Abstract
Rhodopsin has developed mechanisms to optimize its sensitivity to light by suppressing dark noise and enhancing quantum yield. We propose that an intramolecular hydrogen-bonding network formed by ∼20 water molecules, the hydrophilic residues, and peptide backbones in the transmembrane region is essential to restrain thermal isomerization, the source of dark noise. We studied the thermal stability of rhodopsin at 55 °C with single point mutations (E181Q and S186A) that perturb the hydrogen-bonding network at the active site. We found that the rate of thermal isomerization increased by 1–2 orders of magnitude in the mutants. Our results illustrate the importance of the intact hydrogen-bonding network for dim-light detection, revealing the functional roles of water molecules in rhodopsin. We also show that thermal isomerization of 11-cis-retinal in solution can be catalyzed by wild-type opsin and that this catalytic property is not affected by the mutations. We characterize the catalytic effect and propose that it is due to steric interactions in the retinal-binding site and increases quantum yield by predetermining the trajectory of photoisomerization. Thus, our studies reveal a balancing act between dark noise and quantum yield, which have opposite effects on the thermal isomerization rate. The acquisition of the hydrogen-bonding network and the tuning of the steric interactions at the retinal-binding site are two important factors in the development of dim-light vision.
Keywords: G Protein-coupled Receptors (GPCRs), Membrane Proteins, Protein Stability, Rhodopsin, Vision, Dark Noise, Dim-light Vision, Thermal Isomerization, Thermal Stability
Introduction
Rhodopsin is a widely studied G protein-coupled receptor responsible for generating visual signals in dim-light environments (1–11). It is expressed in the disc membranes in the outer segment of rod photoreceptor cells (1–3). It has a seven-helical transmembrane structure and incorporates the 11-cis-retinal chromophore via a protonated Schiff base (SB)6 in the transmembrane region (3, 8, 9). Upon absorption of a photon, the retinal chromophore isomerizes from the 11-cis- to all-trans-configuration to form the primary photoproduct bathorhodopsin (12, 13), which then evolves into a number of photointermediates on various time scales (14, 15), forming metarhodopsin II (Meta II) in milliseconds. In Meta II, the SB linkage becomes deprotonated, and the absorption changes from the visible to the UV (380 nm) region. Subsequently, Meta II couples to the G protein transducin to induce an exchange of GDP for GTP in the α-subunit of transducin (16). The α-subunit dissociates from the βγ-subunit and activates phosphodiesterase, which catalyzes hydrolysis of cGMP. A decrease in the concentration of cGMP closes the sodium channels of the rod photoreceptor cells and generates hyperpolarization for a visual signal.
We asked what molecular properties allow rhodopsin to gain photosensitivity for dim-light vision; rhodopsin has the ability to handle intensities of light spanning orders of magnitude and yet the capacity to detect single photons (16, 17). These properties must be related to the optimization of amplification, quantum yield, and dark noise level, three important criteria for evaluating the sensitivity of any light detector, both man-made and biological. First, amplification specifies the magnitude of readout per photon detected. In the case of rhodopsin, one photon can activate ∼102 copies of transducin, which each activate ∼102 copies of phosphodiesterase, blocking 107 Na+ ions from crossing the plasma membrane (16). Because downstream signaling amplification is not directly related to the properties of rhodopsin, it is not the focus of this study. Second, quantum yield denotes the fraction of photons being absorbed that can successfully generate a signal. For rhodopsin, the quantum yield is 65%, which is among the highest in photobiological systems (18). Finally, dark noise is the rate of false-positive response generated by a light detector in a completely dark environment. The dark noise of rhodopsin is extremely low: one count in every 420 years for a rhodopsin molecule in primate rod cells at 36 °C (19). Baylor (16), Lagnado and Baylor (20), and Lamb (21) demonstrated that the dark noise originates from the thermal isomerization of 11-cis-retinal in rhodopsin, which generates the same physiological response as photoisomerization.
To investigate the molecular mechanism that allows rhodopsin to achieve high quantum yield and low dark noise, we examined the thermal properties of rhodopsin. First, we hypothesized that quantum yield is associated with the stereochemistry of the retinal-binding site. As early as 1958, Hubbard observed that the rate of isomerization of 11-cis-retinal added externally to the aqueous solution is faster in the presence of opsin, revealing that opsin can catalyze the retinal isomerization (22). However, the catalytic activity of opsin has not been explicitly discussed, and its mechanism has remained obscure. The crystal structures of rhodopsin show that the interactions between the chromophore and the compact binding site result in distortion of the ethylenic chain of the 11-cis-retinal chromophore in the binding pocket (9, 10). This distortion has been postulated to guide the trajectory of photoisomerization of 11-cis-retinal in rhodopsin (23) such that the relaxation from the excited state to the ground state can effectively lead to formation of the primary photoproduct, bathorhodopsin, and subsequently Meta II for transducin activation. We aimed to link the steric interactions in the retinal-binding site to the ability of opsin to catalyze cis- to-trans-isomerization of retinal to explain the high quantum yield of rhodopsin.
