Abstract
Renal-specific oxidoreductase/myo-inositol oxygenase (RSOR/MIOX) catabolizes myo-inositol and is implicated in the pathogenesis of diabetic nephropathy. How high glucose (HG) ambience up-regulates its expression and enzyme activity was investigated. MIOX up-regulation was associated with an increase in enzyme activity, which was reduced to basal levels with phosphatase treatment. Using phosphothreonine, protein kinase A (PKA), and PKC substrate antibodies, analyses of kidney lysates of diabetic animals and LLC-PK1/HK-2 cells subjected to HG ambience indicated MIOX to be a phosphoprotein. Kinase phosphorylated recombinant RSOR/MIOX proteins had increased activity confined to exons 2–5. Mutants with substituted phosphorylation sites had a minimal increase in activity. Treatment of cells with PKC, PKA, and PDK1 kinase activators increased activity, whereas inhibitors reduced it. Inhibitors also reduced the phosphorylation and activity of MIOX induced by HG. Besides HG, exposure of cells to oxidants H2O2 and methylglyoxal up-regulated MIOX expression and its phosphorylation and activity, whereas antioxidants N-acetylcysteine, β-naphthoflavone, and tertiary butyl hydroquinone reduced MIOX expression. Treatment with HG or oxidants or overexpression of MIOX induced nuclear translocation of redox-sensitive transcription factor Nrf2, which binds to antioxidant response elements of various promoters. Promoter analyses revealed an increase in luciferase activity with HG and oxidants. Analyses of antioxidant response elements and carbohydrate response elements revealed an accentuation of DNA-protein interactions with oxidants and under HG ambience. ChIP-PCR and immunofluorescence studies revealed nuclear translocation of carbohydrate response element-binding protein. These findings suggest that phosphorylation of RSOR/MIOX enhances its activity, which is augmented by HG via transcriptional/translational events that are also modulated by diabetes-related pathobiological stresses.
Keywords: Diabetes, Extracellular Matrix Proteins, Glucose Metabolism, Kidney, Protein Kinase A (PKA), Protein Kinase C (PKC), High Glucose Ambience, Oxidant Stress, Oxidoreductase, Renal Tubular Cells
Introduction
Diabetic nephropathy in man is characterized by thickening of basement membranes, mesangial expansion with progression to glomerulosclerosis, tubular atrophy, and interstitial fibrosis, resulting ultimately in renal failure (1–3). A number of mechanisms, involving a complex array of glucose-metabolizing enzymes and various signaling molecules, have been proposed for the pathogenesis of diabetic nephropathy (4–12). One of the molecules whose homeostasis is well known to be adversely affected in diabetic neuropathy is myo-inositol (MI)2 (13, 14); however, its pathogenetic role in diabetic nephropathy is unclear. MI is an essential molecule distributed ubiquitously, and it plays a vital role in signal transduction and osmoregulation, the latter process due to MI having been endowed with unique properties as an organic osmolyte or polyol (15, 16). As MI is also responsible for phosphoinositide signaling, it would be critical for cellular activity and is likely to have a high turnover rate, and thus its regular supplementation would be necessary to maintain various normal physiological processes (17, 18). The major sources by which it could be derived include dietary uptake, cyclic turnover from phosphoinositide signaling pathway, and de novo synthesis from glucose (19, 20), the latter process confined mainly to kidney, brain, liver, and testis. Overall, it seems that MI homeostasis is maintained by a combination of transport, synthesis, and its catabolism. In diabetes, excessive urinary excretion of MI suggests its deranged homeostasis, which in part may be due to the fact that high extracellular glucose inhibits the cellular uptake of MI by compromising its tubular reabsorption (21, 22). On the other hand, high glucose generates excessive intracellular sorbitol via the polyol pathway, which also interferes in the uptake of MI and causes its depletion (15, 21, 22). Besides sorbitol-induced MI depletion and inhibition of cellular MI uptake, there may be further accentuation of MI deficiency related to its catabolism, which is regulated by an enzyme known as myo-inositol oxygenase (MIOX) (23, 24). It catabolizes MI to d-glucuronate, which enters into the pentose phosphate pathway following the interconversion of glucuronate to xylulose (25). Conceivably, this catabolic pathway is extensively utilized by the kidney because MIOX is highly expressed in the kidney, particularly in the proximal tubules (26). The proximal tubular epithelial cells are believed to be involved in the pathogenesis of corticointerstitial fibrosis (27, 28), underscoring the importance in investigating the relevance of the proximal tubule metabolic enzyme MIOX in diabetic nephropathy.
MIOX is a 32-kDa protein, which was originally identified as mouse renal-specific oxidoreductase (RSOR), has affinity for NADPH, and is also known as kidney-specific protein 32 (KSP-32) (23, 29). MIOX expression is up-regulated in diabetes mellitus (23, 28), and conceivably, its up-regulation may be responsible for the concomitant MI depletion and increased urinary excretion. With respect to MI homeostasis, the pathways involving the metabolism of glucose and MI are interdependent (Fig. 1). As indicated in Fig. 1, Step I and II routes are the major events responsible for the urinary excretion of MI and its depletion. Thus, the MI depletion may be a purely osmoregulatory phenomenon that is further exacerbated by the up-regulation of MIOX and the degradation of MI under high glucose ambience. Interestingly, MIOX is also up-regulated to a certain degree by various osmotic stresses (25, 28). The events that follow high glucose ambience or diabetic state include increased flux of glucose intermediaries into various cellular metabolic pathways, increased synthesis of advanced glycation end products, activation of signaling molecules like PKC and PKA, up-regulation of TGF-β, generation of reactive oxygen species (ROS), and ultimately excessive accumulation of extracellular matrix (4–12, 30, 31). Because MIOX is also intimately involved in glucose metabolism and it catabolizes MI, which in turn modulates phosphoinositide signaling (see above), it would be of special interest to explore the mechanism(s) of its up-regulation by high glucose ambience and stresses associated with it, e.g. oxidant stress. Furthermore, recent studies in a large series of patients with renovascular complications demonstrating association of Type I diabetes mellitus in man with polymorphism(s) of MIOX gene further underscore its enormous clinical significance (32). In view of the above considerations, studies were initiated to investigate how glucose regulates de novo synthesis and intracellular MI turnover and to delineate the transcriptional and post-translational events that modulate the functional activity of RSOR/MIOX.
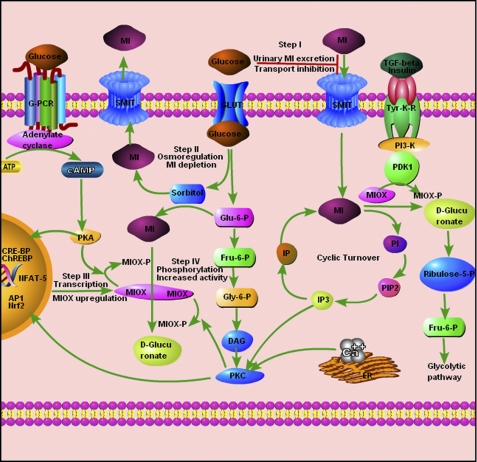
FIGURE 1.
Modulation of MI homeostasis by MIOX in hyperglycemia. The events related to phosphorylation of MIOX by PDK1, PKC, and PKA are depicted along with myo-inositol catabolism and turnover. In the diabetic state, hyperglycemia directly regulates MI homeostasis via various known interdependent pathways. Step I (transport inhibition) and Step II (osmoregulation) are major well established events related to increased MI urinary excretion and its depletion in diabetes mellitus. The high glucose-induced de novo synthesis and intracellular MI turnover are in part controlled by MIOX up-regulation. However, the mechanism(s) of MIOX up-regulation to modulate MI catabolism in hyperglycemia is unclear, and the proposed events are depicted in Step III and Step IV. Conceivably, in Step III, MIOX up-regulation is due to activation of different transcription factors, such as AP1, Nrf2, NFAT-5, cAMP-response element-binding protein (CRE-BP), and ChREBP, by different forms of stress induced by hyperglycemia. In Step IV, the proposed events include post-translational modulation of MIOX with increased enzymatic activity following phosphorylation by kinases, such as PKA, PKC, and PDK1 in the diabetic milieu. G-PCR, G-protein-coupled receptor; GLUT, glucose transporter; Fru, fructose; DAG, diacylglycerol; IP3, inositol 1,4,5-trisphosphate; PIP2, phosphatidylinositol 4,5-bisphosphate; IP, inositol phosphate; PI, phosphoinositide; ER, endoplasmic reticulum; Tyr-K-R, tyrosine kinase receptor; Glu, glucose; Gly, glyceraldehyde; SMIT, sodium-myo-inositol cotransporter.
EXPERIMENTAL PROCEDURES
Animals
A diabetic state was induced in 2-month-old CD1 mice (Harlan) with an intraperitoneal injection of streptozotocin (200 mg/kg of body weight) in normal saline, and the control mice received normal saline only (23). After a week, mice with blood glucose ≥250 mg/dl were selected and maintained for 5 months. Age-matched controls having a glucose level of ∼100 mg/dl were also maintained for a similar period. Six animals were used per variable in various experiments. These animal studies were approved by the Animal Care and Use Committee of our university.
Cell Culture
Renal proximal tubular epithelial cell lines LLC-PK1 and HK-2 were obtained from ATCC. They were maintained in DMEM (Invitrogen) containing 5 mm glucose, 10% FBS, and 1× penicillin-streptomycin, and keratinocyte serum-free medium. One day prior to the experiment, the cells were cultured in 100-mm Petri dishes in DMEM containing 0.05% FBS. The cells were then treated with media containing 5, 20, and 35 mm d-glucose. For electrophile response experiments, cells were treated with d-glucose and oxidants (100 μm H2O2, 1 mm methylglyoxal (MGO), and 200 μm 12-O-tetradecanoylphorbol-13-acetate (TPA)) and antioxidants ((10 mm N-acetylcysteine (NAC), 10 mm reduced glutathione, 100 μm butylated hydroxyanisole (BHA), tertiary butyl hydroquinone (t-BHQ), and 10 μm β-naphthoflavone (β-NF)). To potentiate or dampen the action of different kinases, the cells were also treated, respectively, with the PKA activator forskolin (0.2 μm) and inhibitor H89 (0.2 μm), PKC activator TPA (0.2 μm) and inhibitor calphostin (0.05 μm), phosphoinositide-dependent protein kinase-1 (PDK1)/PI3K inhibitor wortmannin (0.5 μm), TGF-β1 (10 ng/ml), and insulin (0.05 μm).
Phosphosite and Motif Prediction Analyses
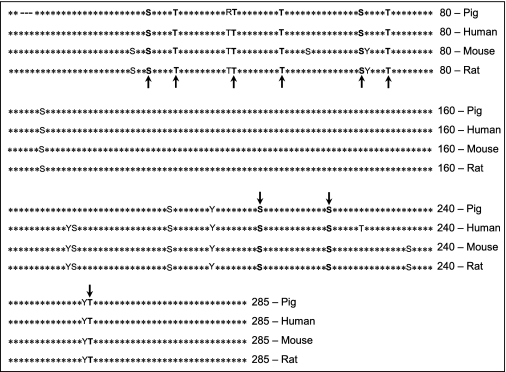
Reference sequences of RSOR/MIOX and its homologue were taken from NCBI (accession numbers NM_019977 (mouse), NM_017584 (human), NM_145771 (rat), and NM_214102 (pig)). Different sequences and structure-based putative protein phosphorylation sites in MIOX were analyzed by Scanprosite, Netphos, Scansite, Predphos, and Predikin as depicted in Table 1. A 3-kb upstream 5′-flanking genomic sequence of RSOR/MIOX was retrieved by using Ensembl bioinformatics, and a search for transcription factor binding motifs was carried out using MOTIF Search.
TABLE 1.
