Abstract
Arabinogalactan proteins are proteoglycans found on the cell surface and in the cell walls of higher plants. The carbohydrate moieties of most arabinogalactan proteins are composed of β-1,3-galactan main chains and β-1,6-galactan side chains, to which other auxiliary sugars are attached. For the present study, an endo-β-1,3-galactanase, designated FvEn3GAL, was first purified and cloned from winter mushroom Flammulina velutipes. The enzyme specifically hydrolyzed β-1,3-galactan, but did not act on β-1,3-glucan, β-1,3:1,4-glucan, xyloglucan, and agarose. It released various β-1,3-galactooligosaccharides together with Gal from β-1,3-galactohexaose in the early phase of the reaction, demonstrating that it acts on β-1,3-galactan in an endo-fashion. Phylogenetic analysis revealed that FvEn3GAL is member of a novel subgroup distinct from known glycoside hydrolases such as endo-β-1,3-glucanase and endo-β-1,3:1,4-glucanase in glycoside hydrolase family 16. Point mutations replacing the putative catalytic Glu residues conserved for enzymes in this family with Asp abolished activity. These results indicate that FvEn3GAL is a highly specific glycoside hydrolase 16 endo-β-1,3-galactanase.
Keywords: Carbohydrate; Cell Wall; Enzyme Purification; Fungi; Hydrolases; Arabinogalactan Protein; Endo-beta-1,3-Galactanase; Glycoside Hydrolase; Winter Mushroom; Beta-1,3-Galactan
Introduction
Arabinogalactan proteins (AGPs)2 are a family of proteoglycans found on the cell surface and in the cell walls of higher plants. AGPs consist of a hydroxyproline-rich core protein and large arabinogalactan (AG) moieties, which generally account for more than 90% of total weight (1–3). AGs lacking a core protein can also be found in several plants. Gum arabic is a commercial mixture of AGPs from the acacia tree and widely used as a food stabilizer. A type of AG has been identified as the active ingredient in the herbal medicine Juzen-Taiho-To (4, 5). Although AGPs are differentiated by their core proteins, the AG moieties have a common overall structure, that is, β-1,3-galactan main chains to which β-1,6-galactan side chains are attached through O-6. The β-1,6-galactan side chains are further substituted with l-Ara and lesser amounts of other auxiliary sugars such as glucuronic acid (GlcUA), 4-O-methyl-glucuronic acid, l-rhamnose, and l-fucose (1–3).3
AGPs are implicated in various physiological phenomena including cell adhesion, stress resistance, programmed cell death, and the growth of pollen tubes in higher plants (6–10). Several lines of evidence have demonstrated the importance of the AG moieties for the functions of AGPs. For instance, β-glycosyl-Yariv reagent (chemical name, 1,3,5-tri(p-glycosyloxyphenylazo)-2,4,6-trihydroxybenzene), which selectively binds to the carbohydrate moieties of AGPs, perturbs development and growth in plants (11–15). An AGP of tobacco stylar transmitting tissue, named TTS protein, undergoes degradation of its carbohydrate moieties in pollen tubes, stimulating the elongation of the pollen tubes (9, 10). In addition, xylogen, which is a non-classical AGP from Zinnia (Zinnia elegans) mesophyll cells, induces the differentiation to tracheary elements. Importantly, the inductive activity is lost when the carbohydrate moieties are removed from xylogen by chemical treatment (16). The specific carbohydrate structures that are crucial for the functioning of these AGPs, however, have not been identified.
AG moieties can be degraded, modified, and investigated by using specific glycoside hydrolases (GHs). Endo-β-1,6-galactanase (EC 3.2.1.164) isolated from Trichoderma viride specifically acts on β-1,6-galactan side chains (17, 18). Based on hydrophobic cluster analysis, this endo-β-1,6-galactanase has been categorized into GH family 5 (19–21). Exo-β-1,3-galactanase (EC 3.2.1.145), belonging to GH family 43, hydrolyzes β-1,3-galactan main chains, bypassing β-1,6-galactan side chains. The enzyme can release Gal, β-1,6-galactooligosaccharides (β-1,6-Galn), and their derivatives substituted with l-arabinofuranosyl, l-fucosyl, and/or uronosyl residues from AGPs (22, 23). On the other hand, no endo-β-1,3-galactanase has so far been identified and satisfactorily described. Arabinogalactanase found in Rhizopus niveus was once labeled as an endo-β-1,3-galactanase (EC 3.2.1.90) (24–26) but never characterized, and the EC entry for endo-β-1,3-galactanase was deleted in 2001.
For the present study, an endo-β-1,3-galactanase, designated FvEn3GAL, was isolated and cloned from winter mushroom Flammulina velutipes. The enzyme specifically hydrolyzed β-1,3-galactan in an endo-fashion. Phylogenetic analysis including hypothetical bacterial fungal proteins indicated that the enzyme belongs to an unknown subgroup of GH family 16 and stands apart from known enzymes.
