Abstract
Differentiation of preadipocytes into mature adipocytes capable of efficiently storing lipids is an important regulatory mechanism in obesity. Here, we examined the involvement of histone deacetylases (HDACs) and histone acetyltransferases (HATs) in the regulation of adipogenesis. We find that among the various members of the HDAC and HAT families, only HDAC9 exhibited dramatic down-regulation preceding adipogenic differentiation. Preadipocytes from HDAC9 gene knock-out mice exhibited accelerated adipogenic differentiation, whereas HDAC9 overexpression in 3T3-L1 preadipocytes suppressed adipogenic differentiation, demonstrating its direct role as a negative regulator of adipogenesis. HDAC9 expression was higher in visceral as compared with subcutaneous preadipocytes, negatively correlating with their potential to undergo adipogenic differentiation in vitro. HDAC9 localized in the nucleus, and its negative regulation of adipogenesis segregates with the N-terminal nuclear targeting domain, whereas the C-terminal deacetylase domain is dispensable for this function. HDAC9 co-precipitates with USF1 and is recruited with USF1 at the E-box region of the C/EBPα gene promoter in preadipocytes. Upon induction of adipogenic differentiation, HDAC9 is down-regulated, leading to its dissociation from the USF1 complex, whereas p300 HAT is up-regulated to allow its association with USF1 and accumulation at the E-box site of the C/EBPα promoter in differentiated adipocytes. This reciprocal regulation of HDAC9 and p300 HAT in the USF1 complex is associated with increased C/EBPα expression, a master regulator of adipogenic differentiation. These findings provide new insights into mechanisms of adipogenic differentiation and document a critical regulatory role for HDAC9 in adipogenic differentiation through a deacetylase-independent mechanism.
Keywords: Adipocyte, Adipose Tissue, Adipose Tissue Metabolism, C/EBP Transcription Factor, Cell Differentiation, Chromatin Immunoprecipitation (ChIP), Gene Expression, Gene Knock-out, Gene Transcription, Histone Acetylase
Introduction
Adipose tissue expansion is a fundamental compensatory mechanism to facilitate storage of excess energy as triglycerides during times of caloric excess. Adipose tissue expands by enhancing lipid loading of existing adipocytes (hypertrophy) and/or by increased differentiation of preadipocytes to mature adipocytes (hyperplasia). Regional adipose depots respond differently to caloric excess; visceral adipose tissue expands primarily by hypertrophy, whereas subcutaneous adipose tissue can expand by hyperplasia (1). Hypertrophic expansion of visceral adipose tissue is associated with adverse metabolic consequences, whereas hyperplasia of subcutaneous adipose tissue (which is commonly observed after thiazolidinedione drug treatment of type II diabetic patients) is associated with improved insulin sensitivity and metabolic parameters (2–6). Conceptually, hyperplastic expansion of adipose tissue is metabolically beneficial because it permits increased lipid storing capacity of the adipose tissue without overloading the individual adipocytes. Excessive enlargement of adipocytes due to lipid overloading promotes mechanical stress and inflammation, alters secretory function, and induces necrotic cell death (7–11). Thus, obese individuals with many large adipocytes exhibit greater degrees of glucose intolerance and insulin resistance than individuals with the same degree of obesity but many small adipocytes (12–16).
Efficient differentiation of preadipocytes to mature adipocytes is central to hyperplastic adipose tissue expansion and thus could favorably modulate whole body glucose and lipid homeostasis in obesity states. Adipogenic differentiation, like other forms of cellular differentiation, is a complex process orchestrated by a concerted action of various transcription factors under the control of transcriptional activators and repressors, leading to stable changes in cellular gene expression and acquisition of a mature terminally differentiated phenotype (17, 18). Histone deacetylases (HDACs)2 and histone acetyltransferases (HATs) are chromatin modifying enzymes known to play pivotal roles as transcriptional suppressors and activators, respectively, in cellular differentiation (19–22). However, the role of HDACs and HATs in adipogenic differentiation, particularly in humans, is largely unknown.
In the present study we examined the contribution of HDACs and HATs in differentiation of human preadipocytes. Our data show that HDAC9 is differentially expressed in omental versus subcutaneous adipocytes and functions as a novel negative regulator of adipogenic differentiation. The mechanism of HDAC9 inhibition of adipogenesis is independent of its deacetylase activity and could be related at least in part to its recruitment with USF1 at the E-box site of the C/EBPα gene promoter. During adipogenic differentiation, HDAC9 expression is down-regulated, leading to its dissociation from the USF1 complex. Concomitantly, p300 HAT is up-regulated to allow its association with the USF1 complex. This reciprocal regulation of HDAC9 and p300 appears to act as a switch to turn on expression of C/EBPα, a master regulator of adipogenic differentiation. Indeed, forced HDAC9 overexpression by transient transfection in 3T3-L1 preadipocytes blunts C/EBPα expression and adipogenesis, whereas preadipocytes from HDAC9 knock-out mice exhibit enhanced C/EBPα expression and accelerated adipogenesis, demonstrating a direct role of this HDAC in adipogenic differentiation. Our findings provide new insight into mechanisms of adipogenic differentiation and document a critical regulatory role of HDAC9 in such differentiation independent of its deacetylase activity.
EXPERIMENTAL PROCEDURES
Isolation of Preadipocytes and Adipogenic Differentiation
Human adipose tissues were collected from subcutaneous and omental depots of non-obese and non-diabetic patients undergoing abdominal surgeries. The institutional review board at the University of Cincinnati approved our protocol. Within 2 h after collection, adipose tissues were minced thoroughly and digested with collagenase as we described previously (23). Digests were centrifuged to separate mature floating adipocytes from the pelleted stromovascular cells. The floating mature adipocyte fractions were collected, washed twice with PBS, and stored frozen before use. Pellets were suspended in red blood cell lysis buffer (0.82% NH4Cl, 0.1% KHCO3, and 0.0037% EDTA, pH 7.4) and incubated at room temperature for 10 min. Pellets were suspended again in PBS and filtered through 70 μm followed by 40 μm membranes. Cells were pelleted again and resuspended in DMEM/F-12 medium containing 10% FBS. Resuspended cells (preadipocyte-enriched stromovascular fraction) were plated, grown, and expanded in DMEM/F-12 medium containing 10% FBS. Preadipocytes were utilized for in vitro adipogenic differentiation within two to four passages in culture. Cells were allowed to differentiate in the presence of adipocyte differentiation medium (Cell Application), which was replaced with fresh medium every fourth day (23). In some cases adipose tissues were flash-frozen in liquid nitrogen immediately after collection for subsequent analysis of RNA and protein expression.
Male C57BL/6J or HDAC9 gene knock-out (in C57BL/6J and 129 SvEv background) mice 18–20 weeks of age, maintained on chow diet and water ad libitum, were used in the present study. Subcutaneous (inguinal) and epididymal adipose tissues were dissected out, processed for preadipocyte isolation, and cultured and differentiated according to the methods described for human cells (23).
A model adipogenic cell line, 3T3-L1 cells, were obtained from ATCC and grown in DMEM/F-12 medium containing 10% FBS. Cells were allowed to differentiate in the presence of adipocyte differentiation medium (Cell Application) as described for human preadipocytes.
Transient Transfection of 3T3-L1 Cells
3T3-L1 cells were transfected with Lipofectamine in a 6-well or 10-cm dish according to the manufacturer's suggestions. FLAG-tagged full-length HDAC9 cDNA construct (amino acids 1–1069) (a generous gift of Dr. Arthur Zelent) (24), FLAG-tagged N-terminal HDAC9 construct (amino acids 1–636), FLAG-tagged C-terminal HDAC9 construct (amino acids 637–1069), all cloned in pCMV 3Tag 6 vector (Agilent Technologies), and empty vector were used in the transfection study. Using anti-FLAG antibody, we detected nearly 3–4-fold higher expression of FLAG-tagged HDAC9 constructs compared with endogenous HDAC9 at 3-day post-transfection. Under our experimental conditions, we achieved ∼30–45% transfection efficiency, as judged by microscopic visualization of cells expressing FLAG-tagged HDAC9 constructs. One day after transfection, adipogenic differentiation medium was added, and cells were allowed to differentiate for 3–7 days. Cells were treated with either Qiazol to extract RNA with RNeasy lipid mini kit (Qiagen) or with radioimmune precipitation assay lysis buffer to extract protein for Western blot analysis.
