Abstract
Prion diseases are fatal neurodegenerative diseases caused by the accumulation of the misfolded isoform (PrPSc) of the prion protein (PrPC). Cell-based screens have identified several compounds that induce a reduction in PrPSc levels in infected cultured cells. However, the molecular targets of most antiprion compounds remain unknown. We undertook a large-scale, unbiased, cell-based screen for antiprion compounds and then investigated whether a representative subset of the active molecules had measurable affinity for PrP, increased the susceptibility of PrPSc to proteolysis, or altered the cellular localization or expression level of PrPC. None of the antiprion compounds showed in vitro affinity for PrP or had the ability to disaggregate PrPSc in infected brain homogenates. These observations suggest that most antiprion compounds identified in cell-based screens deploy their activity via non-PrP targets in the cell. Our findings indicate that in comparison to PrP conformers themselves, proteins that play auxiliary roles in prion propagation may be more effective targets for future drug discovery efforts.
Keywords: Circular Dichroism (CD), High throughput screening (HTS), Neurodegeneration, Prions, Protein Drug Interactions, Antiprion, Assay, Differential Scanning Fluorimetry, Isothermal Titration Calorimetry, Molecular Target
Introduction
Misfolding and aggregation of endogenously expressed proteins cause several neurodegenerative disorders (1–3). Prion diseases belong to this class of proteinopathies and result from the misfolding of the α-helix-rich, cellular prion protein (PrPC) into a β-sheet-rich, disease-causing, infectious isoform termed PrPSc (3–6). Unlike other neurodegenerative disorders, prion diseases are readily transmissible to laboratory animals and cultured cells. The availability of laboratory models harboring the infectious aggregate has enabled the development of an empirical drug discovery strategy against prion diseases. Typically, prion-infected, neuronally derived cell lines that accumulate and stably propagate PrPSc (7, 8) are used as a primary screen for the identification of compounds that reduce prion levels in culture. Subsequently, the in vivo efficacy of putative antiprion compounds is assessed by analyzing their ability to prolong disease incubation periods in prion-infected rodents. Using this approach, numerous antiprion compounds have been identified, including pentosan polysulfate, dextran sulfate, HPA-23, Congo red, suramin, dendritic polyamines, 2-aminothiazoles, and quinacrine (9). However, none of these compounds have been shown to be effective against a variety of prion strains in animal models when administered at a late, post-symptomatic stage, and none have been shown to have significant disease-modifying properties in human clinical studies.
Although measuring PrPSc levels in infected cultured cells can be used to assess antiprion activity, this method does not elucidate the molecular targets of active compounds. As a result, the mechanisms of action of most antiprion compounds remain unknown. In principle, a compound can reduce the prion load in a cell by interacting with a number of molecular targets. The most direct mechanism is through direct binding to PrPC and stabilization of its native conformation (10, 11). Alternatively, a drug may directly interact with PrPSc, leading to its disaggregation (12), or may target auxiliary factors or proteins that play a role in PrPC expression, localization, or conversion to PrPSc (13).
To investigate whether antiprion compounds identified in prion-infected neuronal cell lines have a tendency to interact with PrPC, PrPSc, or other targets, we screened a library of 2,160 known drugs and natural products and identified 206 compounds that cleared PrPSc in neuroblastoma (N2a) cell lines at a concentration of less than 1 μm. Of these initial hits, we validated the activity of 16 compounds and assessed their ability to bind to recombinant PrP, directly disaggregate PrPSc, reduce the expression level of PrPC, and alter the localization of PrPC. Taken together, the results suggest that the antiprion activity of these compounds is mainly mediated by non-PrP targets.
EXPERIMENTAL PROCEDURES
Chemical Library
The chemical library of 2,160 compounds screened in both cell-based and direct-binding assays was obtained from the MicroSource Discovery System (MSDI, Gaylordsville, CT), and includes known drugs, bioactives, and natural products. Compounds were solubilized at 10 mm in dimethyl sulfoxide (DMSO)2 and stored in a 96-well format by the Small Molecule Discovery Center at the University of California San Francisco.
Cell-based Antiprion Activity and Toxicity
A mouse neuroblastoma (N2a) cell line was infected with the Rocky Mountain Laboratory (RML) strain of scrapie prions to produce ScN2a cells (14). Screening the chemical library for antiprion activity was performed in a high-throughput ELISA. Briefly, 4 × 104 ScN2a cells were treated with the compound of interest for 5 days at 1 μm final concentration. Untreated ScN2a cells were used as negative controls; ScN2a cells treated with quinacrine (1 μm) were used as positive controls (15, 16). A toxicity screen was conducted in parallel at the same compound concentration and time of exposure in a 96-well format using an acetomethoxy derivative of calcein (calcein-AM) assay. Untreated ScN2a cells were used as negative controls. Both of these methods have been described previously (17, 18).
Antiprion Activity by Immunoblotting
ScN2a cells (5 × 105) were propagated in a 10-cm plate and treated for 5 days with the compound of interest at 50, 20, 10, or 1 μm, depending on cellular toxicity. Negative controls were performed by treating cells with DMSO alone. As a positive control, cells were treated with 1 μm quinacrine. Cells were lysed with lysis buffer (0.5% Nonidet P-40, 0.5% deoxycholate, 10 mm Tris-HCl, pH 8, 100 mm NaCl) and protein concentration was normalized to 1 mg/ml using the BCA assay. Samples were incubated with 20 μg/ml of proteinase K for 1 h at 37 °C. Digestions were stopped with 2 mm phenylmethylsulfonyl fluoride (PMSF), and samples were centrifuged at 100,000 × g for 1 h at 4 °C. Supernatants were discarded, and pellets were resuspended in reducing SDS sample buffer for SDS-PAGE. Western blotting was performed according to standard procedures. PrP was detected by using D13 antibody Fab fragment conjugated (19) with horseradish peroxidase (HRP) (Rockland Immunochemicals Inc.).
Protein Purification and Preparation
Truncated recombinant mouse (Mo) PrP(89–230) and full-length recombinant MoPrP(23–230) were overexpressed and purified as previously described (20, 21). Lyophilized pellets were dissolved in a denaturation buffer (6.4 m GdnHCl, room temperature, 30 min) at 1 or 5 mg/ml for truncated or full-length proteins, respectively. The protein was then diluted 4-fold with cold refolding buffer (25 mm Tris-HCl, pH 8.0, 5 mm EDTA) and dialyzed 2 times against the screening buffer (20 mm sodium acetate, pH 5.1, 150 mm NaCl) at 4 °C.
