Abstract
Leucine zippers are oligomerization domains used in a wide range of proteins. Their structure is based on a highly conserved heptad repeat sequence in which two key positions are occupied by leucines. The leucine zipper of the cell cycle-regulated Nek2 kinase is important for its dimerization and activation. However, the sequence of this leucine zipper is most unusual in that leucines occupy only one of the two hydrophobic positions. The other position, depending on the register of the heptad repeat, is occupied by either acidic or basic residues. Using NMR spectroscopy, we show that this leucine zipper exists in two conformations of almost equal population that exchange with a rate of 17 s−1. We propose that the two conformations correspond to the two possible registers of the heptad repeat. This hypothesis is supported by a cysteine mutant that locks the protein in one of the two conformations. NMR spectra of this mutant showed the predicted 2-fold reduction of peaks in the 15N HSQC spectrum and the complete removal of cross peaks in exchange spectra. It is possible that interconversion of these two conformations may be triggered by external signals in a manner similar to that proposed recently for the microtubule binding domain of dynein and the HAMP domain. As a result, the leucine zipper of Nek2 kinase is the first example where the frameshift of coiled-coil heptad repeats has been directly observed experimentally.
Keywords: NMR, Protein Assembly, Protein Conformation, Protein Domains, Signal Transduction, Chemical Exchange, Coiled Coils, Leucine Zippers, Protein Dynamics
Introduction
Intracellular signaling pathways that regulate processes such as cell cycle control rely on formation of specific protein complexes at the right time and place. As a result, a wide range of conserved interaction motifs have evolved among which the leucine zipper is one of the most common and versatile. Leucine zippers were first identified as dimerization domains in bZIP transcription factors with a sequence motif consisting of leucines repeated every 7 amino acids (1). The relevance of the repeating heptad sequence was clarified when it was shown that leucine zippers assume a coiled-coil fold (2–4). In this structure, the first and fourth residues (i.e. positions A and D in the heptad sequence, ABCDEFG) of each helix point toward each other and thus form a hydrophobic core. Residues in positions E and G flank the hydrophobic core residues and are often occupied by charged residues that can form salt bridges. The latter are of particular significance in heterodimeric leucine zippers as they help to determine specificity. Residues in positions B, C, and F are usually not of importance as their side-chains point away from the coiled-coil interface. Leucine zippers show great versatility as they can exist as dimers, trimers, or tetramers, can be homo- or hetero-oligomers and can form parallel or anti-parallel complexes (5–7).
Although leucine zippers have been best characterized in transcription factors, they also exist in many other signaling proteins including protein kinases (8). Protein kinase activation often involves a trans-autophosphorylation step that is facilitated by the physical proximity of two kinase molecules. In the case of receptor tyrosine kinases, this may be brought about by crosslinking of two receptors as a result of extracellular ligand binding. Some cytoplasmic kinases on the other hand contain their own oligomerization domain. One example is the cell cycle-regulated kinase, Nek2, which consists of an N-terminal catalytic domain and a C-terminal region that contains multiple regulatory motifs, including a leucine zipper (9) (Fig. 1A). For this kinase, oligomerization via the leucine zipper is essential for full activation both in vitro and in vivo, most likely as a result of it promoting trans-autophosphorylation (10, 11).
FIGURE 1.
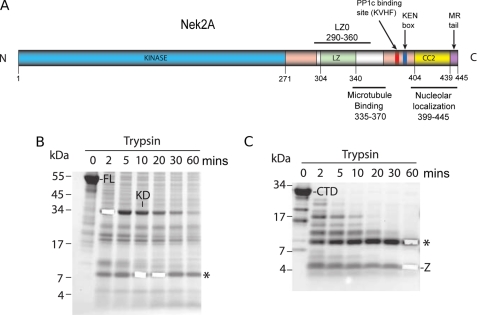
Nek2 domain organization and limited proteolysis. A, scheme of full-length Nek2A kinase annotated with functional and structural motifs indicated with their position in the sequence. B, SDS-PAGE and Coomassie Blue analysis of full-length Nek2A kinase subjected to limited proteolysis for the times indicated (mins) FL, full-length protein; KD, kinase domain; *, 8 kDa fragment. C, as for B, but using the C-terminal non-catalytic region in the proteolysis assay. CTD, C-terminal domain; *, 8 kDa fragment; Z, nonspecific fragment corresponding to a region of Nek2A lacking tryptic sites.
In a previous study on the role of the Nek2 leucine zipper, we noted that the sequence, in terms of the distribution of hydrophobic and charged residues, is somewhat unusual (10) (Fig. 2). However, no structural studies were undertaken at the time. Here, we now show that the Nek2 leucine zipper does indeed display highly atypical biophysical properties with NMR spectroscopy clearly showing that the leucine zipper exists in two conformations, which exchange on a relatively slow timescale. This raises the intriguing possibility that the dimerization and, as a result, activation of the Nek2 kinase may be subject to specific regulation. It is also the first time that the exchange of a coiled-coil domain, indirectly inferred for several completely unrelated proteins, has been directly observed experimentally.
FIGURE 2.
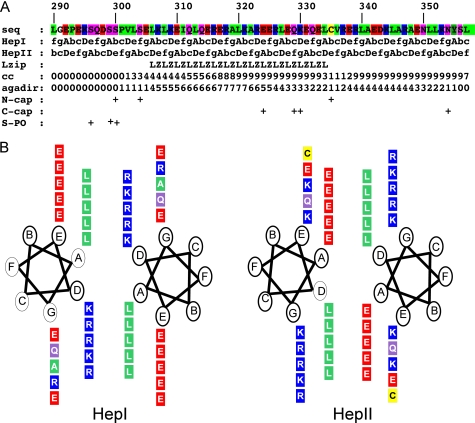
Sequence analysis of the Nek2 leucine zipper. A, sequence of the Nek2 leucine zipper is shown color coded by properties (green, hydrophobic; red, negatively charged; blue, positively charged; magenta, polar uncharged; yellow, cysteine). Below the sequence, the two heptad repeats, HepI and HepII, are shown with the key hydrophobic positions, A and D, marked in uppercase. The leucine zipper prediction by the program 2ZIP is then shown (Lzip) followed by the coiled-coil prediction by the program COILS (cc). The next three lines are provided by the program AGADIR: probability for folding as isolated α-helix (agadir) as well as putative N- and C-caps and finally predicted serine phosphorylation sites (S-PO). B, helical wheel plots of the core leucine zipper residues of Nek2. Amino acids shown in boxes are colored as in A. Heptad repeat positions are indicated in circles for the two possible conformations, HepI and HepII.
EXPERIMENTAL PROCEDURES
Sequence Analysis
Domain definitions for Nek2 kinase were taken from annotations of entry P51955 from the Uniprot database with small modifications for Fig. 1A. Leucine zipper prediction was performed with the 2ZIP server (12). The coiled-coil prediction was performed with the COILS server (13). Single helix preference as well as N- and C-caps were calculated with the web-based version of AGADIR (14). Pattern searches were performed with the program pattinprot (15). Phosphorylation sites around the Nek2 leucine zipper were taken from the literature (11, 16). The helical wheel in Fig. 2B was initially generated using the EMBOSS (17) application pepwheel and then adapted to incorporate the two heptad repeats.