EXPERIMENTAL PROCEDURES
Culture of Native F. velutipes
F. velutipes (NBRC 7663) was distributed from NITE Biological Resource Center (Kisarazu, Japan). The mycelia were cultured in a liquid medium containing 2% (w/v) gum arabic as the carbon source, 0.6% (w/v) Tryptone, 0.3% (w/v) yeast extract, 1% (w/v) potassium phosphate, and 0.05% (w/v) magnesium sulfate at 30 °C for 20 days.
Purification of the Native Enzyme
Native FvEn3GAL was purified from the culture medium of F. velutipes mycelia. The culture medium was filtered through nylon mesh to remove cell debris. The pH of the medium was adjusted to 4.0 with acetic acid. The medium was adsorbed onto a carboxymethyl (CM)-Sepharose fast flow column (2.8 × 84 cm, GE Healthcare, Tokyo, Japan) that had been equilibrated with 20 mm acetate buffer (pH 4.0) and eluted with a linear gradient of 0–500 mm NaCl in the buffer. The active fraction was concentrated and then applied to a Sephacryl S-200 column (2.5 × 145 cm, GE Healthcare) equilibrated and eluted with the buffer. The active fraction eluted was collected and used for the analysis of enzyme properties.
cDNA Cloning
The purified native enzyme was separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (27), blotted onto a PVDF-Plus membrane (Osmonics, Moers, Germany), and subjected to an N-terminal amino acid sequence analysis with a protein sequencer (HP G1000A, Hewlett Packard, Palo Alto, CA). The corresponding partial cDNA was searched for in the cDNA database that was constructed for mycelia of F. velutipes by Beckman Coulter Genomics. For the determination of full-length cDNA sequence, rapid amplification of 5′ cDNA was performed with a GeneRacer kit (Invitrogen) using an internal specific primer (5′-AGTACTAGTCGGCGGGGATGGGT-3′).
Bioinformatics
The secondary structure of FvEn3GAL was predicted with the Phyre program (28). The alignment and phylogenetic analysis were performed by the ClustalW program using sequences of characterized GH 16 enzymes collected at the CAZy web site (29, 30). Information for the sequences including enzyme names, origins, and accession numbers is given in the supplemental Materials and Methods.
Expression of rFvEn3GAL in Pichia pastoris
FvEn3GAL cDNA corresponding to the mature form of the enzyme (Ala-22 to Asn-249) was amplified by PCR and subcloned into pPICzαC (Invitrogen). For the preparation of rPM1 and rPM2, point mutations E138D and E143D, respectively, were introduced into FvEn3GAL cDNA by PCR using the following sets of primers: PM1 forward (5′-GTTGATATCGGTGAATGGAAGGGC-3′) and PM1 reverse (5′-GTCGGGGGGCCATCCAGAC-3′) and PM2 forward (5′-TGGAAGGGCACCCCCGAC-3′) and PM2 reverse (5′-GTCACCGATATCTTCGGGG-3′), respectively, with KOD-Plus (Toyobo, Tokyo, Japan). The nucleotide sequences of the mutated fragments were confirmed with an ABI PRISM 310 genetic analyzer. The methylotrophic yeast P. pastoris strain KM71 was transformed with the linearized plasmid constructs with a multicopy Pichia expression kit (Invitrogen). The transformants were cultured for 2 days, and then the recombinant proteins were induced by 1% (v/v) methanol according to the manufacturer's instructions. The recombinant proteins were purified by the same conventional chromatography procedure that was used for the native enzyme.
Oligo- and Polysaccharides
Preparation of β-1,3-galactan and β-galactan I and II was from gum arabic (Sigma) by Smith degradation (22); β-1,3-galactobiose (Gal2) and β-1,3-galactotriose (Gal3) were prepared from larch wood arabinogalactan (22). To obtain β-1,3-galactotetraose (Gal4), β-1,3-galactopentaose (Gal5), and β-1,3-galactohexaose (Gal6), β-1,3-galactan was partially hydrolyzed with 40 mm trifluoroacetic acid at 100 °C for 60 min. The β-1,3-Galns were purified on a Bio-Gel P-2 (Bio-Rad) column. AGPs from radish roots and leaves and β-1,4- and β-1,6-Galns were prepared as described previously (17, 22, 31, 32). CM-cellulose 4M, β-1,4-galactan from lupin, and β-1,3:1,4-glucan from barley were purchased from Megazyme (Wicklow, Ireland). β-1,3:1,6-Glucan from Laminaria digitata, κ-carrageenan, and p-nitrophenyl (PNP)-glycosides were from Sigma. Keratan sulfate was from Seikagaku Co. (Tokyo, Japan). Agarose and CM-curdlan were from Wako (Osaka, Japan). Xyloglucan was prepared from tamarind as described (33).
Enzyme Assay
The activity of the native enzyme toward polysaccharides was measured using reaction mixtures (total volume, 0.1 ml) consisting of 0.1% (w/v) polysaccharide, 50 mm acetate buffer (pH 5.0), and the enzyme. The liberated sugars were determined reductometrically by the method of Nelson (34) and Somogyi (35). One unit of enzyme activity liberates 1 μmol of reducing sugar/min. The enzymatic activity toward Galn was measured using reaction mixtures (total volume, 8 μl) consisting of the enzyme, 2.5 mm oligosaccharides, and 10 mm acetate buffer (pH 5.0). Mono- and oligosaccharides in enzymatic hydrolysates were separated by thin layer chromatography (TLC) on Silica gel 60 F254 (Merck, Darmstadt, Germany) using 7:1:2 (v/v/v) 1-propanol/ethanol/water as solvent and detected by charring after spraying TLC plates with 20% (v/v) H2SO4-methanol (17, 18).