Isothermal Titration Calorimetry
ITC measurements were performed at 37 °C using the ITC200 Microcalorimeter (GE Healthcare). To counteract the backlash effect observed during the first injection (22), ITC titrations were carried out with one 0.2-μl injection of ligand (compound), followed by 10 consecutive injections of 3.8 μl of the compound with injection durations of 4- and 150-s intervals between injections. The amount of energy released was measured following each injection. Prior to each measurement, DMSO was added to the protein sample to reach a final concentration of 1%. The sample chamber was filled with 15 μm recombinant MoPrP(89–230), which had been dialyzed previously in the screening buffer (see protein preparation). Solution stocks of compounds were prepared at 20 mm in 100% DMSO. Solutions of titrants were freshly prepared by diluting them with the dialyzed buffer to reach a 200 μm final concentration (1% final DMSO). Titrant solutions were centrifuged for 5 min at 14,000 × g, and supernatants were loaded into the syringe. The final molar ratio of ligand:protein exceeded 2.5. Isotherm data were analyzed with Origin7 software (MicroCal) supplied by the manufacturer.
Circular Dichroism
CD spectra of both truncated and full-length recombinant MoPrP were recorded at 10 μm, with a 0.1-cm cuvette using a Jasco J-715 spectrometer with a Jasco PTC-348WI Peltier-effect temperature control device, where the temperature reported reflects that of the heating block. Scans were acquired at 50 nm/min, with a bandwidth of 2 nm and data spacing of 0.1 nm. Thermal unfolding curves were measured at 10 μm recombinant MoPrP(89–230) and MoPrP(23–230). All compound solutions were made fresh from 100 mm DMSO stock solutions. Prior to each run, samples were equilibrated with the compound of interest for 15 min at 20 °C, then a temperature ramp rate of 2 °C/min was applied from 20 to 90 °C. To attenuate DMSO noise background, a wavelength of 230 nm was used to monitor protein unfolding. The melting point was determined from the thermal unfolding curve fit by the two-state folding EXAM algorithm (23, 24). A Δu Cp value of 4.76 kJ mol−1 K−1 was set in the fitting of the van't Hoff equation to determine the melting temperature (Tm) and the ΔH of the transition.
A significant thermal upshift was defined as ΔTm exceeding the standard deviation of the technique (σ = 0.4 °C) by a factor of three. The standard deviation was calculated based on three Tm measurements of native PrP.
Thermal Shift Monitoring by Differential Scanning Fluorimetry (DSF)
Protein stability was assessed in a 96-well format using an MxPro3005P qRT-PCR Detection System (Stratagene, Agilent Technologies). Sypro-Orange dye (Invitrogen) was used to monitor the fluorescence by applying ROX filter for the fluorescence emission (610 nm) and FAM filter for the fluorescence excitation (492 nm). Optimal conditions to perform the assay were determined by varying protein concentrations, Sypro-Orange dilutions, buffers, and thermal ramp parameters.
To experimentally screen compounds, recombinant MoPrP(89–230) and Sypro-Orange dye were plated manually. Stock solutions of 10 mm compounds (100% DMSO) were freshly diluted in the screening buffer (1:10 ratio), then added to the protein by using a 96/384 pipettors robot (Apricot Designs) to achieve a final compound concentration of 1 mm (1% final DMSO).
DSF spectra of 10 μm recombinant MoPrP(89–230) with a 1:2000 Sypro-Orange dilution in the screening buffer were recorded. Samples (150 μl final) were heated at 2 °C/min, from 40 to 90 °C, and the fluorescence values were recorded after every 1 °C increase. Approximations of the melting temperature (Tm) were assessed by using the maximum value of the first derivative generated by the qPCR software (MxPro QPCR software, Stratagene, Agilent Technologies). To evaluate changes in Tm, for each run, first derivatives of the melting curves were generated from the MxPro software, exported as a text file and imported in Mathematica software. The first derivative of the curves was fitted with a polynomial function. Tm was approximated by determining the maximum value of the fitted first derivative plot.
PrPC Expression
To analyze the effects of the compounds on PrPC expression, uninfected N2a cells were incubated with the selected molecules for 5 days. Negative control cells were incubated with DMSO (0.5% final concentration). After treatment and cell lysis, normalized crude extracts were analyzed by Western immunoblotting using conjugated D13-HRP antibody to detect PrP. PrPC expression levels were normalized with respect to actin controls (Fig. 6). For dose-response analysis of amcinonide, N2a cells were treated for 3 days and quantified by Western immunoblotting as described above.
FIGURE 6.
PrPC expression levels in uninfected N2a cells treated with the 16 validated antiprion compounds at the indicated concentrations for 5 days. Conjugated D13-HRP was used to detect PrP in Western immunoblots. Lane numbers correspond to compound IDs in Table 1. As a negative control, N2a cells were treated with DMSO.
Western Blot Quantification
Western blots were quantified using ImageJ software. PrPSc and PrPC levels in compound-treated cells were normalized against the levels in untreated control cells.
In Vitro Susceptibility of PrPSc to Protease
Brain homogenates (10% w/v) were prepared from terminal CD-1 mice infected with RML prions, then diluted 10-fold using an acidic sodium acetate buffer supplemented by detergents (5 mm sodium acetate, pH 3.5, 1% Nonidet P-40). Ninety μl of 1% brain homogenates (0.6 mg/ml) were incubated with 10 μl of test compounds or polyamidoamine (PAMAM) generation 4.0 and 4.5 to reach final concentrations of 100 μm or 100 μg/ml, respectively. Samples were incubated for 2 h with constant shaking at 37 °C. After neutralization with 100 μl of a freshly prepared buffer containing 0.2 m HEPES, pH 7.5, 0.3 m NaCl, and 4% Sarkosyl, samples were subjected to PK digestion (20 μl/ml, 1 h of incubation at 37 °C). Proteolytic digestions were stopped by the addition of PMSF (2 mm final concentration), and samples were analyzed by Western immunoblotting using conjugated D13-HRP antibody to detect PrP.
Lipid Raft Isolation
For lipid raft isolation, N2a cells were treated with the compounds for 3 days in 6-well plates. Plates were placed on ice, cells were rinsed twice with chilled PBS and incubated for 20 min with ice-cold Triton X-100 lysis buffer made with Mes-buffered saline (25 mm Mes, 150 mm NaCl, pH 6.5) containing 1% (v/v) Triton X-100. The lysates were then homogenized by passing through a Luer 22-gauge needle and centrifuged at 500 × g for 5 min at 4 °C to pellet cell debris. Cold supernatants were harvested, normalized to 0.5 mg/ml, and mixed with an equal volume of 80% (v/v) sucrose to obtain a 40% (v/v) sucrose solution. A 1-ml aliquot of the sample was then transferred to the bottom of a SW-60 centrifuge tube. Two ml of 30% sucrose was then added to the top, followed by the addition of 1 ml of 5% sucrose to create a discontinuous sucrose gradient. Tubes were centrifuged at 4 °C for 18 h at 140,000 × g in a SW-60 rotor (Beckman Instruments). Fractions (1–8; 500 μl) were collected from the top and analyzed by Western immunoblotting using D13-HRP antibody (see the immunoblotting section). Flotillin-1 (Sigma) was probed by immunoblot and used as a marker for detergent-resistant membranes.
