Abstract
The ∼2-megadalton evolutionarily conserved histone deacetylase-associated Rpd3L/Sin3L complex plays critical roles in altering the histone code and repressing transcription of a broad range of genes involved in many aspects of cellular physiology. Targeting of this complex to specific regions of the genome is presumed to rely on interactions involving one or more of at least 10 distinct subunits in the complex. Here we describe the solution structure of the complex formed by the interacting domains of two constitutively associated subunits, mSin3A and SAP30. The mSin3A paired amphipathic helix 3 (PAH3) domain in the complex adopts the left-handed four-helix bundle structure characteristic of PAH domains. The SAP30 Sin3 interaction domain (SID) binds to PAH3 via a tripartite structural motif, including a C-terminal helix that targets the canonical PAH hydrophobic cleft while two other helices and an N-terminal extension target a discrete surface formed largely by the PAH3 α2, α3, and α3′ helices. The protein-protein interface is extensive (∼1400 Å2), accounting for the high affinity of the interaction and the constitutive association of the SAP30 subunit with the Rpd3L/Sin3L complex. We further show using NMR that the mSin3A PAH3-SAP30 SID complex can bind to nucleic acids, hinting at a role for a nucleolar localization sequence in the SID αA helix in targeting the Rpd3L/Sin3L complex for silencing ribosomal RNA genes.
Keywords: Corepressor Transcription, Gene Silencing, NMR, Protein Assembly, Protein-Protein Interactions, Transcription Regulation, Transcription Repressor, Chromatin-modifying Complex, Nucleolar Targeting, Protein-Nucleic Acid Interactions
Introduction
The transcriptional output at any given locus in eukaryotes is determined in part by the pattern of post-translational histone modifications, broadly defined as the histone code (1, 2). The reversible nature of the modifications allows site-specific alterations to the code with concomitant changes in the levels of gene expression. Histone deacetylases (HDACs)3 comprise an important class of enzymes that effect transcriptional repression by reversing the acetylation status of histones. Nuclear HDACs are commonly found in large, multisubunit complexes with proteins that target the enzymes to specific sites on the genome, lending specificity to the deacetylation process.
HDAC1 and HDAC2, the mammalian homologues of yeast Rpd3, are found in at least five biochemically distinct complexes, two of which share several subunits, including the scaffolding protein and transcriptional corepressor Sin3 (3–6). These complexes are found in a broad range of plant and animal species, although the two Rpd3/Sin3 complexes are particularly widespread, also being found in yeast. The two Rpd3/Sin3 complexes differ in size and subunit composition and have distinct transcriptional roles. The smaller ∼0.6-MDa Rpd3S/Sin3S complex is targeted to the intragenic regions of actively transcribed genes to mitigate RNA polymerase progression and suppress aberrant transcription initiation (7–12). In contrast, the larger 1.2–2-MDa Rpd3L/Sin3L complex is recruited to promoter regions to effect transcriptional repression of a wide variety of genes involved in cell cycle control, differentiation, DNA replication and repair, mitochondrial metabolism, and apoptosis (13–20). Consistent with the broad impact of these Rpd3/Sin3 complexes on cellular physiology, sin3 or hdac1/hdac2 knockouts in mammals have been shown to lead to embryonic lethality or developmental defects (14, 15, 21–25).
Besides the paralogous proteins, including HDAC1/HDAC2, mSin3A/mSin3B, and the histone-interacting RbAp46/RbAp48 proteins, the mammalian Rpd3L/Sin3L complex comprises at least five other subunits, including SAP30, Sds3, SAP180/RBP1, SAP130, and ING1b/ING2, whose precise roles at the molecular level are poorly understood but most likely involve targeting the complex to specific genomic loci via one or more interaction surfaces (16–20, 26). The SAP30 subunit was among the first subunits of the complex to be identified and biochemically characterized (17, 20) and has since then been consistently detected in fractionation experiments involving this complex (13, 16, 26–28). The constitutive association of the SAP30 subunit with the Rpd3L/Sin3L alludes to a key role for the protein in stabilizing the complex, as has been suggested by sap30 genetic knockouts in yeast that yield similar effects as those involving sin3 and rpd3 (20, 29–31). SAP30 also appears to play a role in complex assembly because the protein has been proposed to interact with the ING1b/ING2 and SAP180/RBP1 subunits of the complex (26, 32–34).
At the molecular level, SAP30 harbors a poorly conserved N-terminal region of low sequence complexity, a highly conserved central region with a zinc finger motif found in organisms ranging from flies to humans, and a C-terminal Sin3 interaction domain (SID) that is well conserved from yeast to humans. Belying its small size, SAP30 appears to be targeted by a remarkably large and diverse array of cellular transcription factors and viral proteins, including CIR (CBF1-interacting factor of the Notch pathway (35)), yin yang 1 (YY1 (36)), human papillomavirus-binding factor (37), non-structural NSs protein of the Rift Valley fever virus (38), and the HTRP protein of the herpes simplex virus type 1 (HSV-1 (39)). Aside from the novel zinc finger motif that has been implicated in nonspecific interactions with nucleic acids (40, 41), little is known structurally regarding how SAP30 interacted with its targets.
Here, we structurally and functionally characterize the interaction between the SAP30 SID and the mSin3A PAH3 domains. Structures of Sin3 PAH1 and PAH2 domains in complex with diverse targets have been described, providing insights into how these four-helix bundles bind to relatively short sequence motifs embedded within isolated helical segments of the SIDs (42–48). The Sin3 PAH3 domains share relatively low levels of sequence identity with the PAH1 and PAH2 domains (∼25 and ∼16%, respectively). Here we show that the mSin3A PAH3-SAP30 SID complex shares several underlying themes with previously characterized PAH-SID complexes, but there are additional structural and functional elaborations on these themes that could not have been predicted solely from sequence analysis.