Subcellular Fractionation and Western Blot Analysis
Nuclear and cytoplasmic fractions of 3T3-L1 and human omental cells were prepared using NE-PER nuclear and cytoplasmic extraction reagent (Thermo Scientific) according to the manufacturer's suggestions. Authenticity of nuclear fraction was verified by Western blotting for enrichment of NuMA (nuclear mitotic apparatus protein; Santa Cruz) nuclear protein. Western blot was performed by separating proteins on SDS-PAGE, transferring to nitrocellulose membrane, and probing with the appropriate antibody, and subsequently blots were developed using ECL system (Amersham Biosciences). Antibodies were obtained from Santa Cruz (USF1, USF2, p300, Tip60, GCN5, and GAPDH), Cell Signaling (P/CAF, HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC7, HDAC8), -FLAG antibody M2 (Sigma) and HDAC9 (Abcam). Other chemicals used in this study were obtained from Sigma.
Quantitative PCR Quantification of mRNA Levels
Total RNA was extracted from tissues or cells with Qiazol and purified with RNeasy lipid mini kits (Qiagen). qRT-PCR quantification of mRNA levels of the genes of interest was performed using SYBR green qRT-PCR kits (Agilent Technologies). Normalized Ct values were subjected to statistical analysis, and -fold difference was calculated by ΔΔCt method as described previously (23).
Immunoprecipitation
Cells were lysed with immunoprecipitation lysis buffer (Thermo Scientific), clarified by centrifugation, and immunoprecipitated by overnight incubation at 4 °C with the appropriate antibody and Dynabeads Protein G (Invitrogen). FLAG-tagged HDAC9 proteins were precipitated from transfected 3T3-L1 cell lysates by anti-FLAG M2 magnetic beads (Sigma). In some experiments cells were cross-linked with dithiobis(succinimidyl propionate) (Thermo Scientific) in PBS for 30 min at room temperature before cell lysis and immunoprecipitation.
Immunocytochemistry
Transfected 3T3-L1 cells were grown on two-chambered glass slides (Lab-Tek), fixed with 4% paraformaldehyde, permeabilized by treatment with 0.1% Triton X-100, and labeled with Alexa 488-conjugated-FLAG antibody (Cell Signaling) to detect expression of FLAG-tagged HDAC9 proteins. Cells were visualized under a confocal microscope (Zeiss LSM710), with the nucleus stained blue with DAPI. Untransfected 3T3-L1 cells were fixed with acetone/methanol (1:1) at −20 °C and incubated with HDAC9 antibody (Abcam) followed by treatment with FITC-conjugated secondary antibody (Sigma) to label endogenously expressed HDAC9 (green) in these cells, whereas the cell nucleus was stained with DAPI (blue).
Chromatin Immunoprecipitation (ChIP)
Cells were first fixed with 2 mm disuccinimidyl glutarate (Thermo Scientific) for 45 min at room temperature followed by cross-linking with 1% formaldehyde (Sigma) at room temperature for 10–15 min. Reactions were quenched with glycine, and DNA was sheared by sonication and by digestion with endonuclease (Active Motif). DNA was precipitated with the appropriate antibody and Dynabeads Protein G overnight at 4 °C. Precipitated DNA was reversed cross-linked, purified by phenol-chloroform extraction, and subjected to SYBR Green ChIP- quantitative PCR quantification (Invitrogen) using primer pairs (sequence information will be provided upon request) from the E-box region of the C/EBPα gene promoter. This primer combination amplified a 121-bp fragment. IgG-precipitated samples were used as a negative control.
RESULTS
Down-regulation of HDAC9 during in Vitro Adipogenic Differentiation of Human and Mouse Preadipocytes
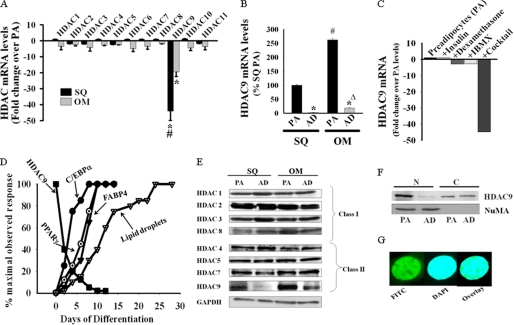
Members of the HDAC family of proteins are emerging as critical regulators of cellular differentiation processes. Here, we explored the possible involvement of this family of proteins in the regulation of adipogenic differentiation. We initiated our study by examining the mRNA expression of members of the HDAC family of genes in preadipocytes and in vitro differentiated adipocytes from human subcutaneous and omental adipose tissues. All 11 members of the HDAC family of genes (class I, II, and IV) are expressed in preadipocytes from these two regional human adipose depots. Among the 11 HDAC genes, only HDAC9 mRNA levels showed progressive down-regulation during adipogenic differentiation of preadipocytes, and differentiated cells exhibited markedly reduced HDAC9 mRNA levels. Fig. 1A depicts -fold decreases in the mRNA levels of members of the HDAC family of genes in preadipocytes versus 7-day in vitro differentiated adipocytes from human subcutaneous and omental adipose tissues. In contrast to other members of the HDAC gene family, HDAC9 mRNA levels were reduced by nearly 50-fold in subcutaneous preadipocytes and by nearly 20-fold in omental preadipocytes after their in vitro adipogenic differentiation. This down-regulation of HDAC9 transcripts is not an artifact of the in vitro differentiation process because mature adipocytes isolated from human subcutaneous and omental adipose tissues demonstrate similarly reduced HDAC9 mRNA levels compared with their corresponding stromal preadipocyte-enriched fractions (15–40-fold; Fig. 1B). Interestingly, HDAC9 mRNA levels are relatively higher in freshly isolated preadipocytes and mature adipocytes from omental adipose tissue as compared with their subcutaneous counterparts (∼3–8-fold).
FIGURE 1.
Down-regulation of HDAC9 during in vitro adipogenic differentiation of human subcutaneous (SQ) and omental (OM) preadipocytes. A, in vitro adipogenic differentiation of freshly isolated human SQ and OM preadipocytes for 7 days demonstrates selective down-regulation of HDAC9 mRNA levels as determined by SYBR Green qRT-PCR. Values are expressed as -fold changes from HDAC mRNA levels in undifferentiated preadipocytes (PA) and normalized to Homo sapiens ribosomal protein, large, P0 (RPLP0). Results are expressed as the mean ± S.E. from three different donors (*, p < 0.001 as compared with HDAC9 expression in corresponding undifferentiated preadipocytes; #, p < 0.001 as compared with HDAC9 expression in differentiated OM cells). B, mature adipocytes (AD) isolated from human SQ and OM adipose tissues show reduced HDAC9 mRNA levels compared with corresponding stromal preadipocytes (PA). Data are the mean ± S.E. from three different donors (*, p < 0.001 as compared with HDAC9 expression in corresponding stromal preadipocytes). Also, HDAC9 mRNA levels are significantly higher in omental preadipocytes (#, p < 0.001) and adipocytes (Δ, p < 0.001) compared with that from SQ. IBMX, isobutylmethylxanthine. C, HDAC9 down-regulation is induced strictly under conditions that trigger in vitro adipogenic differentiation of human SQ preadipocytes. Values are expressed as described above, and data are representative of two independent experiments. Similar results were obtained with human OM and 3T3-L1 preadipocytes. D, HDAC9 mRNA down-regulation precedes adipocyte differentiation-specific genes expression and cytoplasmic lipid droplets accumulation in differentiating SQ preadipocytes in vitro. Values are representative finding from four different donors. Adipocyte differentiation-specific genes expression were determined by SYBR Green qRT-PCR and normalized for RPLPO. E, HDAC9 protein is selectively down-regulated during in vitro adipogenic differentiation (7 days) of human SQ and OM preadipocytes. Among members of the class II HDACs (HDAC4, -5, -7, and -9), only HDAC9 shows significant down-regulation with adipogenic differentiation, whereas none of the class I HDACs (HDAC1, -2, -3, and -8) shows any appreciable change in their levels. GAPDH is shown as the loading control. The figure is representative of four different donors. F, HDAC9 protein exhibits predominant localization in the nuclear fraction of 3T3-L1 preadipocytes; adipogenic differentiation of these cells promotes down-regulation of nuclear HDAC9 levels. The figure is from a representative experiment, replicated with similar results in three independent experiments. Similar findings are also noted with cellular subfractions from human OM and mouse SQ preadipocytes. G, confocal microscopic picture shows constitutive nuclear localization of endogenous HDAC9 protein in 3T3-L1 cells. HDAC9 protein was detected in fixed cells employing HDAC9 antibody and FITC-conjugated secondary antibody (green), whereas the cell nucleus was stained with DAPI (blue). The overlay image demonstrates overlapping green and blue fluorescence.