Limited Proteolysis
SDS-PAGE analysis was performed following a 1:500 trypsin:target (w/w) incubation at 25 °C. Aliquots were withdrawn at 2, 5, 10, 20, 30, and 60 min. The reaction for each time point was immediately halted by the addition of Pefabloc SC (2 mm final) and PSC protector solution (5% v/v final) from Roche. Sequencing grade modified trypsin was obtained from Promega. To identify the protected fragments, a 60-min limited digest was performed, as above, and the products separated by reverse-phase chromatography using a JASCO HPLC and a 4.5 ml Zorbex stable bond 300 C3 column at 1 ml/min heated to 55 °C. The column was developed with 5–50% acetonitrile, 0.05% TFA (trifluoroacetic acid) pH 1.8 at 1% acetonitrile/min. Peaks were collected in 0.5-ml fractions and monitored at 280, 220, and 210 nm wavelengths. The fractions were analyzed by standard electrospray MS procedures.
Mass Spectrometry
Protein molecular weight was determined using a stand-alone syringe pump (Perkin Elmer, Foster City, CA) coupled to a Platform electrospray mass spectrometer (Micromass, Manchester, UK). Samples were desalted on-line using a 2 × 10 mm guard column (Upchurch Scientific, Oak Harbor, WA) packed with 50 micron Poros RII resin (Perseptive Biosystems, Framingham) inserted in place of the sample loop on a rheodyne 7125 valve. Proteins were injected onto the column in 10% acetonitrile, 0.10% formic acid, washed with the same solvent and then step-eluted into the mass spectrometer in 70% acetonitrile, 0.1% formic acid at a flow rate of 10 μl/min. The mass spectrometer was calibrated using myoglobin. Standard samples comprised of 100 pmol of protein at a minimum concentration of 1 μm.
Construct Generation
All protein expression constructs were cloned into pETM-11 vectors obtained from the Protein Expression Laboratory at EMBL Heidelberg. Inserts were generated by polymerase chain reaction using 2.5 units of Platinum® Pfx DNA Polymerase (Roche) with 200 ng of template, 500 nm of each primer, 1.2 mm dNTP mix, and 1 mm MgSO4 on a Techne TC-312 thermocycler. Amplification was done by initial denaturation for 2′ at 94 °C followed by 30 cycles of 15″ melting at 94 °C, 60″ at 60 °C and 30″ extension at 68 °C. Primers for constructs LZ0 and LZ5 were LZ05′: GGAGCGCCCATGGCGCGACAATTAGGAGAG; LZ03′: GGATCCTTATAGCAAGCTGTAGTTCTTCACAGATTTTCTGC; LZ55′: GCGCCCATGGCGGTATTGAGTGAGCTGAAACTG; LZ53′: GGATCCTTAGTCCTCTGCTAGTCTCTCACG, respectively. PCR products were purified with QIAquick PCR purification kit according to the manufacturer's protocol. Purified PCR products and pETM-11 target vector DNA were double digested with NcoI and BamHI. The product of the vector digestion was purified by electrophoresis on a 1% agarose gel (analytical quality, Melford Labs). DNA was extracted from excised bands using the QIAquick gel extraction kit. 50 ng of gel purified digestion product from the vector and 150 ng of digested PCR product were mixed and ligated using the rapid DNA ligation kit (Roche). 1/10 of the ligation reaction was transformed into 100 μl DH5a-T1R chemical competent cells (Invitrogen). Transformed cells were checked for inserts by colony PCR.
Site-directed Mutagenesis
Point mutations were generated using the QuikChange kit (Stratagene) using the manufacturer's protocol. Mutagenesis primers were C335A: CAGAAAGAACAGGAGCTTGCAGTTCGTGAGAGACTAG and GTCTCTCACGAACTGCAAGCTCCTGTTCTTTCTG; K309C: CTGTATTGAGTGAGCTGAAACTGTGTGAAATTCAGTTACAGGAGCGAGA and TCTCGCTCCTGTAACTGAATTTCACACAGTTTCAGCTCACTCAATACAG; E310C: ATTGAGTGAGCTGAAACTGAAGTGTATTCAGTTACAGGAGCGAGAGC and GCTCTCGCTCCTGTAACTGAATACACTTCAGTTTCAGCTCACTCAAT (mutated codon indicated in bold). For all constructs and mutants, small scale cultures were grown for several positive clones, and DNA was prepared with the Qiagen miniprep kit. Accuracy of vector and insert was checked by DNA sequencing.
Protein Expression and Purification
For protein expression, miniprep DNA was transformed into BL21* cells (Invitrogen). Transformed cells were grown up in LB medium to OD ∼0.8 when they were induced with 0.75 mm IPTG for 4 h. For 15N isotope labeling the protocol was modified as suggested (18). Cells were opened using a French Press cell at 1000 psi. The proteins were purified on fast flow 6 (GE Healthcare) columns of 2 ml resin, equilibrated as per the manufacturer's instructions. After loading the samples, the columns were washed with 30 ml of wash buffer (20 mm phosphate pH 7.5, 500 mm NaCl, 10 mm imidazole, 1 mm β-ME, 0.02% NaN3) before elution with 10 ml of elution buffer (as wash buffer, but with 500 mm imidazole). The eluted protein was incubated with AcTEV protease (Invitrogen) for 2 h at room temperature followed by dialysis 3× against 1 liter of fast flow 6 wash buffer. The protein solution was applied a second time to the affinity column to remove nonspecific binding proteins. Where required, a final polishing step using a Sephadex 16/70 gel filtration column (GE Healthcare) was performed using an AKTA purification system. Fractions containing the pure protein were pooled and concentrated in Vivaspin concentrators (Sartorius) with 3 kDa molecular mass cut-off. Protein concentrations were determined using the Qbit fluorescence assay (Invitrogen). Protein samples were exchanged into NMR buffer (20 mm sodium phosphate, 50 mm NaCl, pH 7.0, 2 mm DTT, 0.02% NaN3) using PD10 or Nap5 columns (GE Healthcare), which were also used for all other experiments. The only exceptions were samples of disulfide locked LZ5 K309C/C335A, LZ5 E310C/C335A, and LZ0 K309C/C335A for which DTT was omitted from the buffer.
CD spectroscopy. All CD experiments were recorded on a JASCO700 instrument fitted with a Peltier temperature control system. Square cuvettes with 0.1 or 1 mm path length were used with protein concentrations ranging from 20–200 μm. Spectra were calibrated using software provided by the manufacturer. Secondary structure content was estimated using home written Mathematica (Wolfram Research) macros by comparison to standard curves for α-helix, β-sheet, and random coil. To measure thermal unfolding curves, samples were heated at 1 °C/min while the CD signal at a constant wavelength of 222 nm was measured. Unfolding curves were fitted to the equation for a two-state unfolding reaction using a home written Mathematica macro to extract the melting temperature.