Consensus sequences for kinase-specific (bold arrows) and non-kinase-specific phosphorylation sites in pig MIOX as surveyed by Scanprosite, NetPhos, Scansite, and Predphos
The PKC sites were Thr-40, Thr-49, Ser-64, and Thr-253; the PKA sites were Ser-24, Ser-205, and Ser-218; and the PDK1/PKC sites were Ser-24, Ser-64, and Thr-253. The kinase-specific phosphorylation sites seem to be highly conserved across all the mammalian species.
Plasmid Construction and Generation of Recombinant Proteins
Mouse full-length MIOX cDNA was cloned in pET15b prokaryotic expression vector and expressed in Escherichia coli as described previously (23). For cloning of porcine cDNA, MIOX was amplified from the cDNA of the LLC-PK1 cell line using the sense and antisense primers 5′-GGGGATCCGATGAAGGACCCAGACCCTTCC-3′ and 5′-GGGGATCCTCACCAGCACAGGACACCGGG-3′, respectively (Underline, Restriction sites; Bold, open reading frame). The amplicon was first cloned in pCRII vector, sequence-confirmed, then subcloned into pET15b vector at the BamHI site, and expressed in E. coli.
Site-directed Mutagenesis and Generation of Exon Deletion Constructs
Mutagenesis was carried out for constructing mutants using a QuikChange multisite-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All the putative PKA-directed phosphorylation sites at serine (Ser-24, Ser-205, and Ser-218) and threonine (Thr-29 and Thr-69) residues were substituted by either alanine or glycine using antisense primers. Similarly, putative PKC phosphorylation sites (Ser-64, Thr-40, Thr-49, Thr-69, and Thr-253) were substituted by alanine using antisense primers. Another mutant was constructed using all PKA and PKC phosphorylation sites and then used for generating PDK1 constructs. The sequences of various antisense primers used to generate pET15 MIOX plasmid constructs with substituted nucleotides italicized and underlined are as follows: S24G (70A→G), 5′-GTTTCGGAAGCCGCCCTTGTCTTTGG-3′; T29A (85A→G), 5′-GCGGGCCAGACGCGTAGTTTCGGAA-3′; T40A (118A→G), 5′-CATCAGCTTGTAGGCGCGGAAGACCCG-3′; T49A (145A→G), 5′-CGAAGTCCACGGCCTGCCACGTGTG-3′; S64A (190T→G), 5′-CATTCTCTTGTAGGCGAAGCCCCCGAA-3′; T69A (205A→G), 5′-CCTCCAAGACAGCCATTCTCTTGTA-3′; S205A (613T→G), 5′-CTCCTGGGCGGGCGAATTTGTTGAA-3′; S218A (652T→G), 5′-ACGGGTAGAAGGCGTGGCAGCGGAT-3′; and T253A (757A→G), 5′-CGGAGCCCTTGGCGTAGAGATCGAA-3′.
Various exon deletion constructs encompassing different regions of mouse RSOR/MIOX and pET15 B vector were generated using sense and antisense primers as follows: RSOR nt 1–858 (exons 1–10): NdeI (sense), 5′-GGCATATGATGAAGGTCGATGTGGG-3′; XhoI (antisense), 5′GGCTCGAGTCACCAGCTCAGGGTGC-3′; RSOR nt 1–636 (exons 1–8): NdeI (sense), 5′-GGCATATGATGAAGGTCGATGTGGG-3′; XhoI (antisense), 5′GGCTCGAGCTCTGAAGGCAGGGAGA-3′; RSOR nt 1–408 (exons 1–5): NdeI (sense), 5′-GGCATATGATGAAGGTCGATGTGGG-3′; XhoI (antisense), 5′GGCTCGAGCTGAGGTTCCCCCCACA-3′; RSOR nt 16–858 (exons 2–10): Nde1 (sense), 5′-GGCATATGGGCCCAGACCCTTCCCT-3′; XhoI (antisense), 5′GGCTCGAGTCACCAGCTCAGGGTGC-3′; and RSOR nt 409–858 (exons 6–10): Nde1 (sense), 5′-GGCATATGTGGGCTGTTGTTGGAGA-3′; XhoI (antisense), 5′GGCTCGAGTCACCAGCTCAGGGTGC-3′.
These constructs were subcloned into pET15b vector and used as templates for generating in vitro translated products using the TnT reticulocyte system. The restriction sites of various enzymes within the primers are italicized and underlined.
Orthophosphate Labeling
Phosphorylation studies were carried out in LLC-PK1 cells maintained in 5 ml of DMEM with 5–35 mm concentrations of d-glucose for 36 h in 60-mm Petri dishes, and the cells were transfected with pcDNA-RSOR/MIOX using Lipofectamine 2000. Cells treated with l-glucose served as control. For orthophosphate labeling, 36 h after glucose treatment the transfected cells were washed with phosphate-deficient DMEM containing low glucose and incubated for 1 h in 1 ml of the same medium. Cells were then labeled with 250 μCi of [32P]orthophosphate (Amersham Biosciences) in 1 ml of deficient medium for 4 h at 37 °C in a CO2 incubator. The cells were washed twice with 5 ml of ice-cold Tris-buffered saline (TBS) and then lysed with 1 ml of radioimmunoprecipitation assay buffer (Pierce) with 200 μm sodium orthovanadate and 50 mm NaF. The lysate was then subjected to immunoprecipitation with anti-RSOR/MIOX antibody followed by SDS-PAGE and autoradiography (23, 28).
In Vitro Phosphorylation with PKC, PKA, and PDK1
For in vitro phosphorylation, first a prokaryotic (bacterially expressed purified protein in pET15B) system was used. The phosphorylation was performed using different kinases, i.e. cAMP/cGMP-dependent protein kinases, protein kinase C, casein kinase, and PDK1. The radioactive phosphorylation of recombinant RSOR/MIOX (1 μg/reaction) was carried out by protein kinase C (10 ng/μl; Promega) using 1× kinase buffer (20 mm HEPES, 10 mm MgCl2, 17 mm CaCl2, 1 mm DTT), phosphatidylserine (600 ng/μl), and [γ-32P]ATP (2 μCi/μl). For the non-radioactive phosphorylation, the radioisotope was replaced with 150 μm cold ATP. For the phosphorylation with cAMP-dependent protein kinase (Promega), the reaction was carried out in 1× buffer (40 mm Tris·HCl, pH 7.4, 20 mm magnesium acetate), [γ-32P]ATP (2 μCi/μl), and cAMP (2 μCi/μl). For the non-radioactive phosphorylation, the isotope was replaced with 200 μm ATP. The phosphorylation of 1 μg of RSOR/MIOX with PDK1 (Cell Signaling Technology) was carried out with 1× reaction buffer (25 mm Tris·HCl, pH 7.5, 1 mm MgCl2, 5 mm β-glycerophosphate, 0.1 mm sodium orthovanadate, 2 mm DTT), ATP (100 μm), and [γ-32P]ATP (2 μCi/μl). All phosphorylation reactions were carried out in a total volume of 50 μl for 30 min at 30 °C.
In Vitro Translation
As an alternative to the eukaryotic expression system, in vitro translation was carried out by using TnT® T7 coupled rabbit reticulocyte lysate system. RSOR/MIOX and its mutants were expressed and radiolabeled at 30 °C for 90 min in a 25-μl reaction mixture containing 12.5 μl of rabbit reticulocyte, 1× TnT buffer, 0.5 μl of amino acid mixture minus methionine, 1 μl of [35S]methionine (10 mCi/ml), 1 μl of T7 RNA polymerase, 0.5 μl of RNasin, and 1 μg of normal and mutant RSOR/MIOX plasmid DNA. For non-radioisotopic labeling, 0.5 μl of amino acid mixture minus leucine and cysteine was used in place of the amino acid mixture minus methionine and [35S]methionine. For in vitro phosphorylation of non-radiolabeled TnT product, the reaction was carried out for 60 min followed by [γ-32P]ATP labeling by different kinases for another 30 min. The expressed and phosphorylated products were subjected to SDS-PAGE followed by autoradiography. Aliquots were also saved for measuring the MIOX activity before and after phosphatase treatment.
Phosphatase Treatment
Protein λ-phosphatase (λ-PPase; Calbiochem, EMD Biosciences) was used. The MIOX protein, translated and phosphorylated in vitro by different kinases, and cortical kidney tissue lysates were used in the absence of phosphatase inhibitor. The phosphatase reaction mixture included 50 mm Tris-HCl, pH 7.8, 5 mm DTT, 2 mm MnCl2, 100 μg/ml BSA, 100 units of λ-PPase, and 50 μg of protein in a total volume of 500 μl. The reaction was carried out for 30 min.
MIOX Assay
Kidney cortices or LLC-PK1 cells were homogenized in a buffer containing 50 mm sodium acetate, 1 mm ferrous ammonium sulfate, 2 mm l-cysteine, 1 mm glutathione, 1 mm PMSF, 0.2 mm sodium orthovanadate, and 50 mm sodium fluoride followed by a brief sonication. MIOX activity was also determined in the kidney cortices following phosphatase treatment. They were homogenized in 1× phosphatase buffer containing 50 mm Tris·HCl, pH 8.0, 5 mm DTT, 2 mm MnCl2, and 100 μg/ml BSA without any phosphatase inhibitor. The MIOX assays were carried out at 30 °C for 30 min in a 500-μl reaction volume containing 50 mm sodium acetate, 1 mm ferrous ammonium sulfate, 2 mm l-cysteine, and 60 mm myo-inositol. Fifty microliters (100 μg/ml) of LLC-PK1 cells or kidney homogenates, recombinant proteins, or in vitro translated MIOX protein was added into the reaction mixture for the MIOX activity assay. The reaction was terminated by boiling followed by precipitation with 3% TCA. Following a centrifugation at 1,000 × g for 5 min, d-glucuronate was determined in the supernatant by the addition of a double volume of freshly prepared orcinol reagent (40 mg of orcinol and 9 mg of FeCl3·6H2O dissolved in 10 ml of concentrated HCl) (33). Colorimetric readings were made at an A660 nm. The MIOX activity was averaged from five different experiments.
Immunoprecipitation and Western Blotting with Phosphoamino Acid-specific Antibodies
Mouse kidney cortices were homogenized and lysed in radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors. Equal amounts (50 μg) of the tissue lysates from control and diabetic mouse kidneys were used for immunoprecipitation with the following antibodies: 1) anti-phosphoserine/threonine-specific PKA substrate antibody, 2) anti-phosphoserine-specific PKC substrate antibody, 3) anti-phosphothreonine antibody, and 4) anti-phosphotyrosine antibody (Cell Signaling Technology). The immunoprecipitates were utilized for 10% SDS-PAGE, and then the Western blots were probed with anti-RSOR/MIOX antibody.
Immunoblot Analysis
For cellular expression analysis of RSOR or NF-E2-related factor 2 (Nrf2), a transcription factor that regulates redox-sensitive genes, 20 μg of cytoplasmic extracts of oxidant- or antioxidant-treated cells was subjected to 10% SDS-PAGE and blotted onto nitrocellulose membrane. For nuclear expression of Nrf2, 40 μg of nuclear extracts was prepared as previously described (34) and subjected to 10% SDS-PAGE. The blots were prepared and individually treated with various primary antibodies (rabbit anti-RSOR or anti-Nrf2; Cell Signaling Technology) at a dilution of 1:1,000 for 12 h at 4 °C followed by treatment with secondary antibody (anti-rabbit or -goat) conjugated with horseradish peroxidase (HRP). The autoradiograms were prepared using the ECL detection system (Amersham Biosciences).