EXPERIMENTAL PROCEDURES
Production of SAP30 SID and mSin3A PAH3
The coding sequences of mouse SAP30 SID (residues 130–220) and mammalian Sin3A PAH3 (residues 457–528) were amplified by PCR and inserted into the pMCSG23 and pMCSG7 expression vectors, for expression as His6-maltose-binding protein (MBP) and His6-tagged fusion proteins, respectively. The cloned gene segments were confirmed by DNA sequencing. Escherichia coli BL21(DE3) cells (Novagen) co-transformed with both plasmids were grown at 37 °C in LB broth (EMD Chemicals). The growth temperature was shifted to 20 °C when the A600 nm reached about 0.6–0.8. Protein expression was induced using 1 mm isopropyl β-d-thiogalactopyranoside, and the cells were harvested 16–18 h thereafter. Cell pellets were suspended in 50 mm Tris-HCl buffer (pH 8.0) containing 0.15 m NaCl, 2 mm tris(2-carboxyethyl)phosphine hydrochloride, 1 mm phenylmethylsulfonyl (PMSF), 1 μm leupeptin, 1 mm pepstatin, and 0.1% Triton X-100 and lysed via sonication followed by DNase I treatment for 15 min at room temperature and centrifugation at 12,000 rpm for 30 min. The supernatant was incubated with His-Select Ni2+-resin (Sigma-Aldrich) for 40 min, followed by extensive washing with 50 mm Tris buffer (pH 8) containing 0.15 m NaCl and 2 mm tris(2-carboxyethyl)phosphine hydrochloride and subsequent incubation with tobacco etch virus protease at room temperature for 4 h and then at 4 °C overnight. The cleaved proteins were collected and purified to homogeneity via reversed-phase HPLC using a C18 or C8 column (Grace Vydac) and a linear gradient of 0.1% trifluoroacetic acid (TFA) and 0.1% TFA in 80% acetonitrile. The mSin3A PAH3 and SAP30 SID polypeptides eluted with distinct retention times, and fractions containing each protein were pooled and lyophilized. Uniformly 15N- and/or 13C-labeled proteins were produced using the same procedure except that the cells were grown in M9 minimal medium containing 15N-labeled ammonium sulfate and/or 13C-labeled d-glucose (Cambridge Isotopes), respectively. The identity and integrity of the proteins and the extent of isotope enrichment were established by electrospray ionization mass spectrometry and SDS-PAGE.
SAP30 SID mutants V148E, F186E, F200E, V148E/F186E, V148E/F200E, F186E/F200E, and V148E/F186E/F200E for in vitro binding assays were generated using the QuikChange site-directed mutagenesis protocol (Agilent). All mutations were confirmed by DNA sequencing. Wild-type and mutant SAP30 SID proteins for these assays were expressed as His6-MBP fusion proteins and purified in an analogous manner to when they were co-expressed with mSin3A PAH3, except the proteins were neither cleaved by tobacco etch virus protease nor subjected to HPLC.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) experiments were performed on a BIAcore 3000 instrument (Biacore) at 25 °C in HBS-P buffer (0.01 m HEPES (pH 7.4), 0.15 m NaCl, 0.005% (v/v) surfactant P20) at a flow rate of 25 μl/min. The mSin3A PAH3 polypeptide was immobilized on a CM5 sensor chip, whereas the wild-type and mutant forms of the SAP30 SID were injected at different concentrations. Protein-protein association was monitored for 200 s, whereas the protein-protein dissociation was monitored for 450 and 250 s for the wild-type and the mutant proteins, respectively. The chip was regenerated using 0.01 n sodium hydroxide (NaOH) for 10 s at a flow rate of 100 μl/min between runs.
SAP30 SID-mSin3A PAH3 Complex Generation and NMR Sample Preparation
The complex was generated by dissolving 15N,13C-labeled SAP30 SID or mSin3A PAH3A and an equivalent amount of the unlabeled mSin3A PAH3A or SAP30 SID in 20 mm sodium phosphate buffer (pH 6.0) containing 2 mm DTT and 8 m urea, followed by dialysis against the same buffer but without any urea using a Spectrapor DispoDialyzer dialysis unit with a 1000 Da cut-off (Spectrum Laboratories). The sample was exchanged into NMR buffer containing 20 mm sodium phosphate (pH 6.5), 2 mm DTT-d10, and 0.2% (w/v) NaN3 using a YM-3 Centricon unit with a 3000 Da cut-off (Millipore). Protein concentrations were determined spectrophotometrically. Samples of the protein-protein complex were lyophilized and redissolved in 99.996% D2O for experiments performed in D2O (Sigma-Aldrich).
NMR Titrations
A 25-mer single-stranded DNA oligonucleotide with the sequence 5′-GCGTTACGGCTTTTTGCCGTAACGC-3′ was purchased in lyophilized form from Integrated DNA Technologies (Coralville, IA). The DNA was used for the titrations without any additional purification. The DNA was dissolved in 20 mm sodium phosphate buffer (pH 6.5) containing 2 mm dithiothreitol-d10, 10% D2O, and 0.2% (w/v) NaN3. The DNA duplex in hairpin conformation was obtained by heating the sample to 95 °C for 15 min and rapidly cooling it on ice. Hairpin formation was confirmed by one-dimensional 1H NMR; the spectrum revealed no evidence of sample heterogeneity, implying a reasonably pure sample preparation. The complex of 15N-labeled SAP30 SID and unlabeled mSin3A PAH3 was generated as described above; complex formation was confirmed by recording a two-dimensional 1H-15N HSQC spectrum. The protein-DNA complex was generated by slowly titrating the 15N-labeled SAP30 SID-mSin3A PAH3 complex with the hairpin duplex. Titrations were performed at 0.05 mm protein concentration and terminated when five equivalents of DNA had been added.
Transcription Repression Assays
SAP30 cDNA encoding various mutations was cloned into the pCMV-BD plasmid (Agilent) to create GAL4-DBD fusions. Luciferase reporter under the control of the 14D promoter was a kind gift from D. Ayer (University of Utah). LacZ in pcDNA 3.1 plasmid (Invitrogen) was used as a transfection control in repression experiments. Repression assays were performed as described previously by Viiri et al. (49). Two independent experiments with three replicates were performed showing comparable results. Human embryonal kidney epithelial cells (HEK 293T) were cultured in DMEM (Invitrogen) supplemented with penicillin/streptomycin antibiotics, 5% fetal bovine serum, 1 mm sodium pyruvate, and 50 μg/ml uridine. Cells were plated to a density of 20,000 cells/cm2 24 h prior to transfection. HEK293T cells were transfected with 250 ng of pCMV-BD, 200 ng of the 14D luciferase reporter, and 50 ng of pcDNA-LacZ using Fugene 6 or Fugene HD reagents (Roche Applied Science) as instructed by the manufacturer.
NMR Spectroscopy and Structure Determination
All NMR data were acquired on a Varian Inova 600-MHz spectrometer equipped with a pulsed field gradient triple resonance cold probe at 35 °C. NMR data processing and analysis were performed using Felix 98.0 (Accelrys) and Sparky. Sample concentrations used for the structure determination were ∼1 mm. Backbone and side chain 1H, 15N, and 13C resonance assignments for each of the interacting proteins in the SAP30 SID-mSin3A PAH3 complex were obtained by analyzing three-dimensional HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, C(CO)NH-TOCSY, HNCO, HCACO, HCCH-COSY, and HCCH-TOCSY spectra (50, 51). Aromatic resonances were assigned based on a careful analysis of two-dimensional 1H-13C aromatic HSQC, three-dimensional 13C-edited NOESY, and 15N,13C-double half-filtered NOESY spectra (52).
For structure determination, backbone ϕ and ψ torsion angle restraints were derived from analysis of 1Hα, 13Cα, 13Cβ, 13C′, and backbone 15N chemical shifts using TALOS+ (53). Restraints were imposed only for those residues with TALOS+ reliability scores of 10. NOE-based distance restraints for each protein in the complex were derived from three-dimensional 15N-edited NOESY (mixing time (τm) = 75 ms) and three-dimensional 15N,13C-filtered, 15N,13C-edited NOESY (τm = 120 ms (54)) spectra recorded in H2O, three-dimensional 13C-edited aliphatic NOESY (τm = 60 ms), and two-dimensional 15N,13C-double half-filtered NOESY (τm = 60 ms (52)) spectra recorded in D2O.
