Abstract
The Collaborative Cross (CC) is a mouse recombinant inbred strain panel that is being developed as a resource for mammalian systems genetics. Here we describe an experiment that uses partially inbred CC lines to evaluate the genetic properties and utility of this emerging resource. Genome-wide analysis of the incipient strains reveals high genetic diversity, balanced allele frequencies, and dense, evenly distributed recombination sites—all ideal qualities for a systems genetics resource. We map discrete, complex, and biomolecular traits and contrast two quantitative trait locus (QTL) mapping approaches. Analysis based on inferred haplotypes improves power, reduces false discovery, and provides information to identify and prioritize candidate genes that is unique to multifounder crosses like the CC. The number of expression QTLs discovered here exceeds all previous efforts at eQTL mapping in mice, and we map local eQTL at 1-Mb resolution. We demonstrate that the genetic diversity of the CC, which derives from random mixing of eight founder strains, results in high phenotypic diversity and enhances our ability to map causative loci underlying complex disease-related traits.
Mouse models have made significant contributions to our understanding of human biology and disease. Analogous physiology and anatomy between humans and mice enables the study of human diseases in an organism that is amenable to laboratory investigation. Moreover, we can manipulate the mouse experimentally, and as a result, characterize disease mechanisms in ways that are impossible in humans. Mouse models allow application of powerful genomic techniques to identify and validate causative genes underlying human diseases with complex etiologies.
Recombinant inbred lines (RILs) are one of the many mouse resources that have been developed to advance complex disease research. RILs are well suited for modeling variable susceptibility to disease among individuals, particularly for common diseases that can be attributed to the combined effects of multiple genetic and environmental factors. RIL panels are genetic reference populations that can be studied across a range of perturbations, and have the added benefit of known population structures (Churchill 2007). However, existing RIL panels have limitations such as large regions of the genome lacking genetic variation and low statistical power due to small panel sizes that make them suboptimal for genome-wide analyses with contemporary “-omic” technologies. This study demonstrates the utility of a next-generation RIL resource specifically designed to overcome limitations of current mouse resources.
The Collaborative Cross
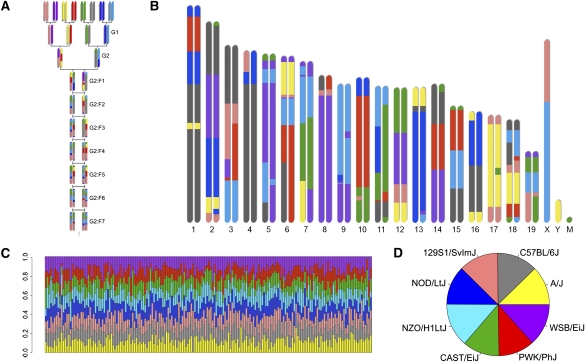
The Collaborative Cross (CC) is currently under development as a next-generation platform for mammalian systems genetics (Threadgill et al. 2002; Churchill et al. 2004). The CC will consist of a panel of RILs derived from eight diverse founder strains, including five classical inbred strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, and NZO/H1LtJ) and three wild-derived strains representing the three Mus musculus subspecies (CAST/EiJ, PWK/PhJ, and WSB/EiJ). The eight founder strains capture a level of genetic diversity unsurpassed by any existing RIL panel or other extant mouse resource (Roberts et al. 2007). The CC will comprise substantially more lines than existing RIL panels and, thus, it will provide more statistical power. Because the captured genetic variation will be randomly distributed among the lines, the CC will minimize spurious associations in mapping studies. In the CC RIL breeding scheme (Fig. 1A), founder strains contribute equally to each of the independently bred lines. Two generations of outcrossing are required to produce generation G2:F1 mice, whose genomes include balanced contributions from the eight founder strains. After this generation, CC strains become inbred by brother–sister mating. The rate of inbreeding is rapid at first and slows with each subsequent generation.
Figure 1.
Genetic properties of the Pre-CC panel. (A) Eight founder's inbred strains were bred according to the original funnel design (Churchill et al. 2004). The G2:F1 generation has contributions from all eight founders. Siblings are mated in this and all subsequent generations until the lines are isogenic. (B) An example of a pre-CC genome from a single G2:F7 animal. Ancestry was inferred by comparing pre-CC and founder genotypes using an HMM. (C) Founder contributions to any particular line (columns) vary from 1.3% to 30.7%. (D) Each of the eight founders contributes between 11.4% and 13.5% to 184 lines from the exercise behavior and metabolism phenotyping arm.
Study design
We performed an experiment, referred to as the pre-CC study, to investigate the genetic architecture and phenotypic characteristics of the incipient CC strains. We observed variation in a broad range of phenotypes, examined the genetic properties of the panel, and developed and tested data analysis strategies that will also apply to the finished CC strains.
The pre-CC study used partially inbred mice from 220 CC lines that were initiated at the Oak Ridge National Laboratory (ORNL) (Chesler et al. 2008). Mice ranging from generation G2:F5 to G2:F12 were distributed to four distinct phenotyping arms (Supplemental Fig. 1; Supplemental Table 1). The phenotyping arms focused on (1) exercise behavior and metabolism; (2) H1N1 influenza susceptibility; (3) SARS-CoV susceptibility; and (4) allergic airway disease. Phenotyping for the first three arms was conducted at the University of North Carolina–Chapel Hill (UNC), while the fourth arm was conducted at the National Human Genome Research Institute (NHGRI). To the extent possible, littermates from each CC line were distributed across the four arms of the study. Due to the complex logistics of breeding and shipping mice, the sample sizes varied among study arms with the largest arm containing 184 mice. Tissues from terminal dissection of the mice were sent to National Jewish Health (NJH) for characterization of innate immune response and to The Jackson Laboratory (JAX) for gene expression analysis. In addition to these four study arms, phenotyping efforts were also performed using different sets of mice at the ORNL and Tel Aviv University (TAU) breeding facilities.
