Cell Proliferative Responses to IL-27
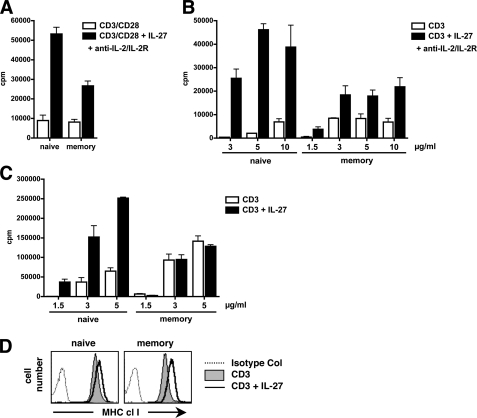
In the IL-27 princeps work, IL-27 was shown to increase proliferation, assessed by thymidine incorporation, of naive, but not memory, human and murine CD4+ T cells, when cells were stimulated with plate-bound CD3 and soluble CD28 mAbs in the presence of neutralizing anti-IL-2 and IL-2R mAbs (2). Using similar experimental conditions, we observed that IL-27 was able to increase the proliferation of naive CD4+ T cells, and also that of memory CD4+ T cells to some extent. However, consistent with the prior findings, IL-27 proliferative effect on memory CD4+ T cells, when observed, was consistently 2–3-fold lower than that observed on naive CD4+ T cells (Fig. 1A). The differential proliferative response of naive versus memory CD4+ T cells to IL-27 was particularly pronounced when suboptimal conditions of stimulation were used, i.e. low doses (1.5 and 3 μg/ml) of plate-coated CD3 mAb only. Under these conditions, IL-27 proliferative effect on memory CD4+ T cells was null or up to over 30-fold lower than that observed in naive CD4+ T cells (Fig. 1, B and C). Upon CD3 stimulation, the selective proliferative effect of IL-27 on naive CD4+ T cells could be observed in the absence of neutralizing anti-IL-2/IL-2R Abs (Fig. 1C).
FIGURE 1.
A–C, effect of IL-27 on naive and memory CD4+ T cell proliferation. Purified naive and memory CD4+ T cells were stimulated for 4 days with plate-coated CD3 mAb (3 μg/ml) and soluble CD28 mAb (0.5 μg/ml) (A) or various concentrations of plate-coated CD3 mAb only (B and C), with (A and B) or without (C) neutralizing anti-IL-2 and IL-2R mAbs, and their proliferation, in the absence or presence of IL-27, was measured by [3H]thymidine incorporation. Results are expressed in cpm (mean of triplicates ± S.D.) and are representative of three independent experiments performed with different donors. D, effect of IL-27 on MHC class I induction in naive and memory CD4+ T cells stimulated for 36 h with CD3-coated beads. Control staining and staining for MHC class I are represented as indicated (x axis, log fluorescence intensity; y axis: cell number). Data are representative of three different donors.
Of note, biological responses to IL-27 were not systematically more robust in naive than memory human CD4+ T cells. For example, up-regulation of MHC class I expression by IL-27 in cells stimulated in suboptimal condition (CD3 stimulation only) was not lower in memory than naive CD4+ T cells (Fig. 1D).
The Poor Proliferative Response of Memory CD4+ T Cells to the Addition of IL-27 Is Not Due to Autocrine Effect of IL-27 in These Cells
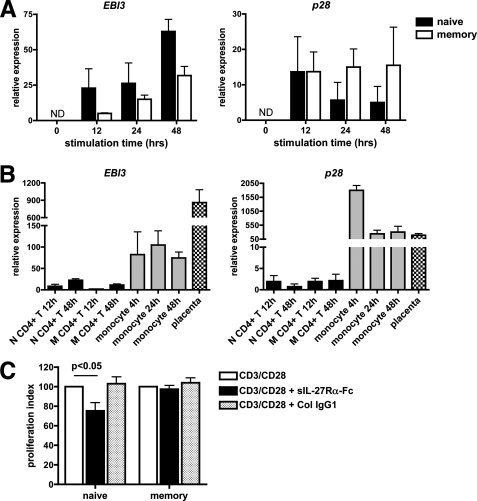
A recent study demonstrated that autocrine production of IL-27 is involved in the proliferation of murine total CD8+ T cells (27). Therefore, we investigated whether the lower proliferative response of human memory CD4+ T cells compared with naive CD4+ T cells to exogenous IL-27 could be due to autocrine production of IL-27 by memory CD4+ T cells specifically. Previously, we had shown by Western blot that naive and memory CD4+ T cells stimulated with CD3/CD28 mAbs express low but detectable levels of EBI3, but their production of IL-27 was not investigated (26). Therefore, we analyzed by RTqPCR the expression of both subunits of IL-27, EBI3 and p28, at different time points upon CD3 or CD3/CD28 stimulation. Upon CD3 stimulation, induction of EBI3 was detected in both naive and memory CD4+ T cells, but was associated with barely detectable levels of p28 in both subsets (not shown). Upon CD3/CD28 stimulation, the two subunits of IL-27 could be detected in both naive and memory CD4+ T cells, at levels that were not significantly higher in the latters (Fig. 2A). These levels were however much lower than those observed in other cell types or tissues known to express high amounts of IL-27, such as LPS-stimulated monocytes or placental tissues (Fig. 2B, when compared with monocytes, EBI3 levels in CD4+ T cells were on average 3–58-fold lower, and p28 levels over 100-fold lower). The expression of IL-27 protein in activated CD4+ T cells could not be unambiguously measured by intracellular staining with commercial anti-IL-27 Ab, which in our hands failed to prove specificity for IL-27 (not shown).
FIGURE 2.
Analysis of IL-27 expression by naive and memory CD4+ T cells and of its role in their proliferation. A, EBI3 and p28 expression in naive and memory CD4+ T cells stimulated for various times with plate-coated CD3 mAb and soluble CD28 mAb was analyzed by RTqPCR. Data from three different donors (mean ± S.E.) are shown. ND, not detected. B, expression levels of EBI3 and p28 determined by RTqPCR in 12 or 48-h CD3/CD28-stimulated naive (N) or memory (M) CD4+ T cells (mean ± S.E. of 4 donors) were compared with those observed in monocytes stimulated for various times with LPS (3 donors) or placenta tissues (4 donors, 9 to 33 weeks of pregnancy). C, purified naive and memory CD4+ T cells were stimulated for 4 days with plate-coated CD3 mAb and soluble CD28 mAb in the absence or presence of sIL-27Rα-Fc or an irrelevant human IgG1 control protein, and their proliferation was measured by [3H]thymidine incorporation. The percentage of proliferation (mean ± S.E. of 7 different donors) observed in the presence of sIL-27Rα-Fc or a control protein, relative to that observed in their absence is represented.