Analysis of Mode of Action
The mode of hydrolysis action was analyzed with a reaction mixture (total volume, 6 μl) containing the native enzyme, 1 mm β-1,3-Gal6, and 50 mm acetate buffer (pH 5.0). For the analysis of transglycosylation activity, 10 mm β-1,3-Gal3 was used as the substrate. After incubation at 37 °C, the reducing sugars in the reaction mixtures were coupled at their reducing terminals with p-aminobenzoic acid ethyl ester (ABEE) according to the method of Matsuura and Imaoka (36). The ABEE-derivatized sugars were analyzed on a high-performance liquid chromatography (HPLC) system equipped with a TSKgel Amide-80 (4.6 × 250 mm; Tosoh, Tokyo, Japan) column as described previously (18).
RESULTS
Purification of the Enzyme from the Culture Medium of F. velutipes
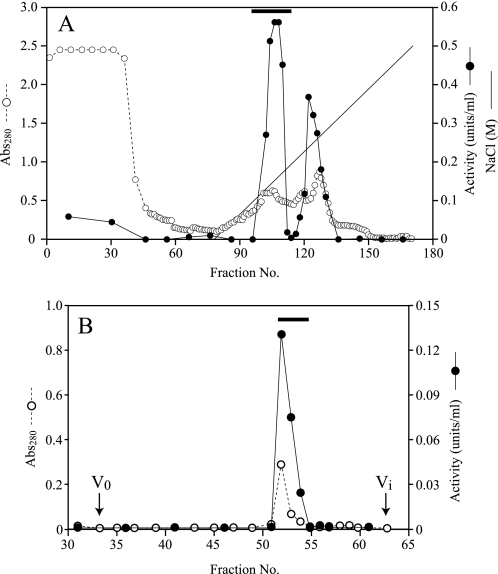
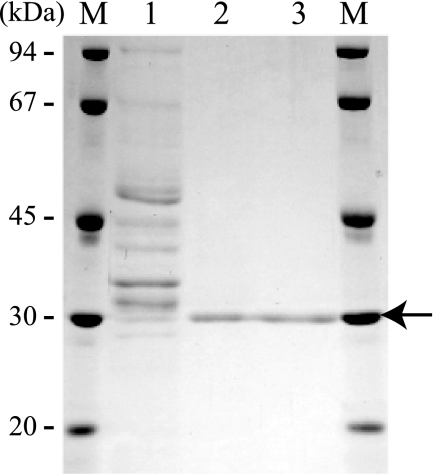
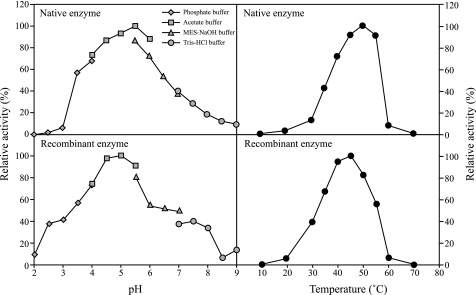
In preliminary experiments, the culture medium of F. velutipes mycelia showed high hydrolytic activity toward β-1,3-galactan that was prepared from gum arabic by Smith degradation when mycelia were grown with 2% (w/v) gum arabic as the carbon source. For the present study, the enzyme hydrolyzing β-1,3-galactan was purified by ion-exchange and gel-permeation chromatography from the culture medium (Fig. 1, supplemental Table S1). The purified enzyme showed high activity toward β-1,3-galactan (specific activity, 12.8 units/mg of protein) but failed to act on β-1,3-Gal2 and PNP-β-galactoside (β-Gal). Based on these observations, the enzyme was inferred to be an endo-β-1,3-galactanase and designated FvEn3GAL. The purified FvEn3GAL appeared as a single protein with a relative molecular mass of 30 kDa on SDS-PAGE (Fig. 2). The enzyme is probably N-glycosylated because the relative molecular mass apparently decreased when it was treated with endoglycosidase H (supplemental Fig. S1). The enzyme showed relatively high activity between pH 4 and pH 6. The optimal temperature for the enzyme action was 50 °C (Fig. 3).
FIGURE 1.
Purification of native enzyme from culture medium of F. velutipes. A, the culture medium (400 ml) was first applied onto a CM-Sepharose fast flow column. Bound proteins were eluted with a linear gradient of 0–500 mm NaCl. The fractions were assayed for endo-β-1,3-galactanase activity (closed circles) using β-1,3-galactan as the substrate and monitored for absorbance at 280 nm (Abs280, open circles). The solid line represents the concentration of NaCl. The fraction eluted with 150 mm NaCl represented by a horizontal bar was pooled, and the second active fraction eluted with 250 mm NaCl was not purified further in the present study. B, the active fraction from CM-Sepharose fast flow was concentrated and applied to gel-permeation chromatography on a Sephacryl S-200 column. V0 and Vt indicate void and inner volume of the column, respectively.
