Abstract
Identification of physiologically relevant substrates is still the most challenging part in protease research for understanding the biological activity of these enzymes. The zinc-dependent metalloprotease meprin β is known to be expressed in many tissues with functions in health and disease. Here, we demonstrate unique interactions between meprin β and the amyloid precursor protein (APP). Although APP is intensively studied as a ubiquitously expressed cell surface protein, which is involved in Alzheimer disease, its precise physiological role and relevance remain elusive. Based on a novel proteomics technique termed terminal amine isotopic labeling of substrates (TAILS), APP was identified as a substrate for meprin β. Processing of APP by meprin β was subsequently validated using in vitro and in vivo approaches. N-terminal APP fragments of about 11 and 20 kDa were found in human and mouse brain lysates but not in meprin β−/− mouse brain lysates. Although these APP fragments were in the range of those responsible for caspase-induced neurodegeneration, we did not detect cytotoxicity to primary neurons treated by these fragments. Our data demonstrate that meprin β is a physiologically relevant enzyme in APP processing.
Keywords: Cell Surface Enzymes, Metalloprotease, Neuroscience, Protein Processing, Proteomics, Astacin, Metzincin
Introduction
All proteins undergo post-translational modification by proteolysis, demonstrating the importance of proteases in health and disease. About 2% of all human genes encode for proteolytic enzymes, which account for 569 representatives in humans (1, 2).
Meprin α and β are members of the astacin family of zinc endopeptidases belonging to the metzincin superfamily (3). Both enzymes exhibit unique characteristics among the seven astacin proteases in humans. Meprin α for instance is capable of forming homo-oligomers up to a size of 6 megadaltons, hence being the largest secreted protease known (4, 5). Meprin β is predominantly membrane-bound but can be released from the cell surface by ectodomain shedding (6). The protease displays a wide expression pattern in humans, including brain, skin, intestine, and kidney (7–10). To date, only a few potential in vivo substrates of meprin β have been described, e.g. interleukin-1β (11), interleukin-18 (12), pro-collagen III (10), tumor growth factor α (TGF-α) (13), epithelial sodium channel (14), and vascular endothelial growth factor A (VEGF-A) (15). Other reports have revealed an involvement of meprin β in immunological mechanisms due to its expression in intestinal leukocytes of the lamina propria (16). Based on the processing of these substrates, certain cleavage specificities have been unraveled for meprins. Although meprin α prefers small aliphatic residues in P1′ (nomenclature by Schechter and Berger (17)), meprin β favors acidic amino acid residues (18).
In this study, based on the proteomics terminal amine isotopic labeling of substrates technique (19), the amyloid precursor protein (APP)4 was identified as a candidate substrate for meprin β. APP is a ubiquitously expressed glycoprotein closely related to the pathogenesis of Alzheimer disease (AD). Together with amyloid precursor-like protein (APLP) 1 and APLP2, APP composes the small APP gene family. All three multidomain proteins are type I integral membrane proteins with large extracellular domains and a transmembrane region (20). Alternative splicing of the APP gene leads to three differently processed APP isoforms as follows: APP770, APP751, and APP695.
Before reaching the cell surface, APP undergoes several post-translational modifications in the secretory pathway. Since the discovery of APP in 1987 (21), multiple physiological functions have been assigned to this protein, e.g. growth factor, cell adhesion, signal transduction, etc. (22–24). However, the actual physiological role of APP is still elusive. The generation of the Aβ peptide from APP involves sequential cleavage by two proteases termed β- and γ-secretase (25). The initial β-secretase cleavage by BACE1 (26, 27) generates the N-terminal soluble domain (APPsβ) and a membrane-tethered C-terminal fragment (βCTF or C99). Subsequently, Aβ is released into the luminal space by γ-secretase cleavage within the transmembrane domain of APP (28, 29). However, only a small percentage of APP is cleaved by β-secretase. A larger portion of APP is alternatively shed within the Aβ peptide by an α-secretase (30). The α-secretase activity has been demonstrated to be mainly associated with ADAM10 (a disintegrin and metalloprotease 10) (31, 32). This α-secretase-dependent processing of APP liberates the longer sAPPα and a shorter CTF, termed αCTF or C83. APP and also meprin β have been shown to be expressed in mice brains (33). Gene expression databases confirm an overexpression of meprin β in murine and human hippocampi (34). Together, these data highlight that the regulation of APP is complex and that there may be putative diverse effects of APP in addition to the pivotal role of APP in AD. This study provides evidence that the metalloprotease meprin β is physiologically relevant for extracellular processing of the amyloid precursor protein, demonstrated in vitro, in cellulo and in vivo.
EXPERIMENTAL PROCEDURES
Terminal Amine Isotopic Labeling of Substrates (TAILS)
For proteomic analysis, HaCaT cells (human adult low calcium high temperature keratinocytes; kindly provided by Dirk Breitkreutz) were grown in DMEM, 5% calf serum, to 70% confluency. Cells were then washed intensively to remove serum proteins and grown overnight serum-free. Cells were washed again, incubated in phenol red-free, serum-free medium, and incubated with recombinant human meprin α or β (5, 9, 35). Conditioned medium proteins were harvested at 48 h when the cells were between 80 and 90% confluent, protease inhibitors (1 mm EDTA, 1 mm PMSF) immediately added, and clarified by centrifugation (5 min, 500 × g), filtration (0.22 μm), and additional centrifugation (30 min, 8,000 × g). The proteins were concentrated ×100 by ultrafiltration using Amicon Ultra-15 centrifugal filter units (3-kDa cutoff, Millipore). The sample buffer was exchanged to 50 mm HEPES, 150 mm NaCl, 10 mm CaCl2 by five cycles of dilution and concentration within the same concentrating device. Protein concentration was determined by bicinchoninic acid assay (Pierce) and Bradford assay (Bio-Rad). Secretome collection and concentration, tryptic digestion, amine-terminal blocked peptide enrichment, liquid chromatography-MS/MS, data analysis, and peptide abundance ratio were performed as described previously (36, 37).
Heterologous Protein Expression, Purification, and Cleavage of APPs by Meprin β
Cloning was performed following standard procedures (38), using human APP751 and APP695 cDNA templates. A truncated version of APP was generated, termed HisAPP, lacking the APP signal peptide, the transmembrane domain, and the intracellular part by using the following primers: sense, 5′-catgccatggctggaggtacccactgatggtaat-3′, and antisense, 5′-acatgcatgcctatttttgatgatgaacttcatat-3′.