Here we present the first genetic data and analysis from the incipient CC lines: 33.4 million genotypes, two phenotypes, and 23,176 gene expression measurements from one arm of the pre-CC study. These data enabled us to illustrate the properties of the study design and to provide an overview of the analysis techniques and strategies developed for the CC. High-density genotype data allowed us to ask questions about the breeding process and compare our expectations with empirical observations. We estimated ancestry across each pre-CC genome, and these analyses indicated that the incipient CC strains are genetically diverse and include balanced contributions from each founder strain. Additionally, we mapped quantitative trait loci (QTL) for white head-spotting, a discrete character observed in one of the eight founder strains; baseline body weight, a trait known to be highly polygenic; and liver mRNA expression profile. Thus, we highlight results from three types of traits–Mendelian, complex, and molecular. An ancestry-based approach proved superior to marker-based methods for mapping QTL in the pre-CC population, and facilitated new approaches to downstream analysis that are introduced here. These genetic analysis strategies and the population genetic architecture are the common elements shared across the four pre-CC study arms.
Results
PreCC genomes have balanced founder contributions
Male mice from the exercise behavior and metabolism study arm (n = 184) were genotyped at UNC using a high-density genotyping platform (Yang et al. 2009). Marker density far exceeds recombination in the panel, making it possible to observe effectively every transition in the ancestry of each mouse. On average, these mice had undergone 6.7 generations of inbreeding and were 84.1% homozygous. We applied a hidden Markov model (HMM) to the genotypes to infer from which of the eight CC founder strains each allele was inherited (Fig. 1B; Mott et al. 2000; Liu et al. 2010) and used these results to assess the properties of the panel as a whole. Genome segments were defined as the regions between inferred recombination breakpoints. Pre-CC autosomal genomes had 142.3 segments on average (st dev. = 21.8). Simulated CC genomes (Broman 2005) had 134 segments on average, which is significantly fewer (one sample t-test, t = 5.16, P ≤ 0.0001). This is consistent with a model in which some of the observed segments will be lost during the process of inbreeding. Segment sizes were exponentially distributed with a median of 10.46 Mb, which is smaller than the simulated median segment size of ∼17 Mb (8.5 cM).
Founder contributions to individual lines varied from 1.3% to 30.7% (Fig. 1C), with one exception. A small number of lines that were determined to include breeding errors were excluded from the study (see Supplemental material). When averaged across all pre-CC lines, each of the eight founders contributed uniformly to the population (11.4%–13.5%) (Fig. 1D). Local allele frequencies in each genomic segment ranged from 4.1% to 27.3%, demonstrating that genetic variation from each of the eight founder strains was present at all loci (Fig. 2). Allele frequencies deviated significantly from the expected 12.5% only in extremely short intervals (spikes on Chr 2 and Chr 15 in Fig. 2), most likely due to uncertainty in haplotype inference.
Figure 2.
Allele frequencies. Genome-wide founder allele frequencies range from 4.1% to 27.3%. The color corresponding to each founder strain is the same as in Figure 1D.
QTL mapping indicates white head-spotting in the pre-CC is a Mendelian recessive trait linked to a region on Chr 10
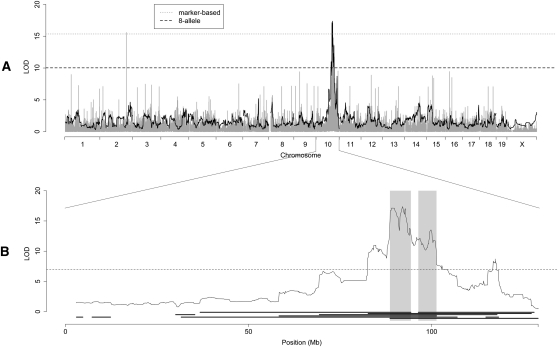
Two methods for mapping QTL were applied to the study data. A marker-based association method regresses phenotypic values on observed marker genotypes. This method does not take advantage of our ability to infer the founder origin of inherited haplotypes. In addition, we used an eight-allele linkage model in the manner previously described for multifounder crosses (Mott et al. 2000; Valdar et al. 2006b; Kover et al. 2009). Specifically, phenotypic values were regressed on estimated founder haplotype probabilities in each marker interval. An allele descended from the A/J founder was considered distinct from an allele descended from C57BL/6J, even though individual marker genotypes in that interval may be identical. In this way we accounted for unobserved variation unique to each founder strain. However, in this partially inbred panel, eight founder alleles can result in 28 unique heterozygous genotypes. Any particular heterozygote genotype was rare enough that we had no power to evaluate its relationship to a QTL. Instead, we made the simplifying assumption that a heterozygote has a phenotype that is intermediate between the phenotypes of the two associated homozygote genotypes. We applied both methods in the pre-CC population. Here we use the white head-spotting trait to illustrate the differences.