Immunocytochemistry
N2a cells (2.5 × 105) were plated on a coverslip (Fisher Scientific, Circles No. 1.5) placed in a 24-well plate format, and treated for 3 days with the selected compounds. Cells were washed with warm (37 °C) PBS and fixed with 4% paraformaldehyde solution for 20 min at room temperature. Cells were rinsed 3 times for 5 min with room temperature PBS, permeabilized with 0.3% Triton X-100 in PBS for 5 min, and then rinsed 3 times with PBS buffer. Cells were blocked with 10% normal goat serum in PBS for 30 min at room temperature, then incubated with primary D18 antibody (5 μg/ml) in 10% normal goat serum overnight at 4 °C. Cells were washed successively 3 times with PBS for 10 min, and incubated in the dark at room temperature for 2 h with a FITC-conjugated goat anti-human IgG (H&L) polyclonal antibody (5 μg/ml diluted in 10% normal goat serum, Jackson ImmunoResearch). Samples were rinsed 3 times with PBS for 10 min in the dark. Coverslips were rinsed briefly with water, then mounted on Superfrost Plus microscope slides (Electron Microscopy Sciences, Hatfield, PA) with Vectashield with the counterstain DAPI (Vector Laboratories), and sealed with Cytoseal. Slides were analyzed at the QB3-UCSF Nikon Imaging Center on a Nikon Eclipse Ti-E Motorized Inverted Microscope. Images represent individual Z-slices taken from the middle of the cell.
Total RNA Purification and Prnp Quantitative PCR
Uninfected N2a cells were incubated with amcinonide at 50 μm or 0.5% DMSO (untreated control cells) for 6, 24, 48, or 72 h. Total RNA was isolated with TRIzol reagent. cDNA was synthesized from 1 g of total RNA using the SuperScript II First Strand Synthesis System for RT-PCR at 42 °C for 60 min (Invitrogen), and then diluted 10-fold in water. Two microliters were used in duplicate for quantitative PCR amplification of Prnp, and actin as an internal control, using the MxPro3005P qRT-PCR apparatus (Stratagene, Agilent Technologies). The following program was used: denaturation step at 95 °C for 10 min, 40 cycles of PCR (denaturation at 95 °C for 10 s, annealing at 55 °C for 8 s, elongation at 72 °C for 15 s). Primers were as follows: Actin, forward, gatcattgctcctcctgagc-5′; reverse, ctcatcgtactcctgcttgc-3′; Prnp: forward, cgagaccgatgtgaagatga-5′; reverse, atcccacgatcaggaagatg-3′. The curves of amplification were read with MxPro 3005P software using the comparative cycle threshold method. Relative quantifications of the target mRNAs were calculated after normalization of cycle thresholds with respect to actin levels. Values are expressed as fold-change compared with untreated cells (0.5% DMSO).
PrPC Degradation Kinetics
N2a cells were preincubated with amcinonide at 20 μm or 0.5% DMSO (untreated control cells) for 3 days. Subsequently, 30 μg/ml of cycloheximide (Sigma) was added to the culture to inhibit protein synthesis, and cells were incubated at 37 °C for various durations. Cells were then lysed and residual PrPC levels were evaluated by Western immunoblotting.
RESULTS
Identification of Compounds with Antiprion Activity
We screened a chemical library of 2,160 compounds containing known drugs and natural products (Microsource) for antiprion activity in ScN2a cells. The cell-based assay was carried out in a multiwell format and PrPSc levels were measured using an ELISA-based assay (18).
Cells treated with DMSO (carrier) and quinacrine (a known antiprion drug (15)) were used as negative and positive controls, respectively. These measurements were used as reference to calculate the normalized percentage change in PrPSc levels following treatment with experimental compounds. ScN2a cells were treated with compounds for 5 days at a concentration of 1 μm. In parallel, we analyzed the cytotoxicity of all compounds at this concentration by employing the calcein AM assay for membrane integrity (25). The level of cytotoxicity was normalized with respect to untreated cells.
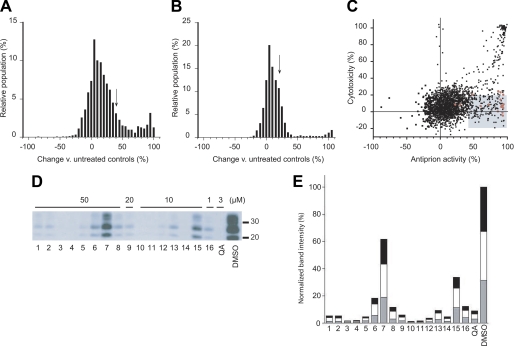
Based on the distribution of activity and cytotoxicity measurements (Fig. 1, A and B), we defined a “hit” as a compound that reduced PrPSc load by 40% with less than 20% cytotoxicity as measured by the calcein AM assay (Fig. 1C). Using these criteria, we identified 206 nontoxic compounds with antiprion activity. A previous screen of the MicroSource compound library also identified active compounds (26), most of which were included in our hit set as well (Fig. 1C, red dots).
FIGURE 1.
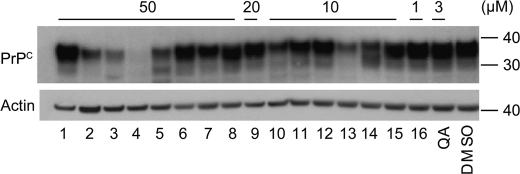
Identification of compounds from the Microsource library with antiprion activity. A, the distribution of induced changes in PrPSc levels in ScN2a cells following 5 days of treatment with 1 μm of each compound. PrPSc levels were measured by ELISA following PK digestion, then quantified relative to untreated control cells. Compounds resulting in ≥40% PrPSc decrease (arrow) were considered hits for antiprion activity. B, the distribution of cytotoxic effects of compounds measured by the calcein AM assay. Compounds decreasing cell viability by ≤20% (arrow) were considered nontoxic hits. C, the hit set (n = 206, shaded box) that fulfilled antiprion activity and cytotoxicity criteria. Red dots correspond to compounds with antiprion activities previously identified by Kocisko et al. (26). D, secondary validation of the antiprion activity for a subset of hits. Compounds were added to ScN2a cells at the indicated concentrations for 5 days, and the resulting PK-resistant PrPSc levels were analyzed by Western blotting using conjugated D13-HRP antibody to detect PrP. Lane numbers (bottom) correspond to compound ID indicated in Table 1. Quinacrine (QA) and DMSO alone were analyzed as positive and negative controls, respectively. Apparent molecular mass markers are indicated in kilodaltons. E, quantification of PrPSc band intensities relative to untreated controls. The gray, white, and black bars indicate the relative intensities of the unglycosylated, monoglycosylated, and diglycosylated bands, respectively.