Constructs were ligated into pFastBac (Invitrogen) containing the meprin β signal peptide, followed by a His6 tag (5), resulting in the expression of soluble APP. Primers were synthesized by Invitrogen, and sequences of constructs were verified by DNA sequencing (Genterprise GmbH). Recombinant protein was expressed using the Bac-to-Bac expression system (Invitrogen) following the manufacturer's instructions. All media and supplements were obtained from Invitrogen. Recombinant baculoviruses were amplified in adherently growing Spodoptera frugiperda (Sf)9 insect cells at 27 °C in Grace's insect medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. Protein expression was performed in 500 ml of suspension cultures of BTI-TN-5B1-4 (HighFive) insect cells growing in Express Five serum-free media supplemented with 4 mm glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin in Fernbach flasks using a Multitron orbital shaker (Infors AG). Cells were infected at a density of 2 × 106 cells/ml with an amplified viral stock at a multiplicity of infection of ∼10. Protein expression was stopped after 72 h; media were stored at −20 °C until further use.
HisAPP was further purified from the media by ammonium sulfate precipitation (60% saturation), stirring overnight at 6 °C, followed by centrifugation at 11,000 × g for 2 h at 4 °C. Pellets were dissolved in 1/10 volume of 50 mm NaH2PO4, 300 mm NaCl, pH 8.0, and dialyzed against 50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 8.0; afterward the protein solution was loaded on a Ni-NTA column. After a washing step using 50 mm NaH2PO4, 300 mm NaCl, 25 mm imidazole, pH 8.0, protein was eluted with the same buffer containing 50 mm imidazole, pH 8.0.
Human APP770 (R & D Systems), APP751, and APP695 were incubated with meprin β for given periods of time at 37 °C in a 100:1 molar ratio. Proteins were analyzed by SDS-PAGE and Western blot. For N-terminal sequencing, proteins were blotted onto PVDF membranes, stained with Coomassie Brilliant Blue, and then sequenced at the protein micro-sequencing center of the Institut Fédératif de Recherche 128 (Lyon, France).
SDS-PAGE and Western Blot Analysis
SDS-PAGE was performed according to standard procedures in 10% polyacrylamide gels. Coomassie Brilliant Blue was used for background free gel staining (39). For immunoblot analysis, proteins were subjected to electrophoresis under reducing conditions and afterward transferred onto a PVDF membrane (Immobilon P; Millipore) by Western blotting. For detection with polyclonal antibodies, the membrane was saturated with 5% dry milk in TBS for 1 h, incubated with the first antibody (polyclonal anti-N-APP, 1:500; Thermo Scientific) for 1 h, and subsequently with horseradish peroxidase-conjugated anti-rabbit IgG (1:10,000) for 1 h at room temperature. Detection was performed using Rotilumin (Roth) following the manufacturer's instructions using x-ray film (hyperfilm enhanced chemiluminescence; Amersham Biosciences). For detection with monoclonal antibodies (Penta-His, 22C11 (40), all diluted 1:1,000), the membrane was blocked with 3% bovine serum albumin (BSA) incubated with the appropriate antibody. The secondary antibody (anti-mouse, coupled with horseradish peroxidase) was added for 1 h, and subsequent detection was performed with Rotilumin.
Transient Transfections of HEK293 Cells with APP and Meprin β cDNAs
HEK293 cells were grown in 6-well cell culture plates until they reached ∼70–80% confluent monolayers. HEK293 cells were transiently transfected with the following cDNAs, by using GeneJuice transfection reagent (Novagen), according to the manufacturer's instructions as follows: 1 μg of pcDNA3 (Invitrogen) and 1 μg of APP751WT-pPCI-neo; 1 μg of pcDNA3 and 1 μg of meprin β-pIRES2-EGFP; 1 μg of APP751WT-pPCI-neo and 1 μg of meprin β-pIRES2-EGFP. The transfections were carried out for 48 h. The wells were then washed with 1 ml of serum-free medium. 800 μl of fresh serum-free medium was then added to the wells, and the medium was conditioned for 5 h. Cell medium was then collected, and cell lysates were prepared in 1% Triton X-100 (Sigma) in phosphate-buffered saline (PBS) lysis buffer containing a mixture of protease inhibitors (Complete, Roche Applied Science). Samples containing cell medium were used for detection of total soluble APP (sAPP) and other sAPP fragments and soluble meprin β. Cell lysates were used to detect membrane-bound meprin β and actin. Total sAPP and other sAPP fragments were detected with anti-N-APP antibody, 22C11 (40). Soluble meprin β was detected with an anti-meprin β antibody, MEP1B (R & D Systems). Actin was detected with an anti-actin antibody (Sigma). Signal detection in all Western blotting experiments was carried out with the enhanced chemiluminescence assay solutions (Millipore).
Preparation of Human Brain Lysates
Brain tissue samples were pulverized into powder in a liquid nitrogen-cooled pestle and mortar. Brain lysates were prepared by adding a 6-fold volume to weight ratio of radioimmunoprecipitation buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% IGEPAL, 0.1% SDS, 0.5% deoxycholate) containing 10 mm NaF, 1 mm β-glycerophosphate, protease inhibitor mixture (Complete, Roche Applied Science), phosphatase inhibitor mixture (PhosSTOP, Roche Applied Science) and by homogenizing the brain tissue powder in a glass homogenizer. Brain homogenates were incubated on ice for 30 min and then centrifuged at 100,000 × g at 4 °C for 30 min. Tissue supernatants were aliquoted and stored at −70 °C. This study was approved by the Institutional Review Board of the University Hospital of Mainz. The experiments were undertaken with the understanding and written consent of each subject or their family member and in compliance with national legislation and the Code of Ethical Principles for Medical Research Involving Human Subjects of the World Medical Association (Declaration of Helsinki).
Animals and Tissue Preparation
Mature congenic Mep1β−/− mice on a C57Bl/6 background, as described previously (41), were maintained on a 12-h light-dark cycle with food and water ad libitum. Control (n = 3) and Mep1β−/− animals (n = 3) were anesthetized by sodium pentobarbital overdose and sacrificed by cervical dislocation. Entire brains were removed and subdissected into cerebellum, frontal cortex, temporal cortex, hippocampus, and “the rest” of the brain prior to further analyses. Individual brain tissue samples were used for generation of a PBS-soluble fraction (S1) and a detergent-soluble fraction (S2). S1 fraction was obtained by adding a 6-fold volume to weight ratio of PBS, containing protease inhibitor mixture (Complete, Roche Applied Science), 10 mm NaF, 1 mm β-glycerophosphate, and phosphatase inhibitor mixture (PhosSTOP, Roche Applied Science). Tissues were homogenized by using a glass homogenizer and incubated on ice for 30 min. PBS-soluble fractions were centrifuged at 100,000 × g at 4 °C for 30 min. S2 fractions were generated by resuspending the remaining pellet in the same original volume of 1% Triton/PBS buffer, containing protease inhibitor mixture (Complete, Roche Applied Science), 10 mm NaF, 1 mm β-glycerophosphate, and phosphatase inhibitor mixture (PhosSTOP, Roche Applied Science). As previously, S2 fractions were incubated on ice for 30 min before being centrifuged at 100,000 × g at 4 °C for 30 min. Tissue supernatants were aliquoted in fresh tubes and stored at −70 °C.
