Abstract
In Escherichia coli, a multicomponent BAM (β-barrel assembly machinery) complex is responsible for recognition and assembly of outer membrane β-barrel proteins. The functionality of BAM in protein biogenesis is mainly orchestrated through the presence of two essential components, BamA and BamD. Here, we present crystal structures of four lipoproteins (BamB–E). Monomeric BamB and BamD proteins display scaffold architectures typically implied in transient protein interactions. BamB is a β-propeller protein comprising eight WD40 repeats. BamD shows an elongated fold on the basis of five tetratricopeptide repeats, three of which form the scaffold for protein recognition. The rod-shaped BamC protein has evolved through the gene duplication of two conserved domains known to mediate protein interactions in structurally related complexes. By contrast, the dimeric BamE is formed through a domain swap and indicates fold similarity to the β-lactamase inhibitor protein family, possibly integrating cell wall stability in BAM function. Structural and biochemical data show evidence for the specific recognition of amphipathic sequences through the tetratricopeptide repeat architecture of BamD. Collectively, our data advance the understanding of the BAM complex and highlight the functional importance of BamD in amphipathic outer membrane β-barrel protein motif recognition and protein delivery.
Keywords: Lipoprotein, Lipoprotein Structure, Membrane Biogenesis, Protein Cross-linking, Protein Structure
Introduction
Gram-negative bacteria are surrounded by two membranes, an inner membrane and the protective outer membrane (OM)2 layer. The outer membrane architecture is highly asymmetric and composed by integral outer membrane β-barrel proteins, lipoproteins, lipopolysaccharides, and phospholipids (1). These components are entirely synthesized in the cytoplasm, translocated over the inner membrane, and finally delivered through the periplasm to the outer membrane by specific shuttle factors that transport their cargo to membrane receptor complexes (1, 2). In Escherichia coli, lipoproteins are targeted to the OM through the LolABCDE complex system via a series of membrane and periplasmic transfer steps (3, 4). Integral OMPs are translocated by the secretion (Sec) machinery and stabilized against premature precipitation or mislocalization in the periplasm by the three major chaperones PpiD, Skp, and SurA (4–7). These chaperones presumably act sequentially with PpiD at an early stage, early after the OMP release into the periplasm (5). This initial rescue event is followed by Skp and SurA chaperoning at a later stage (8). Various experimental studies describing OMP folding through SurA show the particular importance of this chaperone. Phage display combined with peptide binding studies indicated SurA interactions with amphipathic OMP peptides (9, 10). Furthermore, the importance of SurA in vivo is underlined by the observation that E. coli cells being deleted in surA exhibit reduced levels of folded OmpA, OmpC, OmpF, and LamB porins (11).
After the unfolded outer membrane protein is delivered through the periplasmic space, the final steps are the tethering to the BAM complex followed by the catalyzed insertion into the OM. The BAM complex of E. coli is formed by five proteins: the integral BamA receptor (see a model in Fig. 1A) and four accessory lipoproteins, BamB–E of divergent fold and sequences (1, 12). Mutational analysis of the BAM family proteins through entire or partial deletion has demonstrated the essential role of BamA and BamD for cell survival, whereas the remaining three lipoproteins, BamB, BamC, and BamE, are non-essential. These lipoproteins express a rather unspecific influence on the stability of the complex (BamE) and the kinetic folding of selected OMP proteins (BamB, e.g. on the monomer/trimer transition of LamB) (13–15).
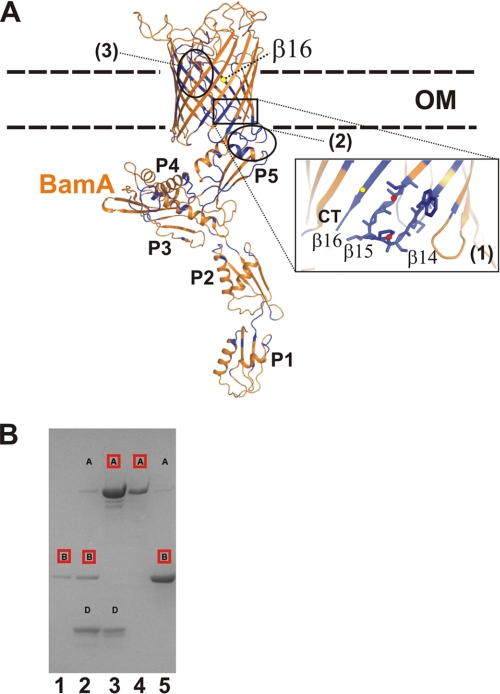
FIGURE 1.
Structure model of BamA from E. coli and BamA complex formation. A, the model of full-length BamA from E. coli is shown inserted in a schematically depicted OM. The model was prepared on the basis of the FhaC transporter structure (PDB code 2QDZ) comprising a β-barrel domain and two N-terminal POTRA domains (18). The membrane spanning the β-barrel part of BamA was modeled using FhaC coordinates and the Modeler program (63). Further positioning of BamA POTRA domains (P4 and P5, PDB code 3OG5) was accomplished by the superposition onto the FhaC POTRA domains (33, 56). This initial model was then extended using the BamA crystal structure comprising the POTRA P1-P4 domains (PDB code 3EFC) by a structure alignment of the overlapping P4 domains (56). The full-length model of BamA (orange) was subsequently used to localize conserved residues that were marked in blue to detect a possible accumulation of residues (e.g. (1) in strands β16, β17, and β18). This part of the structure is highlighted by a close-up view, with the two proline residues emphasized by red dots. Additional parts of the protein are conserved: (2), the junction between POTRA and the barrel domain and (3), the longest loop extending into the barrel domain. B, SDS-PAGE analysis demonstrating the complex formation of BamA (A), BamB (B), and BamD (D). In lane 1, BamB and BamD were applied to Ni-NTA chromatography. Lane 2 shows the elution of a mixture containing BamA, BamB, and BamD, with BamB comprising the His tag. In lane 3, the mixture of BamA and BamD was applied, with BamA as the His-tagged protein. Lane 4 shows a mixture of partially unfolded but soluble and His-tagged BamA in octyl-polyoxyethylene detergent incubated with BamB. A mixture of folded BamA and BamB was applied after Ni-NTA separation. Proteins comprising the His tag are marked in red.
BamA comprises a two-domain architecture: five polypeptide transport-associated (POTRA) subdomains form the N-terminal part followed by the C-terminal OMP domain comprising 16 β-strands (16–18). For proper function, only the C-terminal POTRA domain P5, located proximal to the barrel domain, is essential (Fig. 1A) (16). Recently it has been shown that the BAM complex can also be assembled in a functional form from overexpressed and purified proteins (19). Biochemical and mutational studies provided further evidence for the direct interactions between BamA and BamB or BamD, respectively. Interactions of BamB and BamA are mediated through residues originally identified by a combined approach using bioinformatic and mutational studies (20). Another investigation provided deeper insights into the interaction of BamD with BamA, presumably mediated via the last ∼30 BamD residues in a direct or indirect way (14).
The very C-terminal residues of bacterial OMPs and β-barrel proteins from the mitochondrial outer membrane are important for their recognition and assembly. It has long been reported that the C-proximal residue (‘0′) of most OMPs in E. coli almost invariably carries the aromatic residue Phe and, to a significantly lesser extent, additional aromatic residues (mostly Trp) (21, 22). Additional positions being conserved are at −2, which often harbors a Tyr residue, the −4 position (hydrophobic residue), and the −5 position (often a Gly residue) (22). The distribution of these residues is not only important for the recognition by the BAM complex but also fulfils the archetypical amphipathic pattern criterion of all transmembrane segments, the underlying structure principle for OMP architecture. Because of the evolutionary relationship of Gram-negative bacteria and mitochondria, a similar C-terminal fingerprint has been observed in mitochondrial outer membrane proteins of the β-barrel architecture and is important for proper processing by the sorting and assembly machinery (23). Here, the principle deviation from the bacterial system comprises the presence of a terminal residue (‘0′) being negatively charged. Similar to OMPs, the −1 residue is mostly aromatic but strictly hydrophobic, the −3 residue is hydrophobic (mostly Val, Leu, or Phe), and the −6 residue position is almost invariably Gly (23). Although the SAM and BAM components Sam50 and BamA express a clear sequence similarity, this has not been recognized for the accessory components of the two complexes. Neither Sam37 nor Sam35 of the SAM complex show a strong sequence similarity to the BamB-BamE lipoproteins (24).