White head-spotting results from the absence of pigment from an area on the crown of the head in WSB/EiJ mice. Mutations in numerous genes have been previously identified that result in the absence of pigment-producing melanocytes in mice. However, the genetic basis of the WSB/EiJ white head spot has not been determined. Spotting was not observed in any F1 hybrid crosses with WSB/EiJ, suggesting that head-spotting is due to a single locus recessive allele or to complex inheritance involving multiple loci. Since the white head spot cannot be observed in albino mice, those animals were removed from the mapping population and the trait was mapped using 111 nonalbino mice. We observed six animals with white head spots, which is less than expected for a single locus recessive (expectation = 11.7, see Supplemental material).
Association mapping with the marker-based model revealed several markers with high LOD scores throughout the genome, with two significant loci on Chr 2 (147.4 Mb, LOD = 15.59) and Chr 10 (89.9 Mb, LOD = 15.78) that exceed the 5% genome-wide LOD threshold of 15.35 (Fig. 3A). In contrast, the eight-allele model produced a LOD profile with a single highly significant peak located at 92.0 Mb on Chr 10 (LOD = 17.36, 5% genome-wide threshold = 10.03). To explain this discrepancy, we looked at the genotypes and allele effects underlying each locus (Supplemental Fig. 3). At the Chr 2 locus, two of 111 samples had heterozygous genotypes, and both of these were white-spotted animals. The other four spotted animals did not share the WSB/EiJ allele. Given our observation of white head-spotting in the WSB/EiJ strain and absence of head-spotting in F1 hybrids, the Chr 2 locus is an implausible genetic explanation for white head-spotting. The significant test statistic resulted from the small number of head-spotted samples combined with the small number of heterozygous animals and the chance occurrence of two white-spotted animals being heterozygous at the same locus. In contrast to these results, all six white head-spotted samples share the allele that is private to WSB/EiJ at the Chr 10 locus. We conclude that the Chr 2 locus is spurious and that white spotting is driven by a single QTL on Chr 10.
Figure 3.
White head-spot genome scan. (A) Marker-based (light gray) and eight-allele (black) models implicate an allele on Chr 10. (B) Superimposing WSB/EiJ homozygous regions from white head-spotted samples reveals two candidate regions from 88.6 to 94.3 Mb and from 96.4 to 101.3 Mb.
We identified the founder haplotypes and assigned boundaries to the genomic regions contributed by each founder in each of the six mice with a white head spot. The overlap of homozygous WSB/EiJ haplotypes concordant with the LOD peak on Chr 10 supports a recessive Mendelian mode of inheritance for the white head-spot phenotype. Specifically, the white head-spotted animals share two regions of WSB/EiJ homozygosity, from 88.6 to 94.3 Mb and from 96.4 to 101.3 Mb (Fig. 3B, one animal had a region descended from the A/J strain from 94.3 to 96.4 Mb). We used the Gene Ontology (GO) and Mammalian Phenotype Ontology to query whether any of the 52 genes in this region were known to be associated with white spotting. The kit ligand (Kitl, stem-cell factor, steel factor) was associated with phenotypes including white spotting, head spot, and head blaze, and with the GO biological process positive regulation of melanocyte differentiation. None of the other 51 genes in the region were associated with these or similar terms in either ontology. Numerous alleles of Kitl and its target receptor Kit have been reported to result in white-spotting phenotypes (Sarvella 1956; Rhim et al. 2000). This wealth of evidence makes Kitl a strong candidate for the white-spotting gene in the WSB/EiJ inbred strain and the pre-CC population.
Allele effect patterns for a complex trait QTL provide a means for data integration and candidate gene discovery
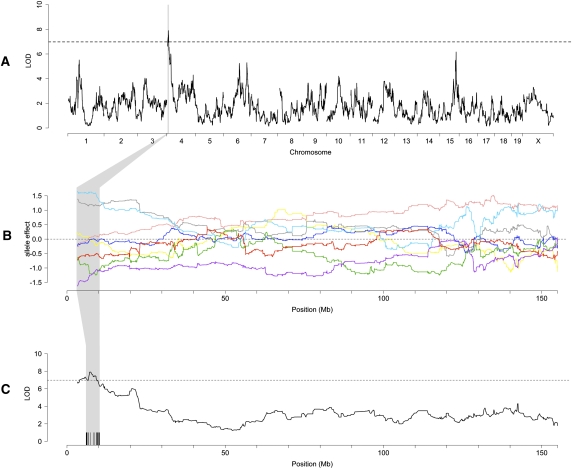
Body weight shows a continuous range of variation that is consistent with polygenic genetic architecture in the pre-CC mice. The founder strains vary widely in body weight. The obese NZO/HlLtJ mice weigh 45.7 g on average (n = 8), while the three wild-derived inbred strains averaged only 16.2 g (n = 28) between 11 and 14 wk of age. We used the eight-allele model to map QTL for baseline body weight (Fig. 4A). One QTL was detected on Chr 4 (LOD = 7.90, 5% genome-wide error rate threshold = 6.98), which explained 18.6% of the observed phenotypic variation. This QTL was designated Bwq14 and spans the region from 3.03 to 10.34 Mb (1.5 LOD drop confidence interval), which contains 42 genes (Supplemental Table 2). Several additional peaks were distinct, but not significant, at a 5% threshold. Of these, peaks on Chr 10 and Chr 15 are in the same location as previously reported QTLs for relative fat content (Purcell-Huynh et al. 1995; Keightley et al. 1998; Brockmann et al. 2000). In contrast, no QTLs were detected using the marker-based approach.
Figure 4.