Structures were determined using ARIA 1.2 in conjunction with CNS 1.1 starting from an initial structure with extended backbone conformations (55–57). All NOEs were calibrated automatically and assigned iteratively by ARIA; the assignments were checked manually for errors after every run. Eighty conformers were calculated in the final iteration, of which the 40 conformers with the lowest restraint energies were refined in a shell of explicit water, and the 20 conformers with the lowest restraint energies, restraint violations, and r.m.s. deviations from the ideal covalent geometry were selected for further analysis. The final conformers were analyzed using CNS (57), PROCHECK (58), MONSTER (59), DALI (60), and DeepView (61). Molecular images were generated using CHIMERA (62).
RESULTS
Only the mSin3A PAH3-SAP30 SID Complex, Not the Isolated Domains, Is Amenable to Structural Studies
Previous studies localized the SAP30-mSin3A interaction to a 91-residue SID at the C terminus of SAP30 and the 74-residue PAH3 domain of mSin3A. Although SAP30 SID could be readily expressed in E. coli, the mSin3A PAH3 domain could not be stably expressed in this host unless when co-expressed with the SAP30 SID. Co-purification and subsequent dissociation of the complex was straightforward, but the individual polypeptides either formed soluble aggregates (SAP30 SID) or were not sufficiently soluble (mSin3A PAH3) at near physiological pH, precluding NMR studies. On the other hand, the mSin3A PAH3-SAP30 SID complex was readily soluble and yielded high quality NMR spectra for the individual proteins to facilitate resonance assignments using standard double and triple resonance approaches and structure determination using traditional approaches.
Both mSin3A PAH3 and SAP30 SID Exhibit Long Range Interactions Characteristic of Globular Domains in the Complex
The solution NMR structure of the mSin3A PAH3-SAP30 SID complex was determined using 4079 NOE-based distance restraints, including 617 intermolecular distance restraints, and 168 chemical shift-based torsion angle restraints (Table 1). The final ensemble of 20 conformers exhibited reasonable precision and good covalent geometry and was in good agreement with NMR data, including no distance violations of >0.5 Å or torsion angle violations of >5° (Fig. 1A and Table 1). The vast majority of the residues of both mSin3A PAH3 and SAP30 SID adopt well defined conformations, although the four N-terminal and two C-terminal residues of mSin3A PAH3 as well as the 13 N-terminal and 11 C-terminal residues of SAP30 SID are disordered in the complex.
TABLE 1.
NMR structure determination statistics for mSin3A PAH3-SAP30 SID complex
| Parameters | Values |
|---|---|
| Restraint statistics | |
| NOE-based distance restraintsa | 4079 |
| Unambiguous NOE-based restraints | 3640 |
| Intraresidue | 1558 |
| Sequential (|i − j| = 1) | 700 |
| Medium range (1 < |i − j| ≤ 4) | 555 |
| Intramolecular long range (|i − j| > 4) | 210 |
| Intermolecular restraints | 617 |
| Ambiguous NOE-based restraints | 439 |
| Hydrogen bonding distance restraints | 106 |
| Torsion angle restraints | 168 (84 φ, 84 ψ) |
| Structure quality of NMR ensemble | |
| Restraint satisfaction | |
| r.m.s. differences for distances (Å) | 0.015 ± 0.001 |
| r.m.s. differences for torsion angles (degrees) | 0.175 ± 0.078 |
| Deviations from ideal covalent geometry | |
| Bond lengths (Å) | 0.003 ± 0.000 |
| Bond angles (degrees) | 0.433 ± 0.010 |
| Impropers (degrees) | 1.077 ± 0.050 |
| Ramachandran plot statistics (%) | |
| Residues in most favored regions | 84 (90.8)b |
| Residues in allowed regions | 14.6 (8.4)b |
| Residues in disallowed regions | 1.4 (0.8)b |
| Average atomic r.m.s. deviations from average structure (Å) | |
| All atoms | 5.54 |
| All atoms in ordered regionsb | 1.26 |
| Backbone atoms (N, Cα, C′) | |
| All residues | 5.88 |
| All residues in ordered regionsb | 0.63 |
a All statistics for NOE restraints correspond to unique or non-redundant restraints.
b Ordered regions include residues 461–526 of mSin3A and residues 143–209 of SAP30.
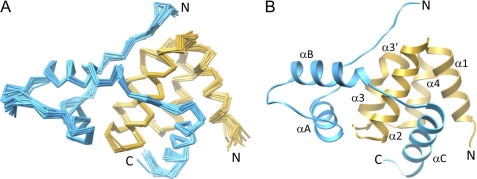
FIGURE 1.
Solution structure of the mSin3A PAH3-SAP30 SID complex. A, backbone Cα traces of the ensemble of 20 structures following a best fit superposition of the polypeptide segments deemed to be in ordered regions (residues 461–526 of mSin3A and residues 143–209 of SAP30). The disordered regions are not shown for clarity. B, a ribbon diagram of the representative structure of the ensemble. The mSin3A PAH3 and SAP30 SIDs are shown in gold and blue, respectively, in both A and B.
The mSin3A PAH3 domain comprises five helices, with the α1, α2, α3, α3′, and α4 helices spanning residues Thr463–Leu475, Ala478–Asn492, Arg498–Leu504, Ser506–Leu509, and Pro513–Leu523, respectively (Fig. 1B). Notwithstanding the premature termination of α3 caused by Pro507 that results in an additional short helix (α3′) immediately following α3, the overall fold of PAH3 is reminiscent of the left-handed four-helix bundle arrangement described for Sin3 PAH1 and PAH2 domains; indeed, the PAH3 domain exhibits best fit backbone r.m.s. deviations of 1.27–1.63 Å with these domains.
The core SAP30 SID comprises three helices, with the αA, αB, and αC helices spanning residues Val155–His164, Lys175–Phe186, and Glu193–Asn206, respectively (Fig. 1B). The αA and αB helices pack against each other and also partially against a segment that is largely in an extended conformation N-terminal to the αA helix forming a globular subdomain (Fig. 2A). In contrast, the αC helix makes no contacts with the rest of the SID.