We next examined whether any one specific component of the adipogenic differentiation medium (dexamethasone, isobutylmethylxanthine, and insulin) is sufficient to promote HDAC9 mRNA down-regulation in preadipocytes. As shown in Fig. 1C, none of these agents was effective individually at down-regulating HDAC9 mRNA levels in human subcutaneous preadipocytes. Marked HDAC9 down-regulation was demonstrable only when all agents were combined together, which is likewise a requisite for adipogenic differentiation. Similar findings were also observed in preadipocytes from human omental adipose tissues as well as in adipogenic 3T3-L1 preadipocytes (data not shown). These results suggest a possible interdependency between down-regulation of HDAC9 and the induction of adipogenesis.
Evaluation of the temporal kinetics of HDAC9 down-regulation with respect to expression of adipocyte differentiation-specific genes during in vitro differentiation of human subcutaneous preadipocytes revealed that HDAC9 down-regulation is an early event, preceding up-regulation of adipocyte differentiation-specific genes (Fig. 1D). After application of adipogenic differentiation medium, HDAC9 levels exhibited progressive down-regulation, reaching 10% of the base-line values by day 6, whereas expression of adipocyte differentiation-specific genes (C/EBPα, PPARγ, and FABP4) exhibited progressive up-regulation to reach at the maximal levels by day 6. Lipid droplet accumulation, lagging behind, reached maximum at day 20–25 post stimulation.
Similar to preadipocytes from human adipose depots, preadipocytes from mouse subcutaneous and epididymal adipose tissues showed selective down-regulation of HDAC9 mRNA during in vitro adipogenic differentiation, and this down-regulation was more marked in the subcutaneous compared with epididymal preadipocytes (37- versus 12-fold) (supplemental Fig. 1A). Murine 3T3-L1 preadipocytes likewise exhibited HDAC9 mRNA down-regulation (nearly 90-fold) during adipogenic differentiation (supplemental Fig. 1B). Thus, HDAC9 down-regulation consistently parallels adipogenic differentiation of preadipocytes from human and mouse regional adipose tissues and in 3T3-L1 cells, a model adipogenic cell line.
We next performed Western blot analysis to examine the relative levels of class I (HDAC1–3 and -8) and class IIa (HDAC4, -5, -7, and -9) HDAC proteins in preadipocytes versus 7-day in vitro differentiated adipocytes from human subcutaneous and omental adipose tissues (Fig. 1E). In agreement with mRNA levels, HDAC9 protein levels showed a marked down-regulation after in vitro adipogenic differentiation of preadipocytes (2–6-fold). In contrast, other members of these classes of HDAC proteins did not exhibit consistent changes in their expression levels after adipogenic differentiation. Further examination revealed that HDAC9 protein is localized predominantly in the cell nucleus in preadipocytes (Fig. 1, F and G), and adipogenic differentiation promotes down-regulation of HDAC9 from the nuclear compartment (Fig. 1F). Thus, HDAC9 is constitutively a nuclear protein in preadipocytes, and down-regulation of HDAC9 protein within the cell nucleus parallels adipogenic differentiation.
Differential HDAC9 Expression in Visceral versus Subcutaneous Preadipocytes Negatively Correlates with Their Adipogenic Differentiation Potential
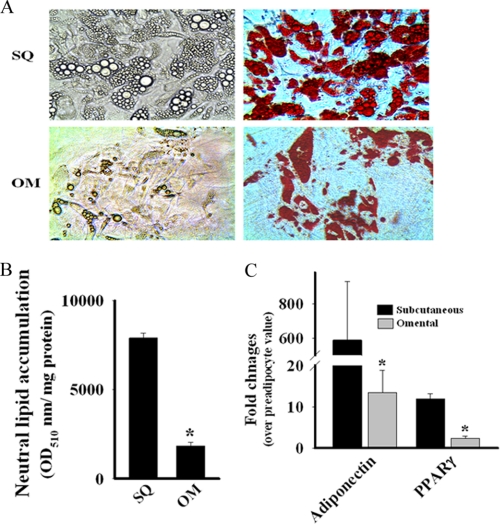
The mRNA data suggested that HDAC9 expression levels differ in preadipocytes isolated from subcutaneous versus visceral adipose tissues. We, therefore, compared HDAC9 protein levels in preadipocytes isolated from human subcutaneous versus omental adipose tissues. The omental preadipocytes invariably exhibited more abundant expression of HDAC9 protein (∼2–3-fold) compared with subcutaneous preadipocytes (Fig. 1E). Furthermore, in vitro differentiation of omental preadipocytes was associated with diminished down-regulation (2–4-fold) of HDAC9 protein compared with that observed in subcutaneous preadipocytes (4–6-fold). Similar characteristics were also displayed by preadipocytes isolated from subcutaneous versus epididymal adipose tissues of C57/BL6 mice, with preadipocytes from the epididymal adipose tissue exhibiting higher base-line levels of HDAC9 protein and diminished down-regulation in response to adipogenic differentiation compared with subcutaneous preadipocytes (data not shown). In parallel with higher HDAC9 levels, omental preadipocytes exhibited attenuated in vitro adipogenic differentiation compared with corresponding subcutaneous preadipocytes. Thus, both cytoplasmic lipid droplet accumulation (Fig. 2, A and B) and adipocyte differentiation-specific gene expression (Fig. 2C) were substantially lower in differentiated human omental preadipocytes compared with their subcutaneous counterparts. Thus, the elevated levels of HDAC9 expression in omental preadipocytes correlate negatively with their capacity to undergo adipogenic differentiation.
FIGURE 2.
SQ preadipocytes exhibit higher adipogenic differentiation potential than OM preadipocytes. A, reduced accumulation of cytoplasmic lipid droplets in differentiated (14 days) human OM compared with human SQ adipocytes are visualized by phase contrast microscopy in live cells (left panel) and further identified by labeling neutral lipids with oil red-O (right panel). B, the levels of oil red-O-labeled cytoplasmic lipid droplets were quantified spectrophotometrically, and the data are expressed as A510 nm/mg of protein ± S.E. from three different donors. *, p < 0.001 compared with SQ. C, adipocyte differentiation-specific gene expression is much higher in differentiating SQ than OM preadipocytes. The mRNA levels of adiponectin and PPARγ were quantified by qRT-PCR and normalized for the human RPLPO. Data are expressed as the mean ± S.E. of three different donors. *, p < 0.01 compared with SQ.
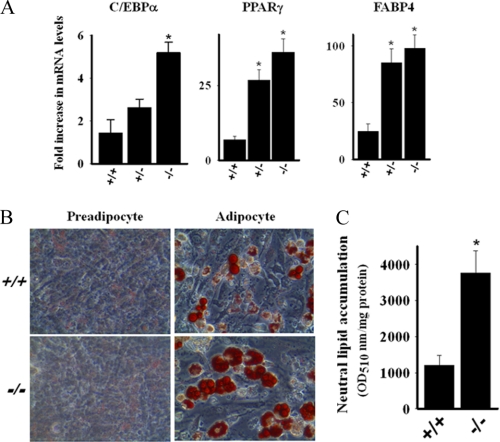
Preadipocytes from HDAC9 Knock-out Mice Exhibit Accelerated in Vitro Adipogenesis
To explore the possibility of a causal relationship between HDAC9 expression and adipogenic differentiation, we compared adipogenic differentiation of preadipocytes from subcutaneous adipose depot of wild-type and HDAC9 gene knock-out mice. Although cells from HDAC9 gene knock-out mice did not undergo spontaneous differentiation, the induction of adipogenesis was potentiated, as demonstrated by enhanced expression of adipocyte differentiation-specific genes and lipid droplet accumulation (Fig. 3, A–C). These findings support the contention that endogenous HDAC9 is a negative regulator of adipogenic differentiation; however, the loss of HDAC9 in the absence of adipogenic stimuli is insufficient to trigger spontaneous adipogenesis.
FIGURE 3.
Preadipocytes from HDAC9 gene knock-out mice exhibit accelerated adipogenic differentiation. A, shown is elevated expression of adipogenic differentiation-specific genes during in vitro adipogenic differentiation (4 days) of preadipocytes from SQ adipose tissue of HDAC9 gene knock-out mice. The mRNA levels of C/EBPα, PPARγ, and FABP4 were quantified by qRT-PCR and normalized for the mouse RPLPO. Data are expressed as the mean ± S.E. of three independent experiments. *, p < 0.005 compared with wild-type (+/+) values. B, microscopic visualization revealed increased oil red-O-labeled lipid droplet accumulation in the cytoplasm of in vitro differentiating (6 days) preadipocytes from SQ adipose tissue of HDAC9 gene knock-out mice. C, the levels of oil red-O-positive materials were quantified spectrophotometrically, and the data are expressed as A510 nm/mg of protein ± S.E. from three independent experiments. *, p < 0.001 compared with wild type (+/+).