To understand the individual contributions of the BamB-BamE lipoproteins in their BAM-associated context for recognition and folding of OMP proteins, we set out to analyze all proteins individually by structure biology methods. The structures, combined with supporting and preexisting biochemical data, provide an advanced picture of their individual function in OMP-biogenesis of E. coli.
EXPERIMENTAL PROCEDURES
The cloning, purification and crystallization of BamC (N- and C-terminal domains), BamD, and BamE have been described (25).
Peptides, Cloning, Purification and Cross-linking: See also supplemental methods.
Protein Cloning and Purification
Genes encoding BamB and full-length BamE (including the signal sequence) were cloned into pET21b, and the TPR1–3 domain of BamD was cloned into pET24b. The proteins were expressed at 37 °C in E. coli BL21 RIL cells (Stratagene) as C-terminally His-tagged fusion products and purified via Ni-NTA chromatography. After cell lysis through a French press in buffer A (see below), cells and membranes were pelleted by centrifugation for 1 h at 160,000 × g. For BamB, the supernatant was directly applied to affinity chromatography. In contrast, BamE was observed in the pellet and was solubilized using buffer A including 1% dodecyl-β-d-maltoside detergent (Glycon). Both proteins were purified via Ni-NTA chromatography using buffer A containing 300 mm NaCl and 10 mm Tris-HCl (pH 8) for equilibration, buffer B (buffer A including 20 mm imidazole) for removal of contaminating proteins, and buffer C (equal to buffer A but including 250 mm imidazole). Samples of BamB or BamE were concentrated and subjected to gel size exclusion chromatography in buffer D (containing 150 mm NaCl, 20 mm Tris (pH 8)) (Superdex200, GE Healthcare). For full-length BamE, all column buffers contained 0.1% dodecyl-β-d-maltoside.
BamA cloned into pET15 was overexpressed as insoluble protein in the E. coli BL21 strain. Cells were disrupted by French press, and inclusion bodies and cell membranes were separated from the soluble fractions by centrifugation. Inclusion bodies were further homogenized by washing steps using buffer A and a potter device for homogenization. Membranes were removed by two additional steps of solubilization using buffer A supplemented by 1% octyl-polyoxyethylene (Bachem). The homogeneity of inclusion bodies was tested using SDS-PAGE. BamA was purified by Ni-NTA under denaturing conditions. Inclusion bodies were solubilized in 6 m guanidine-hydrochloride, Sigma), 20 mm Tris-HCl (pH 8) (buffer E). The protein was applied to Ni-NTA and purified via a two-step gradient using buffer F (6 m guanidine-hydrochloride, 20 mm Tris-HCl, 20 mm imidazole (pH 8)) and buffer G (6 m guanidine-hydrochloride, 20 mm Tris-HCl, 300 mm imidazole (pH 8)) for elution. Refolding of the protein was induced by dropwise addition of the solution into a buffer containing 20 mm Tris-HCl and 0.5% LDAO (Sigma-Aldrich) (pH 8) (buffer H). The protein was further purified by a second Ni-NTA chromatography step using 20 mm Tris and 0.1% LDAO (buffer I) for column equilibration and subsequently buffer J (20 mm Tris, 0.1% LDAO, 20 mm imidazole (pH 8)) and buffer K (20 mm Tris, 0.1% LDAO, 300 mm imidazole (pH 8)) for the washing and elution steps.
Peptides
Peptides Hia1 (GKSMVAIAGSSYQGQNGLAIGYSRIS) and Hia2 (KVIIRLSGTTNSQGKTGVAAGVGYQW), representing C-terminal segments from the Haemophilus influenzae trimeric autotransporter, were purchased from Peptide2.0.
Cross-linking Experiments
For cross-linking of truncated and full-length BamE, concentrated protein solutions were diluted with 10 mm HEPES, 20 mm NaCl (for full-length BamE the buffer also contained 0.1% dodecyl-β-d-maltoside) to approximately 1 mg/ml. Cross-link experiments of BamD and the TPR1–3 domain of BamD with Hia peptides were performed in 100 mm NaHCO3 (pH 7.8). Hia peptides (dissolved in water) were added in 3-fold molar excess. The solutions were diluted to concentrations of about 0.9 mg/ml.
The reactions were performed in a total volume of 50 μl. The solutions were treated with 2.5 μl of 2.5% glutaraldehyde (final concentration, 0.11%) and heated at 37 °C for 5 min. The reactions were interrupted by addition of 10 μl of 1 m Tris (pH 8). After short incubation, SDS gel loading buffer was added, and the reactions were analyzed on 17% SDS-PAGE gels using Coomassie staining.
Protein Crystallization
Crystallization of BamB using the sitting drop vapor diffusion method was performed by mixing the protein solution with the precipitant solution as described earlier for the other lipoproteins (25). BamB crystals appeared 30–60 days after setup from 0.4 μl of BamB at 5.5 mg/ml (in 15 mm Tris (pH 8)) and 0.4 μl of reservoir solutions (3.5 m NH4Cl, 0.1 m Na-acetate (pH 4.6) or 2 m LiCl, 0.1 m Na-acetate (pH 4.6)). A crystal of the (WD40)9 degradation product was observed approximately 110 days after setup from 0.4 μl of BamB at 16 mg/ml and 0.4 μl of ground solution (0.2 m NaCl, 20% PEG 6000, 0.1 m MES (pH 6)).
For data collection in all cases, the initial crystals were used. Crystals were picked using small nylon loops (Hampton Research), briefly immersed in ground solution supplemented with 20–25% glycerol, and immediately flash-frozen in liquid nitrogen. For details see also Ref. 25.
X-ray Data Collection and Structure Solution
Data collections were performed at beamlines PX I and PX II at the Swiss Light Source (SLS, Villigen, Switzerland) at 100 K. Images were recorded on a Pilatus 6 m detector (Dectris). Data were indexed, integrated, and scaled using the XDS and XSCALE programs (26). Data collection and refinement details are given in supplemental Table S1. Structures were solved by the SAD or MIR methods using SHARP for phasing and DM for solvent flattening (27, 28). BamB/(WD40)9 was determined by molecular replacement using the Balbes program (29). Structures were refined using the Refmac program and rebuilt using the Coot program (30). The geometry of the structures was checked with the Rampage program. All crystallographic figures were prepared using PyMOL.
Small-angle X-ray Scattering Measurements
Small-angle x-ray scattering data of BamC were collected at the X33 beamline of the Deutsches Elektronensynchrotron (Hamburg, Germany). Solutions containing different concentrations of full-length BamC in 20 mm HEPES, 150 mm NaCl, 5 mm MgCl2 (pH 7.5) were measured at 283 K. Before and after each sample measurement, scattering data from the corresponding buffer solution were collected and subtracted. Scattering was recorded at a wavelength of 1.5 Å and using a sample-detector distance of 2.7 m, with a MAR345 image plate detector. Fourier transformation was performed using the autoGNOM program (31). Reconstruction of the molecular shape of BamC was calculated using DAMMIN (31). Models were placed into the reconstructed envelopes using the SUPCOMP program (31, 32).