Quantitative Real Time PCR
RNA samples were transcribed into cDNA using random primers and SuperScript II reverse transcriptase according to the manufacturer's instructions (Invitrogen). The obtained cDNA was subjected to quantitative real time PCR measurement using the StepOnePlus RT-PCR system (Applied Biosystems, Darmstadt, Germany). Measurement of the PCR product was enabled by incorporation of SYBR Green®. Amplification reaction consisted of a hold of 10 min at 95 °C and 40 cycles (15 s at 95 °C and 60 s at 60 °C) with subsequent recording of primer melting curves. The primer sequences for amplification of the target gene Mep1b were as follows: meprin_fwd, tgctgatcatcacccttgtc, and meprin_rev, cggagtcaaatttggtcgat. The transcript level was normalized to the transcript level of ARF1 (ADP-ribosylation factor 1). As samples, we used commercially available adult human renal RNA (Agilent Technologies, Waldbronn, Germany), adult human brain RNA (BD Biosciences), and Stratagene universal human reference RNA (Agilent Technologies, Cedar Creek, TX). ΔΔCT values were used to calculate the relative expression for each data point.
Generation and Purification of N-APP20 and N-APP11
8.5·10−7 m APP695 was incubated with 1·10−8 m meprin β for 2 h at 37 °C. Total APP695 fragments were then loaded on a Ni-NTA column as described under “Experimental Procedures.” N-APP11 was purified with a 20 mm imidazole-containing buffer and N-APP20 with 500 mm imidazole.
Preparation of Primary Rat Neurons
Hippocampal or cortical neurons were prepared from Sprague-Dawley rat embryos at day 18 as described before (42). In brief, hippocampi or cortexes were collected in Hanks' balanced saline solution (Invitrogen), cut in pieces, and trypsinized (0.05% trypsin, 0.02% EDTA in PBS) for 15 min. After a mechanical dissociation in Neurobasal medium (Invitrogen) containing B-27 supplement and 2-fold Glutamax (both from Invitrogen), cells were centrifuged at 500 × g for 4 min and resuspended again in Neurobasal medium for counting. The cells were then plated onto poly-l-ornithine (100 μg/ml; Sigma)-coated 24-well plates at a density of 100,000 or 150,000 cells/well (for hippocampus and cortex, respectively). The medium was replaced the next day, and cells were cultured for another 11 days (hippocampus) or 4 days (cortex) at 37 °C in a humidified 5% CO2 incubator.
Live Cell Imaging with Tubulin Tracker
Primary hippocampal cells were grown to days in vitro (div) 11 and pooled hippocampal and cortical cells to div4 in Neurobasal media (Invitrogen) with 0.5 mm glutamine at 37 °C and 5% CO2 in 24-well plates. Cells were washed three times with DPBS and subsequently incubated with 100 nm N-APP20, 100 nm APP695, and 0.1 nm meprin β for 48 h. APP695 fragments were generated by preincubating 100 nm APP695 with 0.1 nm meprin β overnight at 37 °C. Microtubules were immunostained with 250 nm Tubulin Tracker (Invitrogen) according to the manufacturer's instructions. Fluorescence of the cells was monitored by fluorescence microscopy (wDM IRBE microscope, Leica, Wetzlar, Germany). Densitometric analysis was performed with ImageJ, Version A5e.
RESULTS
APP Is a Substrate for Meprin β Identified by Mass Spectrometry-based Degradomics
Degradomic approaches employ quantitative proteomic techniques to identify protease cleavage sites in a cellular context. The proteome-wide coverage prevents bias toward particular substrate classes, hence representing true profiling for potential substrates (43).
In a cell culture-based degradomics approach, named TAILS (19, 36), allowing for identification of newly derived N termini caused by proteases of interest, we identified 96 novel extracellular substrates for human meprin β (Fig. 1 and supplemental Fig. S1). This technique identifies potential substrates in the natural cellular context of the target protein indicated also by their relative abundance ratios (37). Therefore, we were able to identify cleavage of APP by meprin β between Ser124/Asp125, Glu380/Thr381, and Gly383/Asp384 (Fig. 1).
FIGURE 1.
Proteomic identification of APP cleavage products produced by meprin β in cultured keratinocytes. a, proteome was harvested from cultured keratinocytes (HaCaT) with (+) and without (−) meprin β. Isotopic labeling of amine groups and MS/MS analysis were performed as described previously (36, 37). b, APP peptides identified by the degradomics approach TAILS (a) and meprin β cleavage sites in APP isoforms. High probability values calculated by the iProphet algorithm indicate high confidence in spectrum to peptide assignments (for spectrum peptide assignments see supplemental Fig. S1). Meprin β versus control (ctrl) abundance ratios of >15 identify meprin β-generated neo-N termini as high confidence cleavage products (36, 37). Sprot, Swiss-Prot accession numbers.
Meprin β Processing of Recombinant APP Isoforms in Vitro
To further investigate the relevance of this APP processing by meprin β, recombinant forms of APP (APP695, -751, and -770) were incubated with meprin β. APP695 and APP751 were subcloned in a Bac-to-Bac expression system for heterologous expression in baculovirus-infected insect cells and subsequently purified by Ni-NTA chromatography. Expression and secretion of the purified proteins were verified by Western blotting, which revealed signals for a protein species with a molecular mass of about 110 kDa for the 751 variant and 100 kDa for APP695 (Fig. 2b). The yield of affinity chromatography-purified APP was 15 mg/liter cell culture medium for each isoform. Both APP695 and -751 were expressed with a truncated C terminus to maintain solubility of the proteins (Fig. 2, a and b). The identity of the purified APP isoforms was additionally verified by MALDI-TOF analysis.