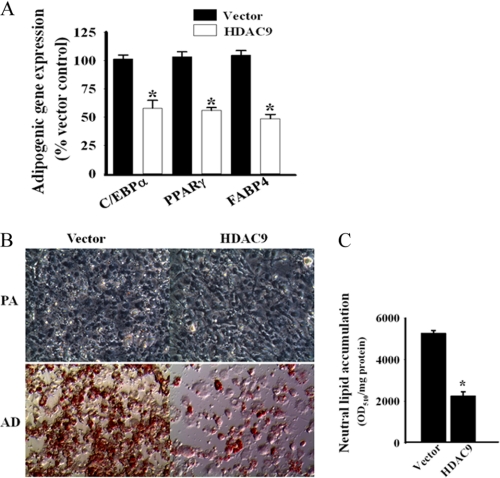
HDAC9 Overexpression by Transient Transfection Inhibits Adipogenic Differentiation of 3T3-L1 Preadipocytes
To further establish the role of HDAC9 as a negative regulator of adipogenic differentiation, we overexpressed HDAC9 in 3T3-L1 cells by transient transfection and subsequently examined adipogenic differentiation of these transfected cells. Overexpression of HDAC9 blunted adipogenic differentiation, as demonstrated by reduced expression of adipocyte differentiation-specific genes (C/EBPα, PPARγ, and FABP4) and reduced cytoplasmic lipid droplet accumulation (Fig. 4, A–C). These data confirm that HDAC9 functions as a negative regulator of the adipogenic differentiation process.
FIGURE 4.
HDAC9 overexpression by transient transfection blunts adipogenic differentiation of 3T3-L1 preadipocytes. A, HDAC9 overexpression by transfection in 3T3-L1 preadipocytes blunts their adipogenic differentiation, as evidenced by reduced expression of adipocyte differentiation-specific genes. The mRNA levels of C/EBPα, PPARγ, and FABP4 were determined by qRT-PCR and normalized for the mouse RPLPO. Empty vector-transfected cells are used as controls. Data are the mean ± S.E. from three independent experiments. *, p < 0.001 compared with corresponding vector-transfected control. HDAC9 overexpression in 3T3-L1 preadipocytes (PA) blunts adipogenic (AD) differentiation of these cells with reduced accumulation of cytoplasmic lipid droplets, as observed by microscopic visualization of oil red-O-stained cells (B), and by spectrophotometric quantification of oil red-O positive materials as described above (C). Data are expressed as the mean ± S.E. of three independent experiments. *, p < 0.001 compare with vector transfected control.
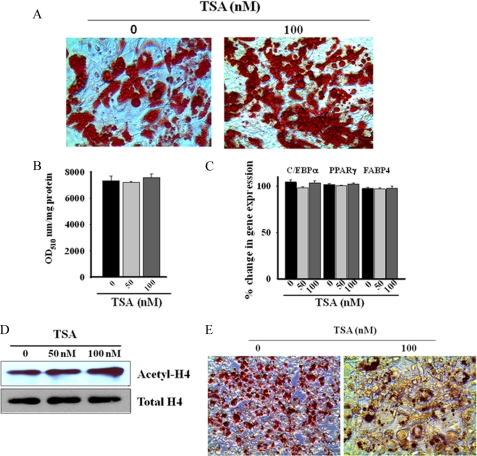
Inhibition of HDAC Activity Is Insufficient to Promote Adipogenic Differentiation of Human Preadipocytes
In some prior studies, inhibitors of HDAC enzymatic activity were reported to modulate adipogenic differentiation. We, therefore, tested the effects of several HDAC inhibitors on the temporal kinetics of adipogenic differentiation in human subcutaneous preadipocytes. We found that the pan-HDAC inhibitor trichostatin A (TSA) did not influence cytoplasmic lipid droplet accumulation (Fig. 5, A and B) or adipocyte differentiation-specific gene expression (Fig. 5C), suggesting a lack of an effect of this deacetylase inhibitor on adipogenic differentiation. TSA, although ineffective to influence adipogenic differentiation, induced significant stimulation of endogenous histone acetylation (Fig. 5D, ∼2-fold stimulation of H4 acetylation within 24 h), suggesting that the compound effectively inhibited histone deacetylase activity in these cells. Likewise, butyrate, a short chain fatty acid inhibitor of HDAC, failed to influence adipogenic differentiation, whereas the nonspecific agent valproic acid markedly inhibited adipogenic differentiation of human subcutaneous preadipocytes (supplemental Fig. 2).
FIGURE 5.
HDAC inhibitor TSA fails to suppress adipogenic differentiation of human SQ preadipocytes. A, the presence of pan-HDAC inhibitor trichostatin A (100 nm) during the course of adipogenic differentiation of human SQ preadipocytes for 14 days produces no appreciable effect on the extent of cytoplasmic lipid droplets accumulation, as observed by microscopic visualization of oil red-O-stained cells. The figure is a representative finding of six different determinations with cells from three different donors. B, spectrophotometric quantification similarly shows no significant effect of trichostatin A on lipid droplets accumulation by differentiated (14 days) human SQ preadipocytes. Values are the mean ± S.E. of three separate experiments from three different donors. C, trichostatin A is similarly without effect on adipogenic gene expression (C/EBPα, PPARγ, and FABP4) during in vitro differentiation of human SQ preadipocytes, as determined by qRT-PCR. Values are the mean ± S.E. of three separate experiments from three different donors. D, Western blot analysis shows a nearly 2-fold stimulation of histone 4 (H4) acetylation in SQ preadipocytes after 24 h of treatment with 100 nm trichostatin A. Total H4 levels are shown as the loading control. The figure is representative of three independent experiments. E, trichostatin A (100 nm) treatment markedly attenuates cytoplasmic lipid droplets accumulation in differentiated (10 days) 3T3-L1 cells, as observed by microscopic visualization of oil red-O-stained cells.
Although adipogenic differentiation of human subcutaneous preadipocytes was insensitive to TSA, this inhibitor blocked adipogenic differentiation of 3T3-L1 cells. Fig. 5E shows that TSA (100 nm) markedly reduced cytoplasmic lipid droplet accumulation in differentiating 3T3-L1 cells, a finding consistent with that of Haberland et al. (25).
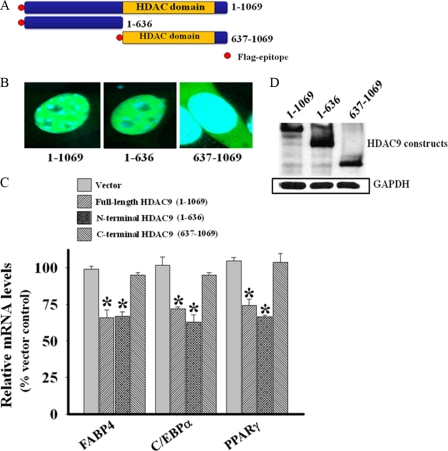
Deacetylase Domain of HDAC9 Is Dispensable for Its Negative Regulatory Effect on Adipogenesis
To test whether the deacetylase domain of HDAC9 is required to suppress adipogenic differentiation, we overexpressed full-length HDAC9, an N-terminal segment of HDAC9 lacking the deacetylase domain, and a C-terminal segment of HDAC9 containing the deacetylase domain (Fig. 6A) in 3T3-L1 preadipocytes. Confocal microscopy demonstrated predominant nuclear localization of the full-length HDAC9 construct (Fig. 6B), similar to that observed for the endogenous HDAC9 in these cells (Fig. 1, F and G). Like the full-length protein, the N-terminal HDAC9 protein exhibited predominant nuclear localization, whereas the C-terminal protein was localized in the cytoplasm (Fig. 6B). The full-length and the N-terminal HDAC9 constructs were equally effective at suppressing adipogenic gene expression, whereas the C-terminal HDAC9 construct was ineffective (Fig. 6C). Western blot analysis documented comparable levels of expression of the full-length and the two truncated HDAC9 constructs (Fig. 6D). These data confirm that the deacetylase domain of HDAC9 is not required for the regulation of adipogenesis. Moreover, our data indicate that the N-terminal region of HDAC9 contains its nuclear targeting sequence and suggest in addition a nuclear mechanism of HDAC9 action in suppressing adipogenesis.