Next, because dark noise originates from thermal isomerization of 11-cis-retinal, to understand how rhodopsin achieves low dark noise, we need to understand how rhodopsin prevents thermal isomerization. Our investigation was inspired by previous observations. First, Li et al. (9) and Okada et al. (10) reported the crystal structures of rhodopsin, which include water molecules in the transmembrane region of rhodopsin. Palczewski and co-workers (24–26) further studied the distribution of transmembrane water and found conserved contacts between these water molecules and microdomains, which are important in receptor activation. These water molecules participate in an extensive hydrogen-bonding network spanning the entire rhodopsin molecule from the extracellular side through the transmembrane region to the cytoplasmic side. This network has been postulated to be important for stabilizing the dark state of rhodopsin (6, 27, 28). Second, Janz et al. (29, 30) reported that a perturbation to the H-bonding network by mutations at the active site leads to an increase in the rate of thermal decay of rhodopsin, which is defined by a decrease in the 500 nm absorption of rhodopsin. They attributed the thermal decay to the hydrolysis of the SB. Around the same time, Del Valle et al. (31) detected thermal cis,trans-isomerization of retinal using HPLC and proposed that thermal decay was the result of isomerization caused by protein denaturation.
In fact, we previously demonstrated that thermal decay consists of both hydrolysis of the SB in dark-state rhodopsin and thermal isomerization of 11-cis-retinal (32). We compared the thermal properties of WT bovine rhodopsin in H2O and D2O at 59 °C because H-bonds are stronger in D2O compared with H2O (32). We found that the rates of thermal processes become 2–3 times slower in D2O than in H2O, suggesting that the rate-determining steps of both hydrolysis and isomerization involve breaking H-bonds.
In this study, we extended our investigation of the thermal properties of rhodopsin by disrupting the H-bonding network in the retinal active site of rhodopsin using two point mutations, E181Q and S186A (Fig. 1). For the first time, we measured the rate of thermal isomerization and SB hydrolysis simultaneously using rhodopsin mutants, which is expected to provide a more complete mechanistic understanding of the thermal decay process of rhodopsin. We also report the first detailed description of opsin-catalyzed thermal isomerization of 11-cis-retinal. We interpret the results in the context of the roles of both the steric interactions at the active site and the H-bonding network in optimizing the quantum yield and dark noise of rhodopsin, which exert opposing effects that are necessary to modulate photosensitivity. The results give insight into the mechanism of the molecular evolution of vision.
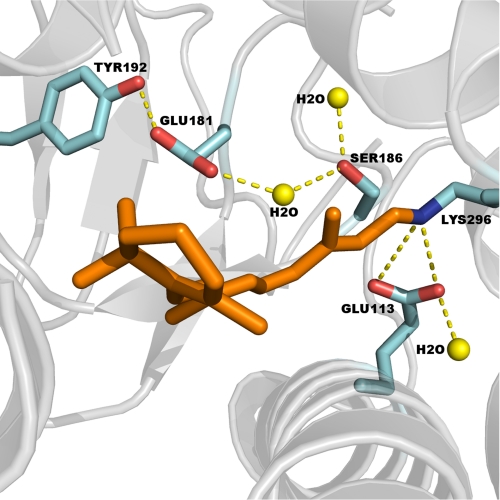
FIGURE 1.
Structure of the 11-cis-retinal chromophore in the binding pocket of rhodopsin (Protein Data Bank code 1GZM): Glu-181 within hydrogen-bonding distance to Tyr-192, Tyr-268, and a water molecule and Ser-186 near the protonated SB counterion complex and within hydrogen-bonding distance to two water molecules.
EXPERIMENTAL PROCEDURES
Preparation of Rhodopsin
Recombinant DNA constructs of WT, E181Q, and S186A opsins in the pACMV-tetO vector (33) were transfected into HEK293T cells using LipofectamineTM. Stable cell lines were created by selecting for transfected cells using the antibiotic Geneticin (300 μg/ml) for 2 weeks (34). Opsin expression was induced with tetracycline (2 μg/ml) and sodium butyrate (5 mm). After 48 h, the cells were harvested, and rhodopsin was regenerated with 5 μm 11-cis-retinal overnight at 4 °C in the dark. All procedures involving the regenerated rhodopsin were conducted under dim red light.