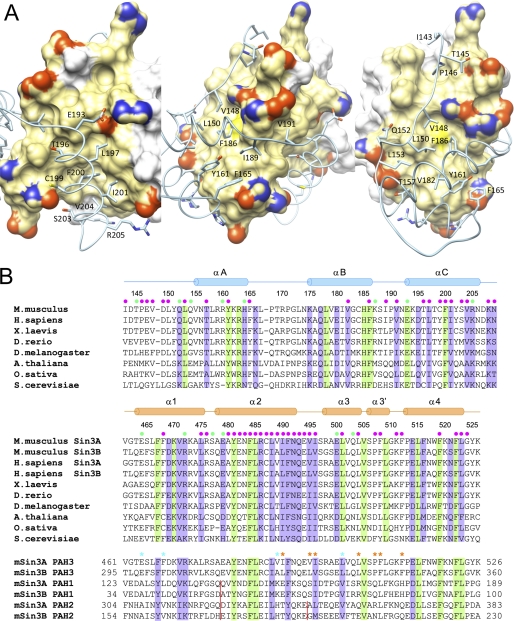
FIGURE 2.
Non-covalent interactions at the mSin3A PAH3-SAP30 SID interface and sequence conservation in the respective domains. A, three views of the protein-protein interface with the PAH3 domain rendered as a molecular surface and the SAP30 SID depicted in a worm-and-sticks representation. Only those SAP30 side chains involved in intermolecular interactions are shown, whereas mSin3A residues engaging in these interactions are colored in gold. Nitrogen, oxygen, and sulfur atoms of interacting residues in both proteins are colored blue, red, and yellow, respectively. B, ClustalW-guided multiple sequence alignments of SAP30 SID orthologs (top), the corresponding mSin3A PAH3 orthologs (middle), and the paralogous Sin3 PAH domains (bottom). Invariant and conserved residues are highlighted in green and purple, respectively. The filled circles in green and magenta denote residues engaging in intermolecular electrostatic/hydrogen bonding and hydrophobic interactions, respectively. Key PAH3 residues that lend hydrophobic character to the secondary surface, which is absent in the PAH1 and PAH2 domains, are denoted by asterisks (in orange) in the bottom panel. The presumed specificity-determining residues in the hydrophobic cleft are denoted by asterisks (in cyan). The red vertical lines represent sites of amino acid insertions in the respective PAH domains.
SAP30 SID Binds via a Tripartite Motif to Two Discrete Surfaces of the mSin3A PAH3 Domain
The mSin3A PAH3 domain features a prominent hydrophobic cleft analogous to those observed previously for the Sin3 PAH1 and PAH2 domains (Fig. 2A (42–48)). A secondary surface, also largely hydrophobic in character, defined by residues in the α2, α3, and α3′ helices as well as in the loops preceding, linking, or following these segments, is unique to the PAH3 domain. The SAP30 SID engages these two discrete surfaces of the PAH3 domain via a tripartite structural motif comprising the N-terminal extension, the globular subdomain, and the αC helix. The latter targets the cleft, whereas the N-terminal extension and the globular subdomain bind to the secondary surface (Fig. 2A). The linker segment connecting the globular subdomain with the αC helix wraps intimately around a hydrophobic segment of the α2 helix. The protein-protein interface in the mSin3A PAH3-SAP30 SID complex is extensive, with the solvent-excluded surface area averaging ∼1440 ± 70 Å2 in each of the 20 conformers of the NMR ensemble.
Non-covalent Contacts across the mSin3A PAH3-SAP30 SID Interface Are Dominated by Hydrophobic Interactions
The mSin3A PAH3-SAP30 SID complex appears to be stabilized largely by hydrophobic interactions, with Ile143, Thr145, Pro146, Val148, Leu150, and Leu153 in the N-terminal segment; Thr157, Tyr161, and Phe165 in the αA helix; and Val182 and Phe186 in the αB helix of SID engaging the secondary surface of PAH3 defined by Leu475, Ala480, Cys487, Ile490, Val495, Ile496, Leu504, Pro507, Phe508, Arg476, and Phe512, with the aliphatic portions of the side chains of Glu479, Glu482, Asn483, Arg486, Gln493, Glu494, Glu500, Gln503, and Lys511 located in the periphery bolstering the hydrophobic character of the surface (Fig. 2, A and B). The hydrophobic cleft of PAH3 is defined by Phe468, Tyr481, Phe484, Leu485, Leu488, Val489, Phe491, Leu501, Phe519, Phe522, and Leu523 and is further bolstered in its character by the aliphatic portions of Glu464, Asn492, and Arg498. The side chains of Glu193, Thr196, Leu197, Cys199, Phe200, Ile201, Ser203, Val204, Arg205, Lys208, and Asn209 of SAP30 SID participate in hydrophobic interactions with the cleft (Fig. 2, A and B). The side chains of Ile189 and Val191 in the loop segment connecting the αB and αC helices of SID also engage in hydrophobic interactions with Leu485, Arg486, Val489, and Ile490 of PAH3.
The aforementioned hydrophobic interactions are buttressed by ionic interactions involving the side chains of Arg160, His164, and Lys187 of SID with those of Glu500, Glu494, and Glu479 of PAH3, respectively; additionally, Glu193 and Arg205 form salt bridges with Arg472 and Glu464, respectively (Fig. 2, A and B). Furthermore, the side chains of Thr145, Gln152, Gln154, and Thr157 of SID engage in hydrogen bonding interactions with those of Ser477, Gln503, Glu500, and Glu500 of PAH3, respectively. Finally, at least two instances of positively charged/amino-aromatic interactions involving Gln493 and Arg486 of PAH3 with Phe165 and Phe186 of SID, respectively, were found that could lend additional stability to the complex.
Residue-specific sequence conservation patterns both within and across species for Sin3 PAH3 and SAP30 SID reveal a strong correlation between conservation of a particular residue and its involvement in stabilizing either the structure of the corresponding domain or that of the PAH3-SID complex (Fig. 2B). Rather surprisingly, the lowest level of sequence conservation is seen for the N-terminal extension of SAP30 SID. However, because this segment adopts a largely extended conformation (Fig. 2A), it is likely to be more malleable to sequence changes than if it were in a regular secondary structure. Predictably, the yeast proteins show the highest level of sequence divergence, although the plant proteins also exhibit high levels of divergence relative to the corresponding proteins from flies to humans, implying that the structures of the Sin3 PAH3-SAP30 SID complex in yeast and plants are likely to show some significant differences (Fig. 2B).
The mSin3A PAH3-SAP30 SID Interaction Is of High Affinity but Can Be Destabilized by Structure-guided Mutagenesis
To evaluate the role of various polypeptide segments in contributing to the stability of the mSin3A PAH3-SAP30 SID complex, we measured the binding affinities of SAP30 SID mutants in in vitro binding assays and compared the results with functional assays conducted in cells. SPR analysis of the mSin3A PAH3-SAP30 SID interaction using the wild-type proteins yielded an equilibrium dissociation constant of ∼9 nm, consistent with the large size of the protein-protein interface.
Given the extensive protein-protein interface and the multitude of interactions stabilizing the complex, we mutated three key residues of SAP30 SID, including Val148, Phe186, and Phe200, either singly or in combination to glutamic acid because all three residues are engaged predominantly in hydrophobic interactions, and replacement with a charged residue is predicted to disrupt these interactions (mSin3A PAH3 mutants could not be studied due to difficulties with protein expression as discussed above). The choice of these three specific sites was guided by our desire to evaluate the contributions of each of the three SAP30 SID structural motifs involved in recognizing mSin3A PAH3. Val148 is located in the N-terminal extension, whereas Phe186 and Phe200 are located in the globular subdomain and the αC helix, respectively (Fig. 2A). Furthermore, all three residues are either invariant or conserved in orthologous proteins (Fig. 2B; Val148 is replaced by aromatic residues in some orthologs).