Analytical Ultracentrifugation
Analytical ultracentrifugation (AUC)3 sedimentation velocity experiments were carried out on a Beckman XL-I centrifuge using an An50-Ti rotor at 4 °C and a speed of 42,000 rpm (LZ0) and an An-60 Ti rotor at 20 °C and a speed of 60,000 rpm (LZ5 and LZ5 mutants). Scans were recorded using the interference optical system until no further sedimentation occurred. Sample concentrations were about 10–200 μm in standard NMR buffers. Protein partial specific volume and buffer density and viscosity were calculated using SEDNTERP (19). The experimental data were analyzed using Sedfit (20) by fitting to the c(s) and c(s,f/f0) models with one discrete component (LZ0) and results for some samples confirmed by two-dimensional spectrum analysis, enhanced van-Holde-Weischet analysis and Genetic Algorithm analysis using UltraScan (21) confirmed by Monte-Carlo analysis.
NMR Spectroscopy
Spectra were recorded at a temperature of 298 K on Bruker Avance 600 and 800 MHz spectrometers fitted with 5 mm cryoprobes. The HSQC spectrum was used as provided by the manufacturer with small modifications to increase safety of the probe. Exchange experiments were as previously described (22), except modified to increase the dispersion of the peaks by changing from a 15N-1H view to a 1H-1H NOESY-like view and also by combining both views into a three-dimensional experiment.4 For qualitative analysis exchange experiments with a mixing time of 64 ms were used, for the quantitative analysis of the exchange rate the following mixing times were used: 8, 16, 24, 32, 64, 96, and 160 ms. Diagonal and cross peak intensities for sufficiently well resolved systems of exchanging amide resonances were extracted using CCPN analysis (23) and fitted to standard equations for slow exchange (22) using a home written Mathematica macro to yield exchange rates and 15N longitudinal relaxation rates. 15N longitudinal (R1) and transversal (R2) were also measured directly using delays of 16, 48, 96, 192, 288, 384, 512, 704, 880, 1120, and 1440 ms for R1 and 5, 10, 15, 20, 31, 41, 61, 82, 102, 133, 154 ms for R2.
Sequence-specific assignment of mutant LZ5 K309C/C335A in non-reducing NMR buffer was based on standard triple resonance three-dimensional experiments (HNCACB, HN(CO)CACB, HBHA(CBCACO)NH) recorded on a 0.6 mm 15N/13C labeled sample on a Bruker Avance 500 MHz spectrometer equipped with a cryoprobe combined with a 3D 15N resolved NOESY spectrum recorded on a 0.8 mm 15N labeled sample on a 700 MHz Bruker Avance spectrometer equipped with a cryoprobe. The assignment was performed with CCPN analysis (23).
RDCs were measured in the presence of 10 mg/ml of pf1 phage (Hyglos GmbH, Germany) in NMR buffer using a standard IPAP 1H-15N correlation experiment (24). The error associated with the measured RDC values is ± 1.5 Hz.
Model Building
A coiled-coil model was generated for the Nek2 leucine zipper residues 299–341 using a program provided by G. Offer (25). This program generates standard coiled-coils based on the definition of the geometry provided as input. No further energy minimization was performed. Parameters used were pitch = 144 Å, helix radius = 4.7 Å, relative rotation of strands = 210°, residue translation for one residue in the helix = 1.495 Å. The same sequence was used as input for models for both heptad repeats. The only difference was the definition of the first A- residue: Leu-306 in HepI and Leu-303 in HepII.
RDCs were calculated for the models using PALES (26) selecting pf1 phage as alignment medium at a concentration of 10 mg/ml, electrostatic mode, a sodium chloride concentration of 50 mm and default settings for all other parameters.
RESULTS
The Nek2 Leucine Zipper Resides within a Larger Proteolytically Resistant Fragment
The Nek2A kinase consists of an N-terminal catalytic domain (residues 8–271) followed by a C-terminal regulatory region (residues 272–445) that encompasses a leucine zipper (residues 305–335) followed immediately by an additional short coiled-coil (residues 340–355) (Fig. 1A). To determine whether the C-terminal region is subdivided into a particular domain organization, limited proteolysis experiments were performed on full-length protein (Fig. 1B) and the complete C-terminal regulatory region (Fig. 1C and supplemental Fig. S1) to identify stable fragments. A ∼8 kDa fragment appeared consistently in both experiments. It was shown by mass spectrometry to cover not only the leucine zipper but all of the following coiled-coil and a short section of the linker connecting it at the N-terminal end to the catalytic domain (residues 290–360). This 8 kDa fragment was subcloned into the pETM-11 expression vector and termed LZ0. For comparison, a series of constructs was generated covering only the predicted core leucine zipper. The best behaved of these, based on a number of criteria including expression yield in bacteria, solubility and stability, was termed LZ5 (residues 299–343). For all biophysical experiments, LZ0 and LZ5 proteins were expressed in bacteria, purified using nickel affinity chromatography followed by removal of the His tag and polished via gel filtration (data not shown).
Sequence Analysis of the Nek2 Leucine Zipper
Meaningful leucine zipper scores provided by the program 2zip (12) start around residue 305 and continue to approximately residue 335 (Fig. 2A). Coiled-coil scores provided by the COILS software (13) cover the leucine zipper, drop to insignificant levels around residue 335, and then return to a significant level from residue 340–355. The leucine zipper is defined by the presence of leucine residues every 7 amino acids. However, in the conventional heptad repeat of a leucine zipper, a second position is also occupied by another leucine or equally compatible hydrophobic residue such that, in the repeating heptad ABCDEFG, positions A and D would normally both be occupied by hydrophobic residues. In Nek2, though, one of these hydrophobic positions is missing, such that the heptad pattern can be positioned in two ways: the leucines can be in the A-position (heptad I) or in the d-position (heptad II). It is traditionally thought that leucine prefers the d-position, but the energetic contribution to coiled-coil stability and the frequency by which leucine is found in either position in leucine zipper sequences are very similar (27). Regardless of the positions of the heptad repeats in Nek2, charged residues would occupy the second conserved position. In heptad I, it would be lysine or arginine in the D position, while in heptad II, it would be glutamate in the A position. Curiously, lysine and arginine in general prefer the A position while glutamate prefers the D position (27, 28). Even though charged residues have been found in the hydrophobic core of other coiled-coils, e.g. myosin, they compromise the stability and make the Nek2 leucine zipper a non-ideal coiled-coil. It is interesting to note the consistent occupation of the normally hydrophobic A and D positions by charged residues in the leucine zipper of Nek2. Of the 6 A positions in heptad II, 5 are occupied by glutamate (the first is occupied by a leucine), while in the case of heptad I, lysine and arginine each take 3 of the 6 D positions (Fig. 2). The high degree of conservation of these destabilizing residues suggests an important function.