Membranes were detergent-solubilized in 50 mm Tris, 100 mm NaCl, 1 mm CaCl2, 1% (w/v) n-dodecyl-β-d-maltoside, and 0.1 mm PMSF (pH 6.8) for 4 h at 4 °C. Solubilized rhodopsin was purified as described previously by immunobinding to anti-rhodopsin C terminus antibody 1D4 coupled to Sepharose beads (34–36). The beads were washed three times with 50 mm Tris, 100 mm NaCl, and 0.1% n-dodecyl-β-d-maltoside (pH 6.8) and three times with Buffer A (50 mm sodium phosphate and 0.1% n-dodecyl-β-d-maltoside (pH 6.5)). Rhodopsin was eluted in Buffer A containing 1D5 peptide (TETSQVAPA) for competitive binding to the antibody. The samples were then concentrated to ∼20 μm in Buffer A.
Thermal Decay
UV-visible spectroscopy was used to monitor thermal decay as described (32). All measurements were made in the dark on a UV-visible spectrophotometer (Shimadzu UV-2450). The temperature of the samples was maintained by a circulating water bath and was monitored by a thermocouple placed at the cuvette holder. Buffer A at a volume of 2.7 ml was equilibrated at 55 °C. At t = 0, a solution of rhodopsin (20 μm) at a volume of 0.3 ml was added. Control experiments suggested that it took <2 s for the mixture to reach 55 °C. UV-visible spectra were taken at various time points, and ∼12 samples (200 μl each) were removed from the cuvette between the acquisition of the spectra. Each sample was immediately frozen in a glass vial precooled by dry ice and stored on ice to quench any thermal process. These samples were divided equally into two parts for measuring the rates of thermal isomerization and hydrolysis of the SB (32).
Thermal Isomerization
The rate of thermal isomerization was obtained by analyzing the isomeric forms of retinal extracted from the samples taken at various time points during the thermal decay experiments. The procedure of the extraction and HPLC analysis has been described previously (32, 37). The retinal extracted in the form of retinal oxime in hexane from each sample was injected into a Beckman analytical silica column (25 cm × 4.6 mm (inner diameter), 5-μm particle size) connected to the HPLC system (Beckman Coulter System Gold®) and detected using UV absorption at 360 nm and a mobile phase of hexane supplemented with 8% diethyl ether and 0.33% ethanol.
Hydrolysis of the SB of Rhodopsin
The second part of the samples taken at various time points during the thermal decay experiments was used to measure the rate of hydrolysis of the SB (37–40). To each 100-μl sample was added 4 μl of 1 m HCl to denature the protein. Retinal covalently linked to the opsin protein via the protonated SB absorbs at 440 nm, whereas free retinal in aqueous solution as a result of hydrolysis of the SB absorbs at 380 nm. Hence, the absorbance at 440 nm of the samples after acidic denaturation reveals the extent of SB hydrolysis. The pH of the solution was measured after the experiments and confirmed to be 1–2.
Opsin-catalyzed Thermal Isomerization
The opsin protein was obtained by photobleaching the purified rhodopsin samples. Buffer A (2.7 ml) was equilibrated in the UV-visible spectrometer at 55 °C. A 0.3-ml concentrated rhodopsin solution (10 μm) was added. The sample was photobleached using a 30-watt fiber optic illuminator at a wavelength of >495 nm to trigger photoisomerization leading to formation of Meta II, which was allowed to decay for 10 min to release all-trans-retinal, yielding opsin protein. Then, a UV-visible spectrum was taken, and a 100-μl sample was taken as a control for the HPLC analysis to confirm that all 11-cis-retinal was converted to all-trans-retinal. Subsequently, 11-cis-retinal solution was added to a final concentration of 1 μm at t = 0. UV-visible spectra were taken to monitor whether rhodopsin regeneration occurred, and at least 10 aliquots (100 μl each) were sampled at various time points for retinal extraction and HPLC analyses to measure the rate of thermal isomerization catalyzed by opsin.