The in vitro mSin3A PAH3 binding activity of each of the SAP30 SID mutants was analyzed by SPR measurements. Association rates as well as complex dissociation rates were measured at multiple concentrations; the equilibrium dissociation constants were inferred from global fits of these rates. The V148E mutant showed a ∼50-fold reduction in binding affinity relative to the wild-type protein, whereas the F186E and F200E mutants exhibited ∼100- and 200-fold reductions in affinity, respectively (Table 2 and supplemental Fig. S1). Each of the three structural motifs involved in mSin3A PAH3 binding thus appears to make significant contributions to the overall affinity. Consistent with this argument, PAH3 binding activity was severely diminished for the double and triple mutants, with the upper limit for the binding affinity estimated to be in the 50 μm range.
TABLE 2.
Kinetic and thermodynamic parameters for mSin3A PAH3-SAP30 SID complexes measured by SPR
| Analytea | Association rate, kab | Dissociation rate (kd)b | Dissociation constant (KD)b |
|---|---|---|---|
| m−1s−1 | s−1 | nm | |
| Wild-type SAP30 SID | 1.07 ± 0.01 × 105 | 9.82 ± 0.33 × 10−4 | 9.2 ± 0.4 × 10−9 |
| SAP30 SID V148E | 2.20 ± 0.04 × 104 | 1.10 ± 0.14 × 10−4 | 5.0 ± 0.2 × 10−7 |
| SAP30 SID F186E | 5.08 ± 0.16 × 103 | 55.4 ± 2.32 × 10−4 | 1.1 ± 0.1 × 10−6 |
| SAP30 SID F200E | 1.49 ± 0.05 × 103 | 31.6 ± 3.00 × 10−4 | 2.1 ± 0.3 × 10−6 |
| SAP30 SID V148E/F186E | NDc | ND | ND |
| SAP30 SID V148E/F200E | ND | ND | ND |
| SAP30 SID V186E/F200E | ND | ND | ND |
| SAP30 SID V148E/F186E/F200E | ND | ND | ND |
a mSin3A PAH3 was the ligand immobilized on the dextran surface in these experiments.
b Uncertainties are from the non-linear least-squares fitting.
c ND, no detectable binding observed at 50 μm analyte concentration.
SAP30 Functional Assays in Cells Qualitatively Mimic the Trends Noted in in Vitro Binding Assays
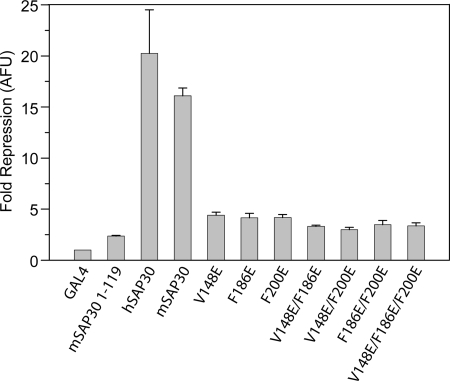
The ability of SAP30 to effect repression of a reporter gene when fused to a heterologous DNA-binding domain has been described previously (17). This repression was shown to be dependent on the interaction with mSin3A because deletion of the C-terminal 91 residues of SAP30 (harboring the SID) abolished both mSin3A binding and the repression activity. Therefore, we analyzed the effect of SAP30 mutations on its transrepression activity by fusing the protein with the Gal4 DNA-binding domain and using a luciferase reporter assay in HEK293 cells. Human as well as mouse full-length SAP30 proteins and a mouse SAP30 mutant lacking the C-terminal 91 residues were used as positive and negative controls, respectively. As expected, both wild-type human and mouse SAP30 potently repressed transcription of the reporter, whereas the C-terminally deleted mutant failed to effect repression (Fig. 3). All three single mutants, including V148E, F186E, and F200E, exhibited diminished repression activity, with ∼4-fold reduction in activity compared with wild-type protein. The double and triple mutants were additionally compromised in their transrepression ability (Fig. 3), although the repression in these cases was not synergistic, which might be a consequence of the limited dynamic range of this assay. Collectively, however, the results from the repression assays implicate all three residues, including Val148, Phe186, and Phe200, in critical roles in Sin3 recruitment with attendant effects on transrepression activity.
FIGURE 3.
Functional analysis of SAP30 mutants analyzed by transcription repression of a 14D luciferase reporter. HEK293 cells were transfected with Gal4DBD fusion plasmids along with the luciferase reporter and pcDNA 3.1.-LacZ control plasmids. The bars in the graph represent the average -fold repression of luciferase activity compared with Gal4DBD alone, whereas the error bars indicate the range of measured values. Results are normalized to β-galactosidase activity from the co-transfected LacZ plasmid. AFU, arbitrary fluorescence units.
SAP30 SID Shares Structural Homology with the SAP Motif, a DNA-binding Motif
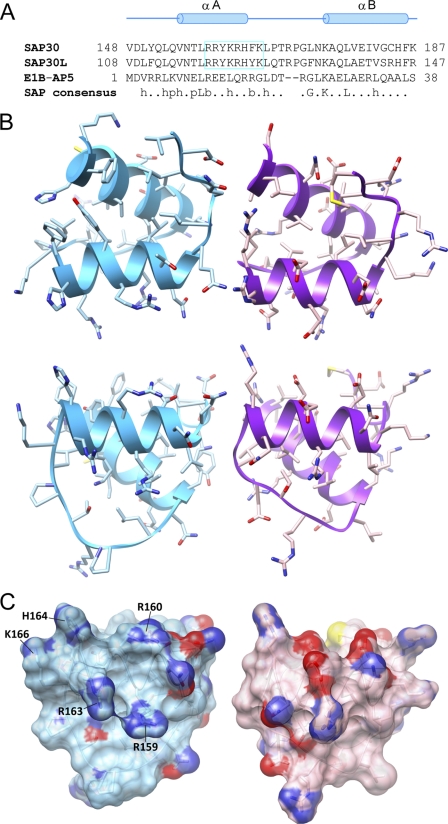
A DALI-based search for potential SAP30 SID structural homologues revealed that the domain shared homology with the SAP (SAF, acinus, PIAS) motif, also known as the SAF-box, which has been implicated in binding nucleic acids (63, 64). A backbone Cα superposition of the SAP domain of E1B-55 kDa-associated protein 5 (E1B-AP5) with the SAP30 SID yields a r.m.s. deviation of 0.87 Å for the segment spanning Asp149–Lys187 of SAP30, encompassing the αA and αB helices (Fig. 4). Although the SAP30 sequence by and large follows the consensus for the SAP motif (Fig. 4A), their relationship at the sequence level could not be established via Pfam and SMART data base searches, in part because of the shortness of the motif and the default settings for E-value cut-offs. We note that the character of the molecular surfaces of the SAP30 SID and E1B-AP5 domains are quite distinct (Fig. 4, B and C), consistent with the two domains sharing only 32% sequence identity and 45% sequence similarity over a 38-residue segment.