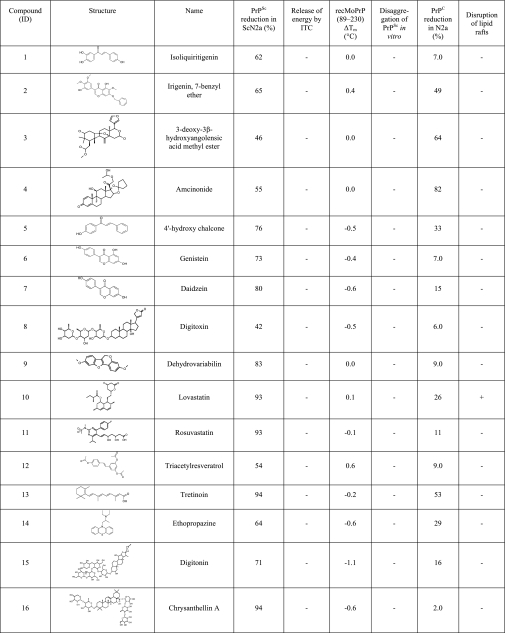
We randomly selected 40 hits for secondary validation. Initially, compounds were added to ScN2a cells at a concentration of 50 μm. For compounds that proved toxic at this concentration, the experiments were repeated at 20, 10, and 1 μm to assess the antiprion activity of the compound at the highest possible nontoxic concentration. Changes in PK-resistant PrPSc levels were analyzed by Western blots. We were able to confirm the antiprion activity of 16 of 40 compounds (Fig. 1D). Some of these 16 compounds belonged to chemical classes previously known to have antiprion properties (statins, flavones, resveratrol, chalcone, quercetin, phenothiazine, and corticosteroid (26, 27)). Several were, as far as we are aware, novel: these include dehydrovariabilin, 3-deoxy-3β-hydroangolensic acid methyl ester, and glycosides (Table 1). Experiments conducted on prion-infected N2a cells in the presence of the 16 compounds revealed a proportional decrease of all three glycoforms (unglycosylated, monoglycosylated, and diglycosylated) of digested PrPSc (Fig. 1E), suggesting that the compounds neither alter PrP glycosylation nor depend on PrP glycosylation for their antiprion activity.
TABLE 1.
Validated antiprion compounds
For each compound, the reduction in PrPC and PrPSc levels in culture, interaction with recPrP detected by ITC, induced ΔTm in recPrP measured by CD, induced disaggregation of PrPSc in vitro, and the ability to disrupt lipid rafts are reported.
Identification of Compounds That Interact with PrP
Isothermal Titration Calorimetry
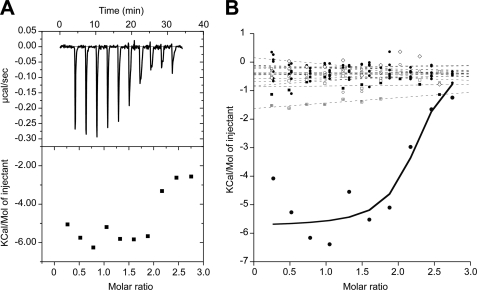
Next, we assessed the ability of these 16 antiprion compounds to interact directly with recombinant MoPrP(89–230) containing the structured domain of PrP found in the proteinase-resistant core of PrPSc (28). ITC was used to measure direct binding between the compounds and recombinant PrP. We first sought to establish the ITC protocol with a control compound that is known to interact with PrP. We used suramin as a positive control as it has been shown to interact nonspecifically with PrP, inducing its aggregation (29, 30). We used ITC to analyze the interaction between suramin and PrP at concentrations of 200 and 15 μm, respectively. In the presence of suramin, we observed a substantial release of energy for the first injections (approximately −6.0 Kcal/mol of injectant), demonstrating an interaction between the partners (Fig. 2A). Using the same experimental parameters, we performed ITC with the 16 confirmed antiprion compounds to detect any significant release of energy upon injection. No substantial release of energy was detected for any of the 16 compounds (Fig. 2B), suggesting that none interacted with recombinant MoPrP(89–230).
FIGURE 2.
Binding of 16 validated compounds to recombinant PrP, measured by ITC. A, peaks and amount of energy released after titration of suramin. B, quantity of energy released after titration of 16 validated antiprion compounds (dashed lines) compared with suramin (solid line).
Thermal Denaturation Upshift Assay
We next sought to measure the binding of compounds to PrP by a thermal-denaturation upshift assay. Ligand binding and protein folding are thermodynamically linked, and the binding of a ligand stabilizes proteins against denaturation (31). We therefore used thermal denaturation to quantify potential binding of antiprion compounds by detecting induced upshifts in the melting temperature of PrP.
We initially assessed the reversibility of the thermal unfolding of recombinant truncated MoPrP(89–230). Circular dichroism (CD) spectra of truncated PrP were measured successively at 20 and 90 °C (supplemental Fig. S1A). As expected, CD spectra indicated a loss of secondary structure at 90 °C. On subsequent cooling, MoPrP(89–230) regained its α-helical signal, indicating that the thermal unfolding of MoPrP(89–230) is reversible. We then determined the melting temperature (Tm) of truncated recombinant PrP by monitoring the change in α-helical content of the protein as a function of temperature. The resulting melting curve was well fit by a two-state unfolding model. The Tm for MoPrP(89–230) was 65.3 ± 0.4 °C and the van't Hoff enthalpy of denaturation was 270.4 ± 15.7 kJ/mol.
We investigated whether changes in protein stability could be robustly detected by this approach. The melting point of PrP was measured in the presence of stabilizing and destabilizing agents. In the presence of 1 m trimethylamine N-oxide (TMAO), a chemical chaperone (11), protein stability increased substantially (Tm = 70.5 ± 0.2 °C; ΔTm = +5.2 °C). Conversely, addition of 1 m of the denaturant GdnHCl decreased the melting temperature (Tm = 62.3 ± 0.4 °C; ΔTm = −3.0 °C) (supplemental Fig. S1B).
Having validated the assay with known stabilizing and destabilizing agents, we proceeded to analyze the potential binding interactions of the 16 cell-active antiprion compounds with the structured domain of PrP. The compounds (100 μm) were added to 10 μm recombinant MoPrP(89–230). Denaturation of truncated PrP in the presence of DMSO alone was used as a negative control. In accordance with the ITC results, none of the 16 compounds significantly increased the Tm of MoPrP(89–230), suggesting that they neither stabilized nor interacted with the folded domain of the protein (Fig. 3, A and B).
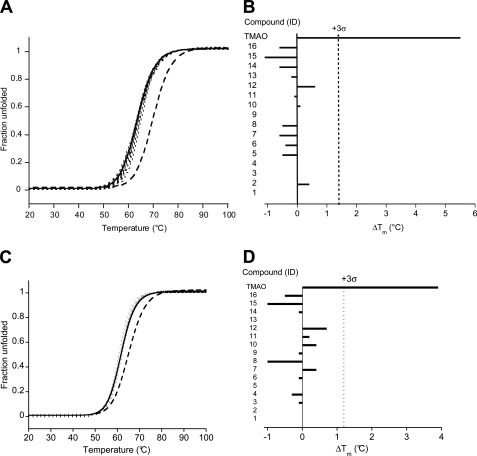
FIGURE 3.