Binding of Retinal into the Active Site of Opsin
The effect of thermal incubation on the retinal-binding activity of opsin proteins was investigated by measuring the entry of 11-cis-retinal into the binding pocket. The intrinsic tryptophan fluorescence at the binding site can be quenched by the presence of retinal (40). The intrinsic fluorescence signal was measured using excitation/emission at 290/330 nm with input and output slit widths of 5 nm. Buffer A (100 μl) was incubated in a Cary Eclipse fluorometer (Varian, Inc.) at 55 °C. The signal was monitored upon the addition of a 100-μl solution of the rhodopsin sample (2 μm). After 30 s, the rhodopsin sample was bleached with light at a wavelength of >495 nm. The bleached sample was kept in the spectrophotometer at 55 °C to allow hydrolysis of the SB and release of all-trans-retinal to yield the opsin samples. After the bleached sample was incubated for ∼10 or 40 min, 2 μl of the concentrated 11-cis-retinal solution in ethanol was added to a final concentration of either 1:1 or 1:10 opsin/retinal. The decay of the fluorescence signal due to the entry of 11-cis-retinal into the active site was monitored.
RESULTS
Sample Preparation
UV-visible spectra of dark-state WT, E181Q, and S186A rhodopsins were recorded at room temperature following purification (supplemental Fig. 1). The peak at 280 nm corresponds to the absorption of opsin, whereas the peak at 500 nm corresponds to rhodopsin. The R values defined as A280/A500 are 1.64 for WT, 1.90 for E181Q, and 1.88 for S186A.
Thermal Decay
Fig. 2 (A–C) shows spectra taken at various time points at 55 °C. Each spectrum was normalized to A280 to account for solvent evaporation. The spectra show that A500 decreases, whereas A380 increases. The peak at 380 nm could be due to 11-cis- or all-trans-retinal free in solution and/or Meta II consisting of all-trans-retinal bound to opsin. Because rhodopsin is the only species that absorbs at 500 nm, the decrease in A500 reveals the thermal decay of rhodopsin. The measured A500 was normalized to the initial value, A500 (t = 0), and plotted as a function of time (Fig. 2D). The half-lives are 70 ± 2, 2.3 ± 0.1, and 0.9 ± 0.1 min for WT, E181Q, and S186A, respectively. Compared with WT, E181Q and S186A decay 30 and 78 times faster, respectively.
FIGURE 2.
Kinetics of the thermal processes of rhodopsin at 55 °C. A–C, UV-visible spectra of the thermal decay of rhodopsin at various time points for WT, E181Q, and S186A. The dark spectra of purified WT and mutant rhodopsins are included as t < 0 min. D, A500 as a function of time. E–G, HPLC chromatograms of the retinal oxime extracts for WT, E181Q, and S186A. H, the fraction of 11-cis-retinal as a function of time. I–K, UV-visible spectra of acid-denatured WT, E181Q, and S186A. L, A440 as a function of time.
Thermal Isomerization
Retinal was extracted as retinal oxime from the sample taken at various time points during the thermal incubation and analyzed by HPLC to determine the extent of thermal isomerization. A total of six isomers were observed in the chromatograms, corresponding to 11-cis-15-syn-, all-trans-15-syn-, 13-cis-15-syn-, 13-cis-15-anti-, 11-cis-15-anti-, and all-trans-15-anti-retinal oxime (32, 37). Because the areas of the peaks for the anti-species are small, only the syn-peaks are shown in Fig. 2 (E–G). During the thermal decay process, the amount of 11-cis-retinal decreases, whereas that of all-trans-retinal increases. This result suggests that the thermal 11-cis- to all-trans-isomerization happens not only in WT, as reported previously (32), but also in E181Q and S186A. To determine the fraction of 11-cis-retinal at each time point, the area of the 11-cis-peak was normalized to the sum of six peaks and the corresponding extinction coefficients for each isomeric form at 360 nm (37). The fraction of 11-cis-retinal was plotted as a function of time (Fig. 2H). The half-lives of thermal isomerization for WT, E181Q, and S186A are 88 ± 10, 3.8 ± 0.6, and 1.3 ± 0.3 min, respectively. The rates for E181Q and S186A are 23 and 68 times faster than that for WT, respectively.
Hydrolysis of the SB
The rate of hydrolysis of the SB was determined by measuring the UV-visible absorption of acid-denatured thermal decay products (Fig. 2, I–K). At the earliest time point (t < 2 min), the predominant peak is at 440 nm, corresponding to retinal covalently attached to denatured opsin via a protonated SB. At later time points, A440 decreases, whereas A380 increases, indicating that the SB is hydrolyzed and the covalent linkage between retinal and opsin is broken. To quantify the extent of hydrolysis of the SB, each absorption spectrum was analyzed by plotting absorbance as a function of frequency and fitting into a sum of two Gaussian functions centered at 380 and 440 nm. The fitted intensity for the 440 nm peak was then plotted as a function of time (Fig. 2L). The half-lives for WT, E181Q, and S186A are 73 ± 3, 2.6 ± 0.3, and 1.4 ± 0.5 min, respectively. The rates for E181Q and S186A are 28 and 52 times faster than that for WT, respectively.