FIGURE 4.
The SAP30 SID is structurally homologous to SAP domains. A, a structure-guided sequence alignment of the two domains along with the consensus sequence for the SAP domains. The consensus sequence follows the convention described in Ref. 63. h, hydrophobic (ACFILMVWY); p, polar (DEHKNQRST); b, bulky (EFIKLMQRWY). SAP30 and SAP30L residues comprising the nucleolar localization sequence are boxed. B, orthogonal views of the structurally homologous regions in the SID (left) and a representative SAP domain (right; Protein Data Bank code 1ZRJ) shown in a ribbon-and-stick representation. The view in the top panel is from the perspective of the secondary hydrophobic surface in the PAH3 domain. C, molecular surface representations of the views in the bottom panel of B. Residues comprising the nucleolar localization sequence are identified.
The Nucleolar Localization Sequence of SAP30 SID Can Mediate Nucleic Acid Binding
SAP30 and SAP30L (SAP30-like protein) have previously been shown to localize to the nucleolus, thereby playing a vital role in targeting the Sin3L complex to this organelle. Nucleolar targeting relies on an intact nucleolar localization signal (NoLS) in both SAP30 and SAP30L that maps to the αA helix and a portion of the succeeding loop in the three-dimensional structure (Fig. 4A). A striking feature of this sequence is its basic character, which is propagated into the molecular surface (Fig. 4, A and C).
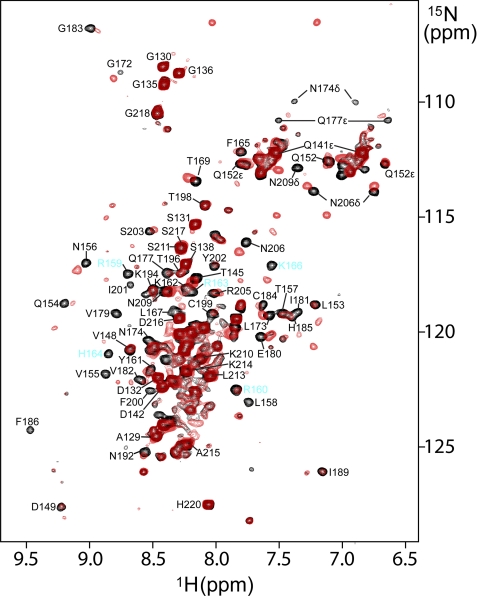
The molecular target of the NoLS is presently unknown. Given the function of the NoLS, the basic character of this sequence, and the structural relationship of the SAP30 SID with the nucleic acid-binding SAP motif, we asked whether SAP30 SID could interact with nucleic acids. To this end, we designed a DNA hairpin duplex as a potential mimic of the natural target of the NoLS and performed NMR titrations with 15N-labeled SAP30 SID prebound to mSin3A PAH3 (an RNA stem-loop structure has been implicated in targeting the nucleolar remodeling complex to the nucleolus (65)). The hairpin duplex induced strong but selective perturbations characterized by shifting and broadening of resonances in the NMR spectra of SAP30 SID (Fig. 5), implicating a role for this domain in nucleic acid binding. Some of the strongest perturbations are observed for the NoLS residues, including Arg159, His164, and Lys166, but similar perturbations are also observed for those in the vicinity at the level of both primary and tertiary structures, including Gln154, Val155, Val179, Gly183, and the side chains of Asn174 and Gln177. In contrast, perturbations are minimal for residues in other regions of the SID, including Asp142, Val148, Thr196, and Thr198. These results thus suggest that the NoLS residues may have a direct role in the recognition of a putative nucleic acid target.
FIGURE 5.
A role for SAP30 SID in nucleic acid binding. Shown is an overlay of the 1H-15N correlated spectra of 15N-labeled SAP30 SID in complex with mSin3A PAH3 recorded in the absence (black) and in the presence of 1.5 equivalents of a 25-mer hairpin DNA duplex (red). NMR data were acquired under identical solution and ambient conditions and processed and displayed with identical parameters. The contour threshold for the experiment conducted in the presence of DNA was adjusted according to the extent of dilution of the sample caused by DNA addition.
DISCUSSION
Unlike the Sin3 PAH1 and PAH2 domains that are broadly available for direct interactions with sequence-specific DNA-binding transcription factors for targeting of the Rpd3L/Sin3L complex to specific gene promoters, the PAH3 domain plays an important role in Rpd3L/Sin3L complex assembly by constitutively associating with the SAP30 subunit. Notwithstanding the extensive structural studies of the Sin3 PAH1 and PAH2 domains in complex with diverse targets (42–48), the remote similarity between these domains and PAH3 precluded reliable predictions of how this domain might interact with SAP30.
The structure of the mSin3A PAH3-SAP30 SID complex reveals significant variations in the themes previously uncovered for PAH domains. For example, the α1 helix is shorter at the N terminus by approximately one to two turns compared with the PAH1 and PAH2 domains, whereas the α3 helix is kinked, formally dividing the helix into two (α3 and α3′). The former along with the drastically bulkier amino acids in the cleft, including Glu464, Phe468, and Leu501, in PAH3 has the effect of dramatically altering the character of the cleft, making it considerably less cavernous than in the other PAH domains (Fig. 2, A and B). This probably explains the contrasting sequence motif (i.e. (D/E)XXφφXXφφXXφ(R/K), where φ represents a hydrophobic residue) preferred by the PAH3 domain and also why the SID mutations that preserve an intact αC helix are still unable to bind PAH3 efficiently (i.e. the single helical motif that was sufficient for high affinity interactions with PAH1 and PAH2 is necessary but not sufficient for high affinity interactions with PAH3).
Perhaps the most striking difference between the PAH3 and the PAH1/2 domains is the involvement of a secondary surface for SID binding. The kinking of the α3 helix and the particular amino acid composition in the vicinity has the effect of creating a narrow hydrophobic groove that is absent in the other PAH domains (Fig. 2, A and B). Additional hydrophobic residues in the α2 and α3 helices, either absent or not in the same structural context in the PAH1/2 domains (Fig. 2B), define a shallow surface that is recognized by the bipartite structural motif of the SIDs that is unique to this complex.
The mSin3A PAH3-SAP30 SID is the highest affinity PAH-SID interaction that has been structurally and functionally characterized thus far. The high affinity of the interaction, which results from the cooperative recognition of the two discrete mSin3A PAH3 surfaces by the SAP30 SID tripartite motif, provides the basis for the constitutive association of this subunit with Sin3 and by extension the Rpd3L/Sin3L complex.