RESULTS
Proteins Involved in the β-Barrel OMP Biogenesis
In E. coli, proteins form the basis for outer membrane protein biogenesis (the integral outer membrane protein BamA with ∼ 85 kDa and four associated lipoproteins (BamB, 40 kDa; BamC, 34 kDa; BamD, 26 kDa; and BamE, 10 kDa). Two proteins of the BAM complex, BamA and BamD, have proven to be essential for the viability of E. coli (14, 16). The outer membrane protein BamA comprises a repetitive N-terminal architecture comprising five POTRA domains, which is followed by the 16-stranded β-barrel for membrane localization, including extended loop structures (see a model of full-length BamA in Fig. 1A and conservation analysis details in supplemental Fig. S1). The protein displays conserved residues which, mapped on the model, cluster, e.g. at the interface between the essential P5 domain and the periplasmic turns of the β-barrel domain (16, 18, 33). Additional conserved areas are located within the last three β-strands (β14-β16) of the barrel walling, with a particular accumulation of residues in the terminal β16-strand representing the C-terminal OMP fingerprint (Fig. 1A, inset).
Assembly of the BamABD Complex
To determine the structures of BAM proteins, we expressed the individual components of the complex. BamA was produced as inclusion bodies and subsequently refolded in LDAO detergent solution. The success of refolding was judged to be ∼80%, using the band shift assay proven to be useful for various OMPs (34, 35). To test whether the proteins employed for structural analysis can reassemble to form the ternary complex, we first tested the coelution from affinity columns and showed that refolded BamA coeluted together with BamD. His-tagged BamB was proven to form a stable complex with BamA and also a mixture of BamA/BamB/BamD, with BamB comprising the only His tag eluted from the column as the expected ternary complex (Fig. 1B). This procedure does, in principle, allow the preparation of BamABD complexes in milligram amounts using the full-length proteins with the N-terminal lipid anchors removed.
Structure of BamB, a WD40 Repeat
To understand the influence of BamB in OMP biogenesis and to identify determinants for BamB-BamA complex formation, we cloned the full-length protein missing the lipid anchor and set out for crystallization. Size determination of the complex by chromatography techniques showed a distribution of oligomeric states resembling monomers, tetramers, and hexamers (Fig. 3B). To our surprise, the crystallization using the oligomeric protein yielded crystals of three BamB fragments that had been processed during the course of crystallization, presumably by limited proteolysis (supplemental Fig. S2A). Interestingly, these fragments consisting of either two or three WD40 repeats (β1/β2/β3, β2/β3, (β5/β6)2) reassembled to yield a regular structure of an unusual novel nine-bladed β-propeller structure with a clear pore shape of ∼1 nm in diameter (supplemental Fig. S2, A and B).
FIGURE 3.
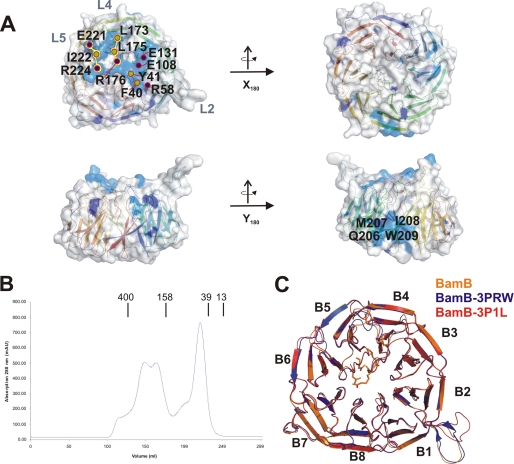
Structure analysis and conservation patterns of BamB. A, analysis of conserved residues using the surface representation of BamB. In the upper panel, the protein surface is shown in top and bottom view (related by a rotation of the molecule around the x axis). Conserved residue patches on the surface are marked in light blue, and important positions facing the surface are marked with numbers. Residue types are color-coded with dots: green, hydrophobic residues; blue, charged residues. In the lower panel, the BamB surface in side view is shown from two different positions that are related by a rotation of the molecule by 180 degrees around the y axis. A second but smaller conserved patch on the outer rim surface appears for residues Gln-206-Met-209. B, size exclusion chromatography of BamB showing the size distribution of the protein as monomers and higher oligomers (1, 4, and 6). C, superposition of BamB structures from this work and recently published structures (38, 40, 54). The three structures show a small structural divergence of 0.6 and 0.5 r.m.s.d., respectively.
Under differing conditions we obtained crystals of a new morphology containing the full-length protein, and the structure to 2.6 Å was solved by molecular replacement using two domains of the nine-bladed propeller as search models (β1/β2/β3 and β5/β6). In line with secondary structure prediction methods, BamB displayed the fold of an eight-bladed β-propeller, and it belongs to the WD40 superfamily of proteins. It also comprised the typical architecture of a Velcro motif, leading to the closure of the propeller ring because of antiparallel pairing of the very N- and C-terminal β-strands (Fig. 2A) (36). While the N-terminal 13 residues were not visible in the electron density, the following 12 residues of the protein were modeled and appear as a random coil structure with only a loose association to the propeller core. In the biological context, these N-terminal structural properties would allow for an enhanced flexibility at the membrane border (Fig. 3A). Most but not all propeller blades show the typical conservation of residues in WD40 motifs, most importantly in β3 and β4 strands (e.g. conserved Trp in β3 and His at the end of β4) (36). In contrast to the nine-bladed and several seven-bladed propeller structures, the eight BamB repeats do not form an open central pore because of the specific folding of extended loops connecting the individual WD40 repeats on the top side (Fig. 2A). In seven-bladed propellers, these loops often form the scaffold for specific protein-protein interactions, although additional interaction sites on the bottom side and the periphery are known (36). This top site has also been proposed based on experimental work done for two WD40 architectures comprising eight repetitive units. One of these is the Skp1-Fbw7-CyclinEdegN complex (FCE), determined by cocrystallization with a substrate peptide (37). The superposition of the FCE complex on BamB indicated a small overall structural deviation (PDB code 2OVR7, r.m.s.d. of 2.5 Å for 316 Cα positions), and the localization of the peptide in the FCE complex is in the vicinity of the putative BamB binding pocket (Fig. 2B) (37). Typical binding motifs recognized by seven- and eight-bladed propellers are often small elongated peptide structures derived from protein sequences often comprising proline residues (36). Arginine residues in loops on the top side of the complex structure are often involved in protein-protein interactions (Fig. 2C) (36, 37). In the superposition of the FCE complex with BamB, it seems possible that BamB also could provide e.g. Arg residues at spatially related positions (e.g. Arg-58, Arg-176, Arg-224) to bind a putative target motif of BamA (20). The advanced analysis of conserved residues in BamB shown here (in comparison to Ref. 20) included more protein sequences mapped on the structure. These data demonstrate a further enhanced accumulation on the top side of the propeller and revealed conserved residues in the neighborhood lining on one surface area of BamB (Fig. 3A). This cluster on the top face more strongly localizes to loops L1, L4, and L5, whereas a small additional cluster was identified at the outside rim (residues Gln-206 to Trp-209, Fig. 3A). Six of these residues on the top side are charged and comprise three arginines (see above) together with glutamates Glu-131, Glu-108, and Glu-221 (Fig. 3A). Substrate peptides of WD40 repeat structures often present one or more proline residues (following a hydrophobic residue) as the typical binding motif of several WD40 structures (36, 37).
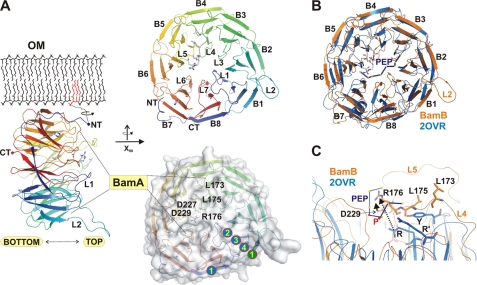
FIGURE 2.