To investigate whether the low IL-27 expression by naive or memory CD4+ T cells could contribute to their proliferation, purified cells were stimulated for 4 days with CD3/CD28 Abs in the absence or presence of a soluble IL-27Rα-Fc chimera known to inhibit IL-27 signaling, but not that of IL-6, another gp130 family cytokine (27, 28). Addition of sIL-27Rα-Fc did not affect proliferation of memory CD4+ T cells in all donors tested (n = 7) (Fig. 2C). However, it partially inhibited naive CD4+ T cell proliferation (25 ± 8% inhibition on average, p < 0.05), suggesting that endogenous IL-27 (or an unknown protein signaling through IL-27Rα) may contribute to naive CD4+ T cell proliferation. Collectively, these data suggested that the lower proliferative response of memory CD4+ T cells to exogenous IL-27 was not due to an autocrine role of IL-27 in these cells, and was most likely due to differential signaling induced by IL-27 in naive versus memory CD4+ T cells.
Human Naive and Memory CD4+ T Cells Are Characterized by Differential Expression Profiles of IL-27 Receptor
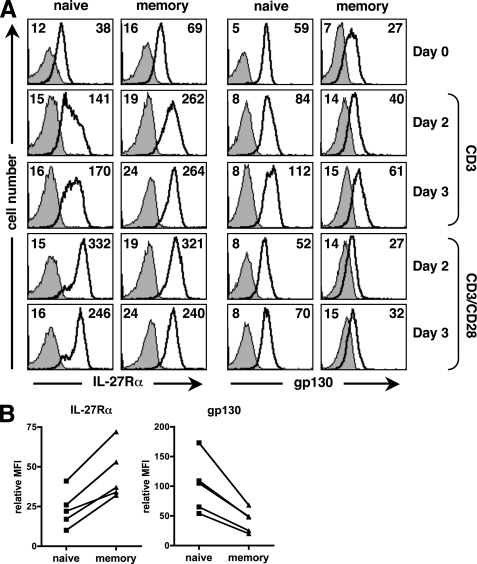
We analyzed the expression profile of both chains of the IL-27 receptor, IL-27Rα and gp130, at the cell surface of resting and activated purified naive and memory CD4+ T cells to evidence possible differences between both subsets (Fig. 3, A and B). FACS analysis showed that constitutive expression of IL-27Rα and gp130 varied between naive and memory CD4+ T cells. Whereas IL-27Rα expression was consistently higher on freshly purified memory than naive CD4+ T cells (MFI 2.5-fold higher on average, p < 0.05), the reverse situation was observed for gp130, whose expression was consistently higher on naive than memory CD4+ T cells (MFI 2.4-fold higher on average, p < 0.05) (Fig. 3B).
FIGURE 3.
Cell surface expression of IL-27 receptor on human naive and memory CD4+ T cells. A, cell surface expression of IL-27Rα and gp130 on naive and memory CD4+ T cells was determined by FACS analysis before stimulation, or after a 2- and 3-day stimulation with plate-coated CD3 mAb, alone or in combination with CD28 mAb. Staining with anti-IL-27Rα mAb and anti-gp130 mAb is indicated by a bold line (MFI indicated at right), while control staining (MFI indicated at left) is represented by a filled gray histogram (x axis, log fluorescence intensity; y axis: cell number). Data from a representative donor out of 3 are shown. B, relative MFI (MFI of specific staining minus MFI of control staining) of IL-27Rα or gp130 staining observed in unstimulated naive and memory CD4+ T cells from 5 donors is shown.
Stimulation with CD3 or CD3/CD28 mAbs for 2 to 3 days induced a substantial increase of IL-27Rα on both naive and memory CD4+ T cells (Fig. 3A). However, upon CD3 stimulation, memory CD4+ T cells still expressed higher levels of IL-27Rα than naive CD4+ T cells. In contrast, CD3/CD28 stimulation induced a strong, but transient, up-regulation of IL-27Rα that resulted in similar levels of IL-27Rα on each CD4+ T cell subset. Gp130 levels were also modulated upon stimulation and for each subset were the highest upon a 3-day CD3 stimulation. Of note, upon either CD3 or CD3/CD28 stimulation, gp130 expression remained higher on naive than memory CD4+ T cells. Addition of IL-27 did not substantially modulate IL-27Rα or gp130 expression in CD3-stimulated human naive or memory CD4+ T cells (not shown). Taken together, these data indicate that while IL-27Rα was expressed at higher levels in resting or CD3-activated memory CD4+ T cells than in their naive counterparts, gp130 was expressed at higher levels in naive CD4+ T cells, both constitutively or upon activation.
Differential Activation of STAT Proteins and of SHP-2 by IL-27 in Naive versus Memory CD4+ T Cells
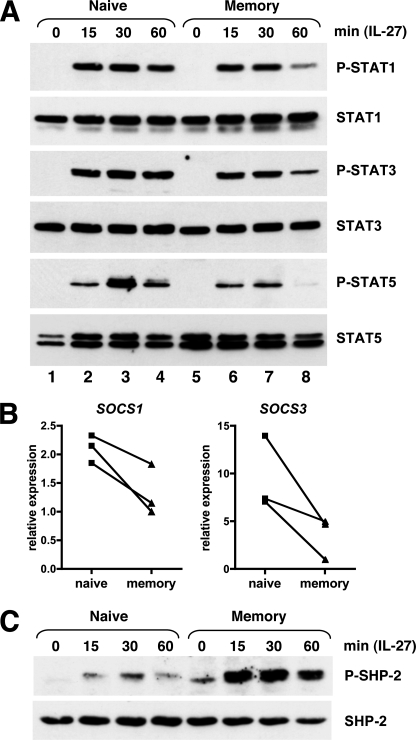
Next, we examined whether IL-27 signaling differs between naive and memory CD4+ T cells. First, we analyzed the activation of STAT1, -3, and -5, that had been previously shown to be activated by IL-27 in human PHA-activated T cells (29). Western blot analysis showed that the kinetic of STAT activation by IL-27 consistently differed between naive and memory CD4+ T cells (Fig. 4A). If after 15 min of stimulation with IL-27, STAT1, -3 and -5 were activated at comparable levels in freshly purified naive or memory human CD4+ T cells, analysis performed at later time points indicated that their activation was more transient in memory than naive CD4+ T cells. Indeed, whereas significant amounts of phospho-STAT1, -3, and -5 were still detected in naive CD4+ T cells after 1 h of IL-27 stimulation, a substantial decrease was observed in memory CD4+ T cells (Fig. 4A, compare lane 4 with lane 8). For STAT5, its activation by IL-27 was not only more sustained, but also stronger in naive than memory CD4+ T cells (compare lane 3 to lane 7). These data indicate that STAT activation by IL-27 differs between naive and memory CD4+ T cells, both in signal magnitude (STAT5) and duration (STAT1, -3, and -5).