FIGURE 2.
a, processing of recombinant human APP isoforms by meprin β. APP wt, schematic structure of APP and alternative spliced variants is shown. Only the full-length APP770 isoform contains both the Kunitz protease inhibitor (KPI) and additionally the Ox2 domain; the latter is absent in APP751. AICD = APP intracellular domain. HisAPP, APP751 and -695 were C-terminally truncated at position 613 or 669, respectively, to generate soluble protein. The insertion of an N-terminal His6 tag (6xHis) allowed affinity purification of the recombinant enzymes by Ni-NTA chromatography. CuBD, copper-binding domain; E2, conserved region of the central APP domain; GFLD, N-terminal growth factor-like domain. b, purified proteins were analyzed by 10% SDS-PAGE and subsequently Coomassie staining (lanes 1–3 and 4–6) as follows: starting material (lanes 1 and 4), washing material (lanes 2 and 5), and the protein sample purified with 50 mm imidazole (lanes 3 and 6). Additionally, proteins were transferred to polyvinylidene fluoride membrane and probed with a monoclonal penta-His (histidine) antibody and a polyclonal antibody specific for the N terminus of APP (anti-N-APP), indicating molecular masses of 100 kDa for APP695 and 110 kDa for APP751. c, proteolytic processing of APP isoforms by meprin β. Recombinant APP770 and purified APP variants 751 and 695 were incubated with meprin β at 37 °C for 1, 5, and 15 min and 30, 60, and 120 min, respectively. After immunoblotting, membranes were exposed to different antibodies (22C11 and Penta His, supplemental Fig. S2, anti-N-APP) specific for certain APP regions as indicated in a. Smaller fragments of processed APP695 about 20 kDa (N-APP20, upper arrow) and 11 kDa (N-APP11, lower arrow) were further analyzed (c). The 11-kDa (N-APP11) fragment starts with the mature N terminus of APP retrieved by Edman degradation.
To investigate whether human meprin β is capable of processing APP in vitro, the two generated isoforms APP695 and -751 as well as APP770 were incubated with meprin β and subsequently examined by Western blot analysis. To demonstrate meprin β-specific activity on APP, we have used several antibodies directed against the N-terminal region of APP (Fig. 2, a and c, and supplemental Fig. S2). Interestingly, compared with the recombinant full-length APP following its incubation with meprin β, we were able to detect various cleavage products (Fig. 2c and supplemental Fig. S2).
Monoclonal 22C11 antibodies revealed the full-length APP751 and -695 protein with apparent molecular masses of 110 and 100 kDa, respectively, whereas the 770 isoform appeared at 120 kDa (Fig. 2c, controls). After incubation of all APP isoforms with meprin β (Fig. 2c, lanes 1, 4, and 7), bands were detected in the range of 20 and 11 kDa. A similar band pattern was detected with the polyclonal antibody anti-N-APP directed against the N terminus of APP (supplemental Fig. S2). Both APP-derived fragments were detectable by the Penta-His antibody, recognizing the N terminus of the generated His-tagged APP695, indicating that these cleavage products share the mature N terminus (Fig. 2c, lanes 10–12).
Calculation of the relative molecular mass of the smallest fragment revealed an average size of 11 kDa. Sequencing of this proteolytic APP695 fragment by Edman degradation revealed indeed the mature N terminus starting with the sequence LEVP. This, in combination with the identified cleavage site by degradomics (Ser124/Asp125), is the exact peptide sequence recently detected in human brain lysates (44, 45).
Meprin β Cleaves APP in Cell Culture-based Assays
To further investigate whether cellular expressed meprin β provides a similar cleavage pattern of APP compared with the applied recombinant proteins, HEK293 cells were transiently transfected with human APP751wt and meprin β. Soluble N-APP fragments of ∼11 and 20 kDa were detected in the medium of HEK293 cells transiently overexpressing APP751 and meprin β (Fig. 3a). This was not observed when meprin β was expressed solely. Additionally, membrane-bound or cellular meprin β (Fig. 3b) as well as soluble meprin β (Fig. 3c) were unaltered by APP overexpression. These results demonstrate that meprin β cleaves APP in vitro and in cellulo, thereby releasing N-terminal metabolites of sAPP.
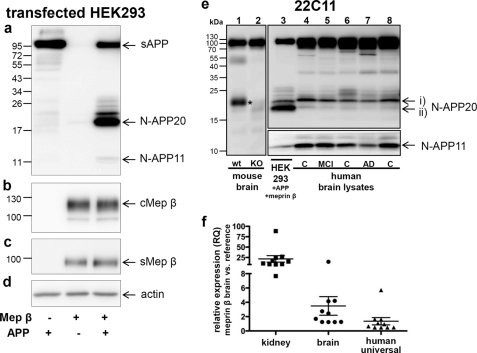
FIGURE 3.
Overexpression of meprin β in human embryonic kidney cells leads to cleavage of N-terminal APP. This processing by meprin β was found in human brain lysates and wild type mouse brain but is missing in meprin β−/− mouse brain. a, human APP was transiently overexpressed in HEK293 cells with or without human meprin β. Cell medium was collected, and total sAPP and other sAPP fragments were detected with anti-N-APP antibody, 22C11. Blots revealed an accumulation of sAPP (N-APP20) bands between the 17- and the 26-kDa markers in media of cells that co-expressed APP and meprin β. Additionally, a N-APP fragment of ∼11 kDa (N-APP11) appeared in the medium of cells that co-expressed APP and meprin β. Levels of membrane-bound or cellular meprin β (cMep β) (b) and soluble meprin β (sMep β) (c) were unaffected by APP overexpression. Equal protein loading was confirmed by blotting for actin protein in cell lysates (d). APP processing, detected with 22C11 antibody, is markedly different between the wild type (wt, lane 1) mice and meprin β knock-out (lane 2) mice (e). The latter showed no N-APP20 (asterisk). Cell medium collected from HEK293 cells transiently transfected with APP and meprin β (lane 3) was loaded on a 15% SDS-PAGE alongside human brain lysates (lane 4, young control; lane 5, mild cognitive impairment (MCI); lane 6, control (C); lane 7, AD; lane 8, control). Different sAPP fragments (N-APP35, -20, and -11) were detected with 22C11 antibody. i and ii indicate subtypes of N-APP20. f, quantitative real time PCR was used to detect human meprin β expression in 10 human brain samples. Three different RNA samples were used as reference for the normalized transcript levels of meprin (see “Experimental Procedures”). Adult human renal total RNA was used as reference because expression of meprin β in mouse and human kidney has been reported previously (46, 47). Expression levels in the brains analyzed were compared with the level of meprin detected in the kidney RNA sample (data were obtained from duplicates of four independent experiments). As additional controls, commercially available RNA samples from human brain and human universal RNA were used as reference samples (data were obtained from duplicates of one experiment). RQ, relative expression.