Baseline body weight genome scan. (A) Eight-allele model (black) indicates a QTL on Chr 4, named here Bwq14, and several suggestive peaks. No peaks reach significance using the marker-based method (light gray). (B) Allele effects plot for Bwq14 suggest a shared NZO/HlLtJ and C57BL/6J allele at this locus is associated with an increase in body weight (dark-gray and light-blue lines). The color corresponding to each founder strain is the same as in Figure 1D. (C) A region of sequence identity between NZO/HlLtJ and C57BL/6J (black bars) reduces the candidate region to 4.69 Mb.
We estimated allele effects at the five highest peaks in order to determine which founder strains contributed high body weight alleles. Two peaks on Chr 6 were associated with different allele effect patterns, indicating that they are indeed separate loci. We found that five of the eight founders (A/J, C57BL/6J, 129S1/SvImJ, NZO/HlLtJ, and CAST/EiJ) contributed high body weight alleles at one or more of these loci. The genomic contribution of the NZO/HlLtJ strain to individual pre-CC mice ranged from 3.7% to 24.9%, but there was no correlation between the proportion of NZO/HlLtJ ancestry and body weight. These observations indicate that variation in body weight is not driven solely by the genetic contributions from the founder with the most extreme body weight. However, the high body weight allele underlying Bwq14 is shared by the NZO/HlLtJ and C57BL/6J strains, the two CC founder strains with the highest body weights (Fig. 4B). We used this allele effects pattern in two separate analyses to refine the QTL location and identify functional candidates for the gene(s) underlying Bwq14.
We performed a founder haplotype analysis using a very high-density SNP resource (Frazer et al. 2007) and genotypes from the Mouse Diversity Array (MDA) (Yang et al. 2009). We identified regions of sequence polymorphism in which NZO/HlLtJ and C57BL/6J share the same allele, but are different from the other six founders. SNPs with this strain diversity pattern (SDP) are densely clustered at 6.01–10.62 Mb (Fig. 4B; Frazer et al. 2007; Szatkiewicz et al. 2008; Yang et al. 2009). This step reduced the candidate region by 37%, to 4.61 Mb. This NZO/HlLtJ-C57BL/6J identical by descent (IBD) region contains 12 genes (71% reduction).
We incorporated liver gene expression data collected from a subset of pre-CC samples (see next section). We performed expression QTL (eQTL) analysis on 11,167 genes that are expressed above background levels among the pre-CC samples. Eighteen of the genes located in the Bwq14 confidence interval had a local eQTL. We compared the allele effects patterns underlying each eQTL with the allele effects pattern of the phenotypic QTL. Only one eQTL, for the gene aspartate-beta-hydroxylase (Asph, cardiac junctin), shared the pattern in which the NZO/HlLtJ and C57BL/6J effects are clustered together and apart from the other six strain effects.
Finally, we used GO and Mammalian Phenotype data to ask whether any of the genes in the region were associated with terms related to body weight. Six genes were linked to relevant GO terms, phenotypes, or both (Supplemental Table 2). The overlap between these three independent analyses determined our highest priority candidates. Four genes had associated ontology terms and fell within the NZO/HlLtJ-C57BL/6J IBD region (Cyp7a1, Chd7, Asph, Gdf6). Of these, only Asph had a local eQTL, and the allele effects pattern for that eQTL matched the effects pattern for Bwq14. This evidence places Asph as the strongest candidate for future functional studies on body weight. We acknowledge that candidates that do not affect mRNA abundance could also be responsible for Bwq14. To further characterize Asph, we examined SNPs present in and around the gene based on complete genome sequences of the eight founder strains (http://www.sanger.ac.uk/resources/mouse/genomes/, see comment in the Methods section). This confirmed that NZO/HlLtJ and C57BL/6J are, in fact, IBD for the entire gene, and all other strains differ by a minimum of seventeen SNPs. The A/J and NOD/LtJ alleles are essentially identical to each other and differ from the NZO/HlLtJ-C57BL/6J allele in the 3′ untranslated region (UTR), which could potentially alter expression levels. The other four strains each harbor distinct haplotypes with variation throughout Asph, including a shared alternative translation stop site.
Abundant local eQTLs reveal high functional diversity and precise mapping
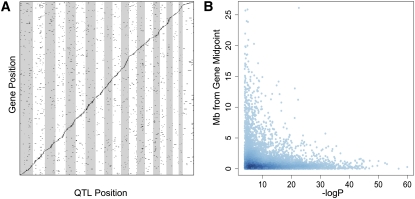
In order to estimate the mapping resolution and to assess functional diversity on a genome-wide scale, we performed gene expression profiling using liver mRNA from 156 available pre-CC samples. We filtered the set of 23,176 probe sets to 11,167 genes that were expressed above background levels in more than 87.5% of the population. A genome-wide eQTL analysis of these transcripts yielded 6182 eQTLs significant above a 5% genome-wide threshold and an additional 1053 eQTLs significant above a 10% threshold. These thresholds correspond to false discovery rates of 2% and 4%, respectively (Storey and Tibshirani 2003). Most eQTLs (5406 or 75%) were local, which we defined as within 10 cM of either end of the gene. Local eQTLs are readily visualized since they appear on the diagonal in a plot of gene location versus eQTL location (Fig. 5A).
Figure 5.
Expression QTL from liver. (A) A total of 7235 eQTLs were detected for 6327 genes. Genes, arranged in order of genomic position along the y-axis, are associated with genetic variation plotted by genomic position on the x-axis. The prominent diagonal band indicates local eQTLs. (B) Local eQTL peaks generally fell within 1 Mb of the gene's genomic location, with the most significant eQTLs also being the most accurate.