FIGURE 4.
Differential IL-27 signaling in naive versus memory CD4+ T cells. A, purified naive and memory CD4+ T cells were incubated in RPMI 1640 containing 2% FBS in the absence or presence of IL-27 for the indicated time (in minutes). Cell lysates were analyzed for STAT activation by Western blot using anti-phospho-STAT1, -3, and -5 Abs. Blots were subsequently analyzed with anti-STAT1, -3, and -5 Abs to monitor STAT expression levels. Data shown are representative of 3 different donors. B, relative levels of SOCS1 and SOCS3 were determined by RTqPCR in purified naive and memory CD4+ T cells from 3 different donors. C, activation of SHP-2 by IL-27 was analyzed in purified naive and memory CD4+ T cells as described in A. Data shown are representative of three different donors.
gp130 signaling is negatively regulated by SOCS1 and SOCS3 (23) and in murine CD8+ T cells, SOCS3 deficiency has been shown to result in prolonged activation of STAT1, -3, and -5 upon IL-27 stimulation (27). However, the prolonged STAT activation observed in naive CD4+ T cells upon IL-27 stimulation was not due to lower SOCS1 or SOCS3 expression levels because both genes were not expressed at lower but higher levels in naive than memory CD4+ T cells in the donors tested (Fig. 4B). In addition to SOCS molecules, gp130 signaling can be negatively regulated by the phosphatase SHP-2. Involvement of SHP-2 in IL-27 signaling had not been previously investigated. Interestingly, we observed that SHP-2 was activated by IL-27 and that its activation was much stronger in memory than naive CD4+ T cells, at all times of stimulation tested (15, 30, and 60 min) (Fig. 4C). Thus, naive CD4+ T cell response to IL-27 was characterized by more sustained STAT1, -3, and -5 activation and conversely lower activation of SHP-2, compared with memory CD4+ T cells.
Analysis of IL-27 Effect on Cell Survival, Cell Division, and Cell Cycle Progression in CD3-stimulated Naive and Memory CD4+ T Cells
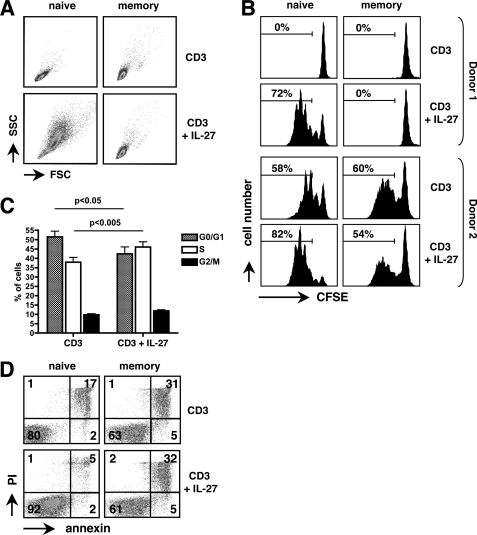
In the experiments described above, proliferation was measured in a [3H]thymidine incorporation assay, that does not discriminate between effects on cell survival, cell division, or cell cycle progression. Therefore, we specifically analyzed IL-27 effect on these events in CD3-stimulated naive and memory CD4+ T cells. In all donors tested (n = 6), IL-27 substantially increased cell division of naive CD4+ T cells assessed by CFSE labeling, an effect most often associated with an increase of cell size (Fig. 5, A and B). Analysis of cell cycle distribution by DNA staining showed that IL-27 induced a statistically significant increase in G0/G1→S transition in these cells (p < 0.05) (Fig. 5C). IL-27 could also affect naive CD4+ T cell survival. Indeed, in the 2 of 6 donors tested for which substantial apoptosis was observed in CD3-stimulated control cells (>15% apoptotic cells), IL-27 increased the percentage of alive naive CD4+ T cells, assessed by IP and annexin staining (Fig. 5D). In contrast, no substantial effect of IL-27 on cell size, cell division, or cell survival was observed in CD3-stimulated memory CD4+ T cells (Fig. 5, A, B, and D).
FIGURE 5.
Effects of IL-27 on cell size, cell division, cell cycle progression, and cell survival in naive and memory CD4+ T cells. A, IL-27 increased the size of CD3-activated naive, but not memory, CD4+ T cells, as assessed by FACS analysis performed at day 5. B, CFSE-labeled naive and memory CD4+ T cells were activated with plate-coated CD3 mAb in the presence or absence of IL-27 and analyzed at day 5 by FACS to monitor cell division. The percentage of cells that underwent over 1 division is indicated. Results from 2 donors, showing either poor (donor 1, top graphs) or strong (donor 2, bottom graphs) proliferation in response to CD3 stimulation, are shown. C, cell cycle distribution of naive CD4+ T cells stimulated for 3 days with plate-coated CD3 mAb in the absence or presence of IL-27 was assayed by FACS analysis after staining with Vybrant® DyeCycle violet stain. Results shown are mean ± S.E. of three independent experiments, each performed on a different donor. D, effect of IL-27 on cell survival was monitored after 4 days of stimulation by staining naive or memory CD4+ T cells with annexin-FITC and IP.