Structure of BamB and comparison with related structures and analysis of conservation. A, structure representation of BamB in side view perspective with the putative membrane-anchoring lipid connected by a flexible linker (dashed lines) to the lipoprotein. The structure is shown in schematic representation, with the N and C terminus (NT/CT) and important loops (L1, L2) labeled. The area interacting with BamA is indicated together with the top and bottom face assignment. Structures in the top panel on the right display the eight WD40 repeats (B1-B8) together with residues known to be essential for BamA interactions (20). These functional residues are almost all located on the top face of the protein and cluster along the central pore of the protein. In the surface representation of BamB, the localization of functional residues is indicated together with the secondary structure assignment of the first WD40 repeat (encircled numbers 1–4). B, superposition of BamB with the Skp-Fbw7-CyclinEdegN complex (PDB code 2OVR), which was cocrystallized with an N-terminal N-degron peptide (PEP, marked in dark blue with the residual C-α position indicated by yellow dots). The peptide is bound on top of the scaffold architecture provided by the extended loop structures (37). C, close-up of the peptide binding site in the FCE complex with structurally important residues emphasized. The localization of amino acids known to be important for function in BamB is assigned (Leu-173, Leu-175, Arg-76), all of which are located in close proximity to residues of the N-degron peptide (here only the residue types are marked with R and R' for arginine residues involved in peptide binding). Notably, the N-degron peptide and related cocrystal structures of WD40 propellers often display proline residues (here specifically marked with P and red dots).
The structure of BamB from E. coli has also been determined recently by other groups (38–40). Our structure essentially resembles the published structures, with only small deviations in the terminal and loop motifs. The r.m.s.d. between the individual BamB structures is ∼0.5–0.7 Å.
Structure of BamC, an Unexpected Repeat Protein
The overall architecture of BamC on the structure level is modular, with a long unstructured N terminus of approximately 70 residues and two structurally related domains connected by an α-helical linker (Fig. 4A). Initially we set out to crystallize the full-length protein but failed to produce any crystals. Therefore we used limited proteolysis of full-length BamC through subtilisin as a tool to define stable fragments (termed BamCND and BamCCD supplemental Fig. S3), which we crystallized separately (25). Both fragments of ∼120 residues in size yielded crystals that diffracted to high resolution (1.6 and 1.3 Å), and domain structures were solved by SAD techniques (see additional details in Ref. 25 and supplemental Table S1).
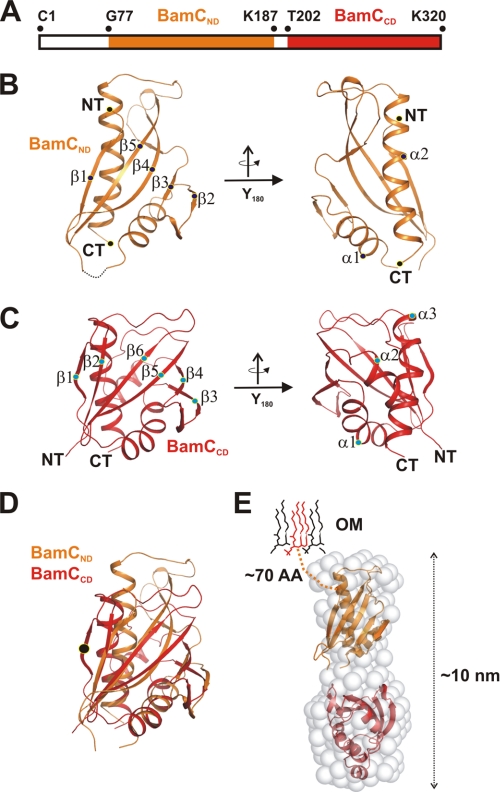
FIGURE 4.
Structure of the BamC scaffold evolved through gene duplication. A, schematic assignment of the N- and C-terminal domains (BamCND and BamCCD) to the BamC sequence. BamCND is color-coded orange, and BamCND is marked in red. These two domains were defined through limited proteolysis, which led to the cleavage of 14 residues in between these domains. B, two structure views of BamCND, both related by a rotation of 180 degrees around the y axis. The structure comprises a α/β-mixed fold arranged as an αβ-sandwich domain. The individual secondary structure elements and domain borders are marked (β1-β5 and α1 and α2). NT, N terminus; CT, C terminus). C, domain architecture of BamCCD with two views related by the rotation of 180 degrees around the y axis. Secondary structure elements are marked as for the N-terminal domain in B. D, superposition of the N- and C-terminal domains. Although the sequence similarity of the domains is low, the structural conservation is clearly visible. The main difference in structure between both domains is the presence of an additional N-terminal β-strand in the BamCCD domain. E, architecture of the full-length protein with the outer membrane delineated. The N-terminal residues of the full-length protein are predicted to be unstructured. The two-domain structure was assembled inside the ∼10-nm-long elongated protein envelope generated by SAXS data analysis. Among all four lipoproteins, BamC shows the longest unstructured N-terminal sequence, approximately 70 residues.
Fold analysis of the BamCND domain indicated a globular compact fold and a mixture of α- and β-secondary structure elements. An antiparallel β-sheet flanks two α-helices with a topology of β1/α1/β2/β3/β4/β5/α2 (Fig. 4B and supplemental Fig. S4B). From the close crystallographic packing of two Bam CND domains we initially anticipated that the BamC protein might be arranged dimeric in an antiparallel manner yielding with a significant interface area of ∼1000 Å2 (supplemental Fig. S4A). Later, however, we noticed that the BamCCD structure was clearly monomeric, and SAXS data of the full-length protein were further supportive in the verification of a monomeric state. The analysis of this domain regarding conserved residues shows two patches of spatially separated residues of aromatic and positively charged nature (supplemental Fig. S4D).
The structure analysis of the C-terminal domain BamCCD proves the significant structure analogy to BamCND, which was not expected from the sequence analysis with respect to repeat recognition (Fig. 4D). Although the r.m.s.d. is ∼3 Å and the identity between the superimposed residues is 12%, secondary structure elements are essentially the same (supplemental Fig. S4, B and C). The topology of this domain is only slightly different, with an additional short N-terminal β-strand and a small α-helical extension between β4 and β5 (Fig. 4C and supplemental Fig. S4, B and C). The domain cores formed by the small β-sheet are structurally much better conserved than the loop or helical elements (see Fig. 4D). The conservation pattern of this BamCCD as deduced from multiple sequence alignments is significantly more pronounced than the BamCND domain. A clear cluster-like appearance along the β5 strand is visible (residues 278 to 286), also including two neighboring aromatic residues (Phe-224 and Trp-228) on α1 (supplemental Fig. S4E). The structure search in the DALI database with both domains revealed the same folds reoccurring in several protein complexes, such as the mammalian AMPK complex or the RocCor domain of the Rab family (41–43). Although these complexes are functionally not related to BamC, the central positioning of these domains within several complexes is appealing (supplemental Fig. S5, A–C).
To determine structure data of the full-length BamC at lower resolution, we performed small-angle x-ray scattering experiments and collected data of the protein, including both domains and part of the anchoring N terminus. These data allowed the determination of the protein envelope structure with approximate dimensions of 5 × 10 nm in diameter. This envelope was used to model two individual BamC domain structures by rigid docking and to estimate their relative orientation within the context of the full-length protein (Fig. 4E).
Activation of BamE through Domain Swapping Events?
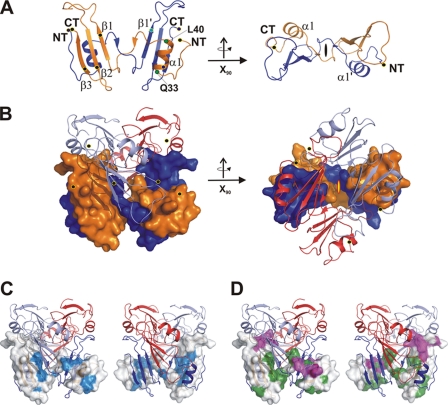
The smallest component of the BAM complex is BamE, with about 10 kDa. The protein was crystallized in an N-terminally truncated form and crystals diffracted to 1.7 Å resolution (see supplemental Table SI). The structure solved by SAD techniques shows a hexameric protein complex that is assembled by three intercalating dimers (Fig. 5, A and B). In the monomer, the very N terminus is followed by a short α-helix (residues Gln-33 to Leu-40), which both interact with the small β-sheet of the adjacent monomer (β1-β3). In the dimer, secondary structure elements arrange to a layer of α/β sandwich molecules formed by an intertwined fold of a pseudoknot-like arrangement (Fig. 5A and supplemental Fig. S6A). Notably, the interface between these two monomers accounts to a significant ∼1200 Å2. The dimer architecture and irreversible monomer-dimer equilibrium are indicative of a three-dimensional domain swapping of the N terminus exchanging with the C-terminal domain (39, 44, 45) (Fig. 5, A and B and supplemental Fig. S6A).