A reason for the abundance of local eQTLs relative to distant eQTLs is that local eQTLs generally have larger effects and are therefore easier to detect. As the threshold is lowered, new distant eQTLs are declared at a rate higher than new local eQTLs. For instance, only 15% of eQTLs declared at the 5% threshold are distant, but they comprise 73% of the additional eQTLs declared at the 10% threshold. Increases in statistical power by using larger numbers of CC lines will therefore likely translate to additional distant eQTLs.
We estimated allele effects for each eQTL peak. Classical inbred strains contributed more alleles with positive effects on expression relative to the population mean than negative effects. The genetically distant PWK/PhJ and CAST/EiJ alleles contributed more extreme effects than the other strains, and the effects were slightly more often negative than positive (Supplemental Fig. 4).
The liver transcriptome data set with an abundance of local eQTLs provided a means to estimate the accuracy and mapping precision in the pre-CC study (Bennett et al. 2010). We computed the distance between the local eQTL peak and the physical midpoint of the corresponding gene based on the assumption that most local eQTLs result from polymorphisms in the cis-regulated genes themselves (Fig. 5B). The median eQTL–gene distance was 0.92 Mb, and distance was correlated with peak height. The most significant eQTLs were also the most accurately located at the gene.
Discussion
The analysis of genetic structure and phenotypes in the pre-CC experiment provides important insights that will apply directly to future CC experiments. We showed that the CC breeding design successfully produced balanced founder contributions to the pre-CC lines. All of the alleles present in the eight founder strains were captured in the pre-CC population. Most allele frequencies were near the expected 1/8 proportion, but ranged from 4% to over 27%. Extreme frequencies could occur by chance, by error in the haplotype inference, or by biological forces such as selection. However, there is no strong evidence for selection in our results. Epistatic incompatibilities could decrease the frequency of alleles, but it was not possible to detect two-locus disequilibrium (64 possible genotype combinations) using such a small number of lines. If selection is occurring, we expect that extreme allele frequencies will be more pronounced in the completed CC lines. When those lines have been genotyped, the pre-CC genotypes will allow a retrospective view of the forces acting on individual alleles across generations.
The pre-CC population has 6% more detectable recombination breakpoints than simulated CC strains (Broman 2005) and segments are of smaller median size. The major differences between the pre-CC lines and the finished CC strains are the fewer inbreeding generations and the resulting residual heterozygosity, and these qualities are reflected by the differences in genome architecture. Recombination breakpoints at the boundary of a heterozygous region are found on only one chromosome, and some portion of these will be lost as inbreeding progresses, resulting in fewer segments overall. It is important to note that new recombination breakpoints will continue to be detected in regions that are still segregating, but on average we expect detectable breakpoints to decrease over the course of inbreeding. As the number of genome segments decreases, the median size of the segments will increase necessarily. Overall, these minor differences are less remarkable than the similarities between our observations in the pre-CC study and our expectations for the CC strains. The genetic composition of the pre-CC lines is a good indicator that the final CC strains will achieve the goal of even distribution of diverse haplotypes, both within lines and at each locus across lines.
Previous studies of multiparent RILs and outcross populations established a framework for the haplotype-based methods used in this study (Valdar et al. 2006b; Macdonald and Long 2007; Kover et al. 2009). We used an HMM to infer genotype probabilities, then segmented the underlying haplotype structure of each pre-CC mouse in order to interrogate allele frequencies and recombination. We also used the haplotype-based method to map QTL, and we contrast this linkage-based approach with association mapping on biallelic marker genotypes. The marker-based method is dependent upon marker loci that share the same strain distribution pattern as the causative alleles. In contrast, the eight-allele model faithfully represents all of the genetic variants in a region with the exception of de-novo mutations that may have arisen in the derivation of strains. Moreover, the haplotype-based method provides a means to assess the local extent of linkage disequilibrium and thus to obtain confidence regions for QTL localization. In the white head-spotting example, the multiple peaks that appear in the QTL region are caused in part by a segment of non-WSB/EiJ genome in one white-spotted animal. This proved to be helpful in localizing the effect.
Aside from these technical considerations, the haplotype-based method allows us to draw conclusions about regions of a specific founder strain genome. This is the essential difference between a linkage study, in which descent is known or can be inferred, and association analysis, which relies solely on the observed genotypes. Our results support theoretical predictions that the linkage approach increases power and accuracy while decreasing false discoveries (Valdar et al. 2006a).
Using the haplotype-based method, we presented three examples demonstrating that the pre-CC population is an excellent resource for QTL mapping. Nevertheless, the pre-CC experiment described here will not be typical of future CC studies. Several limitations resulted from using partially inbred lines that will be ameliorated in the CC. Replication is a key advantage of using RILs, but each arm of the pre-CC study included only a single mouse from each line. In a CC experiment, multiple genetically identical mice can be phenotyped to substantially increase heritability for any trait (Belknap 1998). Further, the fully inbred CC lines will constitute a genetically reproducible population supporting integration of data across experiments, phenotype domains, experimental conditions, and time. Such data integration was problematic with the pre-CC population since littermates used in different arms of the pre-CC experiment were not genetically identical. Sibling data could improve the precision of the haplotype reconstructions, yet, due to the extremely high density of marker genotypes, only marginal improvements would be expected. Additionally, the logistics of the pre-CC experiment were complex. They involved breeding and shipping mice and samples between multiple institutions, genotyping on a newly developed platform, and multistep phenotyping pipelines. The experiment was secondary to animal production at ORNL, so only 138–184 lines were available to any particular study arm. Environmental and seasonal variation was impossible to control.