MAPK Are Not Involved in IL-27-mediated Proliferation of Naive CD4+ T Cells
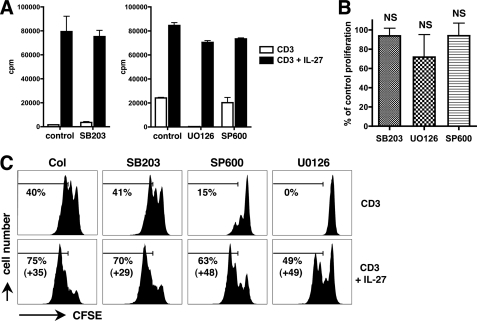
Next, we investigated by which mechanisms IL-27 induces cell proliferation of naive CD4+ T cells. Because MAPKs regulate cell proliferation (30) and can be activated by IL-27 (25), we examined whether they play a role in IL-27-mediated proliferation of human naive CD4+ T cells. Purified naive CD4+ T cells were stimulated for 3 to 4 days with plate-coated CD3 mAb in the absence or presence of IL-27 and of chemical inhibitors of p38 (SB203580), erk1/2 (U0126), or JNK (SP600125), and proliferation was measured in a [3H]thymidine incorporation assay. Consistent with previous observations (31, 32), addition of SB203580 (10 μm) did not affect CD3-mediated naive CD4+ T cell proliferation, whereas addition of U0126 (10 μm) or SP600125 (10 μm) inhibited proliferation by 99 and 54% on average, respectively (Fig. 6A). Importantly, none of the inhibitors abrogated or significantly decreased the proliferative effect of IL-27 (Fig. 6, A and B). Similar results were observed when proliferation of naive CD4+ T cells was measured by CFSE labeling: despite the presence of the inhibitors, a substantial effect of IL-27 on cell division was still observed (Fig. 6C). The lack of effect of the inhibitors on IL-27-mediated cell proliferation was not due to the absence of MAPK activation by IL-27 in these cells. Indeed, when these inhibitors were similarly tested in CD3-stimulated naive CD4+ T cells for their ability to modulate another biological effect of IL-27, i.e. induction of CD95 expression, all three MAPK inhibitors significantly decreased IL-27-mediated induction (p < 0.05) (supplemental Fig. S1). Altogether, these data indicated that neither p38, erk1/2, nor JNK play a substantial role in the proliferative effect of IL-27 on naive CD4+ T cells.
FIGURE 6.
Analysis of the role of MAPK in IL-27-mediated proliferation of naive CD4+ T cells. A, purified naive CD4+ T cells were cultured for 4 days with plate-coated CD3 mAb with or without IL-27, in the absence (control) or presence of U0126, SP600125 (SP600), or SB203580 (SB203) (all at 10 μm). Proliferation was measured by [3H]thymidine incorporation. Results are expressed in cpm (mean of triplicates ± S.D.) and are representative of four independent experiments performed with 3 to 4 different donors. B, increase in cpm induced by IL-27 in the presence of the inhibitors relative to that observed in its absence is shown (mean ± S.E. of four experiments). C, CFSE-labeled naive and memory CD4+ T cells were cultured as described in A and analyzed by FACS to monitor cell division. The percentage of cells that underwent over 1 division and its increase induced by IL-27 are indicated.
IL-27 Induces the Expression of c-Myc, Pim-1, Cyclin D2, Cyclin D3, and CDK4 in Naive but Not Memory CD4+ T Cells
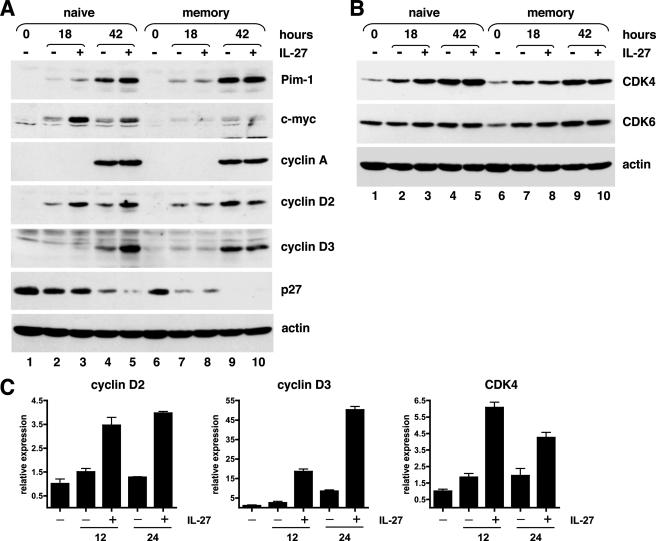
In murine naive CD4+ T cells, IL-27 was recently shown to induce expression of the transcriptional activator c-Myc and of Pim-1 kinase (24), two known regulators of cell proliferation that are induced by gp130 signaling and cooperate to promote cell proliferation by increasing G1 to S transition (33, 34). Consistent with the data obtained in mice, IL-27 induced substantial up-regulation of Pim-1 and c-Myc expression in human naive CD3-stimulated CD4+ T cells (Fig. 7A). Induction of c-Myc was maximal after 18 h of stimulation and rapidly declined thereafter, whereas Pim-1 induction was maximal after 42 h of stimulation. In line with the selective proliferative effect of IL-27 on naive CD4+ T cells, no induction of c-Myc or Pim-1 by IL-27 was observed in CD3-activated memory CD4+ T cells (Fig. 7A).
FIGURE 7.
Effects of IL-27 on the expression of various cell cycle modulators in naive and memory CD4+ T cells. A and B, cell lysates (50 μg/lane) from naive or memory CD4+ T cells, either unstimulated or stimulated for various times with plate-coated CD3 mAb in the absence (−) or presence (+) of IL-27 were analyzed by Western blot with the different indicated Abs and with anti-actin Abs to verify equal total protein loading. Results shown are representative of four independent experiments performed on 3 different donors. C, effect of IL-27 on cyclin D2, cyclin D3, and CDK4 expression was analyzed by RTqPCR in naive CD4+ T cells stimulated with plate-coated CD3 mAb for 12 or 24 h in the absence or presence of IL-27. Data are representative of two donors.
Cell cycle progression is regulated by an interplay between cyclins, CDKs, and CDK inhibitors. D-type cyclins are induced during G1 phase and associate with CDK4 or CDK6, whereas cyclins A and E are induced at the late G1 or at entry in S phase and form complexes with CDK2. Because gp130 signaling had been shown to induce the expression of cyclins A, D2, and D3, and to down-regulate concomitantly the expression of the CDK inhibitors, p21 and p27 (35), we specifically analyzed the effect of IL-27 on the expression of these molecules. In CD3-activated memory CD4+ T cells, IL-27 had no significant effect on the expression of any of the cyclins and CDK analyzed (Fig. 7, A and B). In contrast, in CD3-activated naive CD4+ T cells, IL-27 increased cyclin D2 expression at 18 and 42 h of stimulation (Fig. 7A, lanes 3 and 5) and cyclin D3 expression at 42 h of stimulation (Fig. 7A, lane 5). Among cyclins D2 and D3 partners, only CDK4, but not CDK6, was increased by IL-27 in naive CD4+ T cells at 18 and 42 h of stimulation (Fig. 7B). This induction of cyclin D2, cyclin D3, and CDK4 by IL-27 in naive CD4+ T cells was further confirmed at the mRNA level by RTqPCR. At both time points analyzed (12 and 24 h), an induction of cyclin D2, cyclin D3, and CDK4 gene expression by IL-27 was observed (Fig. 7C). Although IL-27 induced some increase of CDK2 level in naive CD4+ T cells (not shown), it had no consistent effect on the expression of the late G1 cyclins, cyclins A and E, even at later time points (70 h of stimulation) (Fig. 7A and not shown). Of the CDK inhibitors p21 and p27, only p27 was detected in naive and memory CD4+ T cells, and its expression remained unchanged in the presence of IL-27 (Fig. 7A).