Effect of the 16 validated antiprion compounds on the stability of recombinant PrP measured by CD. A and C, thermal denaturation curves (λ = 230 nm) of PrP(89–230) (A) and PrP(23–230) (C) alone (solid lines), in the presence of 1 m TMAO (dashed line), or in the presence of the 16 validated antiprion compounds at 100 μm (dotted lines). B and D, comparison of ΔTm for the 16 validated compounds and the 1 m TMAO control. The dotted line indicates the 3 σ boundary for statistical significance.
To discount the possibility of compounds binding to the N-terminal unstructured region of PrP, we repeated the thermal-denaturation upshift assays using recombinant full-length MoPrP(23–230). The unfolding of MoPrP(23–230) was reversible (supplemental Fig. S1C) and changes in protein stability in the presence of TMAO or GdnHCl could be detected (supplemental Fig. S1D). None of the 16 cell-active antiprion compounds significantly increased the Tm of MoPrP(23–230) (Fig. 3, C and D).
Differential Scanning Fluorimetry
The observation that none of the 16 antiprion compounds directly interacted with recombinant PrP can be interpreted in two ways. First, the analyzed chemical library may have been devoid of compounds that interact with PrP. Indeed, NMR and crystallographic studies indicate that the three-dimensional structure of PrP lacks deep cavities normally required for high-affinity binding interactions with small molecules, and as such, few molecules within an unbiased diversity chemical library are expected to bind PrP (32–36). Alternatively, general binding to PrP may be a poor correlate to antiprion activity and not all molecules that bind to PrP may exhibit antiprion activity. For example, a small molecule may need to bind and locally stabilize specific regions of PrP to inhibit its conversion to PrPSc. To distinguish between these two possibilities, we screened the 2,160 compounds in the chemical library for their ability to interact with PrP using a high-throughput, in vitro binding assay.
We employed an approach based on the DSF assay (37). DSF monitors protein unfolding via a fluorescent dye that interacts preferentially with hydrophobic surfaces exposed during protein unfolding. DSF has been used to detect ligand binding (38), including PrP (39). To measure induced changes in the stability of PrP in a high-throughput fashion, we miniaturized the DSF assay and conducted a screen in a 96-well format (see “Experimental Procedures” and Fig. 4A).
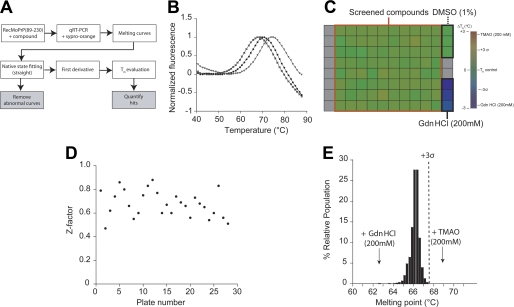
FIGURE 4.
Screening the interaction of the Microsource library with recombinant PrP using DSF. A, the DSF assay strategy; see “Experimental Procedures” for detailed descriptions. B, normalized melting curves of recombinant PrP obtained alone (black circles), with 200 mm TMAO (triangles), or with 200 mm GdnHCl (squares). C, a representative assay plate. The indicated color scale represents the range of Tm values. Gray squares signify empty wells. D, Z-factor measurements plotted for each experimental plate. E, the distribution of Tm values for PrP, after exposure to 100 μm of each compound, is plotted as a percentage of the relative population. The arrows indicate the median value for all GdnHCl and TMAO control measurements. The dotted line indicates the 3 σ boundary for statistical significance.
The 2,160 compounds in the Microsource library were screened at a concentration of 100 μm for direct binding to recombinant MoPrP(89–230). Melting curves of PrP in the presence and absence of compounds were recorded at 1 °C increments from 40 to 90 °C. Typically, the melting curves could be divided into three regimes: the native baseline, the unfolding transition, and the denatured baseline (Fig. 4B). The Tm value indicates the thermodynamic stability of the protein (31). Additionally, the slope and magnitude of the baselines provide data about the aggregation of the native and denatured states of the protein (37). In the absence of compounds, PrP has a stable native baseline, a transition at 65.9 ± 0.4 °C, and a sloped denatured baseline (Fig. 4B). The slope in the denatured baseline likely reflects the aggregation of unfolded PrP at high temperatures. As expected, the addition of 200 mm GdnHCl decreased PrP stability (Tm = 62.8 °C), whereas addition of 200 mm TMAO increased the protein stability (Tm = 68.9 °C) (Fig. 4B).
As controls, each screened plate contained three wells with 200 mm GdnHCl and three wells with 1% DMSO alone (Fig. 4C). To evaluate the experimental noise in the screen, positive and negative controls were used to calculate the Z-factor (40) for each plate (Fig. 4D). Z-factors for all plates were above 0.5, indicating statistical robustness in the screening method. Tm measurement and hit identification were performed by automated analysis of the data (“Experimental Procedures”). The distributions of Tm values were plotted as a percentage of the relative population (Fig. 4E). Hits were defined as those for which the Tm was >67.2 °C, derived from 3 × S.D. The distribution of Tm values from all 2,160 compounds showed that no molecules in the library stabilized PrP (Fig. 4E), indicating none had measurable affinity for recombinant PrP(89–230).
Identification of Compounds That Increase the Susceptibility of PrPSc to Proteolysis in Vitro
Previous studies indicated that the antiprion activity of positively charged branched polyamidoamines could be attributed to their ability to directly interact and disaggregate PrPSc in acidic lysosomal compartments, rendering PrPSc susceptible to proteolysis (12, 41). We therefore analyzed the 16 identified antiprion compounds for this property by measuring their ability to increase the PK sensitivity of PrPSc under acidic conditions in an in vitro degradation assay. Positively and negatively charged polyamidoamines, generation 4.0 and 4.5, were used as positive and negative controls, respectively. Compounds were added to PrPSc-containing brain homogenates from RML-infected mice and incubated for 2 h in acidic buffer (“Experimental Procedures”). Following incubation, the homogenates were digested with PK and the presence of protease-resistant PrPSc was analyzed by Western blots. The results indicate that none of the 16 validated antiprion compounds disaggregated PrPSc and increased the PK sensitivity of the prion-infected mouse brain homogenates in vitro (Fig. 5).
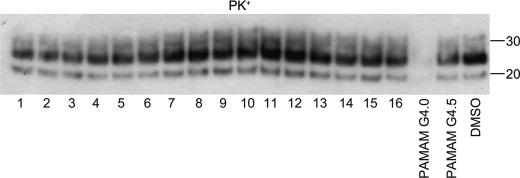
FIGURE 5.
Susceptibility of PrPSc in prion-infected brain extract to protease digestion after exposure to 16 validated antiprion compounds. Brain homogenates of Rocky Mountain Laboratory prion-infected mice were treated with 100 μm of the indicated antiprion compounds for 2 h, then subjected to PK digestion and Western blot analysis. Lane numbers correspond to compound IDs in Table 1. Polyamidoamine (PAMAM) generations 4.0 and 4.5 were used as positive and negative controls, respectively. DMSO was also analyzed as a negative control. Apparent molecular mass markers are indicated in kilodaltons.