Residual heterozygosity presented another analytical challenge for QTL mapping that will not be present in the completed CC lines. This was particularly problematic for the marker-based approach, in which we treated heterozygotes as a separate genotype class, resulting in a false-positive QTL detection for white head-spotting (Fig. 3). For the eight-allele model we used an additive genetic model that treated heterozygous genotypes as having an effect that is intermediate between the estimated homozygous genotype effects, as was done for previous multifounder crosses (Mott et al. 2000; Mott and Flint 2008). Although the additivity assumption was not correct, it did not prevent detection of the recessive white head-spotting phenotype. Over- or underdominant heterozygous loci are exceptions. The success of mapping with simple regression on founder haplotype probabilities provides a baseline for the performance of the more sophisticated analysis methods that are being developed (Durrant and Mott 2010).
Despite these limitations, the pre-CC experiment provided an excellent test bed for analysis strategies and yielded new biological insights that illustrate the potential of the CC. For example, our results revealed Kitl as a strong candidate gene for white head-spotting in the WSB/EiJ strain and pre-CC mice. A variety of Kitl alleles with effects on pigmentation, hematopoiesis, spermatogenesis, and other phenotypes have been characterized over the past half-century (Sarvella 1956; Rhim et al. 2000; Gu et al. 2009; Deshpande et al. 2010). A unique aspect of the CC design is that functional alleles from the eight founders will be fixed on a variety of genetic backgrounds. This will provide opportunities to accurately estimate allele effects in genetically heterogeneous populations and to identify modifier loci.
QTL mapping is only a starting point for genetic analyses, and strategies for incorporating additional data are as important as mapping methods. The eight-way cross design provides a number of unique advantages that enable efficient data integration. The power provided by complete genome sequences of the founder strains that are currently available in draft versions (http://www.sanger.ac.uk/resources/mouse/genomes/) cannot be overstated. When these are combined with haplotype reconstructions such as those presented here, it will be possible to impute the full genome sequence for each CC line, effectively providing a large genetic reference population with completely sequenced genomes. A defining and powerful feature of the eight-allele model is the ability to identify specific patterns of allele effects at a QTL. Balanced allele frequencies in the CC enhance the ability to estimate effects relative to outbred populations. Allele effects can be used to distinguish tight linkage from pleiotropy (Macdonald and Long 2007) and can substantially narrow the list of the candidates for causal polymorphisms in a QTL.
In the genetic analysis of baseline body weight, we used Bwq14 allele effects to integrate our QTL results with high-density SNP genotypes and eQTL data. We successfully narrowed the QTL region and generated a prioritized list of candidate genes with Asph being the top-ranked gene. Asph is involved in calcium (Ca) cycling in cardiomyocytes, and knockout mice exhibit abnormal heart function, morphological defects, and reduced fertility (Dinchuk et al. 2000; Yuan et al. 2007). Although there are no reported effects on body weight in the Asph knockout, Asph has been associated with cell proliferation in the Gene Ontology. Additional analysis is required to determine whether variation in Asph expression contributes to differential body weight.
The Asph example effectively illustrates several points that are relevant to understanding the importance of the CC as a tool for complex trait analysis. The extreme abnormalities reported in the Asph knockout mouse are clearly not relevant to alleles segregating in the CC. This illustrates the stark contrast between the complete ablation of gene function common with reverse genetic approaches and the often-moderate effects of natural allelic variants. Analysis of the founder genome sequences revealed six distinct alleles, underscoring the genetic diversity that will be present in CC strains. Our results demonstrate that sequence variation present in the CC corresponds to functional variation at the molecular and phenotypic level. For example, Asph is expressed in multiple tissues and has 25 reported transcripts and 20 reported protein products (Barrett et al. 2009; Hubbard et al. 2009). The premature stop codon observed in four of the CC founder strains could explain the observed differential gene expression, which could result from strain-specific transcripts not detected on the gene expression platform, differential transcription due to regulatory variation, transcripts targeted for nonsense-mediated decay, differential RNA stability, or a combination of these.
The number and distribution of eQTLs indicate a vast amount of functional diversity. One way of assessing the functional genetic diversity is the number of local eQTLs. The number observed in the pre-CC experiment is more than double the number recently reported in a large panel of classical inbred strains and classical-derived RILs (Bennett et al. 2010), which itself exceeded other currently available resources. One major difference between the CC and these other resources is the genetic variation introduced by the wild-derived founder strains. The CAST/EiJ and PWK/PhJ allele effects were at the extremes of the effect distribution more than twice as often as allele effects from the other strains (Supplemental Fig. 4), supporting the conclusion that much of the functional variation in the pre-CC lines is derived from these strains. These wild-derived alleles had negative effects on expression more often than positive effects. If we assume that each strain should contribute around the same number of negative and positive effect alleles, then we estimate the excess of negative effect wild-derived alleles at around 14%. This excess may result from hybridization errors, in which an undiscovered SNP prevents the wild-derived mRNA from binding to the oligonucleotide expression array. We expect a higher rate of undiscovered SNPs in the wild-derived strains relative to the classical inbred strains, since the expression array platform was developed based on classical inbred strains. When full genome sequence is available for the founder strains, we will be able to test this hypothesis. However, this rate of false positives would not change our conclusions about the abundance of functional diversity relative to other mouse resources or the wild-derived alleles as the source of this diversity (also note that probe effects will undoubtedly also result in false-negative eQTLs, which will lead to an underestimate of their numbers). The genomic distribution of eQTLs is uniform and without gaps across the genome when corrected for local gene density (Supplemental Fig. 5). Furthermore, there are no genomic regions that lack functional genetic variation, in contrast to typical inbred strain crosses (Yang et al. 2007).