To determine whether other proteins contain related leucine zippers which might allow hetero-oligomerization, similarity searches were performed with a pattern search program (15) using the pattern LXXR/KEXX repeated five times. No coiled-coil sequences other than that of Nek2 were found in this search, even allowing for up to two mismatches. Hence, the primary function of this motif in Nek2 would appear to be to promote homodimerization rather than heterodimerization with a different partner molecule.
Circular Dichroism Spectroscopy of the Wild-type Nek2 Leucine Zipper
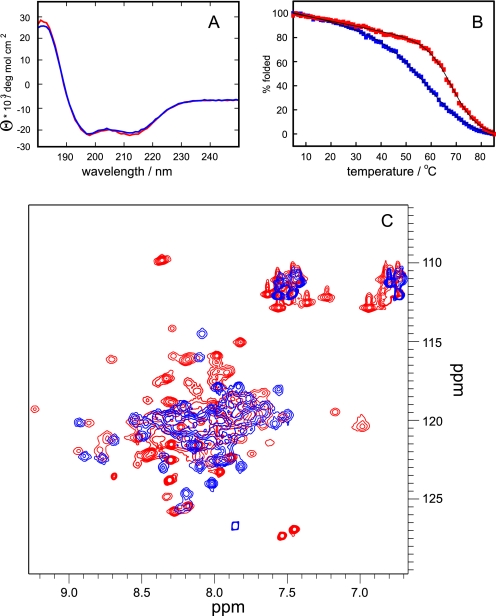
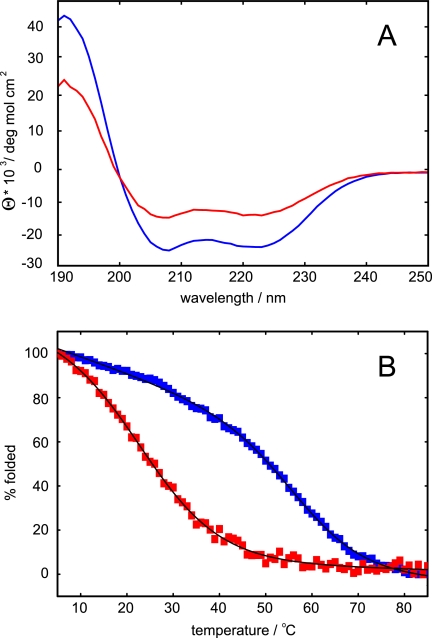
As a first biophysical approach to understand their conformation, CD spectroscopy was performed on the LZ0 and LZ5 polypeptides. The spectra were virtually identical and typical of coiled-coils with minima at 208 and 222 nm and the intensity of the 208 nm peak slightly stronger than the 222 nm peak (Fig. 3A). The molar ellipticities at 222 nm of approximately −22,000 deg mol−1 cm−2 suggested the presence of ∼70% α-helix, in good agreement with expectation. Melting curves showed reasonably cooperative thermal unfolding with relatively high melting temperatures of 57 °C for LZ5 and a slightly higher value of 66 °C for LZ0 (Fig. 3B). These data show that the core leucine zipper sequence alone is sufficient to assume a proper fold and that the additional coiled-coil only adds extra stability.
FIGURE 3.
Comparison of the leucine zipper constructs LZ5 (blue) and LZ0 (red). A, CD spectrum at T = 298 K, concentration 50 μm, path length 1 mm. B, thermal melting curve measured at λ = 222 nm. Melting temperatures obtained from fits to a two state unfolding equation are 57.8 ± 0.2 °C for LZ5 and 66.6 ± 0.2 °C for LZ0. C, superposition of 15N HSQC experiments recorded at 600 MHz and T = 298K.
Analytical Ultracentrifugation
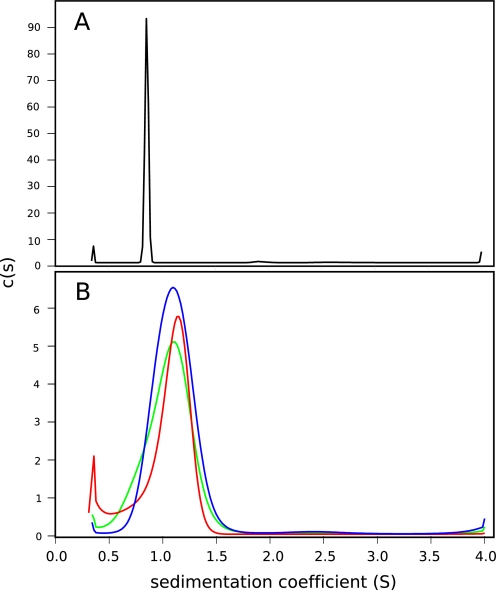
To determine whether the bacterially expressed LZ0 and LZ5 proteins were dimeric as opposed to higher order oligomers, they were subjected to sedimentation velocity analytical ultracentrifugation (AUC). Results strongly supported the conclusion that LZ0 and the LZ5 proteins predominantly formed dimeric molecules that were stable over the relevant concentration range (tested over 10–250 μm). As an example, the sedimentation velocity analysis is shown for LZ0 (Fig. 4A) with the main peak indicating a molecular weight within 500 Da of the calculated molecular mass of the dimer at 17,468 Da. All parameters are listed in Table 1. Although the peaks are not perfectly symmetrical, it can be concluded that the leucine zipper of the Nek2 kinase, with or without the extra coiled-coil portion, forms a dimer as the predominant species. Under the conditions of these experiments there was no sign of higher order oligomers.
FIGURE 4.
Sedimentation velocity analytical ultracentrifugation. c(s) distributions for (A) 228 μM LZ0 (B) LZ5 (blue) and LZ5 K309C/C335A at a concentration of 224 μM in reducing (red) and non-reducing conditions (green). LZ5 wt is shown in blue, LZ5 K309C/C335A in reducing buffer in red and in non-reducing buffer in green. For details on the data analysis see “Experimental Procedures.” For full results see Table 1.
TABLE 1.