Identification of Compounds That Reduce PrPC Expression Levels
We next determined whether the antiprion activity of the 16 validated compounds could be due to induced changes in the expression level of PrPC. Uninfected N2a cells were treated with the compounds, and quantification of Western immunoblots was performed to determine PrPC levels. Of the 16 compounds, four appeared to decrease PrPC expression: irigenin, 7-benzyl ether, 3-deoxy-3β-hydroxyangolensic acid methyl ester, amcinonide, and retinoic acid (Fig. 6). Amcinonide had the most pronounced effect, inducing a substantial decrease (>80%) in PrPC levels (Table 1).
Identification of Compounds That Disrupt Lipid Rafts and PrPC Localization
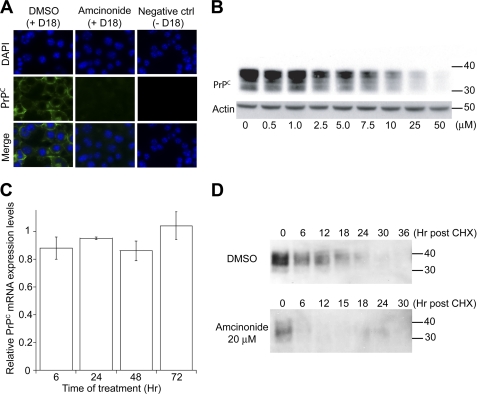
An alternative mechanism for decreasing PrPSc formation is to alter the integrity of lipid rafts, membrane microdomains where PrPC is known to localize (42, 43). We tested the 16 compounds for their ability to disrupt lipid rafts in uninfected N2a cells by detecting PrP and flotillin-1 (a marker of lipid raft integrity (44) in detergent-resistant microdomains) which were isolated by sucrose gradients using a flotation assay (see “Experimental Procedures”) (45). Whereas silver staining of the fractions collected from DMSO-treated cells showed that soluble proteins remained primarily at the bottom of the gradient (fractions 7 and 8), Western blot analysis indicated that PrP colocalized with flotillin-1 (Fig. 7A) at the interface between 5 and 30% sucrose concentrations (Fraction 3). Active compounds were next tested, and all fractions from flotation assays were analyzed by Western immunoblotting for PrP and flotillin-1 (supplemental Fig. S2). To compare directly the effects of the compound set, the levels of PrP and flotillin-1 in fraction 3 of each gradient were compared on the same Western blot (Fig. 7B). Only lovastatin at 10 μm was able to disrupt lipid rafts (Fig. 7B, lane 10). Next, compound-induced changes in PrP localization were analyzed by immunocytochemistry in uninfected N2a cells (Fig. 8A, supplemental Fig. S3). Retinoic acid and lovastatin showed a diffuse signal within the cell that may be attributed to a change in PrP localization. More interestingly, irigenin, 7-benzyl ether, 3-deoxy-3β-hydroxyangolensic acid methyl ester, and amcinonide induced a generalized decrease of intracellular and membrane-localized PrPC. Because amcinonide induced the most substantial decrease in PrPC levels and this effect was dose dependent (Fig. 8B), we decided to further investigate its mechanism of action.
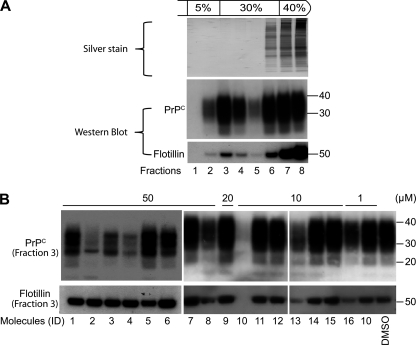
FIGURE 7.
Effects of the 16 antiprion compounds on lipid raft integrity. A, analysis of DMSO-treated cell lysates subjected to the flotation assay. Each fraction (numbered lanes) was analyzed by silver staining (upper panel) or Western immunoblotting with PrPC (middle panel) and flotillin-1 antibodies (lower panel). Detergent-resistant microdomains are concentrated in fraction 3. B, Western blot of fraction 3 of the flotation gradient for cells treated with the 16 antiprion compounds (lanes correspond to compound IDs in Table 1). Blots were probed with anti-PrP (top) and anti-flotillin-1 (bottom) antibodies. Apparent molecular mass markers are indicated in kilodaltons.
FIGURE 8.
Amcinonide reduced PrPC levels by increasing its rate of degradation. A, immunocytochemistry of N2a cells treated with 50 μm amcinonide for 3 days. Cells treated with DMSO were used as control. The negative control indicates DMSO-treated cells without the addition of D18 primary antibody. The green color corresponds to the PrPC (D18) labeling. The blue color locates the nuclei DAPI staining. B, dose-response analysis of the effect of amcinonide on PrPC expression levels. N2a cells were treated for 3 days with the indicated concentrations of amcinonide and PrPC levels were analyzed by Western immunoblotting. Actin staining (bottom) was performed on the same membrane used to detect PrPC expression. C, Prnp mRNA expression levels in N2a cells treated with 50 μm amcinonide for the indicated durations. All mRNAs levels were normalized with respect to actin controls. D, measurement of PrPC half-life after the addition of cycloheximide in cells treated with DMSO (top) or 20 μm amcinonide (bottom). Blots were probed with D18 anti-PrP antibody. For Western blots, apparent molecular mass markers are indicated in kilodaltons.
Effect of Amcinonide on PrPC Expression and Degradation
We investigated whether amcinonide affects PrPC abundance at the level of transcription or protein stability. Cultured mouse N2a cells were treated with control solvent (0.5% DMSO) or amcinonide (50 μm) for 6, 24, 48, and 72 h. Total mRNA was isolated, and quantitative RT-PCR was performed to detect the expression levels of Prnp mRNA. Results indicated that amcinonide did not alter the level of Prnp mRNA (Fig. 8C). Next, we investigated whether amcinonide, added at its half-effective concentration (20 μm), increases the rate of PrPC degradation after the inhibition of translation by cycloheximide. Whereas PrPC in untreated cells had a half-life of 12–18 h, its half-life decreased to less than 6 h with the addition of amcinonide (Fig. 6D). The results indicate that amcinonide alters the PrPC expression levels by increasing its rate of intracellular degradation.
DISCUSSION
In protein aggregation diseases, including prion diseases, a common target for pharmacological intervention has been the aggregating molecule itself (46, 47). Although we were able to identify 16 nontoxic compounds that were active against prion formation in cell culture, none of the 16 molecules interacted with PrPC or PrPSc in vitro, as evaluated by various biophysical techniques. These observations imply that at least one other cellular target (protein X), likely several given the diversity of the active molecules, modulates PrP conversion within the cell. Our results have broad implications for the potential mechanism of known antiprion compounds and the development of novel strategies for the discovery of effective therapeutics against prion diseases.