APP Is Processed by Meprin β in Vivo
To test whether the cleavage of APP by meprin β observed in a proteomics approach, in cell culture and in vitro, is also relevant in vivo, we examined APP processing in human brain lysates from control subjects and AD patients with an N-APP-specific antibody 22C11 (Fig. 3e). N-APP products of about 20 kDa (Fig. 3e, N-APP20, i and ii) and 11 kDa (N-APP11) were observed that correlate with those detected in vitro (Fig. 1c and supplemental Fig. S2) and in cell culture medium (Fig. 3a). The intensities of N-APP20 (Fig. 3e, i and ii) and N-APP11 differed between those identified in cell culture medium and human brain lysates. However, we were unable to detect any statistically significant differences in N-APP fragments between control subjects, mild cognitive impairment, and AD patients (n = 4; one representative is shown) due to the small size of our cohort. The reason for the different amounts of generated N-APP fragments between tissue culture medium and brain lysates might be due to the extensive activity of meprin β in the overexpressing system, probably leading to an enhanced processing of N-APPs. Supporting the physiological relevance of meprin β, we demonstrated that APP processing in the brain of meprin β−/− mice was clearly altered and significantly decreased compared with the wild type (WT) animals (Fig. 3e, lanes 1 and 2). Here, only the full-length sAPP could be detected, although a fragment of about 20 kDa (asterisk, N-APP20) appeared in meprin β-expressing mice only. The N-APP11 fragment was not detected in mice brain, possibly indicating subtle differences in APP processing between mice and human. To quantify the expression level of meprin β in human brain, we performed quantitative real time PCR (Fig. 3f). RNA was extracted from 10 human brain samples and analyzed for meprin β expression. The data were normalized and compared with the expression of meprin β in the kidney, a tissue that is known to highly express the enzyme (46, 47). Interestingly, the expression level of meprin β in the brains analyzed here was on average 21.6 times higher compared with the level of meprin β detected in the kidney RNA sample (Fig. 3f). As further controls, commercially available RNA samples from human brain and human universal RNA were used as reference samples. In comparison with these, meprin β expression in the 10 human brain samples was on average 3.5 and 1.4 times higher, respectively.
Soluble APP695 and APP695 Fragmented by Meprin β Are Not Cytotoxic in Primary Rat Neurons
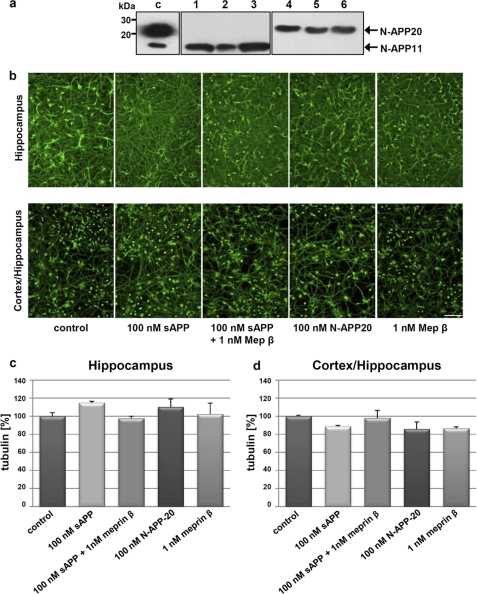
Because small N-terminal fragments of APP have been recently implicated in apoptosis in cells, we analyzed the cytotoxic capacity of meprin β-derived N-terminal APP fragments. To analyze the morphology and viability of isolated embryonic hippocampal and cortical rat neurons, axonal tubulin was stained (Fig. 4b). It has been demonstrated previously that tubulin staining of live cells is an appropriate method to analyze cytotoxicity in neurons (23, 48). Primary neurons at div2 were incubated with sAPP695 and APP695 fragments caused by meprin β activity (Fig. 4b), purified N-APP20 (Fig. 4, a, lanes 4–6, and b), and meprin β solely (Fig. 4b), respectively. To analyze the effects of single N-APP20 in cellulo, N-APP fragments derived from meprin β processing were separated using a Ni-NTA column, with the help of the N-terminal His tag. Isolation of a pure 20-kDa protein from residual N-APPs was attained with 500 mm imidazole (Fig. 4a). After cells were cultured for 48 h, tubulin was stained. Under all conditions, axons and dendrites exhibited a healthy and normal morphology compared with controls (Fig. 4, b–d) indicating that N-terminal APP fragments do not affect neurons cytotoxically. In hippocampal cells, effects of sAPP and N-APP20 rather resulted in an increased tubulin staining indicating a promoting activity of the APP N-terminus. These APP forms just as meprin β cause a marginal decrease of living cells when studying a mixture of cortical and hippocampal neurons (Fig. 4d). However, these observations were not statistically significant. Nevertheless, meprin β is already known to induce apoptosis in keratinocytes thereby reducing the cell number (9). The susceptibility of cortical neurons to APP and meprin β might be higher compared with hippocampal neurons. Previous studies have shown that these two cell cultures exhibit different sensitivities to external impacts (49). Similar effects to the hippocampal cells were observed when the human epithelium-like astrocytic cell line U373-MG was studied under the same conditions as used for rat neurons (data not shown).
FIGURE 4.
N-terminal APP fragments produced by meprin β do not effect morphology of primary neurons. a, separation of N-terminal meprin β-derived APP fragments. After incubation of 3 μm sAPP695 with 10 nm meprin β for 2 h at 37 °C, APP-derived fragments were loaded on a Ni-NTA column. Collected wash (lanes 1–3) and elution fractions (lanes 4–6) were analyzed by Western blotting using the polyclonal N-APP antibody. The arrows indicate the isolated 20- and 11-kDa product derived from buffers containing 20 mm and 500 mm imidazole, respectively. c, control. b, effects of sAPP695 and sAPP695-associated products and meprin β on primary hippocampal and hippocampal/cortex neurons. sAPP695, APP-derived fragments, N-APP20, and meprin β do not influence cell viability, cell morphology, or proliferation of rat neurons. sAPP695-derived fragments were generated by preincubation with 1 nm meprin β. Hippocampal cultures were subjected to proteins at div11 and cortical pooled with hippocampal cells at div4. Tubulin of cell bodies and axons were immunostained after 48 h of incubation for visualization. Scale bar, 100 μm. c and d, densitometric analysis of tubulin staining in primary rat neurons after treatment with sAPP695, sAPP695 fragments, and meprin β (ImageJ version 1.45e). No significant influence on cell morphology could be observed after sAPP and N-APP-20 treatment of hippocampal cells and hippocampal/cortical cultures.