FIGURE 5.
Structure of the oligomeric intertwined BamE complex. A, structure of the dimeric BamE protein unit in side and top view (tilted by 90 degrees around the x axis). Secondary structure elements and termini are marked accordingly. B, structure of the hexameric protein complex, which is formed by intercalation of three dimers (light blue and red). Two monomers (orange and blue) are indicated in surface representation to show the strong knot-like interface. C, representation of the hexameric protein complex from two different orientations, with a monomer represented as surface representation. Conserved residues are plotted onto the surface (cyan) to mark the interface between the two monomers. D, the same surface view as in C, and residues involved in interface stabilization between monomers are marked with different colors depending on the monomer-monomer interface contacts. Taking the conservation fingerprint together with the stabilizing residues of the hexameric complex indicates that the dimeric assembly is of importance, whereas the hexameric complex may assemble under different conditions.
While this monomer-monomer interface is significant and the size and arrangement clearly indicate the potential to form the proper biological interface, interaction areas of the same monomer to additional neighboring chains in the hexameric complex are smaller (Fig. 5C and supplemental Fig. S6, A and B). An analysis of this interface together with a plot of conserved residues on the protein surface indicated the conservation of the large dimeric interface. However, the small additional interfaces leading to hexamerization are not further conserved (Fig. 5D and supplemental Fig. S6A).
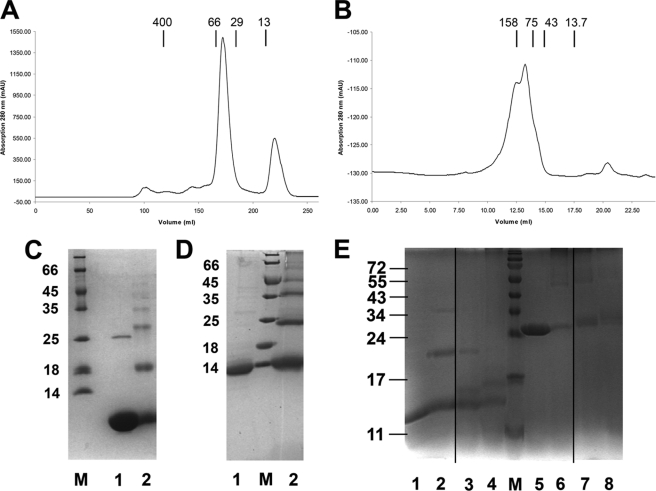
The presence of oligomeric species in solution was also proven by size exclusion chromatography and cross-linking studies of both the full-length protein, including the lipid anchor, and the N-terminally truncated version (Figs. 6, A–D). To confirm that folding and the oligomerization state in the crystal structure were not artificial because of the overexpression of the protein in the cytoplasm, we isolated BamE overexpressed as complete lipoprotein from the natural membrane and analyzed this complex with respect to the oligomeric state. The native complex was isolated based on standard membrane protein extraction procedures using the mild detergent dodecyl-maltoside. Analysis by size exclusion chromatography and cross-linking essentially confirmed the previous finding of a higher molecular weight complex (Figs. 6, C and D). Collectively these data prove the presence of a stable dimeric protein species in the native membrane, which at higher concentrations may assemble to build the hexameric complex.
FIGURE 6.
Protein oligomerization states of BamE and interaction studies of BamD with OMP peptides. A and B, size exclusion chromatography runs of truncated (residues 21–94 of the full-length protein) and the full-length BamE (including the lipid anchor). Molecular masses indicate the oligomeric state (hexameric for the full-length protein and ∼tetrameric for the shortened version) of both species in solution (oligomerization number size is marked with 1, 4, and 6). C and D, further confirmation of the oligomeric state of BamE by glutaraldehyde-mediated cross-linking of truncated BamE (residues 21–94) (C) and full-length BamE (D). Protein samples are shown before (1) and after (2) cross-linking. Samples were subjected to SDS-PAGE and showed a ladder-like appearance, indicating the presence of oligomers (up to hexamers) in solution. E, formation of a BamD-peptide complex as shown by cross-linking experiments. Peptides used for this experiment were derived from the structure of the Hia-autotransporter, and glutaraldehyde has been used a cross-linker (12). The two 26-residue-long peptides comprising β-strands β1, β2, and β3, and β4 were used to be cross-linked with the truncated form of BamD (lanes 1-4) and the full-length version of the protein (lanes 5-8). Lanes 1 and 2 show the protein alone before and after cross-linking. Lanes 3 and 4 demonstrate cross-linking to the two different peptides. The same arrangement is shown in lanes 5 and 6. The full-length protein was cross-linked before the full-length protein was cross-linked to the individual peptides (lanes 7 and 8).
Our search for structurally related folds with the dimer structure led to the identification of another structurally related lipoprotein, OmlA, which shares a similar fold but is monomeric (supplemental Fig. S7A, PDB code 2PXG) (46) with an r.m.s.d. of ∼2.5 Å (for residues aligned to each other within a sphere of 3.5 Å). Although the two structures agree relatively well with each other in the small β-sheet part, the α-helices and loops strongly deviate. The similarity between the BamE crystal structure and those solved by NMR methods shows a r.m.s.d. of ∼2.5 Å, with a reasonable agreement in the β-sheet part but significant deviation in the helix and loop structures (supplemental Fig. S7A) (39, 45).
By contrast, several representatives of β-lactamase inhibitors are also similar in structure and fold. However, in this case, also to the dimeric BamE (supplemental Fig. S7, B–E, PDB code, e.g., 3GMV, 3GMX, or 2G2U) (47, 48). Interestingly, in the model complex, we illustrate that when BamE replaces the β-lactamase inhibitor protein, some conserved residues on the BamE surface point toward the putative β-lactamase complex after superposition of BamE onto the β-lactamase inhibitor in the complex structure of β-lactamase inhibito-SHV-1 (supplemental Fig. S7, E and F) (48).
Structure and Function Analysis of the Essential BamD Lipoprotein
To investigate the second essential protein of BAM, we produced BamD by standard methods. However, purification and increase of the protein concentration of the full-length protein as well as a C-terminally truncated form showed a strong tendency to precipitation. This was reversible if the sample was diluted to a lower concentration. However, the low concentrations of ∼4 mg/ml hampered all crystallization attempts. To stabilize the protein against early precipitation and to be able to increase the protein concentration, urea at small concentrations of 1–2 m was added and indeed stabilized the protein, even at concentrations of 40 mg/ml. We tested the influence of urea on the secondary structure by CD spectroscopy and limited proteolysis but did not find any significant changes compared with the wild-type protein (25).