The set of active antiprion compounds was identified by a high-throughput, ELISA-based assay utilizing ScN2a cells. Sixteen compounds were confirmed by secondary Western blot analysis. The results of three orthogonal, in vitro assays (ITC, CD, and DSF) suggest that none of the compounds directly interacted with recombinant PrP. Indeed, DSF analysis of the entire Microsource library was unable to identify any compounds that stoichiometrically bound to PrP with measurable affinity. The results are consistent with the reported crystal and NMR structures of PrPC, which indicate the lack of prominent structural clefts capable of ligand binding (32–35). Various compounds have been reported to bind PrPC in vitro. However, these interactions have been shown to be nonspecific (e.g. anionic tetrapyrroles (48)), irreproducible in alternative binding assays (e.g. Gn8 (10)), or have millimolar affinities (e.g. quinacrine (49)). More recently, it was reported that a cationic tetrapyrrole (Fe(III)-TMPyP) can specifically interact with the folded domain of human PrP with micromolar affinities, although the large size of the compound may preclude it as an effective brain-permeable clinical therapeutic (50). The results of our study suggest that a drug-discovery screen focused on the identification of specific PrP-interacting molecules, e.g. protein X, is unlikely to identify many efficacious compounds.
Of 16 analyzed compounds, none rendered PrPSc susceptible to proteolysis in vitro. The compounds exert their activity only in the context of intact cells, suggesting that they are acting on other targets involved in the formation and propagation of prions rather than with PrPSc itself. One of the analyzed compounds, amcinonide, substantially reduced the cellular expression level of PrPC, providing a potential mechanism of action for its antiprion activity. Interestingly, a previous screen (26, 39) identified an antiprion compound, budesonide, which is structurally similar to amcinonide. Future studies using an immunoprecipitation approach could be used to identify the molecular targets of this class of compounds and establish a link between their targets and PrPC expression.
Reductions in the expression level of PrPC represent a particularly promising mechanism of action for antiprion compounds. A compound that targets the expression of PrPC has the potential to remove the substrate for all pathogenic misfolded conformations of PrP and be efficacious against a range of prion strains. Several lines of evidence support the idea that reducing the expression level of PrPC can delay or prevent prion disease progression. In fact, one confirmation of the prion hypothesis was the demonstration that mice whose PrP gene has been deleted (Prnp0/0) do not propagate infectious prions and are resistant to prion disease (51). Ablation of the neuronal PrP gene utilizing a cre-lox recombination strategy extended the incubation period of prion-infected mice (52). Using bigenic mice with PrP gene expression under the control of an inducible promoter, it was demonstrated that the down-regulation of PrP can dramatically extend the lifespan of prion-infected mice (53).
Our screen identified two statin drugs: lovastatin and rosuvastatin. By inhibiting the cholesterol synthetic pathway, it had previously been shown that lovastatin alters the integrity of the lipid raft compartments, reducing the quantity of PrPC available for conversion to PrPSc (42). Other hits, including isoliquiritigenin, irigenin, 4′-hydroxychalcone, genistein, daizein, triacetylresveratol, ethopropazine, and chrysanthellin A, belong to chemical classes that had been shown previously to have antiprion activities (15, 26, 27). We also found four structurally novel antiprion compounds: 3-deoxy-3β-hydroxyangolensic acid methyl ester, digitoxin, dehydrovariabilin, and digitonin. Although the mechanisms of action of these compounds are unclear, our in vitro analysis suggests that they are unlikely to interact directly with PrPC.
The key observation to emerge from this study is that none of the antiprion molecules active in cell culture are directly active on PrP itself. Given the presence of previously discovered antiprion compounds among our hits, this may be broadly true of molecules with antiprion properties. From a structural standpoint, this possibility is not unreasonable: the structure of PrPC reveals few, if any, pockets well suited to sequester a small molecule, at least in the native state. Although the molecular targets of the antiprion molecules identified here remain to be determined, the compounds may act on some “druggable” targets, including proteins that modulate PrP expression, localization, and stability in the cell. The identification of these cofactors will advance our understanding of prion and other aggregative disorders as well as enable the optimization of anti-aggregative therapeutics against these debilitating diseases.
Supplementary Material
Acknowledgments
We thank A. Serban for providing recombinant MoPrP(89–230) and MoPrP(23–230) and F. Cohen (TPG Biotech, San Francisco) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants AG02132, AG031220, and AG021601, the Hillblom Foundation, and the Sherman Fairchild Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- DMSO
- dimethyl sulfoxide
- ITC
- isothermal titration calorimetry
- DSF
- differential scanning fluorimetry
- TMAO
- trimethylamine N-oxide
- GdnHCl
- guanidine hydrochloride.
REFERENCES
- 1. Dobson C. M. (1999) Trends Biochem. Sci. 24, 329–332 [DOI] [PubMed] [Google Scholar]
- 2. Rochet J. C., Lansbury P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60–68 [DOI] [PubMed] [Google Scholar]
- 3. Prusiner S. B. (2001) N. Engl. J. Med. 344, 1516–1526 [DOI] [PubMed] [Google Scholar]
- 4. Pan Y. T., Hori H., Saul R., Sanford B. A., Molyneux R. J., Elbein A. D. (1983) Biochemistry 22, 3975–3984 [DOI] [PubMed] [Google Scholar]
- 5. Basler K., Oesch B., Scott M., Westaway D., Wälchli M., Groth D. F., McKinley M. P., Prusiner S. B., Weissmann C. (1986) Cell 46, 417–428 [DOI] [PubMed] [Google Scholar]
- 6. Prusiner S. B. (1987) Annu. Rev. Med. 38, 381–398 [DOI] [PubMed] [Google Scholar]
- 7. Race R. E., Fadness L. H., Chesebro B. (1987) J. Gen. Virol. 68, 1391–1399 [DOI] [PubMed] [Google Scholar]
- 8. Butler D. A., Scott M. R., Bockman J. M., Borchelt D. R., Taraboulos A., Hsiao K. K., Kingsbury D. T., Prusiner S. B. (1988) J. Virol. 62, 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trevitt C. R., Collinge J. (2006) Brain 129, 2241–2265 [DOI] [PubMed] [Google Scholar]