We observed no evidence of eQTL clustering (trans-bands) as reported in previous mouse eQTL studies (Schadt et al. 2003; Chesler et al. 2005). These previous studies have posited that trans-bands are due to a few genes with pleiotropic effects. Others have suggested trans-bands are caused by intersample correlations introduced experimentally that can be statistically corrected (Kang et al. 2008). However, the pre-CC data suggest an alternative origin for their existence in mouse transcriptome analyses. Due to their related origin and breeding histories, existing inbred mouse strains have distinct population substructures, large regions of IBD, and correlated SNP distribution patterns across the genome. eQTL studies using mapping populations with these characteristics (existing RI panels or F2 crosses using common laboratory strains) can erroneously lead to the appearance of trans-bands if any one gene linked to a correlated SNP pattern is under strong trans-regulation. The lack of trans-bands in the pre-CC data suggests that population substructures present in existing mouse resources have been disrupted through the random breeding scheme. Consequently, genetic regulatory variation in the CC is spread throughout the genome, as would be predicted of a random breeding population with genome-wide variation as is present in humans.
The liver transcriptome data was also used to estimate the mapping resolution obtained with as few as 156 pre-CC mice. Local eQTLs are on average <1 Mb from the location of the regulated gene. Extrapolation of this finding to a panel of 500 lines, with replicate measurements to reduce residual variation, implies that mapping resolution of high-effect QTL in the CC could be reduced to <0.25 Mb (Kong and Wright 1994). Local eQTLs typically have larger effects sizes than phenotypic QTLs, but this finding suggests that mapping precision is generally high when there is sufficient power to detect QTLs.
The pre-CC experiment provides strong support for success with future CC experiments. The CC breeding design randomly mixed the genetic diversity of three wild-derived strains with that of five classical inbred strains, and will make vast functional variation accessible in a single resource. The approach used here to precisely map QTLs and prioritize candidate genes will further benefit from having replicate measurements of fully inbred CC strains. As the CC strains are characterized, we expect that a wealth of condition- and tissue-specific transcript, metabolite, and protein-profiling data will become available, essentially at no cost to adopters of the CC platform. The combination of a reproducible genetic reference population with fully imputed genome sequences and high-dimensional molecular phenotypes will open new possibilities for analysis that we have only touched upon, and integrating these data in the context of specific diseases will support human genetic studies by providing new insights to the genetic bases and molecular mechanisms of common human diseases.
Methods
Body weight, white head spot, and gene expression
Pre-CC mice were weaned at ORNL and transported to UNC at 9–13 wk of age. Baseline body weight was measured after acclimating at UNC for 1 wk. Upon completion of the pre-CC experiment at 12–16 wk of age, the presence of a white head spot was noted and liver tissue was dissected and stored in RNAlater (Ambion) before homogenizing in TRIzolPlus (Invitrogen). Total RNA was isolated according to the manufacturer's protocol, and quality was assessed using an Agilent 2100 Bioanalyzer instrument and RNA 6000 Nano LabChip assay (Agilent Technologies). Following reverse transcription with random primers-T7 primers (Affymetrix), double-stranded cDNA was synthesized with the GeneChip WT cDNA Synthesis and Amplification Kit (Affymetrix). In an in vitro transcription (IVT) reaction with T7 RNA polymerase, the cDNA was linearly amplified to generate cRNA. In the second cycle of cDNA synthesis, random primers were used to generate single-stranded DNA in the sense orientation. Incorporation of dUTP in the cDNA synthesis step allowed for the fragmentation of the cDNA strand utilizing uracil DNA glycosylase (UDG) and apurinic/apyrimidinic endonuclease 1 (APE 1) that specifically recognizes the dUTP and allows for breakage at these residues. Labeling was performed with terminal deoxynucleotidyl transferase (TdT), where biotin is added with an Affymetrix Labeling Reagent. Then, 2.3 μg of biotin-labeled and fragmented cDNA were hybridized onto GeneChip Mouse Gene 1.0 ST Arrays (1,102,500 probes; Affymetrix) for 16 h at 45°C. Post-hybridization staining and washing were performed according to the manufacturer's protocols using the Fludics Station 450 instrument (Affymetrix). Finally, the arrays were scanned with a GeneChip(R) Scanner 3000 laser confocal slide scanner.
Feature extraction was performed with GCOS (Affymetrix). CEL files were imported into the R 2.9.2 language/environment for normalization and analysis. Normalization was applied with the rma function in the affy R package from Bioconductor (Gentleman et al. 2004). Probe level data was summarized using a custom probe set definition based on Ensembl genes from the BrainArray website (Sandberg and Larsson 2007; package MoGene10stv1_Mm_ENSG V11.0.1).The minimum number of probes in a probeset was three and the median was 26. Probesets with normalized values <6 in more than 7/8 of samples were removed from further analyses. Probeset level data was transformed using the equation
where φ−1(p) is the quantile with probability p in a normal distribution, ri is the rank of yi with ties resolved by the average rank, and No is the total number of nonmissing observations (Lehmann 1975).