DISCUSSION
In this study, we present a biologically relevant interaction between the metalloprotease meprin β and APP by using a new proteomics approach, knock-out mouse studies, and cell biology experiments. Taken together, the data indicate for the first time that APP is processed in vitro and in vivo by the metalloprotease meprin β resulting in specific N-APP fragments (Fig. 5).
FIGURE 5.
Processing of APP by meprin β in the context of neurodegenerative conditions. The schematic presentation provides an overview of potential interactions of meprin-derived APP fragments. Membrane-bound meprin β (orange) cleaves APP (gray) at distinct sites. N-APP35 that might lead to neuronal death by binding to DR6 might be further cleaved by meprin β into a 20- and 11-kDa fragment sharing the same N terminus, which are unable to bind DR6 and therefore prevent neurotoxicity. Scissors indicate proteolytic cleavage.
To date, APP holds the key role in the hypothesis for the development of Alzheimer disease (25), although the physiological function of APP is poorly understood. However, recently, a novel pathway has been described in mice involving the generation of N-terminal APP peptides binding to the neuronal death receptor DR6, which is required for normal axonal pruning processes (23). Binding of N-APPs to this receptor enables a caspase-dependent cascade to become activated due to nerve growth factor deprivation. Axonal degeneration is then triggered by caspase 6, whereas caspase 3 activation leads to destruction of cell bodies. In this study, we present data that support a role for the metalloprotease meprin β in generation of N-terminal fragments of APPs in vivo.
Meprin β is known to have a clear preference for aspartic and glutamate residues in the P1′ position (18). Two of the three cleavage sites in APP due to meprin β activity identified by TAILS reveal aspartate in P1′ (DALL and DENE), correlating with the specificity of this enzyme. These observations identify meprin β as a promising candidate for the interaction with APP in vivo. By analyzing the GenePaint database, a co-localization of meprin β and APP could be observed throughout the embryonic brain, although with much lower abundance for the metalloprotease. The activity of meprin β, which includes s a transmembrane region and a C-terminal cytosolic part, is dependent on various factors. First, it has to be activated by limited proteolysis, which is done by tryptic proteases like kallikreins (35). Furthermore, it is known that it can be released from the cell surface by ectodomain shedding because of TACE activity (6). In this study, we analyzed the activity of soluble meprin β toward APP. Meprin β was able to cleave all APP isoforms (APP770, -751, and -695) at multiple sites in the N-terminal ectodomain. The full-length APP proteins were converted into fragments of about 20 kDa (N-APP20) and 11 kDa (N-APP11). The physiological relevance of this APP processing by meprin β was demonstrated through the use of meprin β−/− mice. Only faint N-APP fragments could be detected in the knock-out animals, whereas particularly N-APP20 was the most prominent cleavage product in the WT mice. The occurrence of different N-APP peptides in WT animals and their absence in meprin β KO mice clearly indicates a key physiological role of this protease in the turnover of APP.
Most studies in Alzheimer disease research focus on the C-terminal fragments of APP (APP-CTFs), some of which (C99 and C89) serve as substrates for Aβ. Whether the N-terminal processing of APP by meprin β influences the cleavage efficiency toward Aβ by the α-, β-, and γ-secretase is still elusive. Recent evidence suggests that the N terminus of APP is important for the biological role of APP. The copper binding domain, a motif of the N terminus, was shown to build intramolecular disulfide bonds within the APP molecule by oxidizing two cysteines at positions 144 and 158 due to previous copper reduction from Cu(II) to Cu(I), which leads to oxidative stress (50–52). Moreover, copper II induces neurotoxicity in APP−/− neurons to a higher extent than in WT cells (53). Several ligands interacting with sAPP have been characterized, e.g. the low density lipoprotein receptor-related protein (54), fibulin-1 (55), F-spondin (56), and ganglioside GM1 (57), suggesting that the interaction may modulate the activity or function of the protein. Apart from the copper binding domain, the N terminus includes a growth factor-like domain that was reported to be crucial in neurite outgrowth (58). Another function of the N terminus of APP is given by its inhibition capacity that might be critical in regulating proteolytic processing in neurons and soma cells. The Kunitz protease inhibitor-containing APP751 decreases the activity of trypsin significantly in vitro, contrary to APP695 lacking the inhibitory domain (59–61).
Although not proven by N-terminal sequencing, we were able to identify two novel N termini of APP by meprin β between Glu380/Thr381 and Gly383/Asp384 using the proteomics technique. Both cleavage events occur within the E2/carbohydrate binding domain. When an antibody detecting the N-terminal amino acids 1–286 of APP was used, the full-length proteins with molecular masses of 120, 110, and 100 kDa for APP770, -751 and -695, respectively, were processed by meprin β. Signals between 40 and 35 kDa were detected that are in the range of the cleavage sites identified by TAILS. We show that the meprin β-derived N-APP11 fragment identified by the 22C11 antibody and further analyzed by N-terminal sequencing is consistent with a peptide detected in human brain fluids of both AD patients and a control cohort (44). Based on mass spectrometry, this study led to the identification of six novel N-APP fragments starting with the mature N terminus and ending with various amino acids (119, 121–124, and 126). Independent from the amyloid hypothesis, this finding might prove a novel enzymatic mechanism, important for the physiological turnover of APP. Indeed, in our degradomics approach, we could identify meprin β-specific cleavage between Ser124 and Asp125 revealing the exact C terminus of the 11-kDa N-APP fragment observed in vivo (44). Interestingly, the authors could not detect the DR6 ligand in the cerebrospinal fluid (CSF), described by Nikolaev et al. (23). The N-terminal part of this 35-kDa peptide, which is not further characterized yet, potentially serves as a binding partner solely and matches N-terminally with the N-APP peptides of smaller sizes (44). Moreover, the DR6 ligand was not identified in a mouse model either (23). Crossing AD mice with DR6 KO models is necessary to verify this self-destruction mechanism and identify the N-APP ligand in vivo.
Despite the ability of meprin β-releasing N-terminal fragments of APP in vitro and in vivo, these fragments did not affect cytotoxically the primary hippocampal or cortical cells suggesting that the meprin-derived APP fragments are not consistent with those binding to DR6 (23). Supporting our observation, we did not determine significant differences in number and morphology of axons of WT and meprin β−/− cortical sections indicating different physiological functions for meprin β-derived N-APP and the DR6 ligand.