Opsin-catalyzed Thermal Isomerization
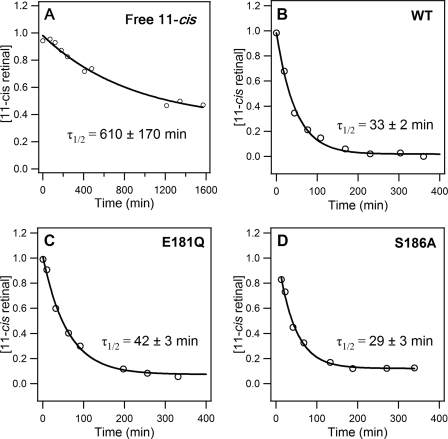
To study the catalytic activity of opsin, defined as the ability of opsin to facilitate thermal isomerization of 11-cis-retinal that is added externally to the solution, the rate of thermal isomerization of 11-cis-retinal in solution was determined in the absence and presence of the WT, E181Q, or S186A opsin at a retinal/opsin ratio of 1:1. Fig. 3 (A–D) shows the time dependence of the fraction of 11-cis-retinal in the sample. The half-lives of thermal isomerization of free 11-cis-retinal were found to be 610 ± 170 min in the absence of opsin protein and 33 ± 2, 42 ± 3, and 29 ± 3 min in the presence of the WT, E181Q, and S186A opsins, respectively. When monitored with UV-visible spectroscopy, no 500 nm pigment appeared during the assay (Fig. 4). These results suggest that opsin catalyzes the 11-cis- to all-trans-isomerization of retinal in solution without formation of the SB at 55 °C. Furthermore, in contrast to the dramatic effect of the E181Q and S186A mutations on the rate of thermal isomerization of rhodopsin, the effect of these mutations on the catalytic rate was much milder (Table 1).
FIGURE 3.
Kinetics of thermal isomerization of 11-cis-retinal free in solution in the absence (A) and presence of WT (B), E181Q (C), and S186A (D) opsins.
FIGURE 4.
UV-visible spectra of 11-cis-retinal in the presence of WT (A), E181Q (B), and S186A (C) opsin proteins. No 500 nm peak was observed over the course of 5–6 h.
TABLE 1.
Half-lives of thermal processes at 55 °C
| τ½ |
|||
|---|---|---|---|
| WT | E181Q | S186A | |
| min | |||
| Thermal stability of rhodopsin | |||
| Thermal decay, A500 | 70 ± 2 | 2.3 ± 0.1 | 0.9 ± 0.1 |
| Thermal isomerization, 11-cis-retinal | 88 ± 10 | 3.8 ± 0.6 | 1.3 ± 0.3 |
| Hydrolysis, A440 | 73 ± 3 | 2.6 ± 0.3 | 1.4 ± 0.5 |
| Catalytic property of opsin protein | |||
| Thermal isomerization of 11-cis-retinal in the presence of opsin protein | 33 ± 2 | 42 ± 3 | 29 ± 3 |
| Thermal isomerization of free 11-cis-retinal in solution | 610 ± 170 | ||
Binding of Retinal to the Active Site of Opsin
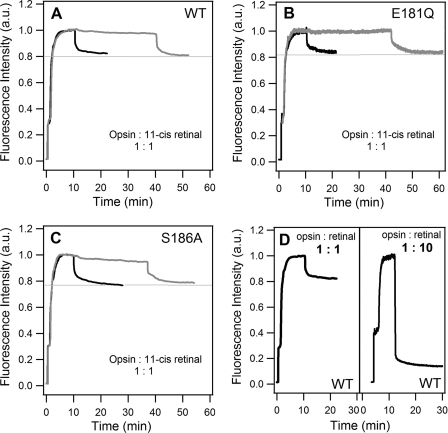
After finding that no SB formation occurs during catalytic isomerization, we performed retinal binding experiments. Fig. 5 (A–C) shows the time dependence of intrinsic tryptophan fluorescence. At t = 0, the sample was buffer alone, and the fluorescence signal was zero. At t ∼ 30 s, the rhodopsin samples were added, which accounts for the first rise of the signal from 0 to 0.2–0.3. Then, the rhodopsin samples were bleached to form Meta II, and all-trans-retinal leaks from the binding pocket. Thus, the fluorescence quenching by retinal in the binding pocket is removed (39), and the signal increases gradually to a maximum at ∼5 min. At this point, opsin proteins are produced. Subsequently, at t = 10 min (black curves) or 40 min (gray curves), 2 μl of concentrated 11-cis-retinal was added to a final concentration of 1:1 opsin/retinal. The addition of 11-cis-retinal triggers a decay of the fluorescence signal, suggesting that 11-cis-retinal enters the binding site and quenches the fluorescence. The magnitude of the decay remains the same regardless of whether 11-cis-retinal is added at t = 10 or 40 min, suggesting that the retinal-binding activity of opsin proteins does not change during incubation in this time period at 55 °C. This also indicates that an equilibrium exists between free and bound 11-cis-retinal, which accounts for the partial occupancy of the binding sites at a 1:1 opsin/retinal ratio. We repeated the measurements with a higher 11-cis-retinal concentration, 1:10 opsin/retinal. The higher concentration of retinal shifts the equilibrium to the bound state such that fluorescence signal drops to a larger extent (Fig. 5D). Moreover, similar results are observed in WT and the E181Q and S186A mutants, revealing that the binding activity is preserved in the mutant opsins.