- 10. Nicoll A. J., Trevitt C. R., Tattum M. H., Risse E., Quarterman E., Ibarra A. A., Wright C., Jackson G. S., Sessions R. B., Farrow M., Waltho J. P., Clarke A. R., Collinge J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17610–17615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tatzelt J., Prusiner S. B., Welch W. J. (1996) EMBO J. 15, 6363–6373 [PMC free article] [PubMed] [Google Scholar]
- 12. Supattapone S., Nguyen H. O., Cohen F. E., Prusiner S. B., Scott M. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14529–14534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perrier V., Wallace A. C., Kaneko K., Safar J., Prusiner S. B., Cohen F. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6073–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bosque P. J., Prusiner S. B. (2000) J. Virol. 74, 4377–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korth C., May B. C., Cohen F. E., Prusiner S. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9836–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. May B. C., Fafarman A. T., Hong S. B., Rogers M., Deady L. W., Prusiner S. B., Cohen F. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3416–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghaemmaghami S., Phuan P. W., Perkins B., Ullman J., May B. C., Cohen F. E., Prusiner S. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17971–17976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghaemmaghami S., May B. C., Renslo A. R., Prusiner S. B. (2010) J. Virol. 84, 3408–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. (1998) J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehlhorn I., Groth D., Stöckel J., Moffat B., Reilly D., Yansura D., Willett W. S., Baldwin M., Fletterick R., Cohen F. E., Vandlen R., Henner D., Prusiner S. B. (1996) Biochemistry 35, 5528–5537 [DOI] [PubMed] [Google Scholar]
- 21. Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B. (2004) Science 305, 673–676 [DOI] [PubMed] [Google Scholar]
- 22. Mizoue L. S., Tellinghuisen J. (2004) Anal. Biochem. 326, 125–127 [DOI] [PubMed] [Google Scholar]
- 23. Morton A., Baase W. A., Matthews B. W. (1995) Biochemistry 34, 8564–8575 [DOI] [PubMed] [Google Scholar]
- 24. Beadle B. M., Shoichet B. K. (2002) J. Mol. Biol. 321, 285–296 [DOI] [PubMed] [Google Scholar]
- 25. Lichtenfels R., Biddison W. E., Schulz H., Vogt A. B., Martin R. (1994) J. Immunol. Methods 172, 227–239 [DOI] [PubMed] [Google Scholar]
- 26. Kocisko D. A., Baron G. S., Rubenstein R., Chen J., Kuizon S., Caughey B. (2003) J. Virol. 77, 10288–10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kocisko D. A., Engel A. L., Harbuck K., Arnold K. M., Olsen E. A., Raymond L. D., Vilette D., Caughey B. (2005) Neurosci. Lett. 388, 106–111 [DOI] [PubMed] [Google Scholar]
- 28. Peretz D., Williamson R. A., Matsunaga Y., Serban H., Pinilla C., Bastidas R. B., Rozenshteyn R., James T. L., Houghten R. A., Cohen F. E., Prusiner S. B., Burton D. R. (1997) J. Mol. Biol. 273, 614–622 [DOI] [PubMed] [Google Scholar]
- 29. Gilch S., Winklhofer K. F., Groschup M. H., Nunziante M., Lucassen R., Spielhaupter C., Muranyi W., Riesner D., Tatzelt J., Schätzl H. M. (2001) EMBO J. 20, 3957–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nunziante M., Kehler C., Maas E., Kassack M. U., Groschup M., Schätzl H. M. (2005) J. Cell Sci. 118, 4959–4973 [DOI] [PubMed] [Google Scholar]
- 31. Matulis D., Kranz J. K., Salemme F. R., Todd M. J. (2005) Biochemistry 44, 5258–5266 [DOI] [PubMed] [Google Scholar]
- 32. Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wüthrich K. (1996) Nature 382, 180–182 [DOI] [PubMed] [Google Scholar]
- 33. James T. L., Liu H., Ulyanov N. B., Farr-Jones S., Zhang H., Donne D. G., Kaneko K., Groth D., Mehlhorn I., Prusiner S. B., Cohen F. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10086–10091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Donne D. G., Viles J. H., Groth D., Mehlhorn I., James T. L., Cohen F. E., Prusiner S. B., Wright P. E., Dyson H. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Knaus K. J., Morillas M., Swietnicki W., Malone M., Surewicz W. K., Yee V. C. (2001) Nat. Struct. Biol. 8, 770–774 [DOI] [PubMed] [Google Scholar]
- 36. Haire L. F., Whyte S. M., Vasisht N., Gill A. C., Verma C., Dodson E. J., Dodson G. G., Bayley P. M. (2004) J. Mol. Biol. 336, 1175–1183 [DOI] [PubMed] [Google Scholar]
- 37. Niesen F. H., Berglund H., Vedadi M. (2007) Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 38. Vedadi M., Arrowsmith C. H., Allali-Hassani A., Senisterra G., Wasney G. A. (2010) J. Struct. Biol. 172, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charvériat M., Reboul M., Wang Q., Picoli C., Lenuzza N., Montagnac A., Nhiri N., Jacquet E., Guéritte F., Lallemand J. Y., Deslys J. P., Mouthon F. (2009) J. Gen. Virol. 90, 1294–1301 [DOI] [PubMed] [Google Scholar]
- 40. Zhang J. H., Chung T. D., Oldenburg K. R. (1999) J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 41. Supattapone S., Wille H., Uyechi L., Safar J., Tremblay P., Szoka F. C., Cohen F. E., Prusiner S. B., Scott M. R. (2001) J. Virol. 75, 3453–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B., Avraham D. (1995) J. Cell Biol. 129, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gilch S., Kehler C., Schätzl H. M. (2006) Mol. Cell. Neurosci. 31, 346–353 [DOI] [PubMed] [Google Scholar]
- 44. Bickel P. E., Scherer P. E., Schnitzer J. E., Oh P., Lisanti M. P., Lodish H. F. (1997) J. Biol. Chem. 272, 13793–13802 [DOI] [PubMed] [Google Scholar]
- 45. Taylor D. R., Whitehouse I. J., Hooper N. M. (2009) PLoS Pathog. 5, e1000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klabunde T., Petrassi H. M., Oza V. B., Raman P., Kelly J. W., Sacchettini J. C. (2000) Nat. Struct. Biol. 7, 312–321 [DOI] [PubMed] [Google Scholar]
- 47. Choi S., Reixach N., Connelly S., Johnson S. M., Wilson I. A., Kelly J. W. (2010) J. Am. Chem. Soc. 132, 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Caughey W. S., Priola S. A., Kocisko D. A., Raymond L. D., Ward A., Caughey B. (2007) Antimicrob. Agents Chemother. 51, 3887–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vogtherr M., Grimme S., Elshorst B., Jacobs D. M., Fiebig K., Griesinger C., Zahn R. (2003) J. Med. Chem. 46, 3563–3564 [DOI] [PubMed] [Google Scholar]
- 50. Lipinski C. A. (2000) J. Pharmacol. Toxicol. Methods 44, 235–249 [DOI] [PubMed] [Google Scholar]
- 51. Büeler H., Aguzzi A., Sailer A., Greiner R. A., Autenried P., Aguet M., Weissmann C. (1993) Cell 73, 1339–1347 [DOI] [PubMed] [Google Scholar]
- 52. Mallucci G., Dickinson A., Linehan J., Klöhn P. C., Brandner S., Collinge J. (2003) Science 302, 871–874 [DOI] [PubMed] [Google Scholar]
- 53. Safar J. G., DeArmond S. J., Kociuba K., Deering C., Didorenko S., Bouzamondo-Bernstein E., Prusiner S. B., Tremblay P. (2005) J. Gen. Virol. 86, 2913–2923 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.