FIGURE 6.
Deacetylase domain of HDAC9 is dispensable for its negative regulatory effect on adipogenic differentiation. A, shown is s schematic presentation of FLAG epitope-tagged constructs of human HDAC9 used in the present study. B, a confocal microscopic overlay image shows subcellular localization of the full-length and the two deletion constructs of HDAC9. HDAC9 proteins appear green (stained with Alexa 488 conjugated FLAG antibody), whereas the nucleus appears blue (stained with DAPI). The full-length and the N-terminal HDAC9 constructs exhibit predominant nuclear localization, whereas the C-terminal HDAC9 construct shows predominant cytoplasmic localization. C, the full-length and the N-terminal HDAC9 constructs were equally active to blunt adipogenic differentiation as evidenced by reduced expression of adipogenic differentiation-specific gene in transfected 3T3-L1 preadipocytes, whereas the C-terminal HDAC9 construct is devoid of such activity. The mRNA levels of FABP4, C/EBPα, and PPARγ were determined by qRT-PCR and normalized for the mouse RPLPO. Empty vector-transfected cells are used as controls. Data are the mean ± S.E. from three independent experiments. *, p < 0.001 compared with corresponding vector transfected control. D, Western blot analysis for the expression of full-length and truncated HDAC9 proteins shows their equivalent expression in transfected 3T3-L1 preadipocytes.
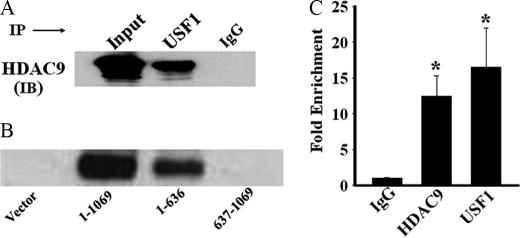
HDAC9 Physically Interacts with USF1 at the Proximal Promoter Site of the C/EBPα Gene in Preadipocytes
HDAC9 has been reported to interact with and repress functional activities of key transcription factors involved in cellular differentiation, including USF1 (26–28). USF1 regulates various genes of the glucose and lipid metabolic pathways (29, 30), and thus, repression of USF1 transcriptional activity by HDAC9 could disrupt adipogenic differentiation. To begin to address this possibility, we first examined whether USF1 could interact with HDAC9 in the context of the adipogenic cells. We immunoprecipitated USF1 from the lysate of human omental preadipocytes and examined by immunoblotting any co-precipitation of HDAC9. Our data demonstrate that HDAC9 co-precipitates with USF1 (Fig. 7A), suggesting a physical interaction between endogenously expressed USF1 and HDAC9 in preadipocytes. In a reciprocal experiment, we found similar co-precipitation of USF1 by HDAC9 from the lysate of omental preadipocytes (data not shown). In differentiated adipocytes, HDAC9 levels are markedly down-regulated, leading to reduced formation of HDAC9-USF1 complex in these cells (supplemental Fig. 3). We further performed co-immunoprecipitation experiments using 3T3-L1 cells transfected with various HDAC9 constructs. Our data show that both the full-length and the N-terminal HDAC9 proteins possess the ability to co-precipitate endogenously expressed USF1 from 3T3-L1 cell lysate, whereas the C-terminal HDAC9 protein lacks such ability (Fig. 7B). These data document that the N-terminal segment of HDAC9 is sufficient for its nuclear localization, USF1 binding, and functional ability to suppress adipogenesis.
FIGURE 7.
HDAC9 physically interacts with USF1 transcription factor and is recruited with USF1 at the E-box site of the C/EBPα gene promoter. A, immunoprecipitation (IP) with USF1 antibody and subsequent immunoblotting (IB) of the precipitates with HDAC9 antibody reveal that HDAC9 co-precipitates with USF1 from the lysate of human OM preadipocytes. IgG antibody replaced USF1 antibody in parallel experiments to serve as negative controls. The figure is representative finding of four independent experiments. Similar results are obtained with lysates from 3T3-L1 preadipocytes. B, the full-length and the N-terminal HDAC9 proteins, but not the C-terminal HDAC9 protein, co-precipitate USF1 from the lysate of transfected 3T3-L1 preadipocytes. Vector-transfected cells were used as a negative control. Data are representative of two separated experiments. C, ChIP analysis demonstrates that HDAC9 and USF1 antibodies promote significant precipitation of the E-box segment of the C/EBPα gene promoter from human OM preadipocytes. Data are the mean ± S.E. of three separate experiments done in duplicate. *, p < 0.001 compared with IgG control.
C/EBPα is a USF1-regulated gene that, in turn, serves as a master regulator of adipogenic differentiation. USF1 binding to the E-box element at the promoter site of the C/EBPα gene induces optimal C/EBPα expression in differentiated adipocytes (31). Because we found that HDAC9 overexpression inhibits C/EBPα expression, we hypothesized that HDAC9 forms an inhibitory complex with USF1 at the E-box site to suppress C/EBPα gene expression in preadipocytes. First, we examined the levels of expression (mRNA and protein) of USF1 and its homolog, USF2, and detected no changes in their levels during the course of adipogenic differentiation of preadipocytes (supplemental Fig. 4, A and B). Additionally, ChIP analysis demonstrated no differences in USF1 binding to the E-box element of the C/EBPα promoter in preadipocytes versus differentiated adipocytes (supplemental Fig. 4C). Together, these data suggest that USF1 is equally expressed and bound to the C/EBPα promoter in preadipocytes and adipocytes, which implies that HDAC9 does not inhibit adipogenesis by disrupting binding of USF1 to the C/EBPα promoter.
Next, we performed ChIP analysis to examine whether HDAC9 is recruited with USF1 to the E-box region of C/EBPα gene promoter. As shown in Fig. 7C, in human omental preadipocytes, both HDAC9 and USF1 antibodies pulled-down the E-box segment of the C/EBPα gene promoter compared with control (IgG). This finding demonstrates that USF1-HDAC9 complex accumulates at the E-box site of the C/EBPα gene promoter in preadipocytes.
HAT Proteins Are Up-regulated during in Vitro Adipogenic Differentiation
Emerging evidence indicates that HDACs and HATs reciprocally regulate gene expression through assembly of repressor and activator complexes, respectively, at the promoter regions of the responsive genes (32, 33). We, therefore, examined whether HAT family members are up-regulated and recruited to the C/EBPα promoter during adipogenic differentiation.
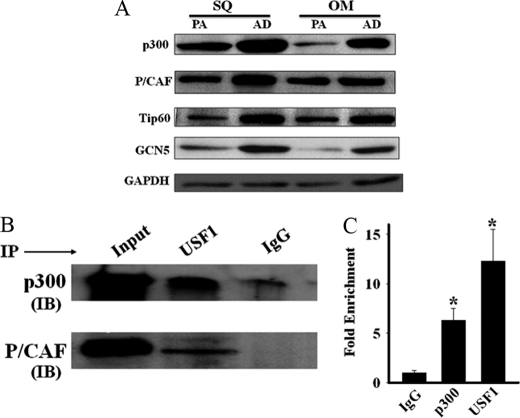
We compared the mRNA levels of members of the HAT family of genes in preadipocytes and 7-day in vitro differentiated adipocytes from human subcutaneous and omental adipose tissues. None of the HAT family members we tested (p300, GCN5, P/CAF, Tip60, CBP, MOF (males-absent on the first), HAT1, and SRC1 (steroid receptor co-activator1)) showed significant changes in mRNA expression during the course of adipogenic differentiation (data not shown). We next examined the protein levels of selected members (P/CAF, Tip60, p300, and GCN5) of this family by Western blot analysis and found that the levels of each of these proteins was up-regulated (2–5-fold) after adipogenic differentiation of preadipocytes (Fig. 8A), a finding consistent with a post-transcriptional mechanism of up-regulation of these HAT family proteins during adipogenic differentiation.
FIGURE 8.
Adipogenic differentiation promotes up-regulation of HAT proteins. A, Western blot analysis shows up-regulation of HAT family of proteins during adipogenic (AD) differentiation of SQ and OM preadipocytes (PA). Data are representative findings from three different donors. B, immunoprecipitation (IP) with USF1 antibody and subsequent immunoblotting (IB) of the precipitates with antibodies against p300 and P/CAF proteins reveal co-precipitation of both p300 and P/CAF with USF1 from the lysates of differentiated human SQ adipocytes. IgG antibody is used in place of USF1 in parallel experiments to serve as negative controls. The figure is a representative finding of three independent experiments. Tip60 and GCN5 antibodies detect no significant precipitation of these proteins with USF1. C, ChIP analysis shows significant precipitation of the E-box segment of the human C/EBPα gene promoter by p300 and USF1 antibodies from the lysates of differentiated human SQ adipocytes. *, p < 0.001 compared with IgG negative control.