Hence, we hypothesize that the most abundant N-terminal APP fragments in vivo do not act as DR6 ligands. Because DR6 is expressed specifically in neuronal tissues and not throughout the body, the N-APP/DR6 interaction appears relevant only to the physiological function of APP in the brain. Because APP is a ubiquitously expressed protein, the activity of meprin β is not restricted to neuronal cells. It is unknown to what extent meprin β is quantitatively involved in N-APP processing by now. Nevertheless, N-APP fragments due to direct activity of meprin β in human and mouse brain were absent in meprin β−/− mice. Involvement and compensation of other proteases due to the lack of meprin β are not excluded, which might lead to different processing forms of APP. It is becoming more evident that the N-terminal processing of APP is important for the elucidation of the biological relevance of this protein. In summary, all experiments described in this study demonstrate that meprin β is a physiologically relevant enzyme for the processing of the extracellular N-terminal region of APP, resulting in proteolytic fragments observed in human brain lysates. Whether meprin β is involved in the pathogenesis of neurodegenerative diseases has to be further elucidated.
Supplementary Material
Acknowledgments
We thank Claudia Broder, Stefan Müller, and Daniel Lottaz for animal care. We thank Walter Stöcker for scientific support, Dominique Mazzocut for N-terminal sequencing, and Norbert Hilger for statistical advice. We thank London Neurodegenerative Diseases Brain Bank at the Institute of Psychiatry, London, United Kingdom, and the Netherlands Brain Bank, Netherlands Institute for Neuroscience, Amsterdam, for providing human brain samples.
This work was supported by the Deutsche Forschungsgemeinschaft Grants DFG BE 4086/1-2 (to C. B.-P.), DFG PI 379/5-1 (to C. U. P.), and SCHI871/2-1 (to O. S.), by the Competence Network Degenerative Dementias of the German Federal Ministry of Education Grants 01GI1004D (to C. U. P.) and 01GI1004B (to S. W.), and a start-up grant of the Johannes Gutenberg-University Mainz, Germany (to C.B.-P.). The meprin β−/− mice were generated with support from National Institutes of Health Grants DK 54625 and DK 19691 from NIDDK (to J. S. B.). The research leading to these results has received funding from the European Community's Seventh Framework Program (FP7) under Grant 200931 (Project IBDase).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- APP
- amyloid precursor protein
- AD
- Alzheimer disease
- TAILS
- terminal amine isotopic labeling of substrates
- Aβ
- amyloid-β
- RIPA
- radioimmunoprecipitation
- NTA
- nitrilotriacetic acid
- s
- soluble
- div
- days in vitro
- DR6
- death receptor 6
- N-APP
- N-terminal APP
- CTF
- C-terminal fragment
- His
- histidine
- div
- days in vitro.
REFERENCES
- 1. Puente X. S., Sánchez L. M., Overall C. M., López-Otín C. (2003) Nat. Rev. Genet. 4, 544–558 [DOI] [PubMed] [Google Scholar]
- 2. López-Otín C., Bond J. S. (2008) J. Biol. Chem. 283, 30433–30437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sterchi E. E., Stöcker W., Bond J. S. (2008) Mol. Aspects Med. 29, 309–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertenshaw G. P., Norcum M. T., Bond J. S. (2003) J. Biol. Chem. 278, 2522–2532 [DOI] [PubMed] [Google Scholar]
- 5. Becker C., Kruse M. N., Slotty K. A., Köhler D., Harris J. R., Rösmann S., Sterchi E. E., Stöcker W. (2003) Biol. Chem. 384, 825–831 [DOI] [PubMed] [Google Scholar]
- 6. Hahn D., Pischitzis A., Roesmann S., Hansen M. K., Leuenberger B., Luginbuehl U., Sterchi E. E. (2003) J. Biol. Chem. 278, 42829–42839 [DOI] [PubMed] [Google Scholar]
- 7. Vizcaíno J. A., Côté R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) Proteomics 9, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schütte A., Lottaz D., Sterchi E. E., Stöcker W., Becker-Pauly C. (2007) Biol. Chem. 388, 523–531 [DOI] [PubMed] [Google Scholar]
- 9. Becker-Pauly C., Höwel M., Walker T., Vlad A., Aufenvenne K., Oji V., Lottaz D., Sterchi E. E., Debela M., Magdolen V., Traupe H., Stöcker W. (2007) J. Invest. Dermatol. 127, 1115–1125 [DOI] [PubMed] [Google Scholar]
- 10. Kronenberg D., Bruns B. C., Moali C., Vadon-Le Goff S., Sterchi E. E., Traupe H., Böhm M., Hulmes D. J., Stöcker W., Becker-Pauly C. (2010) J. Invest. Dermatol. 130, 2727–2735 [DOI] [PubMed] [Google Scholar]
- 11. Herzog C., Haun R. S., Kaushal V., Mayeux P. R., Shah S. V., Kaushal G. P. (2009) Biochem. Biophys. Res. Commun. 379, 904–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banerjee S., Bond J. S. (2008) J. Biol. Chem. 283, 31371–31377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bergin D. A., Greene C. M., Sterchi E. E., Kenna C., Geraghty P., Belaaouaj A., Belaaouaj A., Taggart C. C., O'Neill S. J., McElvaney N. G. (2008) J. Biol. Chem. 283, 31736–31744 [DOI] [PubMed] [Google Scholar]
- 14. Bondarava M., Li T., Endl E., Wehner F. (2009) Pflugers Arch. 458, 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schütte A., Hedrich J., Stöcker W., Becker-Pauly C. (2010) PLoS One 5, e8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crisman J. M., Zhang B., Norman L. P., Bond J. S. (2004) J. Immunol. 172, 4510–4519 [DOI] [PubMed] [Google Scholar]
- 17. Schechter I., Berger A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 18. Bertenshaw G. P., Turk B. E., Hubbard S. J., Matters G. L., Bylander J. E., Crisman J. M., Cantley L. C., Bond J. S. (2001) J. Biol. Chem. 276, 13248–13255 [DOI] [PubMed] [Google Scholar]
- 19. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., Overall C. M. (2010) Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 20. De Strooper B., Annaert W. (2000) J. Cell Sci. 113, 1857–1870 [DOI] [PubMed] [Google Scholar]
- 21. Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., Multhaup G., Beyreuther K., Müller-Hill B. (1987) Nature 325, 733–736 [DOI] [PubMed] [Google Scholar]