FIGURE 5.
Binding of 11-cis-retinal to the active site of opsin. A–C, intensity of intrinsic tryptophan fluorescence upon adding dark-state WT, E181Q, and S186A mutant rhodopsins; bleaching the dark-state samples; and then adding 11-cis-retinal solution at a 1:1 opsin/retinal ratio at either 10 min (black curves) or 40 min (gray curves). D, intensity of intrinsic tryptophan fluorescence upon adding WT rhodopsin, bleaching the dark-state samples, and then adding 11-cis-retinal solution at 1:1 or 1:10 opsin/retinal. a.u., arbitrary units.
DISCUSSION
Low Dark Noise
We focused on the kinetics of thermal processes of rhodopsin to investigate the molecular mechanism of dim-light vision. A motivation of this study was to test the hypothesis that an extensive H-bonding network in rhodopsin is the mechanism for stabilizing the dark-state structure to prevent thermal isomerization; by lowering the rate of thermal isomerization, the dark noise of rhodopsin can be reduced and photosensitivity can be enhanced. We tested the hypothesis by introducing the E181Q and S186A mutations to perturb the H-bonding network at the retinal-binding site and investigated the effect on the kinetics of thermal isomerization of 11-cis-retinal in rhodopsin. The perturbations expedited thermal isomerization by 1–2 orders of magnitude (Fig. 2 and Table 1). These results suggest that an intact H-bonding network at the active site is crucial for preventing thermal isomerization of rhodopsin.
High Quantum Yield
We also investigated the catalytic activity of opsin for thermal isomerization of 11-cis-retinal added externally to solution. The results show that opsin catalyzed thermal cis,trans-retinal isomerization without formation of a protonated SB between retinal and opsin. The rate of thermal isomerization of 11-cis-retinal free in solution was found to become ∼20 times faster in the presence of the WT opsin proteins at 55 °C (Table 1). We also observed this catalytic activity in the E181Q and S186A mutant opsins. We propose that the catalytic activity of opsin originates from the steric interactions between the chromophore and protein at the active site. By constraining the degrees of freedom of 11-cis-retinal, the binding pocket effectively predetermines the trajectory of isomerization to all-trans-retinal (23), which likely enhances the quantum yield of rhodopsin.
Effect of Mutations on Rhodopsin Versus Opsin
We observed a drastic difference between the effects of the E181Q and S186A mutations on the thermal isomerization of rhodopsin and the isomerization of 11-cis-retinal free in solution catalyzed by opsin proteins. We carried out the catalytic measurements by replacing the WT opsin with the E181Q and S186A mutants and found that the rate of catalytic isomerization changed <50%, in contrast to the 1–2 orders of magnitude change in the rate of thermal isomerization in dark-state rhodopsin. Moreover, the fluorescence experiments show that the E181Q and S186A mutant opsins have similar binding activity for 11-cis-retinal (without SB formation) compared with the WT opsin. Hence, the E181Q and S186A mutations have a milder effect on the active state and on the opsin protein than on dark-state rhodopsin.