Intriguingly and belying its simple fold, the globular subdomain of SAP30 SID shows striking structural homology with the SAP motif. The SAP motif was originally discovered in the scaffold attachment factor SAF-A (66), which is involved in attaching chromosomal DNA to the nuclear matrix, but has since been predicted to occur in a number of other proteins, including those involved in DNA repair, RNA splicing, and apoptotic degradation of chromatin (63, 64). Structurally, the SAP motifs, like the globular subdomain of SAP30 SID, harbor two helices separated by a short loop, although isolated SAP motifs as well as those that are part of larger domains comprising four or five helices have been described (67, 68). Although structures of SAP motifs bound to nucleic acids have not yet been described, the N-terminal portions of the two SAP helices have been suggested to bind nucleic acids by NMR titration experiments (67, 68). More rigorous NMR analyses are required to map the precise location of the nucleic acid-binding site on the SAP30 SID when its cognate target is identified. However, the pattern of chemical shift perturbations induced by the surrogate molecule employed in this study suggests that the binding site might not be restricted only to the N termini of the αA and αB helices seen for the SAP motifs.
Given their structural and, to some extent, functional similarities, it would thus appear that the SAP30 SID and the SAP motifs might have shared a common evolutionary history. This is indeed confirmed by a recent phylogenetic analysis of the SAP30 family that also deduced the loss of one of its original functions involving association with the nuclear matrix (69). Our results would suggest that the latter function is somewhat modified to recognize nucleic acids perhaps of a different type and through a different mechanism.
The recent discovery that the SAP30 proteins are involved in the nucleolar targeting of mSin3A (and by extension the Sin3L complex) is exciting (49) because mSin3A, RbAp46, and HDAC1, all members of the mammalian Sin3L complex, have been shown biochemically to be recruited by the nucleolar remodeling complex for the repression of ribosomal gene transcription (70). Furthermore, the repression was dependent on HDAC activity because HDAC inhibitors relieved the repression. The involvement of the mammalian Sin3L complex in the repression of ribosomal gene transcription contrasts with the role of the Rpd3L/Sin3L complex in yeast, where genetic deletions of members of this complex have been implicated in enhanced ribosomal silencing (30). Interestingly, the NoLS is present only in higher organisms from flies to humans in animals (Figs. 2B and 4A), suggesting that this might have been a recently acquired function. Rather strikingly, the SAP30 proteins in the same organisms harbor a novel zinc finger motif N-terminal to the SID; the motif is notably absent in yeast and plants (40, 41). The discovery that the SAP30 zinc finger can also bind to nucleic acids raises the intriguing possibility that the SID and the zinc finger motif might cooperate in the recognition of the same target that promotes their nucleolar localization (40, 41). Additional studies are required to test this exciting possibility.
Supplementary Material
Acknowledgments
We thank Don Ayer for the luciferase reporter used in this study. We thank Philip Page at Reichert Life Sciences and Elena Solomaha at the University of Chicago for useful discussions and advice relating to SPR experiments. We gratefully acknowledge access to resources in the Weinberg College of Arts & Sciences Biological NMR Center and support for structural biology research from the Robert H. Lurie Comprehensive Cancer Center at Northwestern.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM064715 (to I. R.). This work was also supported by the Foundation for Pediatric Research in Finland, the Finnish Medical Foundation, the Competitive Research Funding of Tampere University Hospital (Grant 9K073), and the Nona and Kullervo Väre Foundation (to O. L.) and by Grant P30 CA060553 from the Robert H. Lurie Comprehensive Cancer Center at Northwestern University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 2ld7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The BioMagResBank (BMRB) code for NMR chemical shifts is 17653.
- HDAC
- histone deacetylase
- NOE
- nuclear Overhauser effect
- NOESY
- NOE spectroscopy
- NoLS
- nucleolar localization signal
- PAH
- paired amphipathic helix
- SAF
- scaffold attachment factor
- SID
- Sin3 interaction domain
- SPR
- surface plasmon resonance
- r.m.s.
- root mean square.
REFERENCES
- 1. Gardner K. E., Allis C. D., Strahl B. D. (2011) J. Mol. Biol. 409, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 3. Hayakawa T., Nakayama J. (2011) J. Biomed. Biotechnol. 2011, 129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cunliffe V. T. (2008) Curr. Opin. Genet. Dev. 18, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grzenda A., Lomberk G., Zhang J. S., Urrutia R. (2009) Biochim. Biophys. Acta 1789, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silverstein R. A., Ekwall K. (2005) Curr. Genet. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 7. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 8. Jelinic P., Pellegrino J., David G. (2011) Mol. Cell Biol. 31, 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joshi A. A., Struhl K. (2005) Mol. Cell 20, 971–978 [DOI] [PubMed] [Google Scholar]
- 10. Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J. L. (2007) Science 316, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 11. Li B., Jackson J., Simon M. D., Fleharty B., Gogol M., Seidel C., Workman J. L., Shilatifard A. (2009) J. Biol. Chem. 284, 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicolas E., Yamada T., Cam H. P., Fitzgerald P. C., Kobayashi R., Grewal S. I. (2007) Nat. Struct. Mol. Biol. 14, 372–380 [DOI] [PubMed] [Google Scholar]
- 13. Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Biochim. Biophys. Acta 1731, 77–87; Discussion 75–76 [DOI] [PubMed] [Google Scholar]
- 14. Cowley S. M., Iritani B. M., Mendrysa S. M., Xu T., Cheng P. F., Yada J., Liggitt H. D., Eisenman R. N. (2005) Mol. Cell Biol. 25, 6990–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dannenberg J. H., David G., Zhong S., van der Torre J., Wong W. H., Depinho R. A. (2005) Genes Dev. 19, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fleischer T. C., Yun U. J., Ayer D. E. (2003) Mol. Cell Biol. 23, 3456–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Laherty C. D., Billin A. N., Lavinsky R. M., Yochum G. S., Bush A. C., Sun J. M., Mullen T. M., Davie J. R., Rose D. W., Glass C. K., Rosenfeld M. G., Ayer D. E., Eisenman R. N. (1998) Mol. Cell 2, 33–42 [DOI] [PubMed] [Google Scholar]
- 18. Laherty C. D., Yang W. M., Sun J. M., Davie J. R., Seto E., Eisenman R. N. (1997) Cell 89, 349–356 [DOI] [PubMed] [Google Scholar]