HAT Proteins Interact with USF1 in Differentiated Adipocytes; Role in Switching on Adipogenesis
We investigated whether the up-regulated expression of HAT proteins might foster adipogenic differentiation by converting the HDAC9-USF1 transcriptional repressor complex to a HAT-USF1 transcriptional activator complex. We first examined whether HAT proteins replace HDAC9 as a USF1 binding partner in differentiated adipocytes. We immunoprecipitated USF1 from the lysates of differentiated human subcutaneous adipocytes and examined the immunoprecipitates for the presence of Tip60, GCN5, P/CAF, and p300. We found that both p300 and P/CAF co-precipitated with USF1 in lysates from differentiated adipocytes (Fig. 8B). We did not detect any appreciable precipitation of Tip60 or GCN5 with USF1 from these lysates (data not shown). As expected, HDAC9 protein could not be detected with USF1 in lysates from these differentiated adipocytes (data not shown).
To examine whether p300 and/or P/CAF are present with USF1 at the E-box site of C/EBPα gene promoter of differentiated adipocytes, we performed ChIP analysis with differentiated adipocytes from human subcutaneous adipose tissue. As shown in Fig. 8C, p300 and USF1 antibodies, but not P/CAF antibody (not shown), precipitated the E-box segment of C/EBPα gene promoter in lysates from differentiated human subcutaneous adipocytes, indicating that the USF1-p300 complex occupies the E-box site of C/EBPα promoter in differentiated adipocytes. As expected, the HDAC9 antibody did not precipitate the E-box segment of C/EBPα gene promoter from the lysates of differentiated human subcutaneous adipocytes (data not shown). Together, these findings suggest that during adipogenic differentiation, the binding partner of USF1 is switched from HDAC9 to p300, which in turn may facilitate adipogenic differentiation.
DISCUSSION
The major finding of the present study is the demonstration that HDAC9 functions in a histone deacetylase-independent manner to suppress adipogenic differentiation. We provide evidence to show that HDAC9 forms a complex with USF1, which accumulates at the C/EBPα gene promoter site and represses gene expression. Upon induction of adipogenic differentiation, HDAC9 is rapidly down-regulated. Concomitantly, p300 HAT is up-regulated and recruited to the USF1 complex at the C/EBPα gene promoter, permitting induction of C/EBPα expression and adipogenic differentiation. We propose that reciprocal regulation of HDAC9 and HAT proteins acts as a molecular switch to regulate adipogenic gene expression and differentiation.
HDAC9 is 1 of the 11 members of the HDAC family of proteins (33). Based on their structural characteristics, individual members of the HDAC family of proteins are grouped into classes I, II, and IV. Class I HDACs (HDAC1, -2, -3, and -8) are expressed in a wide range of tissues, present as multiprotein complexes within the cell nucleus, catalytically efficient to deacetylate histones, and functionally involved in transcriptional repression and epigenetic regulation. On the other hand, class IIa HDACs (HDAC4, -5, -7, and -9) exhibit restricted tissue expression, possess little catalytic activity, and are involved in transcriptional regulation of various development and differentiation processes. Class IIb HDACs (HDAC6 and -10) are major cytoplasmic deacetylases, whereas very little is known about the class IV HDAC, HDAC11.
Our study is the first to undertake a systematic examination of HDAC expression during in vitro adipogenic differentiation of freshly isolated human and murine preadipocytes. Among the HDAC family members, HDAC9 was noteworthy for its dramatic and consistent down-regulation both at the mRNA and protein levels during adipogenic differentiation of preadipocytes. Moreover, mature adipocytes isolated from tissue depots exhibited much lower HDAC9 levels compared with corresponding stromal preadipocytes. Interestingly, HDAC9 expression levels also were found to differ among visceral versus subcutaneous adipose depots. Visceral adipose tissues from both human and murine sources (omental and epididymal, respectively) exhibited much higher HDAC9 levels as compared with their subcutaneous counterparts. Similar findings were also observed in preadipocytes isolated from these depots. These differences in HDAC9 expression correlated with the potential of these preadipocytes to undergo in vitro adipogenic differentiation. Thus, when induced to undergo adipogenic differentiation, preadipocytes from the subcutaneous adipose tissues exhibited higher levels of expression of adipocyte differentiation-specific genes and more cytoplasmic lipid droplet accumulation compared with preadipocytes from the visceral sources. These findings suggest that differential HDAC9 expression could play a role in controlling depot-specific differences in adipose tissue function.
HDAC9 down-regulation preceded up-regulation of adipocyte differentiation-specific genes, suggesting that endogenous HDAC9 could inhibit adipogenic differentiation. This notion was reinforced by the finding that preadipocytes isolated from adipose tissue of HDAC9 gene knock-out mice exhibit accelerated in vitro adipogenic differentiation, whereas HDAC9 overexpression by transient transfection blocked adipogenic differentiation of 3T3-L1 preadipocytes. Our findings provide the first evidence to document that HDAC9 is a negative regulator of adipogenic differentiation, in keeping with the emerging view that members of the class IIa HDACs are important regulators of cellular differentiation processes (34). We propose that the elevated HDAC9 levels in preadipocytes function to maintain the undifferentiated state; upon exposure to appropriate adipogenic stimuli, HDAC9 down-regulation removes this block to allow the adipogenic differentiation process to occur. A similar negative regulatory role for HDAC9 in suppressing differentiation of myocytes and Treg cells has previously been proposed (26, 27).
The involvement of HDAC family of proteins in adipogenic differentiation was investigated previously by several laboratories (25, 35). Conflicting evidence supporting both positive and negative regulatory effects of HDAC proteins on adipogenic differentiation of 3T3-L1 preadipocytes were presented. None of these previous studies, however, examined HDAC9 expression during adipogenic differentiation. Most attention has been focused on class I HDACs; one study suggested that HDAC1 is negatively involved in adipogenic differentiation (35), whereas another study reported that both HDAC1 and HDAC2 are required for adipogenic differentiation of 3T3-L1 preadipocytes (25). We did not observe significant changes in HDAC1 or HDAC2 mRNAs or protein levels during adipogenic differentiation of human subcutaneous preadipocytes. Moreover, we found no evidence of mislocalization of class I HDACs in the cytoplasmic compartment during adipogenic differentiation of human preadipocytes (data not shown).
Several prior studies also utilized HDAC inhibitors as tools to examine the involvement of HDACs on adipogenic differentiation. Catalioto et al. (36) showed that adipogenic differentiation of cryopreserved human subcutaneous preadipocytes was blocked by HDAC inhibitors. Similarly, Haberland et al. (25) demonstrated that HDAC inhibitors blocked adipogenic differentiation of 3T3-L1 cells. Likewise, inhibition of adipogenic differentiation of 3T3-L1 cells by trichostatin A and a class I-selective HDAC inhibitor, MS275, was recently reported (37). In contrast, Yoo et al. (35) showed that HDAC1 down-regulation precedes adipogenic differentiation of 3T3-L1 cells, and the HDAC inhibitor butyrate dramatically enhanced adipogenic differentiation of these cells. Haberland et al. (25) proposed that the short chain fatty acid nature of butyrate and valproic acid, but not their HDAC inhibitory activity, contributes to induction of adipogenesis. Such findings are not entirely consistent, as several laboratories, including ours, observed profound inhibitory effects of valproic acid on adipogenic differentiation of human preadipocytes and 3T3-L1 cells (38–40). In contrast, butyrate had no effect on adipogenic differentiation of human preadipocytes in our cells. As both butyrate and valproic acid may have off-target effects, we utilized trichostatin A, a well characterized and selective pan-HDAC inhibitor. Trichostatin A, at a dose that effectively inhibited HDAC enzymatic activity, did not affect adipogenic differentiation of human subcutaneous preadipocytes. Trichostatin A did, however, inhibit (40–50%) adipogenic differentiation of 3T3-L1 cells, similar to results reported by Haberland et al. (25). These conflicting findings that HDAC inhibition suppresses adipogenic differentiation of 3T3-L1 cells, but not that of human preadipocytes, are not surprising. During adipogenic differentiation, growth-arrested 3T3-L1 preadipocytes need to undergo mitotic clonal expansion before adipogenic differentiation, and this clonal expansion process was shown to be sensitive to HDAC inhibitors (25). In contrast, human preadipocytes do not require such clonal expansion for adipogenic differentiation (41, 42) and, therefore, may bypass the step that is sensitive to HDAC inhibitor. Our finding that the N-terminal HDAC9 construct lacking the deacetylase domain was equally competent as the full-length HDAC9 to suppress adipogenesis in 3T3-L1 preadipocytes indicates that the deacetylase activity of HDAC9 is dispensable for its negative regulatory control of adipogenesis also in 3T3-L1 cells.