These results have two implications. First, the results lead to the conclusion that the contribution to the H-bonding network by Glu-181 and Ser-186 plays an important role in maintaining the thermal stability of rhodopsin but a minor role in the catalytic activity of opsin. This conclusion implies that the H-bonding network is structurally and functionally important in the dark-state rhodopsin but not in opsin and Meta II. Second, the results suggest that the H-bonding network is likely coupled to the electrostatic interactions between the protonated SB and the Glu-113 counterion. This electrostatic interaction, absent in both opsin and Meta II, can possibly act as a switch for the H-bonding network. When the counterion Glu-113 interacts with the positively charged protonated SB in the dark state of rhodopsin, the H-bonding network, involving Ser-186 and Glu-181, remains intact for stabilization of the dark state. On the other hand, when the electrostatic interaction between the counterion and the protonated SB is absent, the H-bonding network can be effectively switched off. As another way of testing this model, we measured the rate of thermal isomerization of the dark-state rhodopsin mutant E113A, which eliminates the dark-state counterion, resulting in a deprotonated SB at neutral pH. We expected that the rate of thermal isomerization of the E113A mutant would be very fast, lacking the restraint provided by the H-bonding network. Indeed, we found that the half-life for thermal isomerization is 1.7 ± 0.6 min at 55 °C (supplemental Fig. 2), in support of our hypothesis.
Mutagenesis Studies of Thermal Isomerization of Rhodopsin
This work supplements the previous studies performed by Janz and Farrens (29) on the thermal stability of E181Q and S186A mutants because we have considered an additional process: thermal isomerization. Aside from measuring the rate of SB hydrolysis using acid denaturation, we also measured the rate of thermal isomerization by analyzing the retinal isomers extracted from the thermal decay products of rhodopsin. We argue that it is extremely important to investigate the effect of mutations on the rate of thermal isomerization because thermal isomerization of rhodopsin is the origin of dark noise and thereby is correlated with the ability of rhodopsin to detect photons in a dim-light environment. Although the way we measured the rate of SB hydrolysis differed from the one used by Janz and Farrens, our observations agree with their conclusion that the E181Q and S186A mutations destabilize dark-state rhodopsin and that the effect of E181Q is generally milder than that of S186A. The different effect is likely due to their different roles and energetics contributing to the H-bonding network at the active site.
Molecular Mechanism of Dim-light Vision
We asked what molecular properties account for the extraordinary photosensitivity of rhodopsin. We now propose a balance of forces that is critical for modulating both high quantum yield and low dark noise in photoreceptor function. On the one hand, the stereochemistry at the retinal-binding site increases quantum yield by orienting 11-cis-retinal to isomerization. However, this improved efficiency of rhodopsin's photoresponse would lead to an increase in the rate of thermal isomerization and thus an increase in dark noise, jeopardizing the photosensitivity. To lower the dark noise, an extensive H-bonding network has developed and coupled with the formation of the protonated SB. The H-bonding network stabilizes the dark state of rhodopsin and lowers the dark noise for dim-light detection. It is conceivable that both the optimization of steric interactions in the binding pocket to enhance quantum yield and the acquisition of the H-bonding network to lower the dark noise were important milestones in the divergence from cone to rod pigments in the evolution of dim-light vision. The application of our experimental approach to other rhodopsin mutations and other visual pigments will provide further insights into both the evolution of visual pigments and the mechanism of visual diseases, to which rhodopsin is intimately connected.
Supplementary Material
This work was supported in part by National Science Foundation Career Grant MCB-0955407.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- SB
- Schiff base
- Meta II
- metarhodopsin II.