Preparation of Cytoplasmic and Nuclear Extracts
For preparation of nuclear extract, LLC-PK1 and HK-2 cells were harvested from 100-mm Petri dishes by scraping with a rubber policeman followed by washing with ice-cold PBS three times and pelleted at 500 × g at 4 °C. The pelleted cells were resuspended in 100 μl of sucrose buffer (0.32 m sucrose, 10 mm Tris·HCl, pH 8.0, 3 mm CaCl2, 2 mm MgOAc, 0.1 mm EDTA, 0.5% Nonidet P-40, 1 mm DTT, 0.5 mm PMSF) per 1e+07 cells. The nuclei were spun down for 5 min at 1,500 × g at 4 °C, and cytoplasmic fractions in the supernatants were collected for Western blot analysis. The nuclear pellet was washed with 1 ml of sucrose buffer (without Nonidet P-40) and mixed gently by a pipette with a wide bore tip. The nuclei were recentrifuged at 1,500 × g at 4 °C for 5 min, and the supernatant was aspirated. The nuclei were then resuspended in an appropriate volume (30 μl/1e+07 cells) of Low Salt Buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 20 mm KCl, 0.2 mm EDTA, 25% glycerol (v/v), 0.5 mm DTT, 0.5 mm PMSF) by finger tapping the Eppendorf tube. To this, an equal volume of High Salt Buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 800 mm KCl, 0.2 mm EDTA, 25% glycerol (v/v), 1% Nonidet P-40, 0.5 mm DTT, 0.5 mm PMSF, 4.0 μg/ml leupeptin, 4.0 μg/ml aprotinin, 4.0 μg/ml pepstatin) was added very slowly while mixing with the wide bore pipette. The samples were incubated for 30–45 min at 4 °C while rotating on an orbital shaker. The nuclear extracts and the supernatants were separated by centrifugation at 14,000 × g for 15 min. The concentration of protein was measured, and samples were stored at −70 °C for electrophoretic mobility shift assay (EMSA) studies.
EMSA
EMSA was performed as described previously (26, 31). Briefly, single-stranded oligos were custom synthesized by Integrated DNA Technologies. Their sequences are as follows: −573 murine antioxidant response element (ARE), AGCACATGTGACATCTCTCCTAAG; −673 murine ARE, GATACGTGTGACCTTGGGAGAGGG; −2400 mouse carbohydrate response element (ChRE), TGCCTCACGTGCTAACTCA; and −1380 human ChRE, AGCACGTGACCTACCCGTGTTGGGACACGTGAGG. The motifs are underlined and bold. Both sense and complimentary oligomers (50 pmol) in a volume of 50 μl were annealed in 1× annealing buffer (10 mm Tris, pH 7.5, 100 mm NaCl, 1 mm EDTA) by boiling for 10 min followed by slow cooling to 30 °C for 3 h. The double-stranded probe (10 pmol) was end-labeled by [γ-32P]ATP using T4 polynucleotide kinase (Promega). The unlabeled double-stranded oligonucleotides were used for competition assays. For EMSA, the binding reaction was carried out in a 20-μl reaction volume containing 1× binding buffer (50 mm Tris·HCl, pH 8.0, 750 mm KCl, 2.5 mm EDTA, 0.5% Triton X-100, 62.5% glycerol (v/v), 1 mm DTT), 1–3 μg/μl poly(dI-dC), 1 pmol/μl labeled DNA probe, and 10 μg of nuclear proteins. The latter were prepared as described above from LLC-PK1 and HK-2 cells subjected to various treatments, i.e. oxidant, antioxidant, high and low salts, and activators and inhibitors of PKA and PKC. For specificity competition EMSA, a 50-fold excess of unlabeled oligos was used. The binding reaction was carried out for 30 min at 22 °C. The samples were subjected to 8% non-denaturing PAGE, the gels were dried, and autoradiograms were prepared.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were carried out for the binding of carbohydrate response element-binding protein (ChREBP) to the promoter region of human MIOX. The HK-2 cells were grown for 24 h, serum-starved for 12 h, and then treated with high glucose for 24 h. The ChIP assay was performed by modifying methods described previously (34, 35). Briefly, cross-linking of DNA and protein was carried out by addition of 1% formaldehyde directly to the cell medium and incubation for 30 min. The cross-linking reaction was terminated by the addition of 0.125 m glycine for 5 min. The cells maintained in 60-mm Petri dishes were then washed with ice-cold PBS, scraped with a rubber policeman, and pelleted by centrifugation at 500 × g for 5 min. The pellet was resuspended in 10 volumes of swelling buffer (25 mm HEPES, pH 7.8, 1.5 mm MgCl2, 10m M KCl, 0.1% Nonidet P-40, 1 mm DTT, 0.5 mm PMSF) and incubated on ice for 10 min. The cellular nuclei were pelleted by centrifugation at 1,000 × g for 5 min. The pellet was resuspended in 500 μl of sonication buffer (50 mm HEPES, pH 7.9, 140 mm NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 0.5 mm PMSF), incubated on ice for 10 min, and sonicated to yield DNA fragments of 200–1,000 bp. The sonicated material was centrifuged at 15,000 × g for 15 min to remove the insoluble debris. BSA (1 mg/ml) was added to the supernatant containing the soluble chromatin. The soluble chromatin was precleared by incubation with Protein A-Sepharose/single-stranded DNA for 2 h at 4 °C. The samples were centrifuged, a 50-μl aliquot was kept as input DNA, and the remainder was processed for immunoprecipitation with ChREBP antibodies followed by incubation with Protein A-Sepharose beads. The samples with no antibody were used as a negative control. The beads were centrifuged and washed twice, first with 1 ml of low salt sonication buffer and then with 1 ml of high salt wash buffer (sonication wash buffer A with 500 mm NaCl). The beads were rewashed twice with 1 ml of wash buffer B (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, 250 mm LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 0.5 mm PMSF) and twice with 1 ml of Tris-EDTA buffer. The samples were then eluted by adding 400 μl of elution buffer (50 mm Tris-HCl, pH 8.0, 1 mm EDTA, 1% SDS, 50 mm NaHCO3) and then incubating at 65 °C for 10 min. After addition of 20 μl of 4 m NaCl, the samples were further incubated at 65 °C for 6 h for decross-linking. Similarly, samples of input DNA were decross-linked. One microliter of DNase-free RNase A (10 mg/ml) was added and incubated at 37 °C for 1 h. This was followed by addition of 2 μl of Proteinase K (10 mg/ml) and 4 μl of 0.5 m EDTA, and then samples were incubated at 42 °C for 2 h. After phenol/chloroform extraction and addition of 1 μl of glycogen (20 mg/ml), the DNA was precipitated with 40 μl of 3 m sodium acetate and 1 ml of ethanol. The precipitates of ChIP and input samples were resuspended in 100 μl of 10 mm Tris, pH 7.5 and used for PCR analyses. The primers used in ChIP-PCR for ChREBP motifs encompassing the MIOX nt −1499 to −1256 region were 5′-GTCCCACCTCCTGAACCTAT CCAG-3′ (sense) and 5′-TGCCACTCCACCAC CCACTTATTC-3′ (antisense) with a resulting amplified product of 245 bp.
Immunofluorescence Microscopy
Nuclear localization of transcription factors was assessed by immunofluorescence microscopy in HK-2 cells treated with high glucose or the antioxidant t-BHQ, and cells maintained in low glucose medium served as a control. In addition, the cells transfected with MIOX pcDNA3.1 were also processed for Nrf2 localization. The cell monolayers maintained in Lab-Tek chambers were washed with PBS twice at 5-min intervals followed by fixation with 4% paraformaldehyde at 22 °C for 30 min and three washings with PBS at 5-min intervals. The chambers containing monolayers were placed on ice and incubated with 0.1% saponin in PBS for 20 min. The chambers were then incubated with primary antibodies (goat anti-Nrf2 and rabbit anti-ChREBP) at a dilution of 1:100 in PBS containing 0.5% BSA and 0.1% saponin in a humidified chamber at 37 °C for 1 h. After three washes with PBS, the chambers were incubated with secondary anti-rabbit or -goat conjugated with FITC for another hour. The cells were washed twice with PBS, counterstained with 300 nm DAPI for 20 min at 22 °C, coverslip-mounted, and examined by a Zeiss microscope equipped with UV epi-illumination.
Transfection and Promoter Activity Luciferase Assay
The reporter plasmid construct (pGL3-2512-1) was transfected into exponentially growing LLC-PK1 or HK-2 cells. The cells were seeded onto 30-mm tissue culture Petri dishes at a density of 1 × 106 cells/dish and incubated for ∼18 h to achieve ∼80% confluence for transfection. The transfection was carried out with 10 μl of Lipofectamine 2000 reagent (Invitrogen) and 1 μg of reporter plasmid construct. Co-transfection of 200 ng of pcDNA-rLUC (Renilla luciferase) was used as an optimized equalization control. Assays for both Renilla and firefly luciferase activity were carried out 48 h post-transfection using the commercial Dual-Luciferase kit (Promega) and a TD 20/20 luminometer (Promega) according to the manufacturer's guidelines. Basal promoter activity was determined in cells transfected with reporter construct pGL3-basic and maintained in DMEM containing 5 mm glucose. For comparison among various treatments (oxidants, antioxidants, and kinase activators), 6 h post-transfection of DNA-liposome complex, Opti-MEM I medium was replaced with medium containing either 30 mm glucose or other agents individually (MGO, H2O2, BHA, t-BHQ, NAC, and β-NF) as described above.
Statistical Analyses
Results were expressed as mean ± S.E. following statistical analyses. Student's t test was used to compare the data between two groups. p values less than 0.05 were considered to be statistically significant.
RESULTS
The findings described in this section include the induction of MIOX/RSOR expression and activity in a hyperglycemic state and under high glucose ambience via phosphorylation of its potential PKC-, PKA-, and PDK1-phosphorylating sites and how hyperglycemia, glucose-derived carbonyls (MGO), and various oxidants and antioxidants affect its translational and transcriptional activities, the latter seeming to be largely influenced by unique DNA sites for the binding of certain transcription factors localized in its promoter.
Hyperglycemia and High Glucose Increase MIOX Expression and Activity
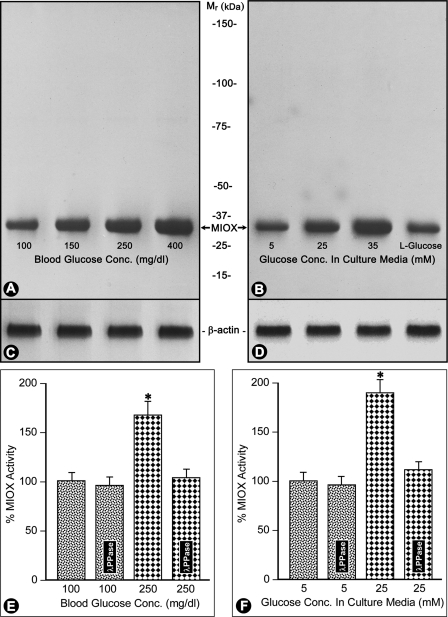
By Western blot analyses, an increased protein expression of RSOR/MIOX was observed in kidneys from streptozotocin-induced diabetic mice compared with the control (Fig. 2A). Also, an increase in enzymatic activity of MIOX was noted in kidney samples of control and diabetic mice with normalized protein concentrations (Fig. 2E). The increased expression or activity was proportional to the degree of hyperglycemia. Similarly, dose-dependent increased expression and activity of MIOX were observed in LLC-PK1 and HK-2 cells treated with various concentrations of d-glucose (Fig. 2, B and F). No increase was observed in control cells treated with l-glucose. The MIOX activity was reduced to almost basal levels with λ-PPase treatment (Fig. 2, E and F). The β-actin expression was unaffected by glucose treatment (Fig. 2, C and D).
FIGURE 2.