FIGURE 2.
SDS-PAGE of rFvEn3GAL at different purification steps. Enzyme fractions obtained after successive purification steps of the endo-β-1,3-galactanase from the culture medium of F. velutipes were analyzed on SDS-PAGE. Lane M, molecular mass markers; lane 1, culture medium of F. velutipes; lane 2, CM-Sepharose fast flow fraction; lane 3, Sephacryl S-200 fraction. The arrow indicates FvEn3GAL. Proteins in the gel were stained with Coomassie Brilliant Blue R-250.
FIGURE 3.
Effects of pH and temperature on actions of native and recombinant enzymes. Activity-pH curves were determined using 50 mm phosphate, acetate, MES-NaOH, and Tris-HCl buffers. Activity-temperature curves were determined by changing the reaction temperature.
Isolation of cDNA Encoding FvEn3GAL
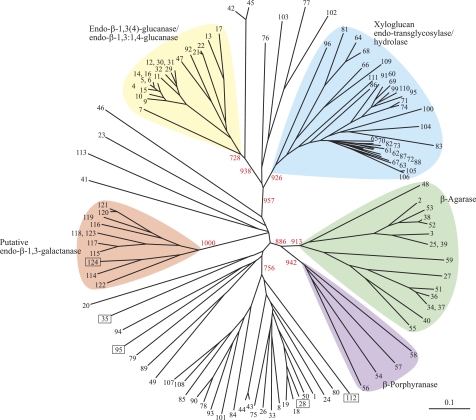
The N-terminal amino acid sequence determined for the purified FvEn3GAL was ATVIPANSFS. With this sequence, full-length cDNA encoding FvEn3GAL (accession number AB610981) was isolated by searching in the cDNA database constructed for F. velutipes mycelia followed by rapid amplification of 5′ cDNA. The cloned FvEn3GAL cDNA appeared to encode a polypeptide of 249 amino acids (molecular mass 26,690 Da) (supplemental Fig. S2). The deduced amino acid sequence showed low but significant similarities to GH 16 enzymes such as endo-β-1,3(4)-glucanase (EC 3.2.1.6) from Phanerochaete chrysosporium (17% identical, accession number BAC67687.1) (37). FvEn3GAL also had relatively high identity to several hypothetical proteins from Streptomyces hygroscopicus (accession number ZP_05512236, 42% identical) and Aspergillus fumigatus (accession number XP_001481519, 41% identical), respectively. Phylogenetic analysis revealed that FvEn3GAL forms a novel subgroup with the highly related sequences, apart from other GH 16 enzymes such as endo-β-1,3-glucanase (EC 3.2.1.39), endo-β-1,3(4)-glucanase, endo-β-1,3:1,4-glucanase (EC 3.2.1.73), and xyloglucan endo-transglycosylase/hydrolase (EC 2.4.1.207/3.2.1.151) (Fig. 4). The Phyre program predicted a secondary structure of FvEn3GAL closely related to an endo-β-1,3-glucanase from Nocardiopsis sp. F96, BglF (NspBglF), which is a member of GH family 16 (2.3 × 10−20) (28, 38). The overall structure of FvEn3GAL proposed by the program was a β-jellyroll fold, which is typical for GH 16 enzymes (39, 40). These results indicate that FvEn3GAL belongs to GH family 16 but is distinct from known enzymes.
FIGURE 4.
Relationships of FvEn3GAL with other members of GH family 16. The phylogenetic relationship of FvEn3GAL with other GH 16 enzymes was analyzed using the ClustalW program. Sequences longer than 500 amino acids are not included. The numbers in boxes are: 28, endo-β-1,3-glucanase NspBglF; 35, κ-carrageenase PcCgkA; 95, endo-β-1,3(4)-glucanase PcLam16A; 112, endo-β-galactosidase/keratanase; 124, FvEn3GAL. 114–123 are uncharacterized proteins. Other information including enzyme names, origins, and accession numbers is given in the supplemental Materials and Methods. The bootstrap values for major branches are shown. The horizontal bar indicates 0.1 substitutions per site.
Properties of the Recombinant Protein
The FvEn3GAL open reading frame, except for the signal peptide, was fused to a yeast secretion signal (α-factor) and heterologously expressed in the methylotrophic yeast P. pastoris. Recombinant FvEn3GAL (rFvEn3GAL) was purified by conventional chromatography (supplemental Table S2). The purified rFvEn3GAL appeared as a single band with a relative molecular mass of 32 kDa (supplemental Fig. S1) and showed hydrolytic activity toward β-1,3-galactan (specific activity, 7.0 units/mg of protein). As is the case for the native enzyme, rFvEn3GAL also appeared to be N-glycosylated (supplemental Fig. S1). The activity of rFvEn3GAL was high between pH 4 and pH 5.5. The optimal temperature for enzyme action was 45 °C (Fig. 3).