It is also important to point out that differences in cell types and culture conditions may be responsible for the divergent experimental results with HDAC inhibitors. We utilized freshly isolated preadipocytes from human adipose tissues and examined these cells at their early passages, and none of the cells was cryo-preserved. Our cells maintained their phenotypes in culture, and their expression profiles closely resembled findings in the native cells. Thus, we believe that our findings are relevant to mechanisms of adipogenic differentiation occurring in vivo.
Our findings suggest that HDAC9 down-regulation is a prerequisite for adipogenic differentiation of human preadipocytes. Because inhibiting deacetylase activity does not affect the differentiation process, the mechanism of HDAC9 inhibition of adipogenesis should be independent of deacetylase activity. We propose that HDAC9 functions as a transcriptional repressor to inhibit adipogenesis through a deacetylase-independent mechanism. Although HDAC9 has no DNA binding activity, it can interact with DNA binding transcription factors to indirectly block transcription. HDAC9 was shown previously to suppress myocyte differentiation by physically interacting with and blocking the transcriptional activity of MEF2 transcription factor at the myogenic gene promoter (26). Similarly, HDAC9 blocks Treg cell differentiation through interacting with and inhibiting FoxP3 (Forkhead box P3) transcriptional activity on cytokine gene promoters (27). Down-regulation of HDAC9, in response to appropriate stimuli, de-represses the transcriptional block to activate the gene expression program supporting differentiation of myocytes or Treg cells. HDAC9 repression of p53 transcriptional activity is recently reported (43), whereas another recent study reported a transcriptional activator role of HDAC9 (44).
We found that HDAC9 physically interacts with USF1 in preadipocytes. USF1 is a ubiquitously expressed transcription factor that regulates numerous genes in the glucose and lipid metabolic pathways (29, 30). USF1 also regulates the expression of C/EBPα (31) and other E-box containing genes critical for adipogenic differentiation (45, 46). Based on our finding that HDAC9 deficiency promotes enhanced expression of C/EBPα in differentiating adipocytes and that HDAC9 overexpression represses C/EBPα gene expression in differentiating 3T3-L1 cells along with the observation that HDAC9 physically interacts with and co-localizes with USF1 at the E-box site of the C/EBPα gene promoter in preadipocytes, we propose that HDAC9 represses C/EBPα gene expression in preadipocytes by inhibiting USF1 transcriptional activity. Consistent with this proposal, elevated C/EBPα expression in differentiated adipocytes parallels HDAC9 loss from the USF1 complex at this promoter site. Previous studies showed that recruitment of HP-1 (heterochromatin protein-1) and CtBP (C-terminal-binding protein) by class II HDACs contributes to their mechanism of transcriptional repression, independent of HDAC enzymatic activity (47, 48). We presume that a similar mechanism is involved in HDAC9 suppression of C/EBPα gene transcription in preadipocytes. Further investigations are needed to fully define the composition of the multiprotein repressor complex assembled by HDAC9 at the C/EBPα gene promoter in human preadipocytes and the critical structural determinants for HDAC9 to interact with USF1 and its other binding partners to repress adipogenic gene expression.
In view of the fact that HDACs and HATs reciprocally regulate various transcriptional processes, we predicted that HAT activation would positively regulate adipogenic differentiation. In support of our conjecture, we found that down-regulation of HDAC9 during adipogenic differentiation is paralleled by the up-regulation of certain members of the HAT family of proteins (GCN5, Tip60, P/CAF, and p300). This up-regulation of HATs is mediated through post-transcriptional mechanisms, because mRNA levels of these genes were unchanged during adipogenic differentiation. Post-transcriptional up-regulation of Tip60 was previously reported to occur during adipogenic differentiation of 3T3-L1 preadipocytes (49). The HAT proteins may, in turn, regulate various transcriptional pathways activated during adipogenesis. For example, Tip60 was shown to regulate PPARγ-dependent gene expression during adipogenic differentiation (49). Similarly, P/CAF was shown to regulate FAS gene expression in the liver (28), and a similar mechanism could also be operative in adipocytes. We probed for P/CAF, Tip60, GCN5, and p300 in the USF1 complex at the E-box region of human C/EBPα gene promoter in differentiated adipocytes and found the presence of only p300 with USF1, suggesting that this complex is functionally active in driving C/EBPα gene expression in differentiated adipocytes. Together, these findings suggest that reciprocal regulation of HDAC9 and p300 levels during adipogenic differentiation regulates C/EBPα gene expression, serving as a switch to repress or activate adipogenic differentiation, respectively. Because preadipocytes from HDAC9 knock-out mice do not undergo spontaneous in vitro adipogenesis, we propose that HDAC9 down-regulation is necessary but not sufficient to initiate the activation of adipogenic differentiation-specific gene expression program.
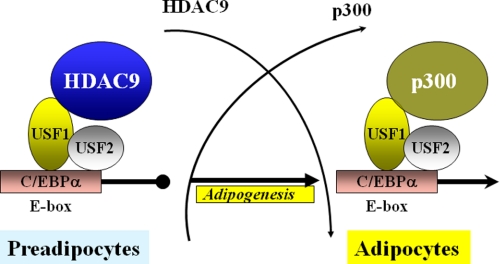
In summary, we have identified HDAC9 as a novel negative regulator of adipogenic differentiation. We propose that HDAC9 inhibits adipogenesis at least in part by inhibiting transcriptional processes independently of its HDAC enzymatic activity. Based on our findings, we propose a working model to suggest that the accumulation of HDAC9-USF1 at the E-box region of C/EBPα gene promoter suppresses adipogenic gene expression in preadipocytes. Upon induction of adipogenic differentiation, HDAC9 is down-regulated and replaced by p300 at the E-box site, which switches on adipogenic gene expression (Fig. 9). Because USF1 functions as a pleiotropic transcriptional regulator of various genes of the adipogenic differentiation pathway, it is conceivable that HDAC9 inhibition of USF1 transcriptional activity will impact multiple genes besides C/EBPα to influence adipogenic differentiation process. Further studies are needed to define the molecular determinants of the interactions between HDAC9, p300, and USF1 and the composition of multiprotein complex involved in regulation of the adipogenic differentiation process.
FIGURE 9.
Schematic model depicts the potential mechanisms of HDAC9 and p300 regulation of USF1-mediated C/EBPα gene expression in preadipocyte and differentiated adipocyte. The model predicts that HDAC9 maintains preadipocytes in the undifferentiated states by binding to and repressing USF1 transcriptional activity at the E-box site of the C/EBPα gene promoter. Induction of adipogenic differentiation promotes HDAC9 down-regulation and replacement by p300 at the E-box site of the C/EBPα gene promoter, thus switching on adipogenic gene expression. This general mechanism could be operative in regulating various USF1-dependent genes in preadipocytes versus differentiated adipocytes.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL076684, HL62984 (to N. L. W.), and ES019480 (to S. H.)

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- HDAC
- histone deacetylase
- HAT
- histone acetyltransferase
- TSA
- trichostatin A
- C/EBPα
- CAAT/enhancer-binding protein α
- P/CAF
- p300/cAMP-response element-binding protein-binding protein (CBP)-associated factor
- Tip60
- Tat-interacting protein 60
- USF
- upstream stimulatory factor
- FABP4
- fatty acid-binding protein 4
- PPARγ
- peroxisome proliferator-activated receptorγ
- Treg
- regulatory T cell
- SQ
- subcutaneous
- OM
- omental
- qRT
- quantitative real-time.