Sedimentation velocity results for the leucine zipper constructs employed in this study
| Construct | Sapp | S20,w | F/F0 | Monomer MW | Apparent MW |
|---|---|---|---|---|---|
| S | S | kDa | kDa | ||
| LZ0, 228 μm | 0.86 | 1.40 | 1.7 | 8.7 | 17.0 |
| LZ0, 29 μm | 0.89 | 1.40 | 1.7 | 8.7 | 17.7 |
| LZ5 | 1.10 | 1.11 | 1.5 | 5.4 | 10.7 |
| LZ5 C335A/K309C reduced | 1.07 | 1.08 | 1.5 | 5.4 | 10.0 |
| LZ5 C335A/K309C oxidized | 1.05 | 1.06 | 1.6 | 5.4 | 10.1 |
NMR Spectroscopy of the Wild-type Nek2 Leucine Zipper
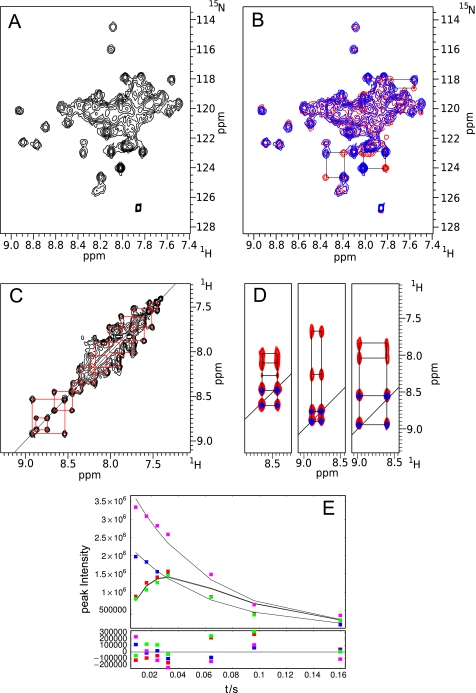
Early NMR studies of constructs of the Nek2 leucine zipper showed unusual “twins” of resonances in homonuclear NOESY and TOCSY spectra suggesting the presence of multiple forms of the protein (data not shown). A more detailed analysis of potential isoforms was therefore performed using a 15N-labeled sample of LZ5. In an HSQC experiment, automatic peak picking with CCPN analysis (23) gave >75 peaks, significantly more than the expected ∼40 peaks, confirming the presence of at least two forms of the protein (Fig. 5A). Very similar spectra were obtained for LZ0, although as the quality of the spectra was much inferior (Fig. 3) all subsequent NMR work was done on LZ5.
FIGURE 5.
NMR spectra of LZ5. A, HSQC spectrum at T = 278K, 600 MHz. B, 15N exchange experiment shown as HSQC (red), mixing time 60 ms, superimposed on HSQC (blue). Connections of exchanging species in the HSQC spectrum via the cross peaks in the exchange spectrum are shown by black boxes for a few residues. C, 15N exchange experiment shown as NOESY, mixing time 60 ms. The exchange for a number of residues is illustrated by red squares. D, superposition of 1H-1H slices of the three-dimensional 15N exchange experiment (64 ms mixing time) with a 15N NOESY-HSQC (100 ms mixing time). Contours for the NOESY are in red, contours for the exchange experiment in blue. E, time dependence of exchange (red, green) and diagonal peak (blue, magenta) intensity of a selected residue (boxes) compared with the results of the fits (continuous lines). Fitting errors are shown below.
To establish if the isoforms were in dynamic equilibrium, two NMR exchange experiments were recorded (22) via transfer to 15N. The resulting two-dimensional spectrum can then take the shape of an HSQC experiment by frequency labeling the nitrogen in t1 (Fig. 5B), or the appearance of a NOESY experiment with frequency labeling of the amide proton in t1 (Fig. 5C).
Both experiments clearly demonstrated that a substantial number of resonances in the Nek2 leucine zipper undergo slow exchange on the chemical shift time scale (Fig. 5, B and C). To further improve the identification of exchange cross peaks, both versions of the exchange experiment were combined into a three-dimensional version that has the appearance of a three-dimensional 15N NOESY-HSQC. A few representative slices are shown in Fig. 5D. Using this three-dimensional exchange spectrum it was possible to identify more than 34 exchanging pairs of resonances suggesting that essentially the entire protein is subject to exchange and that only two isoforms of the protein exist in solution. Comparison of the diagonal peak intensities of selected well-resolved exchange pairs suggests that both forms of the protein exist in almost equal abundance.
A quantitative analysis of the exchange was performed by fitting cross and diagonal peak intensities of pairs of exchanging resonances from a series of exchange experiments with different mixing times to yield the exchange rate (kex) and the 15N longitudinal relaxation rate (R1) (22). An example for the fit is shown in Fig. 5E for one of the four exchanging pairs that were analyzed. The values obtained for kex, 18.2, 17.4, 16.9, 17.2 s−1, were well within the error of the fitting procedure so that they were averaged to give a single rate constant of 17.4 ± 1.7 s−1 assuming a single, cooperative exchange event. 15N R1 values obtained from the same fit for the four systems were 1.5, 1.7, 1.4, and 1.6 s−1, averaged to 1.6 ± 0.3 s−1. Fitting the peak intensities of resolved diagonal peaks directly to an exponential decay to obtain the apparent 15N R1 gave values of 14.9, 16.8, 15.3, 16.1 s−1 which averaged to 15.8 ± 1.6 s−1, approximately ten times the nominal value. These dynamics values were complemented by the direct measurement of 15N R1 and R2 values. For the four well defined resonances R1 values of 14.1, 15.9, 14.6, 15.3 s−1 and R2 values of 29.7, 30.8, 29,9 and 31.1 s−1 were obtained giving averages of 15.0 ± 0.7 s−1 and 30.4 ± 0.8 s−1, respectively.
To ensure that the residues undergoing exchange were not doing so because they are unstructured, a three-dimensional 15N NOESY-HSQC experiment was also recorded and several representative slices are shown superimposed on the exchange spectrum (Fig. 5D). It is apparent that for the majority of resonances with exchange cross peaks there are genuine amide-amide sequential NOE cross peaks, typical for α-helices. Thus, the exchange is not a result of unfolding events.
Probing the Leucine Zipper Conformational Exchange by Site-directed Mutagenesis
The existence of two conformations in all recombinant versions of the leucine zipper made interpretation of NMR spectra very difficult. This was due, firstly, to extensive overlap of peaks: the chemical shift dispersion of coiled-coils is notoriously poor and so was made worse by having two highly similar versions of the same protein in the sample. Furthermore, although the chemical exchange is slow on the chemical shift time scale, it is very close to the transition region toward intermediate exchange. As a result, both longitudinal and transversal relaxation rates of protons and nitrogens are significantly accelerated, making the recording of complex three-dimensional experiments required for sequence specific assignment and structure calculation virtually impossible.
It was therefore decided to employ site-directed mutagenesis to probe the LZ structure and dynamics. For this, we hypothesized that the conformational dynamics might result from exchange between the two alternative heptad repeats described earlier (see Fig. 2). To test this hypothesis, individual arginine/lysine or glutamate positions in the 2nd or 3rd heptad repeats were mutated to cysteine. The intrinsic cysteine of the leucine zipper, Cys-335, was replaced by alanine to avoid interference. Assuming that the Nek2 leucine zipper folds as a parallel coiled-coil, a disulfide bridge can form between cysteines in either the A-position (mutation from glutamate) or the D-position (mutation from lysine/arginine) once the sample is in non-reducing buffer. Disulfide bonds in coiled-coils have been observed to occur naturally, e.g. in myosin (29) and were shown to contribute to the stability of the coiled-coil in synthetic leucine zippers (30).