In general, enzymes belonging to GH family 16 possess a highly conserved catalytic motif, EXDX(X)E, in which the first Glu residue functions as the nucleophile and the last functions as the Brønsted acid/base (Fig. 5) (41, 42). To examine the importance of the residues for the activity of the enzyme, rFvEn3GALs with E138D or E143D mutations, designated rPM1 and rPM2, respectively, were prepared (Fig. 5, supplemental Fig. S1). Neither rPM1 nor rPM2 hydrolyzed β-1,3-galactan at all, demonstrating the importance of these Glu residues for activity.
FIGURE 5.
Replacement of putative catalytic residues of FvEn3GAL. The putative catalytic motif of FvEn3GAL was aligned with those of other GH 16 enzymes by the pairwise method using the ClustalW program. Gaps (–) were introduced to achieve maximum similarity. Amino acid residues identical for all nine sequences are highlighted in black. The mutations E138D and E143D, introduced into rPM1 and rPM2, respectively, are shown.
Substrate Specificity of Native and Recombinant Enzymes
The substrate specificity of the native FvEn3GAL was determined using various polysaccharides as substrates. The native enzyme substantially hydrolyzed β-1,3-galactan prepared from gum arabic by triple Smith degradation but failed to act on β-1,3:1,4-glucan, β-1,3:1,6-glucan, β-1,3-glucan, xyloglucan, keratan sulfate, κ-carrageenan, and agarose, which are substrates for other members of GH family 16 (Table 1). PNP-β-Gal, PNP-β-glucoside, PNP-β-glucuronide, and PNP-β-xyloside did not serve as substrate for the enzyme, and β-1,4-galactan was also resistant to hydrolysis by the enzyme. The rFvEn3GAL exhibited substrate specificity essentially identical to the native enzyme, demonstrating the high specificity of FvEn3GAL toward β-1,3-galactosyl residues (Table 1). The relative activities of the native and recombinant enzymes toward radish AGPs and gum arabic were low when compared with that toward β-1,3-galactan. These observations suggest that the β-1,6-galactosyl branches of AG moieties hinder the access of the enzyme to the β-1,3-galactan main chains. Supporting this suggestion is the fact that removal of β-1,6-galactosyl branches from gum arabic by Smith degradation made it more susceptible to the enzyme, showing that β-galactan II with fewer β-1,6-galactosyl branches is a better substrate than gum arabic and β-galactan I (Table 1) (22, 43). It seems likely that FvEn3GAL acted only on the limited parts of β-1,3-galactan main chains devoid of branches in the AGPs.
TABLE 1.
Substrate specificities of native and recombinant enzymes
| Substrate | Relative activitya |
|
|---|---|---|
| Native enzyme | rFvEn3GAL | |
| % | ||
| β-Galactans | ||
| β-1,3-Galactan prepared from gum arabic | 100 | 100 |
| Gum arabic from acacia tree | Trace | Trace |
| β-Galactan I prepared from gum arabic | 14 | 9 |
| β-Galactan II prepared from gum arabic | 50 | 46 |
| Native AGP from radish leaves | 7 | 8 |
| Native AGP from radish roots | 5 | 5 |
| β-1,4-Galactan from lupin | 0 | 0 |
| Substrates for other GH 16 enzymes | ||
| Agarose | 0 | 0 |
| Curdlan (β-1,3-glucan)b | 0 | 0 |
| CM-curdlan (β-1,3-glucan) | 0 | 0 |
| CM-cellulose (β-1,4-glucan) | 0 | 0 |
| β-1,3:1,4-Glucan from barley | 0 | 0 |
| β-1,3:1,6-Glucan from L. digitata | 0 | 0 |
| Xyloglucan from tamarind | 0 | 0 |
| κ-Carrageenan | 0 | 0 |
| Keratan-sulfate from bovine cornea | 0 | 0 |
a Activity is expressed in percentage of that toward β-1,3-galactan.
b The enzymes were incubated with this polysaccharide at a concentration of 0.5 mg/ml. The concentration of all other polysaccharides was 1.0 mg/ml.
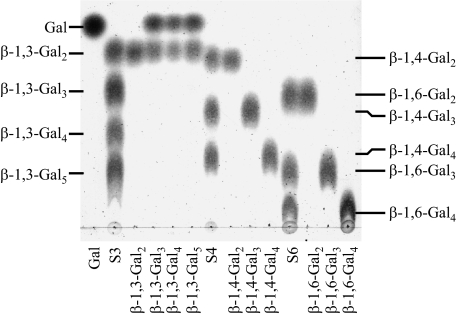
The activity of the native enzyme toward Galn was also examined using β-1,3-, β-1,4-, and β-1,6-Galn. The enzyme hydrolyzed β-1,3-Gal3, β-1,3-Gal4, and β-1,3-Gal5 into Gal and β-1,3-Gal2, whereas β-1,4- and β-1,6-Galn were impervious to the enzyme (Fig. 6). The enzyme probably requires at least three contiguous β-1,3-galactosyl residues for hydrolysis as it failed to act on β-1,3-Gal2. These results confirm the high specificity of the enzyme toward β-1,3-galactosyl residues.
FIGURE 6.
Substrate specificity of FvEn3GAL toward Galns. The products of various oligosaccharides by the action of the native FvEn3GAL were analyzed on TLC. Lane Gal, standard Gal; lane S3, standard β-1,3-Galn with degree of polymerization 2–5; lane S4, standard β-1,4-Galn with degree of polymerization 2–4; lane S6, standard β-1,6-Galn with degree of polymerization 2–4. Susceptibility of these oligomers to the enzyme was examined as indicated below each lane.