- 19. Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. (1997) Cell 89, 357–364 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y., Sun Z. W., Iratni R., Erdjument-Bromage H., Tempst P., Hampsey M., Reinberg D. (1998) Mol. Cell 1, 1021–1031 [DOI] [PubMed] [Google Scholar]
- 21. Pennetta G., Pauli D. (1998) Dev. Genes Evol. 208, 531–536 [DOI] [PubMed] [Google Scholar]
- 22. LeBoeuf M., Terrell A., Trivedi S., Sinha S., Epstein J. A., Olson E. N., Morrisey E. E., Millar S. E. (2010) Dev. Cell 19, 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilting R. H., Yanover E., Heideman M. R., Jacobs H., Horner J., van der Torre J., DePinho R. A., Dannenberg J. H. (2010) EMBO J. 29, 2586–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dovey O. M., Foster C. T., Cowley S. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8242–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montgomery R. L., Davis C. A., Potthoff M. J., Haberland M., Fielitz J., Qi X., Hill J. A., Richardson J. A., Olson E. N. (2007) Genes Dev. 21, 1790–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuzmichev A., Zhang Y., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Mol. Cell Biol. 22, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doyon Y., Cayrou C., Ullah M., Landry A. J., Côté V., Selleck W., Lane W. S., Tan S., Yang X. J., Côté J. (2006) Mol. Cell 21, 51–64 [DOI] [PubMed] [Google Scholar]
- 28. Sif S., Saurin A. J., Imbalzano A. N., Kingston R. E. (2001) Genes Dev. 15, 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bernstein B. E., Tong J. K., Schreiber S. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13708–13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Z. W., Hampsey M. (1999) Genetics 152, 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loewith R., Smith J. S., Meijer M., Williams T. J., Bachman N., Boeke J. D., Young D. (2001) J. Biol. Chem. 276, 24068–24074 [DOI] [PubMed] [Google Scholar]
- 32. Binda O., Roy J. S., Branton P. E. (2006) Mol. Cell Biol. 26, 1917–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai A., Kennedy B. K., Barbie D. A., Bertos N. R., Yang X. J., Theberge M. C., Tsai S. C., Seto E., Zhang Y., Kuzmichev A., Lane W. S., Reinberg D., Harlow E., Branton P. E. (2001) Mol. Cell Biol. 21, 2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsieh J. J., Zhou S., Chen L., Young D. B., Hayward S. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang N. E., Lin C. H., Lin Y. S., Yu W. C. (2003) Biochem. Biophys. Res. Commun. 306, 267–275 [DOI] [PubMed] [Google Scholar]
- 37. Sichtig N., Körfer N., Steger G. (2007) Arch. Biochem. Biophys. 467, 67–75 [DOI] [PubMed] [Google Scholar]
- 38. Le May N., Mansuroglu Z., Léger P., Josse T., Blot G., Billecocq A., Flick R., Jacob Y., Bonnefoy E., Bouloy M. (2008) PLoS Pathog. 4, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen J., Li Y. M., Li J. F., Liu L. D., Liao Y., Na R. X., Wang J. J., Wang L. C., Li Q. H. (2010) Virol. Sin. 25, 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Viiri K. M., Jänis J., Siggers T., Heinonen T. Y., Valjakka J., Bulyk M. L., Mäki M., Lohi O. (2009) Mol. Cell Biol. 29, 342–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He Y., Imhoff R., Sahu A., Radhakrishnan I. (2009) Nucleic Acids Res. 37, 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brubaker K., Cowley S. M., Huang K., Loo L., Yochum G. S., Ayer D. E., Eisenman R. N., Radhakrishnan I. (2000) Cell 103, 655–665 [DOI] [PubMed] [Google Scholar]
- 43. Spronk C. A., Tessari M., Kaan A. M., Jansen J. F., Vermeulen M., Stunnenberg H. G., Vuister G. W. (2000) Nat. Struct. Biol. 7, 1100–1104 [DOI] [PubMed] [Google Scholar]
- 44. Swanson K. A., Knoepfler P. S., Huang K., Kang R. S., Cowley S. M., Laherty C. D., Eisenman R. N., Radhakrishnan I. (2004) Nat. Struct. Mol. Biol. 11, 738–746 [DOI] [PubMed] [Google Scholar]
- 45. van Ingen H., Lasonder E., Jansen J. F., Kaan A. M., Spronk C. A., Stunnenberg H. G., Vuister G. W. (2004) Biochemistry 43, 46–54 [DOI] [PubMed] [Google Scholar]
- 46. Nomura M., Uda-Tochio H., Murai K., Mori N., Nishimura Y. (2005) J. Mol. Biol. 354, 903–915 [DOI] [PubMed] [Google Scholar]
- 47. Sahu S. C., Swanson K. A., Kang R. S., Huang K., Brubaker K., Ratcliff K., Radhakrishnan I. (2008) J. Mol. Biol. 375, 1444–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kumar G. S., Xie T., Zhang Y., Radhakrishnan I. (2011) J. Mol. Biol. 408, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Viiri K. M., Korkeamäki H., Kukkonen M. K., Nieminen L. K., Lindfors K., Peterson P., Mäki M., Kainulainen H., Lohi O. (2006) Nucleic Acids Res. 34, 3288–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ferentz A. E., Wagner G. (2000) Q. Rev. Biophys. 33, 29–65 [DOI] [PubMed] [Google Scholar]
- 51. Bax A., Grzesiek S. (1993) Accounts Chem. Res. 26, 131–138 [Google Scholar]
- 52. Otting G., Wüthrich K. (1990) Q. Rev. Biophys. 23, 39–96 [DOI] [PubMed] [Google Scholar]
- 53. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zwahlen C., Legault P., Vincent S. J. F., Greenblatt J., Konrat R., Kay L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 55. Linge J. P., Habeck M., Rieping W., Nilges M. (2003) Bioinformatics 19, 315–316 [DOI] [PubMed] [Google Scholar]
- 56. Linge J. P., Habeck M., Rieping W., Nilges M. (2004) J. Magn. Reson. 167, 334–342 [DOI] [PubMed] [Google Scholar]
- 57. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 58. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 59. Salerno W. J., Seaver S. M., Armstrong B. R., Radhakrishnan I. (2004) Nucleic Acids Res. 32, W566–W568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 61. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 62. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 63. Aravind L., Koonin E. V. (2000) Trends Biochem. Sci. 25, 112–114 [DOI] [PubMed] [Google Scholar]
- 64. Kipp M., Göhring F., Ostendorp T., van Drunen C. M., van Driel R., Przybylski M., Fackelmayer F. O. (2000) Mol. Cell Biol. 20, 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mayer C., Neubert M., Grummt I. (2008) EMBO Rep. 9, 774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Göhring F., Schwab B. L., Nicotera P., Leist M., Fackelmayer F. O. (1997) EMBO J. 16, 7361–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Okubo S., Hara F., Tsuchida Y., Shimotakahara S., Suzuki S., Hatanaka H., Yokoyama S., Tanaka H., Yasuda H., Shindo H. (2004) J. Biol. Chem. 279, 31455–31461 [DOI] [PubMed] [Google Scholar]
- 68. Suzuki R., Shindo H., Tase A., Kikuchi Y., Shimizu M., Yamazaki T. (2009) Proteins 75, 336–347 [DOI] [PubMed] [Google Scholar]
- 69. Viiri K. M., Heinonen T. Y., Mäki M., Lohi O. (2009) BMC Evol. Biol. 9, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhou Y., Santoro R., Grummt I. (2002) EMBO J. 21, 4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.