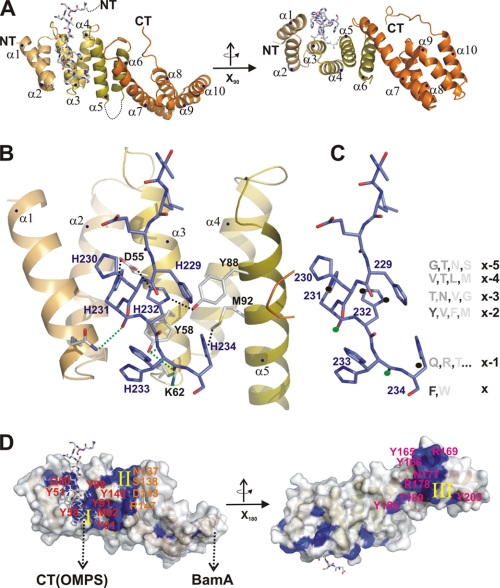
Using the concentrated sample, the protein readily crystallized and yielded crystals that diffracted to a resolution of 1.8 Å. The structure was solved by MIR techniques and showed an entirely α-helical protein fold that was in agreement with secondary structure prediction methods (Fig. 7A). Ten α-helices form an extended protein fold, and all helix pairs (α1/α2 … α9/α10) show the typical fold of TPR (TPR1–5) repeats (Fig. 7A and supplemental Fig. S8, A and B). Interestingly, the TPR3 domain displays an insertion of a 14-residue-long loop (residues 107–120) between helices α5 and α6 that is not visible in the crystal structure (Fig. 7A). The first three of the five TPR domains (TPR1–3) form the structural scaffold known to target C-terminal residues of substrate proteins. In fact, in the crystal lattice, the C-terminal His-tag of a neighboring molecule was localized in the groove of this scaffold (supplemental Fig. S9A, list of interactions). Although this terminus is likely to be non-native in terms of side chain distribution, the binding properties may resemble conditions of the natural substrate protein. Binding of this terminus is mediated by side chain and main chain interactions and is summarized in Fig. 7, B and C. To further investigate the potential of the His tag as a putative model for the C terminus as a potential substrate protein, we hypothesized that residues could originate from the conserved C terminus of OMP proteins. This hypothesis was proven and is underlined by cross-linking experiments of the full-length protein and the TPR1–3-fold with amphipathic peptides and peptides following the C-terminal consensus of OMP proteins (Fig. 7D). In fact, the residues known to be conserved in this terminus would sterically fit with the observed scaffold if the backbone conformation was kept stable and the side chains mutated into the most probable residues (e.g. if the terminal residue is phenylalanine and so forth).
FIGURE 7.
The essential BamD protein represents a TPR scaffold. A, structure representation of BamD shown from two different sides. Secondary structure elements of the exclusively α-helical protein are marked (α1-α10). The C-terminal extension of a second BamD protein is marked in stick representation. B, close-up of the TPR1–3 binding site. The C-terminal peptide is marked in stick representation (blue), and interactions between the peptide and TPR scaffold are marked by dashed lines (important residues are numbered and given in stick presentation). C, the same orientation of the peptide as in B, but additional information of OMPCT sequences is provided. The residues of the peptide carry the nomenclature of x (last residue) to x-5. Conservation of residues as the “OMP fingerprint” for E. coli OMP proteins is given to compare residues supposedly binding at the same site with the actual sequence of the C-terminal BamD portion. D, conservation analysis of BamD sequences. The protein visualized from two sides (identical to A) shows three conserved patches (I-III), residues are numbered and marked in blue), one of which comprises residues located in the TPR domain fold. The second patch is nearby and may be involved in SurA or BamA binding. The third patch is localized on the opposite side of the protein, with a significant number of tyrosine residues involved.
To further examine BamD with respect to conservation, we marked conserved residues on the entire surface of the protein and analyzed them with respect to clusters that would potentially allow interactions to substrate proteins or proteins of the BAM complex (conserved residues are summarized in supplemental Fig. S9B). Three patches can be clearly distinguished because of their distinct localization on the surface. One patch covers the scaffold offered by the TPR1–3 domains. Some of these residues (e.g. Tyr-58, Tyr-88) are also involved in interactions to the polyhistidine extension of the adjacent protein. A second patch comprises several residues within the Asn-137-Arg147 sequence (Fig. 7D, II, and supplemental Fig. S9B). A third patch is located on the opposite side of the protein and involves a significant number of four conserved Tyrosine residues together with two positively charged arginines (Fig. 7D).
Similar structures of TPR proteins have been observed earlier. In particular, the arrangement of the TPR1–3 fold is typical for several representatives of this protein family. Notably, the TPR1–3 fold is reminiscent, e.g., of the Hop protein, which is an adaptor protein of the Hsp chaperones in higher eukaryotic cells (PDB code 1ELR) (49) and which was crystallized with the C-terminal peptide of Hsp90. The strongest structural similarity exists to bacterial proteins YbgF from E. coli and SycD from Shigella flexneri (50, 51), both of which are implicated in protein secretion.
DISCUSSION
The BAM complex comprising the β-barrel OMP BamA and the four lipoproteins BamB-BamE is important for the biogenesis of outer membrane proteins in Gram-negative bacteria. Although the composition of this complex can vary between species, the essential components BamA and BamD are most exclusively present in bacterial genomes. The process of β-barrel biogenesis is mechanistically intriguing, as the unfolded OMP needs to pass several steps from entry into the periplasm until folding in the OM is accomplished. In particular, the delivery step from a chaperone-substrate complex to BAM, the influence of the lipoproteins on the folding step, and the final release into the membrane remain unclear. To examine this problem, we set out to investigate the complex components by structural and biochemical methods and describe the investigation of BamA and the four lipoproteins BamB-BamE.
From sequence analysis of the complex members it has been recognized earlier that three proteins, BamA (POTRA domains), BamB (WD40 domains), and BamD (TPR domains) exhibit the typical features of proteins known to be involved in protein-protein interactions (7). More specifically, the WD40 and TPR domains are among the most common protein-protein interaction domains and often show a variable substrate spectrum for a unique single domain (36). This also applies to the POTRA superfamily which is implicated in the binding of C-terminal motifs of OMPs or amphipathic peptides (52). As a consequence, mutants (partial or full deletions) of these three proteins show the most severe impact on the biogenesis of OMP proteins in mutant strains (14, 16, 53).
The structure analysis of BamB presented here (Fig. 2B) largely resembles the structures published very recently (38, 40, 54). Our interpretation of the structure is, however, more elaborated in terms of sequence analysis, which we mapped onto the surface of the BamB structure to predict an increased number of conserved residues potentially also involved in BamA binding (data are shown in Fig. 3A, alignment not shown). In fact, by mutational studies it has been demonstrated earlier that Arg-176 (together with Leu-175 and Leu-173) is not only conserved but also important for the binding to BamA. Two additional residues, Asp-227 and Asp-229 in BamB, turned out to be equally essential after being mutated. However, these residues are not exposed on the surface and may rather contribute to destabilization by an indirect effect because of a formation of the salt bridge between Asp-227 and Arg-176 (20). Although is are a limited number of conserved areas visible on BamB, we speculated that BamB might be able to bind amphipathic sequences. However, all amphipathic peptides we used for cross-linking studies did not react with the protein, and we hypothesize that BamB may have a different function, possibly in the general alignment of the complex relative to the substrate complex or in support of lateral substrate release. This idea would also be in agreement with the rather small influence of mutants lacking BamB or similar to the phenotype of surA strains, where these factors seem responsible for optimizing the transfer efficiency and kinetics (11, 20).
A search of BamB against the DALI database returned additional eight-bladed propeller structures, some of which as complexes with substrate peptides (37, 55). In combination with the structural repertoire of WD40 structures comprising seven repeats in complex with peptides from protein interaction partners, it is remarkable that most of these short peptides used for cocrystallization contain at least one proline residue (36). Stimulated by this finding, we developed a model of E. coli BamA on the basis of the FhaC homolog and BamA partial structures (Fig. 1A) (18, 33, 56). We analyzed this model with respect to conserved and exposed proline-“rich” sequence stretches. Indeed, we found one segment at the periplasmic side of the β-barrel comprising a loop-like structure between β14 and β15 (Fig. 1A). This part of BamA is obviously among the strongest conserved parts and could contribute to the preselection of the unfolded OMP at a later stage right after the chaperone-OMP complex has been recognized by factors such as BamD. The structure topology around this site is of particular interest, as the lateral release of an OMP monomer may occur by transiently widening the barrel between β1 and β16 to further allow the insertion of the newly folding OMP. An analogous mechanism has been proposed for the SAM machinery of mitochondria (23).