The effect of substrate concentration on the activity of the native enzyme was examined using β-1,3-galactan, β-1,3-Gal3, and β-1,3-Gal4. The resulting Km, kcat, and catalytic efficiency values are listed in Table 2. As the Km value (0.58 mm) toward β-1,3-Gal4 was lower than that (1.25 mm) toward β-1,3-Gal3, the longer β-1,3-Galn probably serves as a better substrate for FvEn3GAL.
TABLE 2.
Kinetics of native FvEn3GAL
| Substrate | Kma | kcata | kcat/Kma |
|---|---|---|---|
| s | m/s | ||
| β-1,3-Galactan | 0.137 mg/ml | 7.56 | —b |
| β-1,3-Gal3 | 1.25 mm | 5.47 | 4.38 × 103 |
| β-1,3-Gal4 | 0.57 mm | 6.50 | 1.14 × 104 |
a To examine the effects of substrates, reactions were carried out with varying concentrations of β-1,3-galactan (0.05–2.0 mg/ml), β-1,3-Gal3 (0.25–2.5 mm), and β-1,3-Gal4 (0.25–2.5 mm).
b The value was not calculated.
Mode of Action
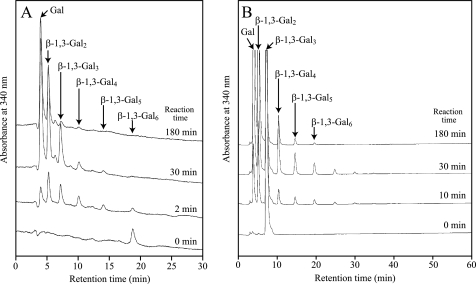
To understand the mode of action of FvEn3GAL, the products released from β-1,3-Gal6 by the action of the native enzyme were analyzed. The enzyme released β-1,3-Gal2, β-1,3-Gal3, β-1,3-Gal4, and β-1,3-Gal5 together with Gal in the initial phase of the hydrolysis reaction (Fig. 7A, supplemental Fig. S3). This is convincing proof that the enzyme acts on the oligosaccharide in an endo-fashion because exo-acting enzymes, including exo-β-1,3-galactanase and β-galactosidase, would accumulate Gal and β-1,3-Gal5 in the initial phase. Upon prolonged incubation, β-1,3-Gal6 was mostly hydrolyzed into Gal and β-1,3-Gal2, although a small amount of β-1,3-Gal3 remained in the reaction mixture. These observations also imply that the enzyme requires at least three contiguous β-1,3-galactosyl residues to act on.
FIGURE 7.
Mode of action of FvEn3GAL. A, the mode of action of the native FvEn3GAL was examined using 1 mm β-1,3-Gal6 as substrate. B, the transglycosylation reaction was performed using 10 mm β-1,3-Gal3 as substrate. Reducing sugars released from each substrate were derivatized with ABEE and analyzed by HPLC. Arrows indicate the elution positions of ABEE-derivatized Gal and β-1,3-Galns with degree of polymerization 2–6.
Through a “retaining” mechanism, GH 16 enzymes may catalyze not just hydrolysis but also transglycosylation, in which glycosyl transfer to carbohydrate occurs. In fact, GH family 16 includes xyloglucan endo-transglycosylases that catalyze transglycosylation rather than hydrolysis (39, 44). We examined the transglycosylation activity of native FvEn3GAL in the presence of a high concentration (10 mm) of β-1,3-Gal3. The native enzyme formed a small amount of the transglycosylation products β-1,3-Gal4, β-1,3-Gal5, and β-1,3-Gal6, as well as the hydrolysis products, Gal and β-1,3-Gal2 (Fig. 7B). We conclude that the linkage formed by the transglycosylation reaction is a β-1,3-galactosidic type linkage because we found that the transglycosylation products were completely hydrolyzed into Gal by the action of exo-β-1,3-galactanase specific to β-1,3-galactosyl residues (supplemental Fig. S4A). Because the transglycosylation reaction was apparently weak when compared with the hydrolysis activity, it is not likely that transglycosylation is the predominant type of reaction catalyzed by this enzyme in nature. As no transglycosylation activity toward 10 mm β-1,6-Gal3 was observed (supplemental Fig. S4B), the transglycosylation activity of the enzyme is presumably specific to β-1,3-galactosyl residues.
DISCUSSION
On the basis of the high specificity toward β-1,3-galactan and its mode of action, FvEn3GAL can be classified as an endo-β-1,3-galactanase. With regard to the arabinogalactanase previously found in R. niveus, we could not find definitive information about its mode of action (24–26). The enzyme has been found to hydrolyze coffee AG into β-1,6-Galn; however, these oligosaccharides could also be released by the action of exo-β-1,3-galactanase (22, 43). We could not find a sequence related to FvEn3GAL in the genomic database of an organism of the Rhizopus genus, Rhizopus oryzae (Rhizopus oryzae Database available from the Broad Institute). Even if we assume that the Rhizopus arabinogalactanase hydrolyzes β-1,3-galactan main chains of coffee AG in an endo-fashion, it would have to be an enzyme distinct from FvEn3GAL.