Expression and activity of MIOX/RSOR in hyperglycemia and under high glucose ambience in vitro. RSOR/MIOX expression as assessed by Western blot analysis (A) and its enzyme activity (E) are increased in proportion to the degree of hyperglycemia in kidneys of mice with streptozotocin-induced diabetes mellitus. Similarly, a dose-dependent increase in RSOR/MIOX expression and activity (B and F) is observed in LLC-PK1 and HK-2 cells subjected to high d-glucose ambience. Treatment with λ-PPase reduced the MIOX activity to basal levels. l-Glucose treatment showed no significant increase in the activity or protein expression. n = 4; (Mean ± S.E.) *, p < 0.01 versus MIOX activity in the controls and following λ-PPase treatment. Panels C and D are β-actin controls.
Evidence for RSOR/MIOX Being a Phosphoprotein
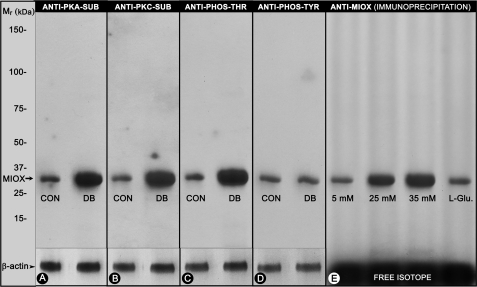
Immunoprecipitation of kidney tissue lysates from control and streptozotocin-induced diabetic mice with substrate-specific phosphoantibodies to PKA (Ser/Thr) and PKC (Ser) and to phosphothreonine and phosphotyrosine residues was carried out. This was followed by Western blot analyses with anti-RSOR/MIOX antibody using equal concentrations of proteins in the immunoprecipitated samples. An increased intensity of bands, indicative of phosphorylation, was observed in kidneys of diabetic mice where samples were immunoprecipitated with phosphoantibodies directed against serine and threonine residues (Fig. 3, A–C). The kidney samples of diabetic mice immunoprecipitated with anti-phosphotyrosine antibody did not yield any significant increase in phosphorylation (Fig. 3D). Also, immunoprecipitation of protein lysates of [32P]orthophosphate-labeled LLC-PK1 cells exposed to various concentrations of d-glucose with RSOR/MIOX antibody revealed an increased intensity of autoradiographic bands in samples subjected to high glucose ambience, suggesting that RSOR/MIOX is a phosphoprotein (Fig. 3E).
FIGURE 3.
Status of phosphorylation of RSOR/MIOX in kidneys of diabetic mice and LLC-PK1 cells subjected to high d-glucose ambience. The PKA and PKC substrate (SUB)-specific and anti-phosphothreonine (PHOS-THR) antibodies revealed increased expression of RSOR/MIOX in diabetic mice (A, B, and C). No significant increase in the phosphorylation/expression was observed as evaluated by anti-phosphotyrosine (PHOS-TYR) (D). Similarly, under high glucose ambience, a dose-dependent increase in the phosphorylation of [32P]orthophosphate-labeled LLC-PK1 cells was observed (E), suggesting that RSOR/MIOX is a phosphoprotein. CON, control; DB, diabetic.
Prediction of Phosphorylation Sites in MIOX/RSOR
Potential serine, threonine, and tyrosine phosphorylation sites of MIOX were identified using the NetPhos 2.0 program. Kinase-specific phosphorylation site prediction by NetPhos1.0, Scanprosite, Predphos, and Scansite revealed multiple potential sites that can be phosphorylated with protein kinases A, G, C, PDK1, casein kinase II, and Ca2+/calmodulin-dependent protein kinase and tyrosine kinases insulin receptor and EGF receptor. Prediction with the program Predikin revealed that MIOX by itself cannot act as a kinase. Potential PKA phosphorylation sites identified were Ser-24, Thr-29, Thr-69, Ser-205, and Ser-218; potential PKC phosphorylation sites were Ser-24, Ser-64, Thr-40, Thr-49, Thr-69, and Thr-253; and potential PDK1 phosphorylation sites were Ser-24, Ser-64, Thr-29, and Thr-253. Most of the kinase-specific phosphorylation sites were conserved across species lines as indicated by the arrows in Table 1. Interestingly, the majority of the potential phosphorylation sites were clustered toward the N-terminal segment of MIOX.
Phosphorylation of Recombinant RSOR/MIOX and Mutant Proteins by Various Kinases and Their Differential Effect on Enzymatic Activity in Vitro
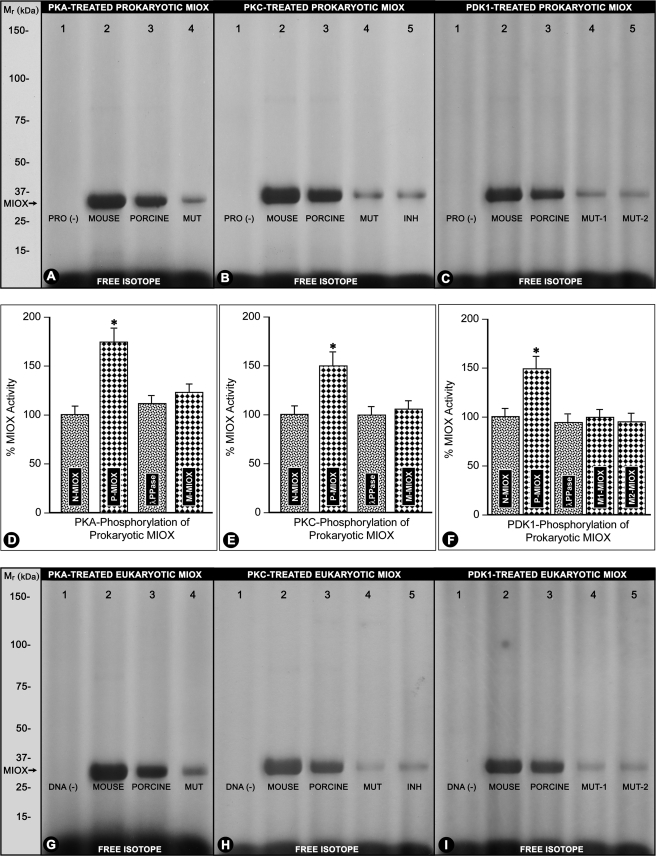
Several protein kinases, including protein kinases A, G, C, casein kinase II, and PDK1, were used for in vitro phosphorylation of recombinant proteins generated by utilizing prokaryotic bacterial and eukaryotic rabbit reticulocyte (TnT) systems using pET15b pig MIOX and RSOR plasmid constructs. Phosphorylation was assessed by [γ-32P]ATP incorporation followed by immunoprecipitation with MIOX antibody, SDS-PAGE, and autoradiography. Casein kinase II and cGMP-dependent protein kinase did not phosphorylate MIOX. Protein kinases A, C, and PDK1 were capable of phosphorylating mouse and pig MIOX in both prokaryotic and eukaryotic systems (Fig. 4, A–C and G–I, lanes 2 and 3). No phosphorylation was observed in samples in which addition of plasmid DNA or recombinant protein was omitted (Fig. 4, A–C and G–I, lane 1). Both the single and double mutants were also phosphorylated by PKA, PKC, or PDK1 but to a minor degree (Fig. 4, A–C and G–I, lanes 4 and 5). Addition of myristoylated PKC inhibitor reduced the phosphorylation of recombinant proteins (Fig. 4, B and H, lane 5).
FIGURE 4.
Phosphorylation of prokaryotic and eukaryotic recombinant RSOR/MIOX and mutant proteins by various kinases and subsequent effect on their enzymatic activities. Protein kinase A, protein kinase C, and PDK1 were able to phosphorylate both mouse and pig MIOX in both prokaryotic and eukaryotic systems (A–C and G–I, lanes 2 and 3). In the absence of DNA or recombinant RSOR/MIOX protein (PRO (−)), no phosphorylated band was observed (A–C and G–I, lane 1). The mutant (MUT) proteins were also phosphorylated but to a minor degree (A–C and G–I, lanes 4 and 5), whereas addition of myristoylated PKC inhibitor (INH) reduced the phosphorylation of recombinant proteins (B and H, lane 5). The activities of recombinant RSOR/MIOX proteins (N-MIOX) increased following phosphorylation (P-MIOX) relative to the basal levels (D–F, column 2). Treatment with λ-PPase reduced the MIOX activity to basal levels (D–F, column 3). Phosphorylated mutant recombinant proteins (M-MIOX) had a mild increase in their enzymatic activity (D–F, column 4). n = 4; *, p < 0.01 versus MIOX activity in recombinant RSOR/MIOX proteins, mutant recombinant proteins, and following λ-PPase treatment.
Phosphorylation of bacterial recombinant RSOR/MIOX proteins in vitro with PKA, PKC, or PDK1 increased their activity by 50–100% over basal levels (Fig. 4, D–F, column 2 versus column 1). The treatment with λ-PPase reduced the MIOX activity almost to the basal levels (Fig. 4, D–F, column 3). The PKA, PKC, or PDK1 mutant recombinant proteins had a marginal increase in their enzymatic activity following in vitro kinase treatments (Fig. 4, D–F, column 4). Phosphorylation of the recombinant proteins generated by the TnT reticulocyte system also increased their MIOX activities (figure not included). Although the activity levels were somewhat higher, they were reduced to basal values with λ-PPase treatment.
RSOR/MIOX Activity Analyses of Recombinant Proteins Generated from Deletion Constructs Encompassing Its Various Exons in Vitro
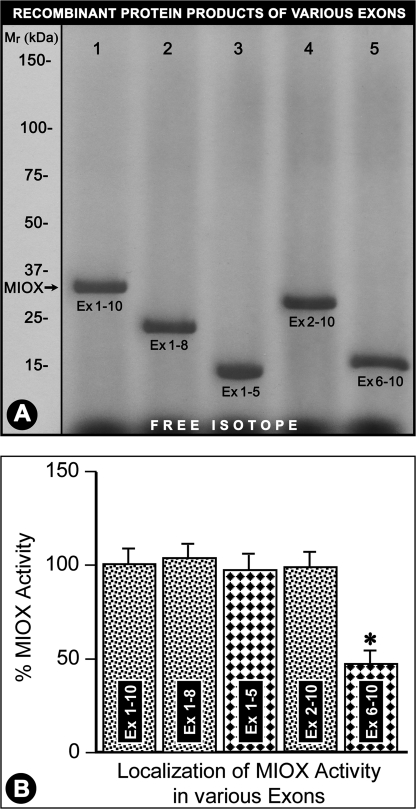
RSOR/MIOX gene includes 10 exons. The DNA constructs inclusive of exons 1–10 (intact), 1–8, 1–5, 2–10, and 6–10 were cloned in pET15b, and truncated proteins were generated using the TnT reticulocyte system. The respective proteins generated from these constructs had approximate molecular masses of 32 (intact), 24, 15, 31, and 17 kDa (Fig. 5A, lanes 1–5). Deletion of exons 1 and 6–10 did not affect the basal MIOX activity compared with the intact protein. However, deletion of exons 2–5 (leaving exons 6–10 intact) markedly reduced the basal activity of RSOR/MIOX in samples with normalized concentration of recombinant protein (Fig. 5, A and B, lane 5 and column 5), suggesting that its N terminus includes the critical phosphorylation sites that are highly relevant to the functionality of this protein.
FIGURE 5.
Delineation of enzyme activity in various RSOR/MIOX exons. Recombinant proteins were generated using pET15b plasmid constructs encompassing exons (Ex) 1–10, 1–8, 1–5, 2–10, and 6–10. The respective proteins generated from these constructs had molecular masses of ∼32 (intact protein), 24, 15, 31, and 17 kDa (A, lanes 1–5). The recombinant protein generated from the plasmid construct having exons 6–10 had markedly reduced activity (B, column 5). Analysis of other truncated proteins suggested that RSOR/MIOX is confined to exons 1–5, the regions where the majority of the phosphorylation sites are clustered (Table 1). n = 4; *, p < 0.01 versus enzyme activity in intact MIOX (exons 1–10) and other truncated recombinant proteins.