Based on this hypothesis, one would predict that a glutamate to cysteine or lysine/arginine to cysteine mutant under non-reducing conditions should exist in only one conformation and should not exchange. Should it be possible to obtain NMR spectra of conformationally “locked” mutants then, at least in general, it should be possible to reconstruct the spectrum of the wild-type protein from the spectra of one pair of cysteine mutants, representing the heptad I or heptad II conformation. Initially, experiments were attempted with an LZ5-E310C/C335A mutant that should lock the protein in the heptad II conformation with the acidic residues in the A position. However, CD spectra showed only a modest amount of α-helix at lower temperatures, regardless of the oxidation state of the buffer, while AUC data of this mutant could not be interpreted and 15N HSQC spectra were even more complex than those obtained with the wild-type protein (data not shown). All of these data suggest that the mutant is unfolded or at best only partially folded.
We therefore prepared an LZ5-K309C/C335A mutant to lock the heptad I conformation with the basic residues in the D position. A preliminary analysis of this mutant by CD showed that oxidation of the cysteines to form the expected disulfide bond led to a substantial increase in the α-helix content (Fig. 6A). This reflects the fact that a reduced cysteine in position D destabilizes the leucine zipper (30). The reduced form also had a much lower stability compared with the oxidized form as illustrated by the thermal unfolding characteristics. The melting temperature of the reduced form was around 27 °C, while the oxidized form melted at 55 °C (Fig. 6B). While the CD experiments did not provide information on the protein dynamics, they did suggest that the mutated protein folds into a parallel coiled-coil. This was supported by AUC sedimentation velocity data indicating the existence of a dimer in non-reducing buffer (Table 1). Fig. 4B shows the c(s) distributions for LZ5 and the double mutant K309C/C335A in reducing and non-reducing conditions. Again, there is one main peak with fitting parameters suggesting a molecular weight close to that of the dimer over the entire concentration range tested.
FIGURE 6.
CD spectroscopy of the LZ5 K309C/C335A mutant. A, CD spectrum at T = 278 K. The spectrum of the reduced protein is shown in red, the spectrum of the oxidized protein is shown in blue. B, thermal unfolding curves. The measured data is shown in red boxes for the reduced sample and in blue boxes for the oxidized sample. The fitted curves are shown for both samples as thin black lines. Melting temperatures obtained from a fit to a two state unfolding equation are 57.7 ± 0.3 °C for the oxidized sample and 24.3 ± 0.3 °C for the reduced sample.
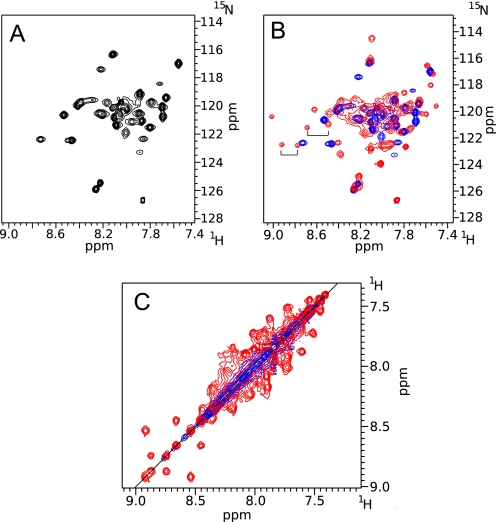
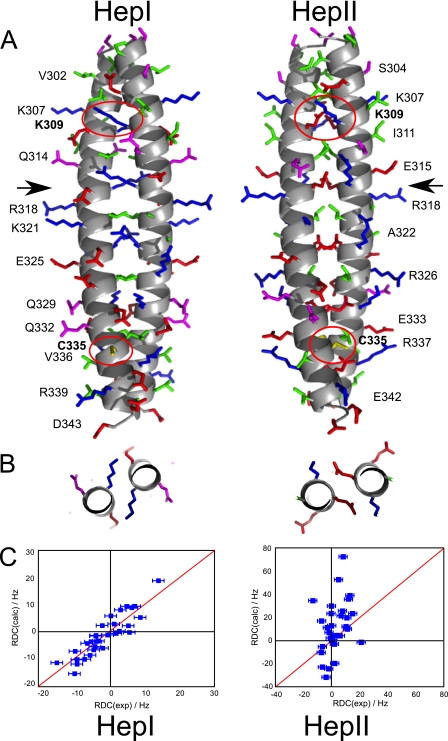
To analyze the effect of disulfide bond formation on the exchange dynamics, a HSQC spectrum of the mutant LZ5 K309C/C335A was recorded in non-reducing buffer (Fig. 7A). It was apparent that the total number of peaks was significantly less than in a spectrum of wild-type LZ5. Automatic peak picking produced a total of just over 40 peaks, very close to the predicted number of 45. This suggests that this sample contained only a single conformation of the leucine zipper. A direct comparison of the C335A and K309C/C335A mutants of LZ5 revealed the similarity of the spectra (Fig. 7B, supplemental Fig. S2). In the resolved region on the left, it was particularly apparent that, as predicted, one peak of an exchanging pair (indicated by brackets) vanished leaving only its partner behind. It was apparent that the peaks were much sharper than in the HSQC of the wild-type protein, also hinting at a significant change in the dynamics of the protein. This observation was confirmed by a NOESY-type exchange experiment which had no exchange cross peaks at all (Fig. 7C). The good quality of the spectra allowed us to obtain an almost complete backbone and partial side chain assignment (deposited with the BMRB, accession code 17417). This was used to collect an extensive set of secondary structure specific proton-proton distances which support the well defined helical structure (supplemental Fig. S6). In addition, 1H-15N residual dipolar couplings were recorded and compared with RDC values calculated for models created for both conformers shown in Fig. 8A. As can be seen in Fig. 8C there is good agreement of the experimental data with those predicted for the HepI model (axial component Da = 9.33 10−4, rhombic component Dr = 1.80 10−4, correlation coefficient r = 0.84, Q-value 0.21) while very little agreement is seen with those predicted for HepII (axial component Da = −3.20 10−3, rhombic component Dr = −2.06 10−3, correlation coefficient r = 0.34, Q-value 37.3). Taken together, these data strongly support the hypothesis that the LZ5 K309C/C335A mutant represents one of the conformers observed for the wild type leucine zipper.
FIGURE 7.
NMR spectra of the LZ5 K309C/C335A mutant. A, 15N HSQC of LZ5-K309C/C335A, oxidized, T = 298K. B, same as in A superimposed on the spectrum of wild-type LZ5. Two exchange pairs are indicated by brackets in the well resolved part of the spectrum of the wild-type protein B: 15N exchange experiment shown in NOESY view of oxidized LZ5 K309C/C335A superimposed on the same experiment for wild-type LZ5. In both B and C the spectrum of wild-type LZ5 is red, for LZ5-K309C/C335A blue.
FIGURE 8.
A, model of the proposed conformations of the Nek2 leucine zipper. The leucine zipper for both conformations is shown with the N terminus at the top and the C terminus at the bottom. The main chain is shown as a helical scheme, the side chains are shown as stick models. No hydrogens are shown. Side chains are colored according to their properties as in Fig. 2. Residues selected for easy visibility are labeled to provide guidance. Arrows indicate the position of Arg-116 in HepI and Glu-317 in HepII where the slices shown in B are taken. Residues Lys-309 and Cys-335, which are mutated, are labeled in bold and the side chains encircled in red. B, packing of charged side chains in the A position of HepI (Arg-316) and the D position in HepII (Glu-317) in a slice through the center of the coiled-coil as marked by arrows in A. C, validation of the model by measurement of 1H-15N RDC values for mutant K309C/C335A of LZ5. Measured RDCs on the x-axis are compared with predicted RDCs (on the y-axis) calculated with PALES for the two models.