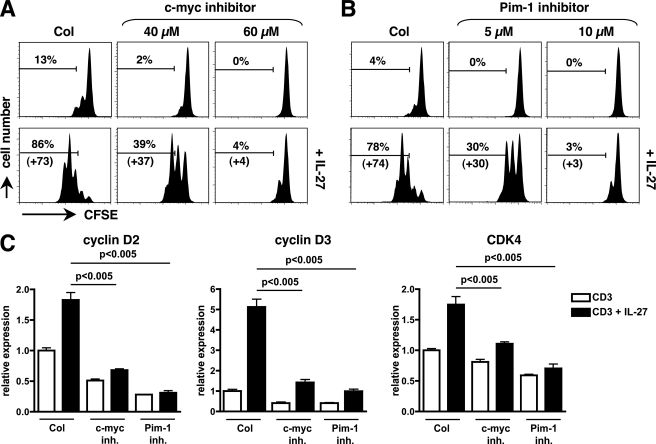
Role of Pim-1 and c-Myc in IL-27-mediated Proliferation of Naive CD4+ T Cells
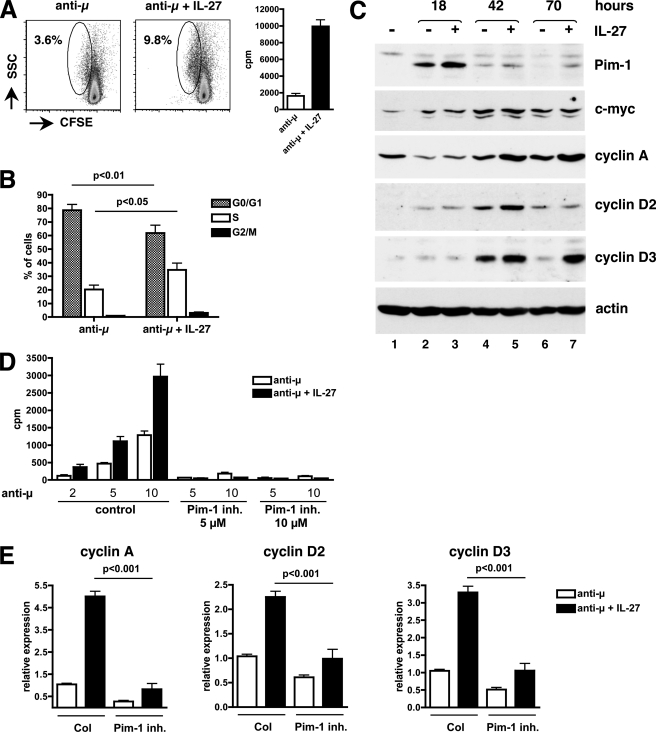
The role of c-Myc and Pim-1 in IL-27-mediated naive CD4+ T cell proliferation was directly assessed by using chemical inhibitors of c-Myc (10058-F4) and of Pim-1 (Pim-1 inhibitor 2) activity. As shown on Fig. 8, A and B, treatment of CD3-stimulated naive CD4+ T cells with c-Myc inhibitor (40 or 60 μm) or Pim-1 inhibitor (5 or 10 μm) significantly inhibited IL-27-mediated proliferation measured by CFSE labeling, in a dose-dependent manner (p < 0.05). This inhibition was associated with decreased induction of cyclin D2, cyclin D3, and CDK4 in IL-27-stimulated naive CD4+ T cells (p < 0.005, Fig. 8C).
FIGURE 8.
Role of c-Myc and Pim-1 in IL-27-mediated cell division and up-regulation of cell cycle regulators in naive CD4+ T cells. A and B, naive CD4+ T cells were labeled with CFSE and stimulated with plate-coated CD3 mAb with or without IL-27, in the absence or presence of various concentrations of specific inhibitors of c-Myc (A) or Pim-1 (B). At day 4, cells were analyzed by FACS to monitor cell division. The percentage of cells that underwent over 1 division and its increase induced by IL-27 are indicated. Results shown are representative of 3 different donors. C, naive CD4+ T cells were stimulated with plate-coated CD3 mAb for 24 h in the presence or absence of 60 μm of c-Myc inhibitor or 10 μm of Pim-1 inhibitor, with or without IL-27, and levels of cyclin D2, cyclin D3, and CDK4 were determined by RTqPCR. Data are expressed as mean (± S.E.) of two independent experiments performed in duplicate cultures.
IL-27 Increases G0/G1→S Transition and Induces the Expression of Pim-1 and Cyclins A, D2, and D3 in BCR-stimulated Naive B Cells
We previously showed that IL-27 increased proliferation, measured by thymidine incorporation, of BCR-activated naive but not memory human tonsillar B cells, through mechanisms that were not investigated (8).
In naive B cells stimulated through their surface IgM via anti-μ Ab stimulation, no effect of IL-27 on either cell survival or cell size was observed (not shown). When monitoring cell division by CFSE labeling, we observed that anti-μ stimulation by itself induced cell division only in a very small fraction of cells (less than 5%), that remained low (∼10% at the best) in the presence of IL-27 (Fig. 9A). However, IL-27 induced a significant increase of the fraction of anti-μ-stimulated naive B cells that left G0/G1 stage and entered the S phase (p < 0.05) (Fig. 9B). Thus, IL-27 proliferative effect observed in BCR-activated naive B cells results essentially from an effect on G0/G1→S transition.
FIGURE 9.