REFERENCES
- 1. Palczewski K. (2006) Annu. Rev. Biochem. 75, 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ridge K. D., Palczewski K. (2007) J. Biol. Chem. 282, 9297–9301 [DOI] [PubMed] [Google Scholar]
- 3. Menon S. T., Han M., Sakmar T. P. (2001) Physiol. Rev. 81, 1659–1688 [DOI] [PubMed] [Google Scholar]
- 4. Birge R. R. (1990) Annu. Rev. Phys. Chem. 41, 683–733 [DOI] [PubMed] [Google Scholar]
- 5. Khorana H. G. (1992) J. Biol. Chem. 267, 1–4 [PubMed] [Google Scholar]
- 6. Sakmar T. P., Menon S. T., Marin E. P., Awad E. S. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 443–484 [DOI] [PubMed] [Google Scholar]
- 7. Okada T., Ernst O. P., Palczewski K., Hofmann K. P. (2001) Trends Biochem. Sci. 26, 318–324 [DOI] [PubMed] [Google Scholar]
- 8. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 9. Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. (2004) J. Mol. Biol. 343, 1409–1438 [DOI] [PubMed] [Google Scholar]
- 10. Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., Buss V. (2004) J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 11. Ahuja S., Hornak V., Yan E. C., Syrett N., Goncalves J. A., Hirshfeld A., Ziliox M., Sakmar T. P., Sheves M., Reeves P. J., Smith S. O., Eilers M. (2009) Nat. Struct. Mol. Biol. 16, 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schoenlein R. W., Peteanu L. A., Mathies R. A., Shank C. V. (1991) Science 254, 412–415 [DOI] [PubMed] [Google Scholar]
- 13. Peteanu L. A., Schoenlein R. W., Wang Q., Mathies R. A., Shank C. V. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11762–11766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hug S. J., Lewis J. W., Einterz C. M., Thorgeirsson T. E., Kliger D. S. (1990) Biochemistry 29, 1475–1485 [DOI] [PubMed] [Google Scholar]
- 15. Randall C. E., Lewis J. W., Hug S. J., Bjorling S. C., Eisnershanas I., Friedman N., Ottolenghi M., Sheves M., Kliger D. S. (1991) J. Am. Chem. Soc. 113, 3473–3485 [Google Scholar]
- 16. Baylor D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baylor D. A., Lamb T. D., Yau K. W. (1979) J. Physiol. 288, 613–634 [PMC free article] [PubMed] [Google Scholar]
- 18. Kandori H., Katsuta Y., Ito M., Sasabe H. (1995) J. Am. Chem. Soc. 117, 2669–2670 [Google Scholar]
- 19. Baylor D. A., Nunn B. J., Schnapf J. L. (1984) J. Physiol. 357, 575–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lagnado L., Baylor D. (1992) Neuron 8, 995–1002 [DOI] [PubMed] [Google Scholar]
- 21. Lamb T. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 566–5708570596 [Google Scholar]
- 22. Hubbard R. (1958) J. Gen. Physiol. 42, 259–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kochendoerfer G. G., Verdegem P. J., van der Hoef I., Lugtenburg J., Mathies R. A. (1996) Biochemistry 35, 16230–16240 [DOI] [PubMed] [Google Scholar]
- 24. Orban T., Gupta S., Palczewski K., Chance M. R. (2010) Biochemistry 49, 827–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Angel T. E., Chance M. R., Palczewski K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8555–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Angel T. E., Gupta S., Jastrzebska B., Palczewski K., Chance M. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rader A. J., Anderson G., Isin B., Khorana H. G., Bahar I., Klein-Seetharaman J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crozier P. S., Stevens M. J., Forrest L. R., Woolf T. B. (2003) J. Mol. Biol. 333, 493–514 [DOI] [PubMed] [Google Scholar]
- 29. Janz J. M., Farrens D. L. (2004) J. Biol. Chem. 279, 55886–55894 [DOI] [PubMed] [Google Scholar]
- 30. Janz J. M., Fay J. F., Farrens D. L. (2003) J. Biol. Chem. 278, 16982–16991 [DOI] [PubMed] [Google Scholar]
- 31. Del Valle L. J., Ramon E., Bosch L., Manyosa J., Garriga P. (2003) Cell. Mol. Life Sci. 60, 2532–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J., Liu M. Y., Nguyen J. B., Bhagat A., Mooney V., Yan E. C. (2009) J. Am. Chem. Soc. 131, 8750–8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reeves P. J., Kim J. M., Khorana H. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yan E. C., Epps J., Lewis J. W., Szundi I., Bhagat A., Sakmar T. P., Kliger D. S. (2007) J. Phys. Chem. C 111, 8843–8848 [Google Scholar]
- 35. Oprian D. D., Molday R. S., Kaufman R. J., Khorana H. G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8874–8878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min K. C., Zvyaga T. A., Cypess A. M., Sakmar T. P. (1993) J. Biol. Chem. 268, 9400–9404 [PubMed] [Google Scholar]
- 37. Tsutsui K., Imai H., Shichida Y. (2007) Biochemistry 46, 6437–6445 [DOI] [PubMed] [Google Scholar]
- 38. Nakagawa M., Iwasa T., Kikkawa S., Tsuda M., Ebrey T. G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6189–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kito Y., Suzuki T., Azuma M., Sekoguti Y. (1968) Nature 218, 955–957 [DOI] [PubMed] [Google Scholar]
- 40. Farrens D. L., Khorana H. G. (1995) J. Biol. Chem. 270, 5073–5076 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.