REFERENCES
- 1. Joe A. W., Yi L., Even Y., Vogl A. W., Rossi F. M. (2009) Stem Cells 27, 2563–2570 [DOI] [PubMed] [Google Scholar]
- 2. Kim J. Y., van de Wall E., Laplante M., Azzara A., Trujillo M. E., Hofmann S. M., Schraw T., Durand J. L., Li H., Li G., Jelicks L. A., Mehler M. F., Hui D. Y., Deshaies Y., Shulman G. I., Schwartz G. J., Scherer P. E. (2007) J. Clin. Invest. 117, 2621–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stefan N., Kantartzis K., Machann J., Schick F., Thamer C., Rittig K., Balletshofer B., Machicao F., Fritsche A., Häring H. U. (2008) Arch. Intern. Med. 168, 1609–1616 [DOI] [PubMed] [Google Scholar]
- 4. Marini M. A., Succurro E., Frontoni S., Hribal M. L., Andreozzi F., Lauro R., Perticone F., Sesti G. (2007) Diabetes Care 30, 2145–2147 [DOI] [PubMed] [Google Scholar]
- 5. Medina-Gomez G., Gray S. L., Yetukuri L., Shimomura K., Virtue S., Campbell M., Curtis R. K., Jimenez-Linan M., Blount M., Yeo G. S., Lopez M., Seppänen-Laakso T., Ashcroft F. M., Oresic M., Vidal-Puig A. (2007) PLoS Genet. 3, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith U., Hammarstedt A. (2010) Biochim. Biophys. Acta 1801, 377–380 [DOI] [PubMed] [Google Scholar]
- 7. Jernås M., Palming J., Sjöholm K., Jennische E., Svensson P. A., Gabrielsson B. G., Levin M., Sjögren A., Rudemo M., Lystig T. C., Carlsson B., Carlsson L. M., Lönn M. (2006) FASEB J. 20, 1540–1542 [DOI] [PubMed] [Google Scholar]
- 8. Monteiro R., de Castro P. M., Calhau C., Azevedo I. (2006) Obes. Surg. 16, 804–806 [DOI] [PubMed] [Google Scholar]
- 9. Skurk T., Alberti-Huber C., Herder C., Hauner H. (2007) J. Clin. Endocrinol. Metab. 92, 1023–1033 [DOI] [PubMed] [Google Scholar]
- 10. Krotkiewski M., Björntorp P., Sjöström L., Smith U. (1983) J. Clin. Invest. 72, 1150–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lundgren M., Svensson M., Lindmark S., Renström F., Ruge T., Eriksson J. W. (2007) Diabetologia 50, 625–633 [DOI] [PubMed] [Google Scholar]
- 12. Weyer C., Foley J. E., Bogardus C., Tataranni P. A., Pratley R. E. (2000) Diabetologia 43, 1498–1506 [DOI] [PubMed] [Google Scholar]
- 13. Brook C. G., Lloyd J. K. (1973) Arch. Dis. Child. 48, 301–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirsch J., Knittle J. L. (1970) Fed. Proc. 29, 1516–1521 [PubMed] [Google Scholar]
- 15. Kissebah A. H., Vydelingum N., Murray R., Evans D. J., Hartz A. J., Kalkhoff R. K., Adams P. W. (1982) J. Clin. Endocrinol. Metab. 54, 254–260 [DOI] [PubMed] [Google Scholar]
- 16. Salans L. B., Knittle J. L., Hirsch J. (1968) J. Clin. Invest. 47, 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lefterova M. I., Lazar M. A. (2009) Trends Endocrinol. Metab. 20, 107–114 [DOI] [PubMed] [Google Scholar]
- 18. Steger D. J., Grant G. R., Schupp M., Tomaru T., Lefterova M. I., Schug J., Manduchi E., Stoeckert C. J., Jr., Lazar M. A. (2010) Genes Dev. 24, 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haumaitre C., Lenoir O., Scharfmann R. (2009) Cell Cycle 8, 536–544 [DOI] [PubMed] [Google Scholar]
- 20. Saladi S. V., de la Serna I. L. (2010) Stem Cell Rev. 6, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vincent A., Van Seuningen I. (2009) Differentiation 78, 99–107 [DOI] [PubMed] [Google Scholar]
- 22. Crepaldi L., Riccio A. (2009) Epigenetics 4, 23–26 [DOI] [PubMed] [Google Scholar]
- 23. Chatterjee T. K., Stoll L. L., Denning G. M., Harrelson A., Blomkalns A. L., Idelman G., Rothenberg F. G., Neltner B., Romig-Martin S. A., Dickson E. W., Rudich S., Weintraub N. L. (2009) Circ. Res. 104, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petrie K., Guidez F., Howell L., Healy L., Waxman S., Greaves M., Zelent A. (2003) J. Biol. Chem. 278, 16059–16072 [DOI] [PubMed] [Google Scholar]
- 25. Haberland M., Carrer M., Mokalled M. H., Montgomery R. L., Olson E. N. (2010) J. Biol. Chem. 285, 14663–14670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haberland M., Arnold M. A., McAnally J., Phan D., Kim Y., Olson E. N. (2007) Mol. Cell. Biol. 27, 518–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L., de Zoeten E. F., Greene M. I., Hancock W. W. (2009) Nat. Rev. Drug Discov. 8, 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong R. H., Chang I., Hudak C. S., Hyun S., Kwan H. Y., Sul H. S. (2009) Cell 136, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu S., Mar-Heyming R., Dugum E. Z., Kolaitis N. A., Qi H., Pajukanta P., Castellani L. W., Lusis A. J., Drake T. A. (2010) Hum. Mol. Genet. 19, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rada-Iglesias A., Ameur A., Kapranov P., Enroth S., Komorowski J., Gingeras T. R., Wadelius C. (2008) Genome Res 18, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J. W., Monila H., Pandey A., Lane M. D. (2007) Biochem. Biophys. Res. Commun. 354, 517–521 [DOI] [PubMed] [Google Scholar]
- 32. Johnsson A., Durand-Dubief M., Xue-Franzén Y., Rönnerblad M., Ekwall K., Wright A. (2009) EMBO Rep. 10, 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haberland M., Montgomery R. L., Olson E. N. (2009) Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin M., Kettmann R., Dequiedt F. (2009) Int. J. Dev. Biol. 53, 291–301 [DOI] [PubMed] [Google Scholar]
- 35. Yoo E. J., Chung J. J., Choe S. S., Kim K. H., Kim J. B. (2006) J. Biol. Chem. 281, 6608–6615 [DOI] [PubMed] [Google Scholar]
- 36. Catalioto R. M., Maggi C. A., Giuliani S. (2009) Exp. Cell Res. 315, 3267–3280 [DOI] [PubMed] [Google Scholar]
- 37. Nebbioso A., Dell'Aversana C., Bugge A., Sarno R., Valente S., Rotili D., Manzo F., Teti D., Mandrup S., Ciana P., Maggi A., Mai A., Gronemeyer H., Altucci L. (2010) J Mol Endocrinol 45, 219–228 [DOI] [PubMed] [Google Scholar]
- 38. Lagace D. C., Nachtigal M. W. (2004) J. Biol. Chem. 279, 18851–18860 [DOI] [PubMed] [Google Scholar]
- 39. Qiao L., Schaack J., Shao J. (2006) Endocrinology 147, 865–874 [DOI] [PubMed] [Google Scholar]
- 40. Brown R., Imran S. A., Ur E., Wilkinson M. (2008) Neuroendocrinology 88, 25–34 [DOI] [PubMed] [Google Scholar]
- 41. Ross A. S., Tsang R., Shewmake K., McGehee R. E., Jr. (2008) Biochem. Biophys. Res. Commun. 366, 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qian S. W., Li X., Zhang Y. Y., Huang H. Y., Liu Y., Sun X., Tang Q. Q. (2010) BMC Dev. Biol. 10, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan Z., Peng L., Radhakrishnan R., Seto E. (2010) J. Biol. Chem. 285, 39329–39338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muralidhar S. A., Ramakrishnan V., Kalra I. S., Li W., Pace B. S. (2011) J. Biol. Chem. 286, 2343–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smih F., Rouet P., Lucas S., Mairal A., Sengenes C., Lafontan M., Vaulont S., Casado M., Langin D. (2002) Diabetes 51, 293–300 [DOI] [PubMed] [Google Scholar]
- 46. Wang D., Sul H. S. (1997) J. Biol. Chem. 272, 26367–26374 [DOI] [PubMed] [Google Scholar]
- 47. Zhang C. L., McKinsey T. A., Olson E. N. (2002) Mol. Cell. Biol. 22, 7302–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang CL, McKinsey TA, Lu JR., Olson EN. (2001) J. Biol. Chem. 276, 35–39 [DOI] [PubMed] [Google Scholar]
- 49. van Beekum O., Brenkman A. B., Grøntved L., Hamers N., van den Broek N. J., Berger R., Mandrup S., Kalkhoven E. (2008) Endocrinology 149, 1840–1849 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.