The lipoprotein BamD is another factor of BAM that is essential for viability of E. coli. A significantly large number of conserved residues in BamD is in line with the essential function of the protein in OMP biogenesis, and those may be assigned to 1) a function in BamA interactions (presumably via the BamD C terminus (supplemental Fig. S8C)) and 2) binding of periplasmic chaperones and OMP substrate recognition (14). Similar to BamB, the TPR fold is typical for adaptor proteins that mediate interactions of a transient nature, e.g. as demonstrated for the cochaperone Hop, which binds the C terminus of Hsp70 or Hsp90. The TPR scaffold of Hop comprising three TPR domains has been cocrystallized with a C-terminal peptide of Hsp90 (supplemental Fig. S8B) (49). In the crystal structure of BamD, we found the C terminus of another BamD protein located in the cavity of the TPR1–3 scaffold, similar to the arrangement obtained for Hop-Hsp90. The terminus appears to be properly clamped by side and main chain interactions. Although being artificial in terms of the presumed motif, we found that an in silico exchange of residues against highly conserved residues from E. coli OMPs at positions of the histidine side chains would still perfectly fit into the binding pocket (Fig. 6C). Supported by cross-linking attempts with two different peptides from the autotransporter Hia, we showed that the TPR domain is necessary for the preselection of substrate OMPs. This hypothesis is further underlined by a comparison with the mitochondrial OMP biogenesis pathway. In mitochondria, adaptor proteins such as Sam35 are known to accomplish the recognition of C-terminal motifs from mitochondrial OMPs (23). In particular, Sam35 plays the decisive role in protein recognition. Using structure prediction tools and TPR repeat prediction servers, it is obvious that Sam35 comprises two TPR domains similar to BamD (data not shown) (57). Interestingly, using the same prediction tool, TPRpred, three of the five TPR domains of BamD are predicted. Consequently, Sam35 and BamD (sequence similarity/homology, 17/55%) may share a similar function in the recognition of C-terminal sequences of OMP proteins and mitochondrial outer membrane proteins, respectively.
A physiological importance of BamC in the BAM-aided folding remains largely unclear from the structural data we obtained. The deletion of BamC also has no physiological effect on the targeting of OMP proteins (11). From previous observations, it appears that this factor may be involved in the response to rifampin (specific for RNA polymerase), tetracycline (ribosome), and ampicillin (β-lactamase) resistance (15). Some of these functions are reoccurring as functional attributes of the BamE protein (see below). Because the BAM complex has also implications in folding and export of autotransporters, it seems possible that BamC carries out a more specialized function involving, e.g., proteins destined for secretion (22). In Serratia marescence, BamC is involved in the swarming behavior but not in the integrity of the cell envelope, which may indicate a potential function in flagella maintenance (58).
The smallest lipoprotein, BamE, shows an interesting equilibrium between a monomer, an intertwined dimer, and a hexamer fold. Recently, this structure was also solved by NMR methods, but a monomer was found, although with a similar topology as the dimeric species (40, 45). Because the equilibrium is irreversible and domain swapping in many proteins was shown to be activating a protein complex, it seems more likely that the dimer is the active form, whereas the monomer is inactive (44). In support of our structural data of the truncated protein, we tested the full-length protein isolated from native membranes toward oligomerization, and both cross-linking and size exclusion chromatography indicated the presence of oligomeric species. In particular, the protein isolated from native membranes should represent the fold most closely with relation to the native conformation. The oligomerization state of this isolated complex is indicating hexamerization, with the protein calculated as full-length protein plus an additional lipid and micelle girdle (Fig. 6B). However, the interface parameters between monomers and the conservation pattern are in favor for a dimeric species. Structure homologs of BamE are found in the class of inhibitors of β-lactamases, a class of proteins that is produced by several strains of bacteria to degrade β-lactam antibiotics, which would otherwise suppress, e.g., cell wall synthesis. Proteins inhibiting β-lactamases typically comprise a tandem fold of (∼70–100)2 residues (59), which is the approximate size of dimeric BamE. The function of natural β-lactamase inhibitors on the basis of proteins is still under debate, but they may be involved in the regulation of cell wall growth (60), a function which could, e.g., reflect a reduced stalk biosynthesis in Caulobacter crescentus (61) and a function in L-form formation of E. coli has been reported recently (62). Together these observations support our view of a BamE function in cell wall maintenance, possibly by a direct or indirect effect involving the β-lactamase AmpC from E. coli.
In summary, the structure and functional data presented here allow new perspectives into the function of the BAM complex-forming proteins (see a model in Fig. 8). Three proteins (BamA, BamB, and BamD) clearly indicate domains that are being used in many protein-protein complex interactions, whereas BamC and BamE may have deviating functions of OMP folding (e.g. β-lactamase resistance). Interestingly, both proteins appear to be located in the periphery of the BAM complex and not directly associated with BamA (7, 13, 14). In contrast, BamB, comprising the WD40 fold, may interact with BamA via a conserved motif at the C terminus of BamA. Studies on BamD in particular indicate amphipathic peptides as a potential substrate of the TPR fold and may represent the linking factor that allows interactions between periplasmic chaperones and the BAM complex by a transfer of the substrate protein to the BAM complex.
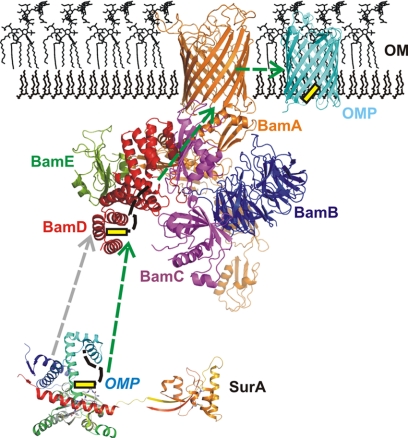
FIGURE 8.
Model of the β-barrel assembly machinery. Proteins destined for the outer membrane are delivered to the BAM complex with the help of periplasmic chaperones. Most importantly, SurA has been shown to bind OMP peptides and to deliver substrates for processing by the BAM complex (6, 9). The entire BAM complex as a model is presented with the five compounds BamA-BamE color-coded (BamA, orange; BamB, blue; BamC, magenta; BamD, red; BamE, green). A possible scenario for OMP biogenesis on the basis of the data presented in this paper is the substrate delivery of a putative OMP (with the OMPCT marked in yellow) from SurA to BamD that can recognize both the BamA protein and OMP peptides. The substrate is presumably transferred to the BAM complex and subsequently inserted into the outer membrane.
Supplementary Material
Acknowledgments
We thank the staff from beamlines PXI and PXII (Swiss Light Source) for excellent support during x-ray data acquisition. We also thank Drs. Manfred Rössle and Dmitri Svergun for fruitful discussions and help during data collection of SAXS data. We thank Dr. Andrei Lupas for continuous support of our work. We also thank several lab rotation students from the University of Tübingen for help.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9, Table S1, methods, and references.
- OM
- outer membrane
- OMP
- outer barrel β-membrane protein
- POTRA
- polypeptide transport-associated
- SAM
- sorting and assembly machinery
- Ni-NTA
- nickel-nitrilotriacetic acid
- LDAO
- lauryl-diaminoxide
- FCE
- Fbw7-CyclinEdegN
- r.m.s.d.
- root mean square deviation
- TPR
- tetratricopeptide
- Sec
- Sec protein secretion system
- SAD
- single anomalous dispersion
- MIR
- multiple isomorphous replacement.