Effect on MIOX/RSOR Phosphorylation and Activity by Various Kinase Activators and Inhibitors in LLC-PK1 Cells
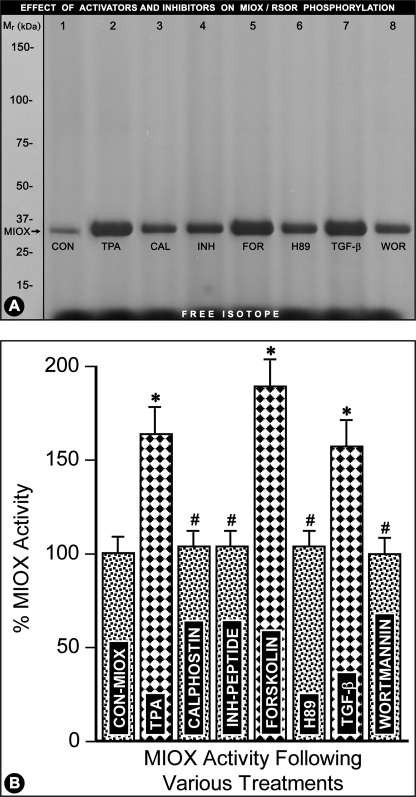
LLC-PK1 cells were treated with activators of PKC (TPA), PKA (forskolin), and PDK1/PI3K (TGF-β). The respective inhibitors included calphostin (PKC), H89 (PKA), and wortmannin (PI3K). Following various treatments, the [γ-32P]ATP-labeled cells were subjected to immunoprecipitation with anti-RSOR antibody and then processed for SDS-PAGE and MIOX activity analyses as described above (Fig. 6, A and B). All the kinase activators, i.e. TPA, forskolin, and TGF-β, were capable of phosphorylating RSOR/MIOX to varying degrees as visualized by distinct bands following autoradiography (Fig. 6A, lanes 2, 5, and 7) compared with the control where no activators were added to the cell lysate (Fig. 6A, lane 1). However, concomitant treatment with kinase inhibitors, i.e. calphostin, H89, and wortmannin (phosphoinositide 3-kinase inhibitor), notably inhibited the phosphorylation (Fig. 6A, lanes 3, 6, and 8 versus lanes 2, 5, and 7). The phosphorylation was also inhibited by the addition of myristoylated PKC peptide inhibitor in the lysate of TPA-treated cells (Fig. 6A, lane 4). With the addition of various activators in various samples with an equal concentration of recombinant protein, the activity was increased 40–60% (Fig. 6B, columns 2, 5, and 7) compared with the basal MIOX activity (Fig. 6B, column 1). Like the phosphorylation, the activity was reduced to basal levels with addition of inhibitors (Fig. 6B, columns 3, 4, 6, and 8).
FIGURE 6.
Modulation of RSOR/MIOX phosphorylation/activity by various kinase activators and inhibitors. Treatment with various activators of PKC (TPA), PKA (forskolin (FOR)), and PDK1/PI3K (TGF-β) increased phosphorylation (A, lanes 2, 5, and 7) and enzyme activity (B, columns 2, 5, and 7). Whereas, the inhibitors, i.e. calphostin (CAL) (PKC), H89 (PKA), and wortmannin (WOR) (PI3K) notably reduced phosphorylation (A, lanes 3, 5, and 6) and the enzyme activity (B, columns 3, 5, and 6) induced by activators almost to the basal levels. The myristoylated PKC peptide inhibitor (INH; A, lane 4) also reduced phosphorylation and RSOR/MIOX activity (B, column 4) to basal control levels. n = 4; *, p < 0.01 versus MIOX activity in the control (CON); #, p < 0.01 MIOX activity in comparison with the corresponding activators (TPA, forskolin, and TGF-β).
Effect on RSOR/MIOX Phosphorylation and Activity by Various Kinase Inhibitors in LLC-PK1 Cells under High Glucose Ambience
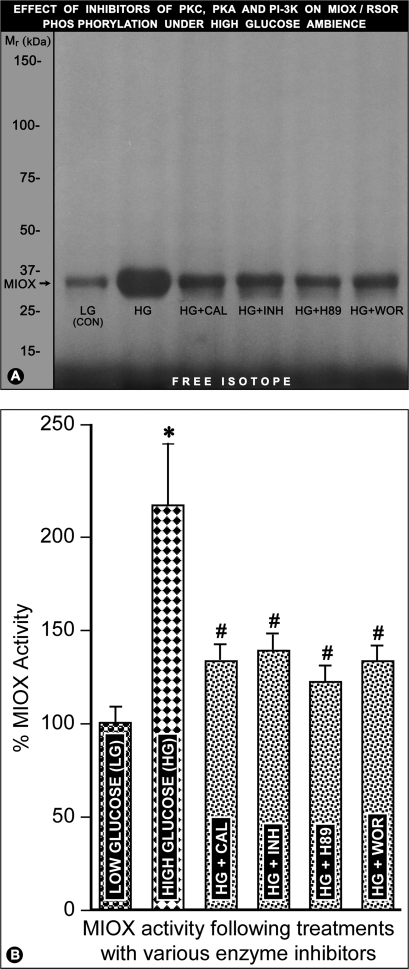
LLC-PK1 cells were subjected to high glucose ambience and treated with various inhibitors, which included calphostin (PKC), H89 (PKA), and wortmannin (PI3K). Following various treatments, the [γ-32P]ATP-labeled cells were subjected to immunoprecipitation with anti-RSOR antibody and then processed for SDS-PAGE and MIOX activity analyses (Fig. 7, A and B). The high glucose increased the phosphorylation and activity of RSOR/MIOX (Fig. 7, A, lane 2, and B, column 2). However, concomitant treatment with various kinase inhibitors substantially reduced the phosphorylation and activity of RSOR/MIOX (Fig. 7, A, lanes 3, 5, and 6, and B, columns 3, 5, and 6). The phosphorylation as well as activity was decreased by the addition of myristoylated PKC peptide inhibitor in the cell lysate (Fig. 7, A, lane 4, and B, column 4). These results suggest that the RSOR/MIOX expression and activity are modulated by various kinases that are known to be involved in the pathogenesis of diabetic nephropathy.
FIGURE 7.
Effect of various kinase inhibitors on RSOR/MIOX phosphorylation and activity in LLC-PK1 cells subjected to high glucose ambience. The high glucose increased RSOR/MIOX phosphorylation and enzyme activity (A, lane 2; B, column 2). Concomitant treatment with various kinase inhibitors notably reduced the phosphorylation and activity of RSOR/MIOX (A, lanes 3, 5, and 6; B, columns 3, 5, and 6). The phosphorylation and activity were decreased by the addition of myristoylated PKC peptide inhibitor (INH) in the cell lysate (A, lane 4; B, column 4). n = 4; *, p < 0.01 versus MIOX activity in the low glucose control (LG (CON)); #, p < 0.01 MIOX activity in comparison with high glucose. CAL, calphostin; WOR, wortmannin.
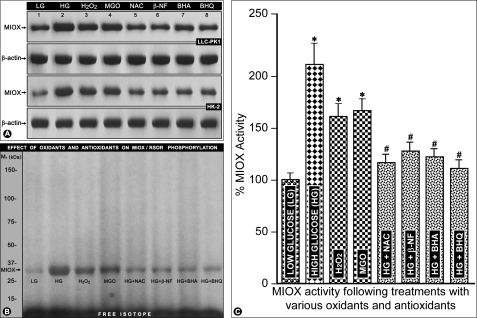
RSOR/MIOX Expression, Phosphorylation, and Activity following Treatment with High Glucose and Various Oxidants and Antioxidants
The LLC-PK1 or HK-2 cells treated with 35 mm d-glucose had an increased expression of RSOR/MIOX as assessed by Western blot analyses (Fig. 8A, lane 2 versus lane 1). A similar increase in RSOR/MIOX expression, although of lesser degree, was noted in cells exposed to oxidants H2O2 (100 μm) and MGO (1 mm) (Fig. 8A, lanes 3 and 4). The expression of RSOR/MIOX was decreased in high glucose-treated cells when exposed to various antioxidants, i.e. NAC (10 mm), β-NF (10 μm), BHA (100 μm), and t-BHQ (100 μm) (Fig. 8A, lanes 5–8). No significant change in the expression of β-actin was observed with treatment with high glucose or various oxidants/antioxidants (Fig. 8A). Similarly, an increased phosphorylation and activity of RSOR/MIOX were observed following treatment with high glucose, H2O2, and MGO (Fig. 8, B, lanes 2–4, and C, columns 2–4). The high glucose-induced phosphorylation and activity were notably decreased following antioxidant treatments (Fig. 8, B, lanes 5–8, and C, columns 5–8), suggesting that RSOR/MIOX is responsive to changes in cellular redox in both porcine- and human-derived cell lines.
FIGURE 8.
Effect of high glucose and various oxidants/antioxidants on RSOR/MIOX expression, phosphorylation, and activity in LLC-PK1 and HK-2 cells. The high glucose increased the expression of RSOR/MIOX, as assessed by Western blot analyses (A, lane 2 versus lane 1), that was associated with increased phosphorylation (B, lane 2 versus lane 1) and activity (C, column 2 versus column 1). A similar increase in its expression, phosphorylation, and activity was noted in cells treated with the oxidants H2O2 and MGO (A, lanes 3 and 4; B, lanes 3 and 4; C, columns 3 and 4). The high glucose-induced RSOR/MIOX expression, phosphorylation, and activity was decreased following treatment with various antioxidants, NAC, β-NF, BHA, and t-BHQ (A, lanes 5–8; B, lanes 5–8; C, columns 5–8), suggesting that RSOR/MIOX is responsive to changes in the cellular redox in both the porcine- and human-derived cell lines. No change in the expression of β-actin was observed with treatment with high glucose or various oxidants/antioxidants (A). n = 4; *, p < 0.01 versus MIOX activity in the low glucose (LG) control; #, p < 0.01 MIOX activity in comparison with high glucose.
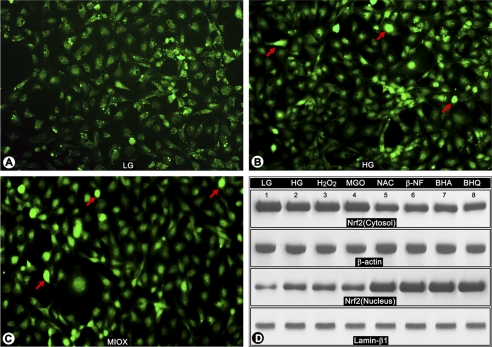
Expression and Cellular Distribution of Redox-sensitive Transcription Factor Nrf2 following Treatment with High Glucose and Various Oxidants/Antioxidants and Transfection of MIOX pcDNA
As shown by immunofluorescence microscopy, high glucose treatment led to an increased translocation of Nrf2 from the cytoplasm into the nucleus (Fig. 9, B versus A, arrows). Also, an increased translocation of Nrf2 was observed with transfection of MIOX pcDNA (Fig. 9C, arrows), thus mimicking the changes seen with high glucose treatment. Conceivably, this up-regulation of MIOX by high glucose may be an adaptive change to modulate cellular redox. To assess whether the translocation of Nrf2 is related to oxidant stress induced by high glucose, the cells were treated with H2O2 and MGO. As shown by Western blot analyses, the high glucose treatment led to an increased expression and translocation of Nrf2 from the cytoplasm into the nucleus (Fig. 9D, lanes 2–4 versus lane 1). Intriguingly, treatment of LLC-PK1 cells exposed to high glucose with various antioxidants led to a further enrichment of Nrf2 in the nuclear compartment (Fig. 9D, lanes 5–8 versus lanes 1–4). These findings led us to explore RSOR/MIOX regulation via oxidant/antioxidant response elements, which may be localized in its promoter.