Signaling events involved in IL-27-mediated proliferation of naive B cells. A, effect of IL-27 on cell division was analyzed by FACS on naive B cells labeled with CFSE and stimulated for 3 days with anti-μ Abs in the absence or presence of IL-27. While IL-27 induced substantial proliferation as measured by [3H]thymidine incorporation (graphs on the right, cpm mean of triplicates ± S.D.), it had no significant effect on cell division (graphs on the left). The percentage of cells that divided is indicated. Results are representative of three different donors. B, effect of IL-27 on cell cycle progression was determined by staining anti-μ-stimulated naive B cells at day 3 with Vybrant® DyeCycle violet stain, and analyzing cell cycle distribution by FACS analysis. Data are shown as mean ± S.E. of three experiments, each performed on a different donor. C, cell lysates (70 μg/lane) from naive B cells, either unstimulated or stimulated with anti-μ Abs for the indicated time in the absence (−) or presence (+) of IL-27 were analyzed by Western blot with the different Abs indicated on the right of each blot. Actin Ab was used to assess equal loading in each lane. Results shown are representative of four experiments performed on 3 different donors. D, purified B cells were stimulated for 3 days with various concentrations of anti-μ Abs (indicated at the bottom of the graphs in μg/ml) with or without IL-27 in the absence or presence of 5 or 10 μm of Pim-1 inhibitor and proliferation was measured by [3H]thymidine incorporation. Results are expressed in cpm (mean of triplicates ± S.D.) and are representative of three independent experiments performed on 2 different donors. Despite a marked effect on cell proliferation, the addition of the inhibitor did not increase mortality (tested at the end of the 3 day culture) by more than 7%. E, anti-μ-stimulated naive B cells were cultured for 64 h in the presence or absence of 5 μm of Pim-1 inhibitor, with or without IL-27, and levels of cyclin A, cyclin D2, and cyclin D3 were determined by RTqPCR. Data are expressed as mean (± S.E.) of three independent experiments.
In anti-μ-stimulated naive B cells, we observed that IL-27 induced the expression of Pim-1 at 18 h of stimulation (Fig. 9C, lane 3) and that of cyclins D2 and D3 at later time points (42 h for cyclin D2, and 42 and 70 h for cyclin D3, Fig. 9C, lanes 5 and 7). It did not induce cyclin E expression (not shown), but induced cyclin A expression at 42 and 70 h of stimulation (Fig. 9C, lanes 5 and 7). No significant induction of c-Myc and of CDK2, 4 or 6 by IL-27 was detected in naive B cells at any time point tested (18, 42, and 70 h) (Fig. 9C and not shown).
As observed for naive CD4+ T cells, Pim-1 was involved in IL-27-mediated proliferation of naive B cells. Addition of Pim-1 inhibitor (5 or 10 μm) to the culture almost completely abolished anti-μ-mediated naive B cell proliferation and also abrogated IL-27 proliferative effect (Fig. 9D). This inhibition was associated with decreased induction of cyclins A, D2, and D3 by IL-27 in anti-μ-stimulated naive B cells (p < 0.001, Fig. 9A). Altogether, these data indicate that IL-27 mediates cell cycle progression in naive CD4+ T cells and naive B cells through induction of both common (Pim-1, cyclins D2, and D3) and distinct (c-Myc, cyclin A, CDK4) sets of mediators.
DISCUSSION
This study further highlights that the response of human CD4+ T cells to IL-27 is differentially regulated at various stages of their differentiation and identifies signaling pathways whose activation by IL-27 differs between naive and memory human CD4+ T cells.
Consistent with a previous study (2), we found that addition of IL-27 to the culture resulted in stronger proliferation of naive than memory human CD4+ T cells, especially when suboptimal conditions of stimulation were used. We also evidenced that endogenous production of IL-27 might contribute to naive, but not memory, human CD4+ T cell proliferation, Of note, not all biological effects of IL-27 differ between human naive and memory CD4+ T cells. Here, we observed that MHC class I induction by IL-27 was comparable in naive and memory CD4+ T cells. Also, a recent study found that IL-27, although it induced IL-10 production in greater folds in naive than memory human CD4+ T cells, induced similar production of IFN-γ in both subsets, and could inhibit IL-17 production and expression of other Th17-associated molecules in both naive and memory CD4+ T cells (36). This heterogeneity of IL-27 responses in naive and memory CD4+ T cells indicates that different mechanisms and signaling pathways are used by IL-27 to mediate the different biological effects.
Little is known on the mechanisms regulating the differential response of naive and memory CD4+ T cells or B cells to IL-27. The expression profile of the IL-27 receptor only partially accounts for this phenomenon. In human B cells, differential response to IL-27 was observed in naive and memory B cells, despite similar levels of IL-27 receptor expression (8). In mice, the differential effect of IL-27 on cytokine production in naive versus effector CD4+ T cells was associated with down-regulation of gp130 and up-regulation of IL-27Rα in effector CD4+ T cells (24). Surprisingly, although IL-27Rα is assumed to contribute preferentially to STAT1 activation and gp130 to STAT3 activation, a stronger activation of STAT3 over STAT1 was observed in effector CD4+ T cells compared with naive CD4+ T cells. The authors suggested that STAT1 activation by IL-27 was specifically inhibited in these cells by an unknown mechanism (24).
Here, we show that human naive and memory CD4+ T cells are characterized by differential expression levels of IL-27Rα and gp130, both constitutively or upon stimulation. The stronger proliferative response of CD3-stimulated naive versus memory CD4+ T cells to IL-27 was associated with lower levels of IL-27Rα and conversely higher levels of gp130 expression. In addition, we evidenced that IL-27 signaling, in particular STAT activation, differed between human naive and memory CD4+ T cells. The difference in STAT activation by IL-27 between both subsets was mainly a difference in signal duration rather than in signal amplitude. Whereas only STAT5 was activated at higher levels in naive than memory CD4+ T cells, activation of STAT1, 3 and 5 as well was more prolonged in naive than memory CD4+ T cells. This sustained activation of STAT molecules, in particular that of STAT3, known to be important for IL-27-mediated proliferation (24), and possibly that of STAT5 also known to regulate T cell proliferation, may be responsible for the stronger proliferative response of naive CD4+ T cells to IL-27.