GH family 16 comprises endo-β-1,3-glucanase, endo-β-1,3(4)-glucanase, endo-β-1,3:1,4-glucanase, β-agarase (EC 3.2.1.81), xyloglucan endo-transglycosylase/hydrolase, keratan-sulfate endo-β-1,4-galactosidase (EC 3.2.1.103), κ-carrageenase (EC 3.2.1.83), and porphyranase (no EC entry) (29, 45). Endo-β-1,3-galactanase will be a new member of GH family 16. Although the substrate specificity of FvEn3GAL was apparently different from these known GH 16 enzymes, the cleavage of β-1,3-galactosyl linkages by FvEn3GAL reminds us of the reaction catalyzed by endo-β-1,3-glucanase. Based on the wide distribution and sequence diversity of endo-β-1,3-glucanase, it has been proposed that the GH 16 enzymes share a common ancestor having laminarinase activity (46, 47). FvEn3GAL has probably evolved from the same ancestor.
Endo-β-1,3-galactanase is likely distributed at least in several fungi and bacteria such as Aspergillus flavus, Neurospora crassa, and S. hygroscopicus, whereas we could not find any closely related sequences in plants. Endo-β-1,3-galactanases are expected to benefit organisms by accelerating the degradation of plant AGPs and AGs, which are utilized as a source of carbon. Because FvEn3GAL alone is restricted to a limited part of β-1,3-galactan main chains of radish AGPs and gum arabic, natural AGPs will be efficiently degraded only by synergistic action with other GHs hydrolyzing the β-1,6-galactan side chains. Indeed, the culture medium of F. velutipes evinced α-l-arabinofuranosidase and β-galactosidase activities beside the endo-β-1,3-galactanase activity investigated here. The degradation by FvEn3GAL of plant material through the release of large AG fragments from cell walls by acting on the exposed parts of β-1,3-galactan main chains of AGPs and AGs may be an important contribution to fungal digestive strategies in nature.
In conclusion, the present study has for the first time isolated and cloned an endo-β-1,3-galactanase. FvEn3GAL from F. velutipes is a member of GH family 16 and seems to belong to a novel subgroup, with a number of hypothetical fungal proteins, that stands apart from known enzymes in family 16. The enzyme, in concert with endo-β-1,6-galactanase and exo-β-1,3-galactanase, should prove a useful tool in efforts to reveal the specific carbohydrate structure of AGPs and AGs and determine their particular functions.
Supplementary Material
Acknowledgment
We are grateful to emeritus Prof. Masaakira Maeda for valuable advice on the preparation of substrate polysaccharides.
This work was supported in part by a grant-in-aid (Development of Biomass Utilization Technologies for Revitalizing Rural Areas) from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AB610981.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Materials and Methods, Tables S1 and S2, and Figs. S1–S4.
Sugars mentioned in this paper belong to the d-series unless otherwise noted.
- AGP
- arabinogalactan protein
- AG
- arabinogalactan
- ABEE
- p-aminobenzoic acid ethyl ester
- DP
- degree of polymerization
- Galn
- galactooligosaccharide
- Gal2
- β-1,3-galactobiose
- Gal3
- β-1,3-galactotriose
- Gal4
- β-1,3-galactotetraose
- Gal5
- β-1,3-galactopentaose
- Gal6
- β-1,3-galactohexaose
- GH
- glycoside hydrolase
- CM
- carboxymethyl
- PNP
- p-nitrophenyl
- r
- recombinant.