FIGURE 9.
Effect of high glucose, various oxidants/antioxidants, and transfection of MIOX pcDNA3.1 on expression and cellular distribution of redox-sensitive transcription factor Nrf2. By immunofluorescence microscopy, an increased translocation of Nrf2 from the cytoplasm into the nucleus was observed in cells under high glucose ambience (B versus A, arrows). Similar results were observed following transfection of MIOX under normal glucose ambience, thus mimicking changes seen with high glucose (C, arrows). Conceivably, the up-regulation of MIOX by high glucose may be an adaptive change to modulate cellular redox. To assess whether the translocation of Nrf2 is related to oxidant stress induced by high glucose, the cells were treated with H2O2 and MGO. As assessed by Western blot analyses, the high glucose treatment led to an increased expression and translocation of Nrf2 from the cytoplasm into the nucleus (D, lanes 2–4 versus lane 1). Intriguingly, treatment of LLC-PK1 cells exposed to high glucose with various antioxidants led to a further enrichment of Nrf2 into the nuclear compartment (D, lanes 5–8 versus lanes 1–4), suggesting that Nrf2 is responsive to both oxidant and antioxidant treatments as has been reported in the literature by Kaspar et al. in other systems (Ref. 48 and supplemental Figs. 1 and 2). LG, low glucose.
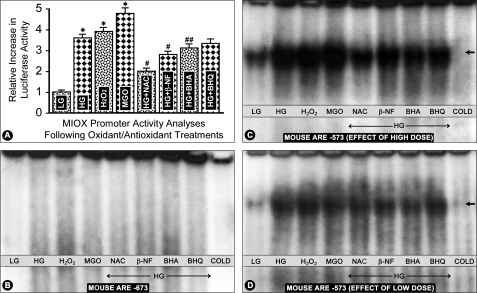
RSOR/MIOX Promoter Activity Analyses following Treatment with High Glucose and Various Oxidants and Antioxidants
For promoter analysis, a DNA fragment of RSOR/MIOX encompassing bp −2521 to −1 was cloned in pGL3-basic promoterless reporter plasmid vector and transfected into LLC-PK1 or HK-2 cells. The cells were treated with high glucose (HG), H2O2, MGO, and HG in combination with antioxidants, i.e. NAC, β-NF, BHA, and BHQ. A 3–5-fold increase in the promoter activity, as assessed by both Renilla and firefly luciferase assays, was observed following treatment with high glucose and oxidants H2O2 and MGO (Fig. 10A, columns 2–4 versus column 1). However, concomitant treatment with HG and antioxidants led to a decrease with antioxidants NAC, β-NF, and BHA in comparison with the cells treated with HG alone (Fig. 10A, columns 5–7). Treatment with BHQ also led to a mild decrease in the luciferase activity, but it was not statistically significant (Fig. 10A, column 8). The promoter activity profiles were similar whether the transfection was carried out in LLC-PK1 or HK-2 cells.
FIGURE 10.
RSOR/MIOX promoter activity analyses and characterization of murine AREs by EMSAs following treatment with high glucose and various oxidants and antioxidants. A 3–5-fold increase in the promoter activity was observed following treatment with high glucose and oxidants H2O2 and MGO (A, columns 2–4 versus column 1). However, concomitant treatment with HG and antioxidants led to a decrease with antioxidants NAC, β-NF, and BHA (A, columns 5–7). A mild decrease with BHQ treatment was also observed, but it was not statistically significant (A, column 8). The murine ARE motif localized to −673 bp of the RSOR/MIOX promoter was found to be non-functional because no distinct band was observed by EMSA following various treatments (B). However, the motif at −573 bp was functional because cells subjected to high glucose ambience or H2O2 and MGO treatments had a marked increase in the intensity of the shifted bands compared with the cells maintained in low glucose (LG) (5 mm) (C, lanes 2–4 versus lane 1, arrow). The intensity of the bands was decreased to a certain extent in cells subjected to high glucose ambience and then treated with antioxidants (C, lanes 5–8 versus lane 2, arrow). The reduction in the intensity of shifted bands by antioxidants was clearly seen when the concentrations of glucose, oxidants, and antioxidants were reduced by 50% intensity (D, lanes 5–8). No distinct band was seen when cold ARE oligo at a 50× concentration was used as a competitor. n = 6; *, p < 0.01 versus luciferase activity in the control low glucose; #, p < 0.01 MIOX activity in comparison with high glucose; ##, p < 0.05 MIOX activity in comparison with high glucose.
RSOR/MIOX Promoter Analyses and Characterization of ARE by EMSA following Treatment with High Glucose, Oxidants, and Antioxidants
Analysis of a 3-kb DNA segment upstream of the 5′-flanking region of mouse and human RSOR/MIOX was carried out to search for various motifs using the TRANSFAC database. It revealed several interesting motifs, including electrophile ARE (or Nrf2) with consensus motif GTGACNNNGC, cAMP response element, ChRE, and AP1 binding sites. The murine ARE motifs were localized to the −673 bp (GTGACCTTGG) and −573 bp (GTGACTGCTC) regions. The murine ARE motif localized to −673 bp of the RSOR/MIOX promoter was found to be non-functional because no distinct band was observed by EMSA (Fig. 10B). However, the motif at −573 bp was functional because shifted bands with marked intensity were seen (Fig. 10C). Cells subjected to high glucose ambience or H2O2 and MGO treatments had a marked increase in the intensity of the shifted bands compared with the cells maintained in low glucose (5 mm) (Fig. 10C, lanes 2–4 versus lane 1, arrow). The intensity of the bands was decreased to a certain extent in cells subjected to high glucose ambience and then treated with antioxidants (Fig. 10C, lanes 5–8 versus lane 2, arrow). In view of the fact that a shifted band could be seen at 5 mm glucose and to confirm the above observations, experiments were performed in which concentrations of glucose as well as those of oxidants and antioxidants in the culture media were reduced to ∼50%, and binding of ARE oligo was assessed by EMSA. The intensity of the shifted band was reduced in cells subjected to various treatments, but the band was still detectable in cells subjected to a much reduced concentration of glucose, i.e. 2.5 mm (Fig. 10D, lane 1). However, the reduction in the intensity of the band was clearly seen with antioxidant treatment in cells subjected to high glucose ambience (Fig. 10D, lanes 5–8). No distinct band was seen when cold ARE oligo at a 50× concentration was used as a competitor.
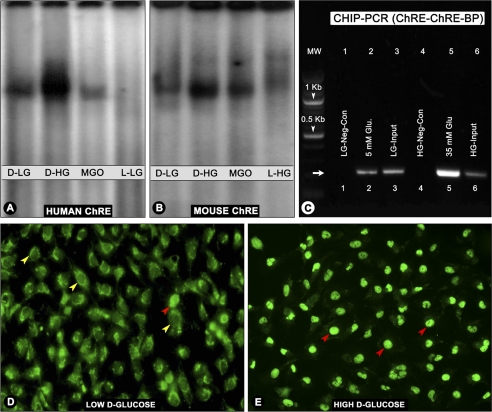
Characterization of ChREs of RSOR/MIOX Promoters
The consensus sequences for ChRE are two repeat E-box (CACGTG) motifs separated by 5 bp, whereas only the CAGTG motif can also be recognized by upstream stimulated factor 1. These consensus sequences were present in both human and murine RSOR/MIOX promoters. In human, the promoter E-box motif (CACGTG) is separated by 15 bp, and a degenerate motif (CCCGTG) with 5-bp spacing is present. The mouse ChRE is located at −2400 bp, whereas that of human is at −1385 bp. Both these motifs were found to be functional as reflected by a notable increase in the band intensity in samples from cells subjected to high glucose ambience (Fig. 11, A and B, lane 2). The samples of cells treated with the oxidant MGO did not show any notable increase in the band intensity relative to those subjected to a basal concentration of 5 mm d-glucose (Fig. 11, A and B, lane 3 versus lane 1), suggesting that ChREs are responsive to high glucose ambience and not to oxidant stress. No distinct binding pattern was observed in cells subjected to 5 or 35 mm l-glucose as controls (Fig. 11, A and B, lane 4).
FIGURE 11.
Characterization of human and murine ChREs following treatment with high glucose and various oxidants/antioxidants by EMSA, ChIP, and morphological analyses. The ChREs of both human (−1380 bp) and mouse (−2400) were functional because a remarkable increase in the band intensity in samples from cells subjected to high d-glucose (d-HG) ambience was observed by EMSA (A and B, lane 2). No significant increase in the band intensity was observed following MGO treatment compared with the low d-glucose (d-LG) control (A and B, lane 3 versus lane 1), suggesting that ChREs are responsive to high glucose ambience and not to oxidant stress. No distinct band was observed in cells subjected to l-glucose (low or high) controls (A and B, lane 4). The ChIP assays confirmed the functionality of ChRE because PCR analyses of the anti-ChREBP antibody-immunoprecipitated nuclear material from cells subjected HG ambience yielded a 245-bp product with strong band intensity (C, lane 5). Faint bands with remarkably reduced intensity were seen in samples from cells subjected to 5 mm d-glucose or from input aliquots (C, lanes 2, 3, and 6). No bands were seen in samples immunoprecipitated with normal IgG (C, lanes 1 and 4). Cellular translocation of ChREBP into the nucleus was readily seen by immunofluorescence microscopy of cells subjected to high d-glucose (E, red arrowheads) compared with low glucose where the ChREBP is seen mainly in the cytoplasm (D, yellow arrowheads). A few cells with ChREBP nuclear localization were also observed (D, red arrowhead). Glu, glucose; Neg-Con, negative control.
To confirm the functionally of ChRE, ChIP assays followed by PCR analyses of −1380 human ChRE region was carried out, yielding a PCR product with the expected size of 245 bp. An antibody directed against ChREBP was used for immunoprecipitation. The PCR analysis of the immunoprecipitated products revealed a band with marked intensity in samples from HK-2 cells treated with high glucose (Fig. 11C, lane 5). Faint bands with significantly less intensity were seen in samples from cells subjected to 5 mm d-glucose or from input aliquots (Fig. 11C, lanes 2, 3, and 6). No band was seen in control negative samples in which normal IgG was substituted for ChREBP antibody (Fig. 11C, lanes 1 and 4).
To delineate the cellular localization of ChREBP, the HK-2 cells were exposed to various concentrations of d-glucose and stained with anti-ChREBP antibody. Under low glucose ambience, ChREBP was mainly seen in the cytoplasmic compartment (Fig. 11D, yellow arrowheads). A few cells with nuclear localization of ChREBP were also seen under basal conditions (Fig. 11D, red arrowheads). However, under high glucose ambience, the majority of the cells showed nuclear localization of ChREBP (Fig. 11E, red arrowheads). Collectively, these three sets of experiments suggest that the ChREs are functional in the RSOR/MIOX promoter.