- 22. Hoareau C., Borrell V., Soriano E., Krebs M. O., Prochiantz A., Allinquant B. (2008) Neurobiol. Aging 29, 542–553 [DOI] [PubMed] [Google Scholar]
- 23. Nikolaev A., McLaughlin T., O'Leary D. D., Tessier-Lavigne M. (2009) Nature 457, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Young-Pearse T. L., Bai J., Chang R., Zheng J. B., LoTurco J. J., Selkoe D. J. (2007) J. Neurosci. 27, 14459–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Selkoe D. J. (2001) Neuron 32, 177–180 [DOI] [PubMed] [Google Scholar]
- 26. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 27. Sinha S. (2002) Med. Clin. N. Am. 86, 629–639 [DOI] [PubMed] [Google Scholar]
- 28. Sastre M., Steiner H., Fuchs K., Capell A., Multhaup G., Condron M. M., Teplow D. B., Haass C. (2001) EMBO Rep. 2, 835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weidemann A., Eggert S., Reinhard F. B., Vogel M., Paliga K., Baier G., Masters C. L., Beyreuther K., Evin G. (2002) Biochemistry 41, 2825–2835 [DOI] [PubMed] [Google Scholar]
- 30. Sisodia S. S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6075–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuhn P. H., Wang H., Dislich B., Colombo A., Zeitschel U., Ellwart J. W., Kremmer E., Rossner S., Lichtenthaler S. F. (2010) EMBO J. 29, 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., Serneels L., Snellinx A., Craessaerts K., Thathiah A., Tesseur I., Bartsch U., Weskamp G., Blobel C. P., Glatzel M., De Strooper B., Saftig P. (2010) J. Neurosci. 30, 4833–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Visel A., Thaller C., Eichele G. (2004) Nucleic Acids Res. 32, D552–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lukk M., Kapushesky M., Nikkilä J., Parkinson H., Goncalves A., Huber W., Ukkonen E., Brazma A. (2010) Nat. Biotechnol. 28, 322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohler A., Debela M., Wagner S., Magdolen V., Becker-Pauly C. (2010) Biol. Chem. 391, 455–460 [DOI] [PubMed] [Google Scholar]
- 36. Prudova A., auf dem Keller U., Butler G. S., Overall C. M. (2010) Mol. Cell. Proteomics 9, 894–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. auf dem Keller U., Prudova A., Gioia M., Butler G. S., Overall C. M. (2010) Mol. Cell. Proteomics 9, 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sambrook J., Gething M. J. (1989) Nature 342, 224–225 [DOI] [PubMed] [Google Scholar]
- 39. Neuhoff V., Stamm R., Pardowitz I., Arold N., Ehrhardt W., Taube D. (1990) Electrophoresis 11, 101–117 [DOI] [PubMed] [Google Scholar]
- 40. Hilbich C., Mönning U., Grund C., Masters C. L., Beyreuther K. (1993) J. Biol. Chem. 268, 26571–26577 [PubMed] [Google Scholar]
- 41. Yura R. E., Bradley S. G., Ramesh G., Reeves W. B., Bond J. S. (2009) Am. J. Physiol. Renal Physiol. 296, F135–F144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin A. M., Kuhlmann C., Trossbach S., Jaeger S., Waldron E., Roebroek A., Luhmann H. J., Laatsch A., Weggen S., Lessmann V., Pietrzik C. U. (2008) J. Biol. Chem. 283, 12004–12013 [DOI] [PubMed] [Google Scholar]
- 43. auf dem Keller U., Schilling O. (2010) Biochimie 92, 1705–1714 [DOI] [PubMed] [Google Scholar]
- 44. Portelius E., Brinkmalm G., Tran A. J., Zetterberg H., Westman-Brinkmalm A., Blennow K. (2009) Neurodegener. Dis. 6, 87–94 [DOI] [PubMed] [Google Scholar]
- 45. Portelius E., Price E., Brinkmalm G., Stiteler M., Olsson M., Persson R., Westman-Brinkmalm A., Zetterberg H., Simon A. J., Blennow K. (1996) Neurobiol. Aging 67, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 46. Bond J. S., Matters G. L., Banerjee S., Dusheck R. E. (2005) FEBS Lett. 579, 3317–3322 [DOI] [PubMed] [Google Scholar]
- 47. Oneda B., Lods N., Lottaz D., Becker-Pauly C., Stöcker W., Pippin J., Huguenin M., Ambort D., Marti H. P., Sterchi E. E. (2008) PLoS One 3, e2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nday C. M., Drever B. D., Salifoglou T., Platt B. (2010) Brain Res. 1352, 265–276 [DOI] [PubMed] [Google Scholar]
- 49. Kusumoto M., Dux E., Paschen W., Hossmann K. A. (1996) J. Neurochem. 67, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 50. Multhaup G., Ruppert T., Schlicksupp A., Hesse L., Bill E., Pipkorn R., Masters C. L., Beyreuther K. (1998) Biochemistry 37, 7224–7230 [DOI] [PubMed] [Google Scholar]
- 51. Multhaup G., Schlicksupp A., Hesse L., Beher D., Ruppert T., Masters C. L., Beyreuther K. (1996) Science 271, 1406–1409 [DOI] [PubMed] [Google Scholar]
- 52. Hesse L., Beher D., Masters C. L., Multhaup G. (1994) FEBS Lett. 349, 109–116 [DOI] [PubMed] [Google Scholar]
- 53. White A. R., Multhaup G., Maher F., Bellingham S., Camakaris J., Zheng H., Bush A. I., Beyreuther K., Masters C. L., Cappai R. (1999) J. Neurosci. 19, 9170–9179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kounnas M. Z., Moir R. D., Rebeck G. W., Bush A. I., Argraves W. S., Tanzi R. E., Hyman B. T., Strickland D. K. (1995) Cell 82, 331–340 [DOI] [PubMed] [Google Scholar]
- 55. Ohsawa I., Takamura C., Kohsaka S. (2001) J. Neurochem. 76, 1411–1420 [DOI] [PubMed] [Google Scholar]
- 56. Ho A., Südhof T. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2548–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang H., Ding J., Tian W., Wang L., Huang L., Ruan Y., Lu T., Sha Y., Zhang D. (2009) Neurobiol. Aging 30, 1245–1253 [DOI] [PubMed] [Google Scholar]
- 58. Small D. H., Nurcombe V., Reed G., Clarris H., Moir R., Beyreuther K., Masters C. L. (1994) J. Neurosci. 14, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kitaguchi N., Takahashi Y., Tokushima Y., Shiojiri S., Ito H. (1988) Nature 331, 530–532 [DOI] [PubMed] [Google Scholar]
- 60. Petersen L. C., Birktoft J. J., Flodgaard H. (1993) Eur. J. Biochem. 214, 271–279 [DOI] [PubMed] [Google Scholar]
- 61. Delvaux A., Van der Elst L., Octave J. N. (1992) FEBS Lett. 297, 124–126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.