REFERENCES
- 1. Fincher G. B., Stone B. A., Clarke A. E. (1983) Annu. Rev. Plant Physiol. 34, 47–70 [Google Scholar]
- 2. Nothnagel E. A. (1997) Int. Rev. Cytol. 174, 195–291 [DOI] [PubMed] [Google Scholar]
- 3. Seifert G. J., Roberts K. (2007) Annu. Rev. Plant Biol. 58, 137–161 [DOI] [PubMed] [Google Scholar]
- 4. Majewska-Sawka A., Nothnagel E. A. (2000) Plant Physiol. 122, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kiyohara H., Matsumoto T., Yamada H. (2002) Phytomedicine 9, 614–624 [DOI] [PubMed] [Google Scholar]
- 6. Showalter A. M. (2001) Cell. Mol. Life Sci. 58, 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shi H., Kim Y., Guo Y., Stevenson B., Zhu J. K. (2003) Plant Cell 15, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao M., Showalter A. M. (1999) Plant J. 19, 321–331 [DOI] [PubMed] [Google Scholar]
- 9. Cheung A. Y., Wang H., Wu H. M. (1995) Cell 82, 383–393 [DOI] [PubMed] [Google Scholar]
- 10. Wu H. M., Wang H., Cheung A. Y. (1995) Cell 82, 395–403 [DOI] [PubMed] [Google Scholar]
- 11. Yariv J., Rapport M. M., Graf L. (1962) Biochem. J. 85, 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komalavilas P., Zhu J. K., Nothnagel E. A. (1991) J. Biol. Chem. 266, 15956–15965 [PubMed] [Google Scholar]
- 13. Willats W. G., Knox J. P. (1996) Plant J. 9, 919–925 [DOI] [PubMed] [Google Scholar]
- 14. Ding L., Zhu J. K. (1997) Planta 203, 289–294 [DOI] [PubMed] [Google Scholar]
- 15. Park M. H., Suzuki Y., Chono M., Knox J. P., Yamaguchi I. (2003) Plant Physiol. 131, 1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Motose H., Sugiyama M., Fukuda H. (2004) Nature 429, 873–878 [DOI] [PubMed] [Google Scholar]
- 17. Okemoto K., Uekita T., Tsumuraya Y., Hashimoto Y., Kasama T. (2003) Carbohydr. Res. 338, 219–230 [DOI] [PubMed] [Google Scholar]
- 18. Kotake T., Kaneko S., Kubomoto A., Haque M. A., Kobayashi H., Tsumuraya Y. (2004) Biochem. J. 377, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henrissat B. (1991) Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henrissat B., Bairoch A. (1993) Biochem. J. 293, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henrissat B., Bairoch A. (1996) Biochem. J. 316, 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsumuraya Y., Mochizuki N., Hashimoto Y., Kovác P. (1990) J. Biol. Chem. 265, 7207–7215 [PubMed] [Google Scholar]
- 23. Ichinose H., Yoshida M., Kotake T., Kuno A., Igarashi K., Tsumuraya Y., Samejima M., Hirabayashi J., Kobayashi H., Kaneko S. (2005) J. Biol. Chem. 280, 25820–25829 [DOI] [PubMed] [Google Scholar]
- 24. Hashimoto Y., Tsujisaka Y., Fukumoto J. (1969) Nippon Nogeikagaku Kaishi 43, 831–836 [Google Scholar]
- 25. Hashimoto Y. (1971) Nippon Nogeikagaku Kaishi 45, 147–150 [Google Scholar]
- 26. Dekker R. F., Richards G. N. (1976) Adv. Carbohydr. Chem. Biochem. 32, 277–352 [DOI] [PubMed] [Google Scholar]
- 27. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 29. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233-D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsumuraya Y., Ogura K., Hashimoto Y., Mukoyama H., Yamamoto S. (1988) Plant Physiol. 86, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsumuraya Y., Hashimoto Y., Yamamoto S., Shibuya N. (1984) Carbohydr. Res. 134, 215–228 [Google Scholar]
- 33. Kooiman P. (1961) Rec. Trav. Chim. Pays-Bas. 80, 849–865 [Google Scholar]
- 34. Nelson N. (1944) J. Biol. Chem. 153, 375–380 [Google Scholar]
- 35. Somogyi M. (1952) J. Biol. Chem. 195, 19–23 [PubMed] [Google Scholar]
- 36. Matsuura F., Imaoka A. (1988) Glycoconj. J. 5, 13–26 [Google Scholar]
- 37. Kawai R., Igarashi K., Yoshida M., Kitaoka M., Samejima M. (2006) Appl. Microbiol. Biotechnol. 71, 898–906 [DOI] [PubMed] [Google Scholar]
- 38. Fibriansah G., Masuda S., Koizumi N., Nakamura S., Kumasaka T. (2007) Proteins 69, 683–690 [DOI] [PubMed] [Google Scholar]
- 39. Baumann M. J., Eklöf J. M., Michel G., Kallas A. M., Teeri T. T., Czjzek M., Brumer H., 3rd (2007) Plant Cell 19, 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vasur J., Kawai R., Andersson E., Igarashi K., Sandgren M., Samejima M., Ståhlberg J. (2009) FEBS J. 276, 3858–3869 [DOI] [PubMed] [Google Scholar]
- 41. Hahn M., Olsen O., Politz O., Borriss R., Heinemann U. (1995) J. Biol. Chem. 270, 3081–3088 [DOI] [PubMed] [Google Scholar]
- 42. Masuda S., Endo K., Koizumi N., Hayami T., Fukazawa T., Yatsunami R., Fukui T., Nakamura S. (2006) Extremophiles 10, 251–255 [DOI] [PubMed] [Google Scholar]
- 43. Kotake T., Kitazawa K., Takata R., Okabe K., Ichinose H., Kaneko S., Tsumuraya Y. (2009) Biosci. Biotechnol. Biochem. 73, 2303–2309 [DOI] [PubMed] [Google Scholar]
- 44. Nishitani K., Tominaga R. (1992) J. Biol. Chem. 267, 21058–21064 [PubMed] [Google Scholar]
- 45. Hehemann J. H., Correc G., Barbeyron T., Helbert W., Czjzek M., Michel G. (2010) Nature 464, 908–912 [DOI] [PubMed] [Google Scholar]
- 46. Barbeyron T., Gerard A., Potin P., Henrissat B., Kloareg B. (1998) Mol. Biol. Evol. 15, 528–537 [DOI] [PubMed] [Google Scholar]
- 47. Michel G., Chantalat L., Duee E., Barbeyron T., Henrissat B., Kloareg B., Dideberg O. (2001) Structure 9, 513–525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.