In naive CD4+ T cells, the prolonged STAT activation was observed despite higher expression of SOCS1 and SOCS3, two negative regulators of gp130 signaling. Interestingly, SHP-2, another regulator of the Jak/STAT pathway, was activated by IL-27 in naive CD4+ T cells, at lower levels compared with memory CD4+ T cells. SHP-2 negatively regulates cytokine signaling by binding to activated receptors and dephosphorylating Jak, in particular Jak1 (23, 37). It binds the gp130 receptor at a specific tyrosine residue, Tyr-759, that is the same as that used by SOCS3, resulting in competition between SOCS3 and SHP-2 for gp130 binding. The lower levels of SOCS3 detected in memory CD4+ T cells may result in greater levels of SHP-2 recruitment and activation by IL-27, leading to more transient STAT activation. SHP-2 is not only involved in the negative regulation of gp130 signaling, but also positively regulates signaling by recruiting growth factor receptor-bound protein (grb2) and MAPK (23, 37). The finding that SHP-2 activation by IL-27 differs between memory and naive CD4+ T cells suggests that the MAPK pathway may be differentially activated by IL-27 in naive versus memory CD4+ T cells. In memory CD4+ T cells, the stronger activation of SHP-2 may contribute to their lower proliferative response to IL-27. Consistent with this hypothesis, we observed that the addition of the SHP-1/SHP-2 inhibitor NSC-87877 to the culture increased the proliferation of CD3-stimulated memory CD4+ T cells in response to IL-27.3
The mechanisms by which IL-27 can modulate proliferation have been so far analyzed only in melanoma cell lines and in murine naive CD4+ T cells (22, 24, 38). In melanoma cell lines, IL-27 was shown to inhibit proliferation through IRF-1 induction (38). In murine naive CD4+ T cells, IL-27 proliferative effect has been shown to be dependent on STAT3 activation by gp130. In these cells, lack of STAT3 resulted in failure of IL-27 to induce c-Myc and Pim-1 expression and to promote cell proliferation (24). In human CD4+ T cells, the pattern of c-Myc and Pim-1 induction by IL-27 correlated with IL-27 ability to induce cell proliferation, in that their induction was observed only in naive, but not memory, CD4+ T cells. In addition, we directly established the role of c-Myc and Pim-1 in IL-27-mediated proliferation, as inhibition of c-Myc or Pim-1 activities inhibited IL-27-induced proliferation of naive CD4+ T cells. Further, we showed that Pim-1 was also induced by IL-27 in anti-μ-stimulated naive B ells and was involved in IL-27-induced proliferation of these cells. Pim-1, that is also involved in cell survival (33), may also play a role in IL-27 effect on naive CD4+ T cell survival.
The precise effect of IL-27 on cell cycle progression and on expression of cyclins and their regulators had not been previously investigated. In this study, we showed that IL-27 increased G0/G1 to S phase transition in both CD3-stimulated naive CD4+ T cells and anti-μ-stimulated naive B cells and increased the expression of cyclins D2 and D3 in both cell types. In addition, an induction of cyclin A and CDK4 expression by IL-27 was detected in activated naive B cells and CD4+ T cells, respectively. Induction of cyclins and CDK expression by IL-27 involves c-Myc and Pim-1, most likely through both direct and indirect mechanisms. In naive CD4+ T cells, the induction of cyclin D2 and CDK4 by IL-27 may be a direct consequence of c-Myc induction, as both genes have been identified as direct c-Myc target genes (39, 40). Pim-1 may play an indirect role, through its ability to regulate c-Myc activity. Indeed, Pim-1 has recently been shown to act as a co-activator of c-Myc and to contribute to the induction of many c-Myc-induced genes (34, 41). Also, the fact that different sets of cell cycle regulators were induced in naive CD4+ T cells and naive B cells, suggests that IL-27 uses cell type-specific factors and/or synergizes with pathways specifically activated by either anti-μ stimulation in naive B cells or CD3 stimulation in naive CD4+ T cells to induce these molecules. Overall, this study provides an insight into the molecular mechanisms involved in IL-27-mediated naive B and CD4+ T cell proliferation and shows that IL-27 signaling is finely regulated according to the cell type, the stage of differentiation and the co-stimuli.
Supplementary Material
Acknowledgments
We thank the Dept. of Otorhinolaryngology of Necker Hospital for tonsils, and Jérôme Mégret and Corinne Cordier for electronic cell sorting. We thank Stefan Pflanz (Schering-Plough Biopharma) for critical reading of the manuscript.
This work was supported by Grant 3733 from the Association de Recherche contre le Cancer.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
O. Devergne, unpublished observations.
- CDK
- cyclin-dependent kinase
- EBI3
- EBV-induced gene 3
- SOCS
- suppressor of cytokine signaling
- RTqPCR
- real-time quantitative PCR analysis
- mAb
- monoclonal antibody.
REFERENCES
- 1. Devergne O., Hummel M., Koeppen H., Le Beau M. M., Nathanson E. C., Kieff E., Birkenbach M. (1996) J. Virol. 70, 1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pflanz S., Timans J. C., Cheung J., Rosales R., Kanzler H., Gilbert J., Hibbert L., Churakova T., Travis M., Vaisberg E., Blumenschein W. M., Mattson J. D., Wagner J. L., To W., Zurawski S., McClanahan T. K., Gorman D. M., Bazan J. F., de Waal Malefyt R., Rennick D., Kastelein R. A. (2002) Immunity 16, 779–790 [DOI] [PubMed] [Google Scholar]
- 3. Devergne O., Coulomb-L'Herminé A., Capel F., Moussa M., Capron F. (2001) Am. J. Pathol. 159, 1763–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coulomb-L'Herminé A., Larousserie F., Pflanz S., Bardel E., Kastelein R. A., Devergne O. (2007) Placenta. 28, 1133–1140 [DOI] [PubMed] [Google Scholar]