REFERENCES
- 1. Knowles T. J., Scott-Tucker A., Overduin M., Henderson I. R. (2009) Nat. Rev. Microbiol. 7, 206–214 [DOI] [PubMed] [Google Scholar]
- 2. Hagan C. L., Silhavy T. J., Kahne D. E. (2010) Annu. Rev. Biochem. [DOI] [PubMed] [Google Scholar]
- 3. Tokuda H., Matsuyama S. (2004) Biochim. Biophys. Acta. 1694, IN1–9 [PubMed] [Google Scholar]
- 4. Tokuda H. (2009) Biosci. Biotechnol. Biochem. 73, 465–473 [DOI] [PubMed] [Google Scholar]
- 5. Antonoaea R., Fürst M., Nishiyama K., Müller M. (2008) Biochemistry 47, 5649–5656 [DOI] [PubMed] [Google Scholar]
- 6. Behrens-Kneip S. (2010) Int. J. Med. Microbiol. 300, 421–428 [DOI] [PubMed] [Google Scholar]
- 7. Gatsos X., Perry A. J., Anwari K., Dolezal P., Wolynec P. P., Likić V. A., Purcell A. W., Buchanan S. K., Lithgow T. (2008) FEMS Microbiol. Rev. 32, 995–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bos M. P., Robert V., Tommassen J. (2007) Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 9. Bitto E., McKay D. B. (2003) J. Biol. Chem. 278, 49316–49322 [DOI] [PubMed] [Google Scholar]
- 10. Xu X., Wang S., Hu Y. X., McKay D. B. (2007) J. Mol. Biol. 373, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ureta A. R., Endres R. G., Wingreen N. S., Silhavy T. J. (2007) J. Bacteriol. 189, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meng G., Surana N. K., St Geme J. W., 3rd, Waksman G. (2006) EMBO J. 25, 2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sklar J. G., Wu T., Gronenberg L. S., Malinverni J. C., Kahne D., Silhavy T. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malinverni J. C., Werner J., Kim S., Sklar J. G., Kahne D., Misra R., Silhavy T. J. (2006) Mol. Microbiol. 61, 151–164 [DOI] [PubMed] [Google Scholar]
- 15. Xu C., Lin X., Ren H., Zhang Y., Wang S., Peng X. (2006) Proteomics 6, 462–473 [DOI] [PubMed] [Google Scholar]
- 16. Kim S., Malinverni J. C., Sliz P., Silhavy T. J., Harrison S. C., Kahne D. (2007) Science 317, 961–964 [DOI] [PubMed] [Google Scholar]
- 17. Jacob-Dubuisson F., Villeret V., Clantin B., Delattre A. S., Saint N. (2009) Biol. Chem. 390, 675–684 [DOI] [PubMed] [Google Scholar]
- 18. Clantin B., Delattre A. S., Rucktooa P., Saint N., Méli A. C., Locht C., Jacob-Dubuisson F., Villeret V. (2007) Science 317, 957–961 [DOI] [PubMed] [Google Scholar]
- 19. Hagan C. L., Kim S., Kahne D. (2010) Science 328, 890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vuong P., Bennion D., Mantei J., Frost D., Misra R. (2008) J. Bacteriol. 190, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Cock H., Struyvé M., Kleerebezem M., van der Krift T., Tommassen J. (1997) J. Mol. Biol. 269, 473–478 [DOI] [PubMed] [Google Scholar]
- 22. Lehr U., Schütz M., Oberhettinger P., Ruiz-Perez F., Donald J. W., Palmer T., Linke D., Henderson I. R., Autenrieth I. B. (2010) Mol. Microbiol. 78, 932–946 [DOI] [PubMed] [Google Scholar]
- 23. Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., Prinz C., Meisinger C., Guiard B., Wagner R., Pfanner N., Wiedemann N. (2008) Cell 132, 1011–1024 [DOI] [PubMed] [Google Scholar]
- 24. Walther D. M., Rapaport D., Tommassen J. (2009) Cell. Mol. Life Sci. 66, 2789–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Albrecht R., Zeth K. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1586–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 28. Cowtan K. D., Main P. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 43–48 [DOI] [PubMed] [Google Scholar]
- 29. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Svergun D. I. (1999) Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Svergun D. I., Barberato C., Koch M. H., Fetler L., Vachette P. (1997) Proteins 27, 110–117 [PubMed] [Google Scholar]
- 33. Gatzeva-Topalova P. Z., Walton T. A., Sousa M. C. (2008) Structure 16, 1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poolman J. T., de Marie S., Zanen H. C. (1980) Infect. Immun. 30, 642–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frasch C. E., Mocca L. F. (1978) J. Bacteriol. 136, 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stirnimann C. U., Petsalaki E., Russell R. B., Müller C. W. (2010) Trends Biochem. Sci. 35, 565–574 [DOI] [PubMed] [Google Scholar]
- 37. Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007) Mol. Cell 26, 131–143 [DOI] [PubMed] [Google Scholar]
- 38. Heuck A., Schleiffer A., Clausen T. (2011) J. Mol. Biol. [DOI] [PubMed] [Google Scholar]
- 39. Kim K. H., Kang H. S., Okon M., Escobar-Cabrera E., McIntosh L. P., Paetzel M. (2011) Biochemistry 50, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 40. Kim K. H., Paetzel M. (2010) J. Mol. Biol. 406, 667–678 [DOI] [PubMed] [Google Scholar]
- 41. Xiao B., Heath R., Saiu P., Leiper F. C., Leone P., Jing C., Walker P. A., Haire L., Eccleston J. F., Davis C. T., Martin S. R., Carling D., Gamblin S. J. (2007) Nature 449, 496–500 [DOI] [PubMed] [Google Scholar]
- 42. Gotthardt K., Weyand M., Kortholt A., Van Haastert P. J., Wittinghofer A. (2008) EMBO J. 27, 2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin X., Townley R., Shapiro L. (2007) Structure 15, 1285–1295 [DOI] [PubMed] [Google Scholar]
- 44. Liu Y., Eisenberg D. (2002) Protein Sci. 11, 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Knowles T. J., Browning D. F., Jeeves M., Maderbocus R., Rajesh S., Sridhar P., Manoli E., Emery D., Sommer U., Spencer A., Leyton D. L., Squire D., Chaudhuri R. R., Viant M. R., Cunningham A. F., Henderson I. R., Overduin M. (2011) EMBO Rep. 12, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vanini M. M., Spisni A., Sforça M. L., Pertinhez T. A., Benedetti C. E. (2008) Proteins 71, 2051–2064 [DOI] [PubMed] [Google Scholar]
- 47. Gretes M., Lim D. C., de Castro L., Jensen S. E., Kang S. G., Lee K. J., Strynadka N. C. (2009) J. Mol. Biol. 389, 289–305 [DOI] [PubMed] [Google Scholar]
- 48. Reynolds K. A., Thomson J. M., Corbett K. D., Bethel C. R., Berger J. M., Kirsch J. F., Bonomo R. A., Handel T. M. (2006) J. Biol. Chem. 281, 26745–26753 [DOI] [PubMed] [Google Scholar]
- 49. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 50. Lunelli M., Lokareddy R. K., Zychlinsky A., Kolbe M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9661–9666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barta M. L., Zhang L., Picking W. L., Geisbrecht B. V. (2010) BMC Struct. Biol. 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gentle I. E., Burri L., Lithgow T. (2005) Mol. Microbiol. 58, 1216–1225 [DOI] [PubMed] [Google Scholar]
- 53. Charlson E. S., Werner J. N., Misra R. (2006) J. Bacteriol. 188, 7186–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Noinaj N., Fairman J. W., Buchanan S. K. (2011) J. Mol. Biol. 407, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gatzeva-Topalova P. Z., Warner L. R., Pardi A., Sousa M. C. (2010) Structure 18, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Karpenahalli M. R., Lupas A. N., Söding J. (2007) BMC Bioinformatics 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Soo P. C., Wei J. R., Horng Y. T., Hsieh S. C., Ho S. W., Lai H. C. (2005) Infect. Immun. 73, 6075–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Strynadka N. C., Jensen S. E., Johns K., Blanchard H., Page M., Matagne A., Frère J. M., James M. N. (1994) Nature 368, 657–660 [DOI] [PubMed] [Google Scholar]
- 60. Doran J. L., Leskiw B. K., Aippersbach S., Jensen S. E. (1990) J. Bacteriol. 172, 4909–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ryan K. R., Taylor J. A., Bowers L. M. (2010) Microbiology 156, 742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Glover W. A., Yang Y., Zhang Y. (2009) PLoS ONE 4, e7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fiser A., Sali A. (2003) Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.