DISCUSSION
Unusual slow chemical exchange between two conformations with a rate of 17 s−1 was observed for the Nek2 leucine zipper in solution. This exchange was observed regardless of construct, sample or any other experimental conditions (supplemental Fig. S4–6) so that it cannot be dismissed as an artifact but has to be seen as a genuine property of the isolated domain that may well hold true for the full-length kinase in vivo.
Examples of dynamics in leucine zippers based on experimental data are few and far between. A modest level of dynamics, albeit fast on the chemical shift timescale, was inferred from a comparison of the NMR data with the crystal structure of the first well characterized leucine zipper, GCN4 (3, 31) and similar observations were made in the leucine zipper of c-Jun (32, 33). These were based on the presence of an asymmetric conformation of a central asparagine in the crystal structure while only a single signal was seen in the NMR spectra suggesting fast averaging in solution.
Slow chemical exchange between folded conformations for a protein involving a leucine zipper was only ever observed once, in the case of the tetramerization domain of the Mnt repressor (34). The tetrameric leucine zipper exists in two different conformations which are generated by a staggered assembly of two coiled-coil dimers that are packed into a distorted four helical coiled-coil. Such a complex assembly is well beyond the capabilities of the small dimeric Nek2 leucine zipper.
The best explanation for the unusual dynamics of the Nek2 leucine zipper would therefore seem to be the existence of two coiled-coils based on a shift in the heptad register (Fig. 8A) where the charged side chains in the A/D positions minimize adverse interactions by extending outside the hydrophobic core with their charged groups (Fig. 8B). Determination of the structure by experimental methods is very challenging and efforts to crystallize the protein may well fail as a result of these inherent dynamics. Also standard NMR methods cannot be used to obtain structural information because of the detrimental effects caused by the exchange rate of 17 s−1. Even though it is slow on the chemical shift timescale, it is still sufficiently fast to have detrimental effects in NMR experiments. As a consequence it is impossible to record triple resonance three-dimensional experiments required for complete assignments.
We were therefore forced to use an indirect approach based on its unusual sequence to investigate the dynamics of the Nek2 leucine zipper. As this sequence contained only one leucine every seven amino acids, there were two ways of positioning the heptad (Figs. 2A and 8A). In essence, both heptads satisfied the definition of a leucine zipper albeit with charged residues in places normally occupied by hydrophobic amino acids. A similar arrangement has been previously described in the myosin coiled-coil (29), where polar residues in these positions have long side chains so that the charged group is at least partly solvent accessible, while the long aliphatic portion can make some contribution to the hydrophobic core as shown in Fig. 8B. The downside of such an arrangement is unfavorable side chain torsion angles that compromise the stability of such leucine zippers.
To provide experimental proof for the two heptad hypothesis we decided to lock the protein in one of the two conformations using a disulfide bond. Two mutants were generated in LZ5, K309C to lock heptad I and E310C to lock heptad II, both as double mutants with C335A to avoid interference. Mutant K309C/C335A in non-reducing buffer performed very much as expected: it stopped the exchange and reduced the number of peaks in the HSQC to that expected for a simple leucine zipper. Moreover, the line widths of the peaks were now more consistent with proteins of this molecular weight. The fact that these peaks did not exactly match the peaks in the wild type spectrum can be explained by the presence of a double mutant and the significant changes in environment introduced by the disulfide bond. A similar, though less apparent, effect of the formation of the disulfide bond in the K309C/C335A double mutant was also seen in LZ0 (supplemental Fig. S6). In addition to supporting our assumption that the K309C/A335C mutant represents the HepI conformation this result provides indirect evidence for the distinct charge distribution on the surface of the two conformers. Their overall shape is virtually identical (Fig. 8A) so that not much of a difference could be expected if their alignment was purely steric. Using pf1 phage as alignment medium, however, means that the alignment is strongly driven by electrostatic interactions, which allows to differentiate the two different conformers as suggested previously (35, 36).
In contrast, mutant E310C/C335A did not give a clear cut result in agreement with previous results. The disulfide bond for residue Cys-310 would be in the A position which was shown to be destabilizing rather than stabilizing unless located right at the N or C terminus (30). As the Nek2 leucine zipper is already less than ideal, the modest destabilization by a disulfide bond in the A position could compromise the entire structure.
While it would be preferable to have at least one mutant to lock each conformation, we believe that these data are sufficient to provide support for the hypothesis that the slow exchange seen in the NMR spectra of the Nek2 leucine zipper results from the interconversion of the two possible heptad arrangements. The actual conformational change would be a simple rotation of each helix by ∼20° about its long axis. The relationship between degree of structural change required and time scale of exchange matches well the two examples in the literature: the rearrangement in the Mnt tetramerization domain is substantial which is reflected in the slow exchange rate (1 s−1) when compared with the Nek2 leucine zipper (17 s−1), which in turn is much slower than the simple rotation of the asparagine side chain in GCN4 and c-Jun, which happens on a fast timescale (>1000 s−1).
The populations of the two conformations as judged by the peak intensities in the HSQC spectra is not far from 50:50, suggesting that the exchange, at least in vitro, does not serve to shift the equilibrium toward one or other of the conformations. Nevertheless, the surfaces, in particular the charge distribution, of both conformations are very different: heptad I has a highly positive charge density in the center and negative charges on the edges, while heptad II has negative charges on the inside and positive charges on the ridges, indirectly supported by the RDC data (Fig. 8A). As a result the conformational exchange could be modulated by the interaction of Nek2 with partner proteins or via phosphorylation, which has been shown to occur on serines 299 and 300 in a cell cycle-dependent manner (16).
An externally triggered shift in the register of a heptad repeat has been suggested recently for two completely different proteins, the microtubule binding domain (MTBD) of dynein (37, 38) and the HAMP domain that is found in a range of transmembrane proteins (39). In the case of MTBD it is hypothesized that binding to microtubules is the initial trigger and that the heptad repeat shift is used to transmit this signal to the distant AAA+ domain ring structure to activate the ATPase activity. For both these proteins indirect data based mainly on site directed mutagenesis was used to support this hypothesis even though only one form of each protein was observed. The latter is possibly due to the fact that the heptad repeat sequence conforms much more to the standard than is the case with Nek2. As a result, it is therefore not surprising to see a heptad repeat shift as a tool for signal transmission within a protein. The leucine zipper of Nek2 kinase is simply a more extreme case than MTBD and HAMP, which makes it the first protein where this shift could be observed experimentally. In eukaryotic cells coiled-coil proteins are often part of highly dynamic structures and we expect that our findings will stimulate new approaches to understanding cellular regulation. This will make the Nek2 leucine zipper a powerful model system for the experimental study of this novel mechanism for the regulation of protein function.