- 5. Pflanz S., Hibbert L., Mattson J., Rosales R., Vaisberg E., Bazan J. F., Phillips J. H., McClanahan T. K., de Waal Malefyt R., Kastelein R. A. (2004) J. Immunol. 172, 2225–2231 [DOI] [PubMed] [Google Scholar]
- 6. Yoshimoto T., Okada K., Morishima N., Kamiya S., Owaki T., Asakawa M., Iwakura Y., Fukai F., Mizuguchi J. (2004) J. Immunol. 173, 2479–2485 [DOI] [PubMed] [Google Scholar]
- 7. Villarino A. V., Larkin J., 3rd, Saris C. J., Caton A. J., Lucas S., Wong T., de Sauvage F., Hunter C. A. (2005) J. Immunol. 174, 7684–7691 [DOI] [PubMed] [Google Scholar]
- 8. Larousserie F., Charlot P., Bardel E., Froger J., Kastelein R. A., Devergne O. (2006) J. Immunol. 176, 5890–5897 [DOI] [PubMed] [Google Scholar]
- 9. Shimizu M., Shimamura M., Owaki T., Asakawa M., Fujita K., Kudo M., Iwakura Y., Takeda Y., Luster A. D., Mizuguchi J., Yoshimoto T. (2006) J. Immunol. 176, 7317–7324 [DOI] [PubMed] [Google Scholar]
- 10. Wang S., Miyazaki Y., Shinozaki Y., Yoshida H. (2007) J. Immunol. 179, 6421–6428 [DOI] [PubMed] [Google Scholar]
- 11. Kalliolas G. D., Ivashkiv L. B. (2008) J. Immunol. 180, 6325–6333 [DOI] [PubMed] [Google Scholar]
- 12. Seita J., Asakawa M., Ooehara J., Takayanagi S., Morita Y., Watanabe N., Fujita K., Kudo M., Mizuguchi J., Ema H., Nakauchi H., Yoshimoto T. (2008) Blood 111, 1903–1912 [DOI] [PubMed] [Google Scholar]
- 13. Kanda N., Watanabe S. (2008) Eur. J. Immunol. 38, 1287–1296 [DOI] [PubMed] [Google Scholar]
- 14. Diveu C., McGeachy M. J., Cua D. J. (2008) Curr. Opin. Immunol. 20, 663–668 [DOI] [PubMed] [Google Scholar]
- 15. Yoshida H., Miyazaki Y., Yoshiyuki M. (2008) Immunol. Rev. 226, 234–247 [DOI] [PubMed] [Google Scholar]
- 16. Stumhofer J. S., Hunter C. A. (2008) Immunol. Lett. 117, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshimura T., Takeda A., Hamano S., Miyazaki Y., Kinjyo I., Ishibashi T., Yoshimura A., Yoshida H. (2006) J. Immunol. 177, 5377–5385 [DOI] [PubMed] [Google Scholar]
- 18. Diveu C., McGeachy M. J., Boniface K., Stumhofer J. S., Sathe M., Joyce-Shaikh B., Chen Y., Tato C. M., McClanahan T. K., de Waal Malefyt R., Hunter C. A., Cua D. J., Kastelein R. A. (2009) J. Immunol. 182, 5748–5756 [DOI] [PubMed] [Google Scholar]
- 19. El-behi M., Ciric B., Yu S., Zhang G. X., Fitzgerald D. C., Rostami A. (2009) J. Immunol. 183, 4957–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeda A., Hamano S., Yamanaka A., Hanada T., Ishibashi T., Mak T. W., Yoshimura A., Yoshida H. (2003) J. Immunol. 170, 4886–4890 [DOI] [PubMed] [Google Scholar]
- 21. Lucas S., Ghilardi N., Li J., de Sauvage F. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15047–15052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kamiya S., Owaki T., Morishima N., Fukai F., Mizuguchi J., Yoshimoto T. (2004) J. Immunol. 173, 3871–3877 [DOI] [PubMed] [Google Scholar]
- 23. Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Muller-Newen G., Schaper F. (2003) Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Owaki T., Asakawa M., Morishima N., Mizoguchi I., Fukai F., Takeda K., Mizuguchi J., Yoshimoto T. (2008) J. Immunol. 180, 2003–2911 [DOI] [PubMed] [Google Scholar]
- 25. Owaki T., Asakawa M., Fukai F., Mizuguchi J., Yoshimoto T. (2006) J. Immunol. 177, 7579–7587 [DOI] [PubMed] [Google Scholar]
- 26. Bardel E., Larousserie F., Charlot-Rabiega P., Coulomb-L'Herminé A., Devergne O. (2008) J. Immunol. 181, 6898–6905 [DOI] [PubMed] [Google Scholar]
- 27. Brender C., Tannahill G. M., Jenkins B. J., Fletcher J., Columbus R., Saris C. J., Ernst M., Nicola N. A., Hilton D. J., Alexander W. S., Starr R. (2007) Blood 110, 2528–2536 [DOI] [PubMed] [Google Scholar]
- 28. Wirtz S., Tubbe I., Galle P. R., Schild H. J., Birkenbach M., Blumberg R. S., Neurath M. F. (2006) J. Exp. Med. 203, 1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hibbert L., Pflanz S., De Waal Malefyt R., Kastelein R. A. (2003) J. Interferon Cytokine Res. 23, 513–522 [DOI] [PubMed] [Google Scholar]
- 30. Wilkinson M. G., Millar J. B. A. (2000) FASEB J. 14, 2147–2157 [DOI] [PubMed] [Google Scholar]
- 31. Bennett B. L., Sasaki D. T., Murray B. W., O'Leary E. C., Sakata S. T., Xu W., Leisten J. C., Motiwala A., Pierce S., Satoh Y., Bhagwat S. S., Manning A. M., Anderson D. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13681–13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Appleman L. J., van Puijenbroek A. A., Shu K. M., Nadler L. M., Boussiotis V. A. (2002) J. Immunol. 168, 2729–2736 [DOI] [PubMed] [Google Scholar]
- 33. Shirogane T., Fukada T., Muller J. M., Shima D. T., Hibi M., Hirano T. (1999) Immunity 11, 709–719 [DOI] [PubMed] [Google Scholar]
- 34. Naud J. F., Eilers M. (2007) Nat. Cell Biol. 9, 873–875 [DOI] [PubMed] [Google Scholar]
- 35. Fukada T., Ohtani T., Yoshida Y., Shirogane T., Nishida K., Nakajima K., Hibi M., Hirano T. (1998) EMBO J. 17, 6670–6677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murugaiyan G., Mittal A., Lopez-Diego R., Maier L. M., Anderson D. E., Weiner H. L. (2009) J. Immunol. 183, 2435–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alexander W. S. (2002) Nat. Rev. Immunol. 2, 1–7 [DOI] [PubMed] [Google Scholar]
- 38. Yoshimoto T., Morishima N., Mizoguchi I., Shimizu M., Nagai H., Oniki S., Oka M., Nishigori C., Mizuguchi J. (2008) J. Immunol. 180, 6527–6535 [DOI] [PubMed] [Google Scholar]
- 39. Bouchard C., Thieke K., Maier A., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M. (1999) EMBO J. 18, 5321–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hermeking H., Rago. C., Schuhmacher M., Li Q., Barrett J. F., Obaya. A. J., O'Connell B. C., Mateyak M. K., Tam W., Kohlhuber F., Dand C. V., Sedivy J. M., Eick D., Vogelstein B., Kinzler K. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nawijn M. C., Alendar A., Berns A. (2011) Nat. Rev. Cancer 11, 23–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.