DISCUSSION
Identification of the enzyme MIOX as an RSOR with relevance to diabetes has given further considerations to investigate the biology of myo-inositol in renal diseases, such as diabetic nephropathy (23, 24). The relevance of RSOR/MIOX in renal pathobiology is further underscored by the fact that this enzyme, which is exclusively expressed in the renal cortical tubules, is markedly down-regulated in acute kidney injury (29). Moreover, currently, the tubulointerstitial compartment is weighted equally to glomerulus in the pathogenesis of diabetic nephropathy because tubulointerstitial injury is known to be well reflected in the degree of compromise in renal functions (27, 28). Another enzyme that is involved in hexose metabolism and is co-expressed with MIOX in proximal tubules is aldehyde reductase (ALR1) (36, 37). The latter enzyme catabolizes d-glucuronate generated from myo-inositol by MIOX to l-gulonate, which through a series of reactions is converted into xylulose and ribulose, which finally enter into the glycolytic pathway (38). Thus, it seems that RSOR/MIOX and MI are apparently vital to the homeostasis of the kidney, especially in the hyperglycemic state. In our previous studies, MIOX was found to be up-regulated in db/db mice and in initial stages of streptozotocin-induced diabetes (23, 26). In the latter, the issue whether the up-regulation was related to hyperglycemia or toxicity of streptozotocin remained unresolved. To address this issue, long term studies in which toxicity concerns can be excluded were initiated. Even after 5 months following the induction of diabetic state there was a sustained up-regulation of MIOX expression associated with the increase in its activity (Fig. 2, A and E). The other issue addressed was cell and species specificity of MIOX because porcine and human homologues lack NADPH binding activity (24). Both porcine (LLC-PK1) and human (HK-2) proximal tubular cells exhibited up-regulation and an increase in the enzyme activity under high glucose ambience (Fig. 2, B and F), suggesting that the basic core structure of MIOX is conserved across species lines as far as its catalytic activity is concerned. Nevertheless, the crystallographic studies have revealed a rather complex structure of this enzyme (39). The spectroscopic analyses indicate that MIOX is a non-heme di-iron oxygenase that utilizes a Fe(II)/Fe(III) binuclear iron center for catalysis (40). Also, the motif structural analyses of the RSOR/MIOX protein revealed multiple putative phosphorylation sites. These sites were clustered toward the N terminus of the protein (Table 1), and the significance of such post-translational modifications was investigated in the present study.
Phosphorylation is a ubiquitous post-translational modification of a given protein that can induce conformational change within the molecule, thereby either increasing or decreasing its activity or altering the activity of the substrate, subcellular localization, binding properties, or association with other proteins (41, 42). This cellular regulatory mechanism occurs through reversible, covalent modification of a protein by addition of phosphate groups via transfer of the terminal phosphate from ATP to an amino acid residue by protein kinases and/or by their removal with protein phosphatases. These kinases and phosphatases specifically target serine, threonine, or tyrosine residues, thereby either phosphorylating or dephosphorylating the protein. They are usually a single molecule that activates many substrate molecules and hence play an important role in signal transduction, transcription, and post-translation modifications (41, 42). Because cAMP-dependent kinases (e.g. PKA) mediate a vast majority of cellular responses, including metabolism and gene expression (43), and in hyperglycemia, the activity of other kinases (e.g. PKC) increases with activation of downstream signaling pathways (5, 6), this led us to investigate the phosphorylation of RSOR/MIOX by different kinases, its significance in diabetes/hyperglycemia, and the mechanisms related to its up-regulation and enzymatic activity. Immunoprecipitation/Western blot analyses revealed increased phosphorylation of renal RSOR/MIOX in diabetic animals as assessed by substrate-specific anti-PKA and -PKC antibodies (Fig. 3, A and B). An increased phosphorylation of threonine residues was also observed, but tyrosine residues seemed to be unaffected (Fig. 3, C and D). Similarly, orthophosphate labeling of LLC-PK1 cells revealed that there is an increased dose-dependent phosphorylation of MIOX (Fig. 3E), suggesting that under high glucose ambience this protein gets phosphorylated in both in vivo and in vitro systems. We then proceeded to assess whether phosphorylation increases the enzymatic activity using in vitro phosphorylation methods and recombinant RSOR/MIOX proteins generated in prokaryotic or eukaryotic systems. Both the prokaryotic and eukaryotic recombinant proteins generated by using murine and porcine MIOX cDNA templates could be phosphorylated by treatment with PKA, PKC, and PDK1 to a variable degree (Fig. 4, A–C and G–I). The mutants were also phosphorylated but to a minor degree. As speculated, phosphorylation was associated with a 50–75% increase in the enzyme activity that could be abolished by phosphatase treatment (Fig. 4, D–F), thus confirming the observation made in in vivo systems (Fig. 2). Because the majority of the serine and threonine phosphorylation sites are clustered in the N-terminal segment of the MIOX protein across all species, one would expect that the catalytic activity would be confined to this region as also deduced from the crystallographic and spectroscopic data (39, 40). This speculation appears to be correct because measuring MIOX activity in the truncated recombinant proteins following exon deletions revealed that the enzyme loses its catalytic properties in vitro in the absence of exons 2–5 (Fig. 5B). The role of phosphorylation in the modulation of MIOX activity was further confirmed by the observation that exposure of LLC-PK1 cells to activators and inhibitors of PKC, PKA, and PDK1 negatively or positively regulated MIOX activity (Fig. 6, A and B). Such modulation by these signaling enzymes lends further credence to the relevance of MIOX in the pathogenesis of diabetic nephropathy, especially in view of the fact that they profoundly modulate various cellular signaling events initiated by hyperglycemia (5). The fact that the inhibitors of PKC, PKA, and PDK1 were capable of reducing the high glucose-induced phosphorylation and activity of RSOR/MIOX would also be supportive of the above notion (Fig. 7, A and B).
In diabetic nephropathy, besides hyperglycemia, the perpetuation of injury also occurs as a result of activation of a multitude of signaling pathways with ROS as a common injurious denominator (8). The ROS influence a wide variety of biological systems, including the biology of the enzyme aldose reductase, which is critically linked to the pathobiology of diabetic nephropathy (44, 45). Unlike RSOR/MIOX, aldose reductase is exclusively expressed in the renal medullary tubules; however, it is transcriptionally up-regulated by high glucose ambience and oxidative stress stemming from the advanced glycation end products (46). In view of certain similarities between RSOR/MIOX and aldose reductase in terms of osmotic regulation (25, 26), the transcriptional/translational responsiveness of MIOX to oxidants (H2O2 and MGO, a potent glycating agent) and antioxidants (NAC, β-NF, BHA, t-BHQ, and reduced glutathione) was assessed. Immunoblot analyses revealed that the exposure of both LLC-PK1 and HK-2 cells to high glucose and oxidants up-regulated the protein expression of MIOX, whereas treatment with antioxidants down-regulated its expression (Fig. 8A). Likewise, phosphorylation and MIOX activity were affected by oxidants and antioxidants under high glucose ambience (Fig. 8, B and C), reinforcing the significance of post-translational modifications that influence the biological properties of this enzyme and ultimately link it to the ROS-initiated cellular redox events described in the literature for the pathogenesis of diabetic nephropathy (8).
One of the well characterized transcription factors that is responsive to changes in the cellular redox is Nrf2 (47). Nrf2 is a member of the Cap “n” Collar subfamily of basic region-leucine zipper (bZip), and it responds to changes in the intracellular redox with increased translocation into the nucleus. The Nrf2 translocation is influenced by a variety of stimuli, including xenobiotics, electrophiles, heavy metals, radiation, and intriguingly both antioxidants and ROS (Refs. 47–49 and supplemental Fig. 1). Because Nrf2 nuclear translocation is known to be affected by both oxidants and antioxidants, its modulation was investigated in both states besides under high glucose ambience. High glucose ambience or overexpression of MIOX led to a translocation of Nrf2 into the nucleus as assessed by immunofluorescence microscopy (Fig. 9, A–C). Also, by immunoblot analyses, a translocation of Nrf2 was observed following high glucose, H2O2, or MGO treatment (Fig. 9D). Intriguingly, concomitant exposure of cells to high glucose and antioxidants enriched the translocation of Nrf2 into the nucleus, suggesting that this transcription factor responds to both oxidant and antioxidant treatments as reported in the literature (48, 49). The fact that Nrf2 was also responsive to the overexpression of MIOX suggests that the biology of oxidant stress and high glucose-induced up-regulation of MIOX are interlinked in the pathogenesis of diabetic nephropathy.
These intriguing results relevant to RSOR/MIOX translation and Nrf2 translocation led us to investigate the transcriptional regulation of this enzyme with respect to cellular redox. The promoter analyses revealed an increased luciferase activity under high glucose ambience and treatment with oxidants (Fig. 10A). The high glucose-induced promoter activity was reduced to a variable degree by various antioxidants, in particular by NAC. NAC, a known antioxidant like reduced glutathione, is used in the treatment of experimental diabetic nephropathy (50, 51). The promoter activity findings were further corroborated by EMSA studies where binding to AREs localized in the murine MIOX promoter was evaluated. Putative motifs for ARE were localized at the −573 and −673 bp regions of the mouse RSOR promoter. No binding was observed with the nuclear extract of treated cells when an oligo encompassing the −673 bp region was used for the EMSA (Fig. 10B), suggesting this motif to be non-functional. However, a strong binding was observed when an oligo of the −573 bp region (GTGACATCTC) was used (Fig. 10C). Interestingly, weak binding was also observed under basal conditions with 5 mm glucose. The intensity of the EMSA bands was decreased to a variable degree in cells subjected to high glucose ambience and then treated with antioxidants (Fig. 10C, lanes 5–8 versus lane 2). The reduction in the intensity of shifted bands by antioxidants could be clearly delineated in EMSA experiments in which half the concentrations of glucose, oxidants, and antioxidants were used (Fig. 10D, lanes 5–8 versus lane 2).
Interestingly, the core sequences of ARE like that of the functional −573 bp oligo have been found be conserved in the promoter regions of several drug-metabolizing enzymes, such as glutathione S-transferase (GST) and NAD(P)H:quinone oxidoreductase (52, 53), and also in antioxidative stress proteins like γ-glutamylcysteine synthetase and heme oxygenase-1 (55, 56), which are known to play a critical role in the maintenance of the intracellular redox state. Nrf2 is one of the major transcription factors involved in redox signaling (51–56). Nrf2 is also modulated by various xenobiotics, including phenolic antioxidants, flavonoids, and isothiocyanate, such as β-NF and 2(3)-tert-butyl-4-hydroxyanisole (57). The fact that high glucose- or oxidant-induced ARE binding could be partially reduced with antioxidants supports the above assertions made in a wide variety of other systems. The results of promoter activity and EMSA binding assays clearly demonstrate transcriptional modulation of MIOX by oxidants, including the oxidant stress that may be stemming from high glucose ambience as has been well reported in the literature.
The next question addressed was whether high glucose can directly modulate transcriptional regulation of RSOR/MIOX, and the answer seems to be affirmative. Such a functionality of the promoter region is usually dependent upon ChREs where the E-box motifs are separated by 5–15 bp. A single ChRE motif was identified in RSOR/MIOX promoter, and it was found to be functional because an increase in the intensity of the shifted band was observed under high d-glucose (35 mm) ambience (Fig. 11, A and B, lane 2). Treatment with MGO or other oxidants failed to increase the band intensity seen under a basal d-glucose concentration of 5 mm in the culture media. Also, no distinct band was observed when the media were substituted with l-glucose (Fig. 11, A and B, last lane). These observations suggest that ChREs are responsive to high glucose challenge somewhat selectively and do not respond to other pathobiological stimuli. Conceivably, such responsiveness is also observed in other metabolizing enzymes, such as liver pyruvate kinase, acetyl-CoA carboxylase, and fatty-acid synthase, because they include E-box and ChRE motifs in their promoters (58). Acetyl-CoA carboxylase and fatty-acid synthase are the two key hepatic liver enzymes whose expression/activity is modulated by the transcription factor ChREBP. Using anti-ChREBP antibody, ChIP-PCR analyses and immunofluorescence studies were carried out. The PCR analysis revealed a band of marked intensity in the nuclear extract isolated from cells exposed to high glucose ambience, suggesting the possibility of strong DNA-protein interactions (Fig. 11C, lane 5). These observations were further strengthened by immunofluorescence studies in which nuclear translocation of ChREBP was clearly observed (Fig. 11, D and E), thus suggesting that high glucose ambience can also directly modulate the transcriptional regulation of RSOR/MIOX.