Supplementary Material
Acknowledgments
We thank the Protein Expression Laboratory at EMBL Heidelberg for a gift of pETM-11 plasmid, Gerald Offer for making his coiled-coil modeling program available to us, Andrew Atkinson for help with the IPAP experiment, and Ragini Gosh for help with the AUC experiments.
This work was supported by grants (to A. M. F.) from the Hope Foundation for Cancer Research, Cancer Research UK, the Association for International Cancer Research, and The Wellcome Trust.
Assignments were deposited with the Biological Magnetic Resonance Data Bank, accession code 17417.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
F. W. Muskett, R. A. Croasdale, A. M. Fry, and M. Pfuhl, unpublished results.
- AUC
- analytical ultracentrifugation
- NOESY
- nuclear Overhauser enhancement spectroscopy
- HSQC
- heteronuclear single quantum coherence
- TOCSY
- total correlation spectroscopy
- RDC
- residual dipolar couplings
- MTBD
- microtubule binding domain
- HAMP
- domain in histidine kinases, adenylyl cyclases, methyl accepting chemotaxis receptors, and phosphatases.
REFERENCES
- 1. Landschulz W. H., Johnson P. F., McKnight S. L. (1988) Science 240, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 2. Oas T. G., McIntosh L. P., O'Shea E. K., Dahlquist F. W., Kim P. S. (1990) Biochemistry 29, 2891–2894 [DOI] [PubMed] [Google Scholar]
- 3. Saudek V., Pastore A., Castiglione M. A., Frank R., Gausepohl H., Gibson T., Weih F., Roesch P. (1990) Protein Eng. 4, 3–10 [DOI] [PubMed] [Google Scholar]
- 4. O'Shea E. K., Klemm J. D., Kim P. S., Alber T. (1991) Science 254, 539–544 [DOI] [PubMed] [Google Scholar]
- 5. Alber T. (1992) Curr. Opin. Genet. Dev. 2, 205–210 [DOI] [PubMed] [Google Scholar]
- 6. Adamson J. G., Zhou N. E., Hodges R. S. (1993) Curr. Opin. Biotechnol. 4, 428–437 [DOI] [PubMed] [Google Scholar]
- 7. Mason J. M., Arndt K. M. (2005) ChemBioChem 5, 170–176 [DOI] [PubMed] [Google Scholar]
- 8. Mukai H., Ono Y. (1994) Biochem. Biophys. Res. Commun. 199, 897–904 [DOI] [PubMed] [Google Scholar]
- 9. Fry A. M. (2002) Oncogene 21, 6184–6194 [DOI] [PubMed] [Google Scholar]
- 10. Fry A. M., Arnaud L., Nigg E. A. (1999) J. Biol. Chem. 274, 16304–16310 [DOI] [PubMed] [Google Scholar]
- 11. Rellos P., Ivins F. J., Baxter J. E., Pike A., Nott T. J., Parkinson D. M., Das S., Howell S., Fedorov O., Shen Q. Y., Fry A. M., Knapp S., Smerdon S. J. (2007) J. Biol. Chem. 282, 6833–6842 [DOI] [PubMed] [Google Scholar]
- 12. Bornberg-Bauer E., Rivals E., Vingron M. (1998) Nucleic Acids Res. 26, 2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lupas A., Van Dyke M., Stock J. (1991) Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 14. Muñoz V., Serrano L. (1994) Nat. Struct. Biol. 1, 399–409 [DOI] [PubMed] [Google Scholar]
- 15. Bucher P., Karplus K., Moeri N., Hofmann K. (1996) Comput. Chem. 20, 3–23 [DOI] [PubMed] [Google Scholar]
- 16. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 17. Rice P., Longden I., Bleasby A. (2000) Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 18. Marley J., Lu M., Bracken C. (2001) J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 19. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton J. C. eds) pp. 90–125, Royal Society of Chemistry, Cambridge, U.K [Google Scholar]
- 20. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Demeler B. (2005) UltraScan A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments, In Modern Analytical Ultracentrifugation: Techniques and Methods, (Scott J., Harding S. E., Rowe A. eds) pp. 210–229, Royal Society of Chemistry (UK) [Google Scholar]
- 22. Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E. (1994) J. Biomol. NMR 4, 727–734 [DOI] [PubMed] [Google Scholar]
- 23. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 24. Ottiger M., Delaglio F., Bax A. (1998) J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 25. Offer G., Sessions R. (1995) J. Mol. Biol. 249, 967–987 [DOI] [PubMed] [Google Scholar]
- 26. Zweckstetter M. (2008) Nat. Protoc. 3, 679–690 [DOI] [PubMed] [Google Scholar]
- 27. Tripet B., Wagschal K., Lavigne P., Mant C. T., Hodges R. S. (2000) J. Mol. Biol. 300, 377–402 [DOI] [PubMed] [Google Scholar]
- 28. Straussman R., Ben-Ya'acov A., Woolfson D. N., Ravid S. (2007) J. Mol. Biol. 366, 1232–1242 [DOI] [PubMed] [Google Scholar]
- 29. Blankenfeldt W., Thomä N. H., Wray J. S., Gautel M., Schlichting I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17713–17717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou N. E., Kay C. M., Hodges R. S. (1993) Biochemistry 32, 3178–3187 [DOI] [PubMed] [Google Scholar]
- 31. Saudek V., Pastore A., Morelli M. A., Frank R., Gausepohl H., Gibson T. (1991) Protein Eng. 4, 519–529 [DOI] [PubMed] [Google Scholar]
- 32. Junius F. K., Mackay J. P., Bubb W. A., Jensen S. A., Weiss A. S., King G. F. (1995) Biochemistry 34, 6164–6174 [DOI] [PubMed] [Google Scholar]
- 33. Junius F. K., O'Donoghue S. I., Nilges M., Weiss A. S., King G. F. (1996) J. Biol. Chem. 271, 13663–13667 [DOI] [PubMed] [Google Scholar]
- 34. Nooren I. M., Kaptein R., Sauer R. T., Boelens R. (1999) Nat. Struct. Biol. 6, 755–759 [DOI] [PubMed] [Google Scholar]
- 35. Zweckstetter M., Schnell J. R., Chou J. J. (2005) J. Am. Chem. Soc. 127, 11918–11919 [DOI] [PubMed] [Google Scholar]
- 36. Schnell J. R., Zhou G., Zweckstetter M., Rigby A. C., Chou J. J. (2005) Protein Sci. 14, 2421–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carter A. P., Garbarino J. E., Wilson-Kubalek E. M., Shipley W. E., Cho C., Milligan R. A., Vale R. D., Gibbons I. R. (2008) Science 322, 1691–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gibbons I. R., Garbarino J. E., Tan C. E., Reck-Peterson S. L., Vale R. D., Carter A. P. (2005) J. Biol. Chem. 280, 23960–23965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M. (2006) Cell 126, 929–940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.