In conclusion, the findings of this study further enhance our understanding of transcriptional and translational regulation of RSOR/MIOX by hyperglycemia and various cellular stresses in which MI homeostasis would be perturbed via different signaling pathways. The proposed mechanism(s) relevant to RSOR/MIOX expression and its activity in a high glucose milieu are depicted schematically in Fig. 1. It is well known in diabetic patients that increased urinary MI excretion is due to glucose-induced MI transport inhibition (Fig. 1, Step I) and glucose intermediary polyol (sorbitol)-induced MI depletion (Fig. 1, Step II). Other mechanisms relevant to increased MIOX expression (Fig. 1, Step III) and enzymatic activity (Fig. 1, Step IV) are proposed on the basis of results of this investigation. Glucose-induced intermediaries and activating kinases can modulate different transcription factors, such as NFAT-5 (26), ChREBP, Nrf-2 (present study), AP1, and cAMP-response element-binding protein (data not included), for increased MIOX expression (Fig. 1, Step III). The central hypothesis is that the post-translational modifications (phosphorylation) modulate RSOR/MIOX activity. Also, different phosphorylating enzymes modulate its transcription and hence its expression and enzyme activity through various kinase-dependent pathways.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK60635.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- MI
- myo-inositol
- MIOX
- myo-inositol oxygenase
- RSOR
- renal-specific oxidoreductase
- MGO
- methylglyoxal
- BHA
- butylated hydroxyanisole
- t-BHQ
- tertiary butyl hydroquinone
- β-NF
- β-naphthoflavone
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- λ-PPase
- λ-phosphatase
- Nrf2
- NF-E2-related factor 2
- ARE
- antioxidant response element
- ChRE
- carbohydrate response element
- ChREBP
- carbohydrate response element-binding protein
- ROS
- reactive oxygen species
- PKA
- protein kinase A
- PDK1
- phosphoinositide-dependent protein kinase-1
- EMSA
- electrophoretic mobility shift assay
- nt
- nucleotides
- HG
- high glucose.
REFERENCES
- 1. Price R. G., Hudson B. G. (1987) in Renal Basement Membranes in Health and Disease, pp. 237–272, Academic Press, Boston, MA [Google Scholar]
- 2. Reddy A. S. (2004) Diabetic Nephropathy: Theory & Practice, College Book Publishers, L.L.C., East Hanover, NJ [Google Scholar]
- 3. Susztak K., Böttinger E. P. (2006) J. Am. Soc. Nephrol. 17, 361–367 [DOI] [PubMed] [Google Scholar]
- 4. Wolf G., Ziyadeh F. N. (1999) Kidney Int. 56, 393–405 [DOI] [PubMed] [Google Scholar]
- 5. Feliers D., Duraisamy S., Faulkner J. L., Duch J., Lee A. V., Abboud H. E., Choudhury G. G., Kasinath B. S. (2001) Kidney Int. 60, 495–504 [DOI] [PubMed] [Google Scholar]
- 6. Haneda M., Koya D., Isono M., Kikkawa R. (2003) J. Am. Soc. Nephrol. 14, 1374–1382 [DOI] [PubMed] [Google Scholar]
- 7. Reusch J. E. (2003) J. Clin. Investig. 112, 986–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brownlee M. (2005) Diabetes 54, 1615–1625 [DOI] [PubMed] [Google Scholar]
- 9. Tan A. L., Forbes J. M., Cooper M. E. (2007) Semin. Nephrol. 27, 130–143 [DOI] [PubMed] [Google Scholar]
- 10. Sanchez A. P., Sharma K. (2009) Expert Rev. Mol. Med. 11, e13. [DOI] [PubMed] [Google Scholar]
- 11. Villeneuve L. M., Natarajan R. (2010) Am. J. Physiol. Renal Physiol. 299, F14–F25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanwar Y. S., Sun L., Xie P., Liu F. Y., Chen S. (2011) Annu. Rev. Pathol. 6, 395–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palmano K. P., Whiting P. H., Hawthorne J. N. (1977) Biochem. J. 167, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whiting P. H., Palmano K. P., Hawthorne J. N. (1979) Biochem. J. 179, 549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Olgemöller B., Schwaabe S., Schleicher E. D., Gerbitz K. D. (1993) Diabetes 42, 1119–1125 [DOI] [PubMed] [Google Scholar]
- 16. Stevens M. J., Lattimer S. A., Kamijo M., Van Huysen C., Sima A. A., Greene D. A. (1993) Diabetologia 36, 608–614 [DOI] [PubMed] [Google Scholar]
- 17. Pendaries C., Tronchère H., Plantavid M., Payrastre B. (2003) FEBS Lett. 546, 25–31 [DOI] [PubMed] [Google Scholar]
- 18. Ooms L. M., Horan K. A., Rahman P., Seaton G., Gurung R., Kethesparan D. S., Mitchell C. A. (2009) Biochem. J. 419, 29–49 [DOI] [PubMed] [Google Scholar]
- 19. Holub B. J. (1986) Annu. Rev. Nutr. 6, 563–597 [DOI] [PubMed] [Google Scholar]
- 20. Colodny L., Hoffman R. L. (1998) Altern. Med. Rev. 3, 432–447 [PubMed] [Google Scholar]
- 21. Gabbay K. H. (1975) Annu. Rev. Med. 26, 521–536 [DOI] [PubMed] [Google Scholar]
- 22. Kawa J. M., Przybylski R., Taylor C. G. (2003) Exp. Biol. Med. 228, 907–914 [DOI] [PubMed] [Google Scholar]
- 23. Yang Q., Dixit B., Wada J., Tian Y., Wallner E. I., Srivastva S. K., Kanwar Y. S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9896–9901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arner R. J., Prabhu K. S., Thompson J. T., Hildenbrandt G. R., Liken A. D., Reddy C. C. (2001) Biochem. J. 360, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prabhu K. S., Arner R. J., Vunta H., Reddy C. C. (2005) J. Biol. Chem. 280, 19895–19901 [DOI] [PubMed] [Google Scholar]
- 26. Nayak B., Xie P., Akagi S., Yang Q., Sun L., Wada J., Thakur A., Danesh F. R., Chugh S. S., Kanwar Y. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17952–17957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phillips A. O., Steadman R. (2002) Histol. Histopathol. 17, 247–252 [DOI] [PubMed] [Google Scholar]
- 28. Phillips A. O. (2003) Curr. Diab. Rep. 3, 491–496 [DOI] [PubMed] [Google Scholar]
- 29. Hu E., Chen Z., Fredrickson T., Gellai M., Jugus M., Contino L., Spurr N., Sims M., Halsey W., Van Horn S., Mao J., Sathe G., Brooks D. (2000) Am. J. Physiol. Renal Physiol. 279, F426–F439 [DOI] [PubMed] [Google Scholar]
- 30. Lauer M. E., Hascall V. C., Wang A. (2007) J. Biol. Chem. 282, 843–852 [DOI] [PubMed] [Google Scholar]
- 31. Ren J., Hascall V. C., Wang A. (2009) J. Biol. Chem. 284, 16621–16632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang B., Hodgkinson A., Millward B. A., Demaine A. G. (2010) J. Diabetes Complications 24, 404–408 [DOI] [PubMed] [Google Scholar]
- 33. Charalampous F. C., Lyras C. (1957) J. Biol. Chem. 228, 1–13 [PubMed] [Google Scholar]
- 34. Xie P., Sun L., Nayak B., Haruna Y., Liu F. Y., Kashihara N., Kanwar Y. S. (2009) J. Am. Soc. Nephrol. 20, 807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soutoglou E., Talianidis I. (2002) Science 295, 1901–1904 [DOI] [PubMed] [Google Scholar]
- 36. Ansari N. H., Srivastava S. K. (1993) Biochem. Mol. Biol. Int. 30, 37–44 [PubMed] [Google Scholar]
- 37. Barski O. A., Papusha V. Z., Ivanova M. M., Rudman D. M., Finegold M. J. (2005) Am. J. Physiol. Renal Physiol. 289, F200–F207 [DOI] [PubMed] [Google Scholar]
- 38. Hankes L. V., Politzer W. M., Touster O., Anderson L. (1969) Ann. N.Y. Acad. Sci. 165, 564–576 [PubMed] [Google Scholar]
- 39. Brown P. M., Caradoc-Davies T. T., Dickson J. M., Cooper G. J., Loomes K. M., Baker E. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15032–15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xing G., Barr E. W., Diao Y., Hoffart L. M., Prabhu K. S., Arner R. J., Reddy C. C., Krebs C., Bollinger J. M., Jr. (2006) Biochemistry 45, 5402–5412 [DOI] [PubMed] [Google Scholar]
- 41. Narayanan A., Jacobson M. P. (2009) Curr. Opin. Struct. Biol. 19, 156–163 [DOI] [PubMed] [Google Scholar]
- 42. Xue Y., Liu Z., Cao J., Ma Q., Gao X., Wang Q., Jin C., Zhou Y., Wen L., Ren J. (2011) Protein Eng. Des. Sel. 24, 255–260 [DOI] [PubMed] [Google Scholar]
- 43. Foretz M., Carling D., Guichard C., Ferré P., Foufelle F. (1998) J. Biol. Chem. 273, 14767–14771 [DOI] [PubMed] [Google Scholar]
- 44. Nishinaka T., Yabe-Nishimura C. (2001) Free Radic. Biol. Med. 31, 205–216 [DOI] [PubMed] [Google Scholar]
- 45. Srivastava S. K., Ramana K. V., Bhatnagar A. (2005) Endocr. Rev. 26, 380–392 [DOI] [PubMed] [Google Scholar]
- 46. Nakamura N., Obayashi H., Fujii M., Fukui M., Yoshimori K., Ogata M., Hasegawa G., Shigeta H., Kitagawa Y., Yoshikawa T., Kondo M., Ohta M., Nishimura M., Nishinaka T., Nishimura C. Y. (2000) Free Radic. Biol. Med. 29, 17–25 [DOI] [PubMed] [Google Scholar]
- 47. Kaspar J. W., Jaiswal A. K. (2010) J. Biol. Chem. 285, 21349–21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaspar J. W., Niture S. K., Jaiswal A. K. (2009) Free Radic. Biol. Med. 47, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Faraonio R., Vergara P., Di Marzo D., Pierantoni M. G., Napolitano M., Russo T., Cimino F. (2006) J. Biol. Chem. 281, 39776–39784 [DOI] [PubMed] [Google Scholar]
- 50. Huang J. S., Chuang L. Y., Guh J. Y., Huang Y. J., Hsu M. S. (2007) Am. J. Physiol. Renal Physiol. 293, F1072–F1082 [DOI] [PubMed] [Google Scholar]
- 51. Zhang Y., Wada J., Hashimoto I., Eguchi J., Yasuhara A., Kanwar Y. S., Shikata K., Makino H. (2006) J. Am. Soc. Nephrol. 17, 1090–1101 [DOI] [PubMed] [Google Scholar]
- 52. Wasserman W. W., Fahl W. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. (1997) Biochem. Biophys. Res. Commun. 236, 313–322 [DOI] [PubMed] [Google Scholar]
- 54. Nguyen T., Huang H. C., Pickett C. B. (2000) J. Biol. Chem. 275, 15466–15473 [DOI] [PubMed] [Google Scholar]
- 55. Moinova H. R., Mulcahy R. T. (1999) Biochem. Biophys. Res. Commun. 261, 661–668 [DOI] [PubMed] [Google Scholar]
- 56. Alam J., Wicks C., Stewart D., Gong P., Touchard C., Otterbein S., Choi A. M., Burow M. E., Tou J. (2000) J. Biol. Chem. 275, 27694–27702 [DOI] [PubMed] [Google Scholar]
- 57. Jiang Z. Q., Chen C., Yang B., Hebbar V., Kong A. N. (2003) Life Sci. 72, 2243–2253 [DOI] [PubMed] [Google Scholar]
- 58. Ishii S., Iizuka K., Miller B. C., Uyeda K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15597–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.