Genotyping
Each mouse in the pre-CC experiment was genotyped using a high-density SNP array. Most of the genotyping was completed using “test” arrays. These arrays were developed as an intermediate step in the process of developing the Mouse Diversity array (Yang et al. 2009). There are two versions of the test array: A-array and B-array. The A-array includes 294,878 SNP assays, and the B-array contains 287,687 additional SNP assays. We determined that 181,752 (A-array) and 180,976 (B-array) SNP assays performed well and targeted loci that are polymorphic among the eight founder strains. There is no overlap between the two arrays, but the genome coverage is complete and uniformly distributed in both. In some cases, animals from the same phenotyping arm were genotyped with different arrays. Integration was achieved by merging the two sets and using an HMM to impute haplotypes at loci with missing genotypes. Due to the high marker density, this procedure was very effective. The exercise behavior and metabolism arm was completely genotyped with the A-array. Genotype data is publically available for download at the CC Status website (http://csbio.unc.edu/CCstatus/index.py).
Haplotype reconstruction
Reconstruction of founder haplotypes in the pre-CC animals is based on analysis of biallelic SNP data from each founder strain. The high density and uniform distribution of phylogenetically informative SNPs (Yang et al. 2009) resulted in highly accurate and robust reconstructions. Haplotype probabilities were computed using HMM (Mott et al. 2000; Liu et al. 2010) with a conservative genotyping error rate of 0.01. The marker density exceeded the total density of recombinations in the cross; thus, it was possible to reduce the exercise behavior and metabolism arm data to ∼16,199 intervals, within which estimated genotypes were essentially constant. Interval boundaries were defined at transitions in highest probability genotype, based on Baum-Welch output from the HMM. In most intervals, haplotype probabilities were near 1 for the inferred states. Exceptions occurred in regions where two or more of the five classical strains in the founder set are identical by descent. One sample was removed from architecture analyses because it was an outlier in two criteria; it had high heterozygosity and a high number of genome segments. To evaluate segregation distortion we used a χ2 goodness-of-fit test on the allele frequencies of homozygotes in each segment and identified extremes using false discovery rate (FDR). Genetic map positions are used for reference purpose only and were based on the integrated mouse genetic map using mouse genome build 37 (Cox et al. 2009).
Genome scans
QTL mapping was performed using a regression model (Mott et al. 2000) with previously described optimization and software (Valdar et al. 2009). We used the BAGPIPE package that consists of an R library and accessory scripts in R and Perl. We regressed each phenotype without covariates on the expected number of haplotypes (i.e., haplotype dosage) at each interval between adjacent genotyped markers. Each test produces an F statistic, which we transformed to a LOD score statistic to evaluate significance. A 1.5 LOD drop interval is considered the best approximation to a 95% confidence interval for QTL mapping (Dupuis and Siegmund 1999). We estimated genome-wide significance thresholds by permutation, which is appropriate in unstructured populations (Churchill and Doerge 1994). Allele effects were estimated using partial correlation coefficients from the same model.
To make eQTL analysis computationally feasible, we reduced the genome to the 16,199 genome segments described above. Within each segment, we averaged the probability matrices to create a new segment-wide matrix. We then regressed the z-transformed expression values on this averaged genotype matrix. Genome scans were essentially identical using ∼182 K complete markers or 16,199 genome segments, and the same eQTLs were declared using segmented and nonsegmented genomes. We performed 5000 permutations on 50 traits to determine 0.05/0.1 error thresholds and used the mean threshold across all expression traits. We used this null distribution to calculate adjusted P-values for each genome scan, and used these to determine the FDR for the eQTL analysis (Storey and Tibshirani 2003). To determine how often each strain was at the extreme end of the allele effects distribution for a given eQTL, we estimated each allele's effect and compared the distribution between strains. These results are summarized in Supplemental Figure 4.
We used three data sources for additional sequence analysis: the Perlegen array-based Mouse resequencing (http://mouse.perlegen.com), the Sanger Mouse Genomes Project (http://www.sanger.ac.uk/resources/mouse/genomes/), and the Mouse Diversity Array (http://cgd.jax.org/datasets/popgen.shtml/). Perlegen data was necessarily used for regional analysis (i.e., in the Bwq14 confidence interval) because the Sanger data is embargoed for analysis on regions larger than a single gene (http://www.sanger.ac.uk/resources/mouse/genomes/datarelease.html).
Acknowledgments
This work was supported by National Institutes of Health grants and training grants GM070683, GM076468, GM067553, GM074244, F32GM090667, T32GM07092, MH090338, T32MH076694, DK076050, U01CA105417, and U01CA134240; Ellison Medical Foundation AG-IA-0202-05, and U.S. Department of Energy under Contract DE-AC05-00OR22725 with UT-Battelle, LLC. Some phenotypes were collected using the Animal Metabolism Phenotyping core facility within UNC's Nutrition Obesity Research Center (funded by NIDDK grant DK056350). Essential support was also provided by the Dean of the UNC School of Medicine, the Lineberger Comprehensive Cancer Center at UNC, and the University Cancer Research Fund from the state of North Carolina. We are particularly grateful for infrastructure support at UNC facilitated by Drs. Bill Marzluff, Terry Magnuson, and Shelley Earp.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE22297.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111310.110.
References
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. 2009. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res 37: D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK 1998. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet 28: 29–38 [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, et al. 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 20: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S 2000. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F2 variance of growth and obesity in DU6i x DBA/2 mice. Genome Res 10: 1941–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet 37: 233–242 [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, et al. 2008. The Collaborative Cross at Oak Ridge National Laboratory: Developing a powerful resource for systems genetics. Mamm Genome 19: 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA 2007. Recombinant inbred strain panels: A tool for systems genetics. Physiol Genomics 31: 174–175 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE, Bult C, Paigen B, Flint J, et al. 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S, Agosti V, Manova K, Moore MA, Hardy MP, Besmer P 2010. Kit ligand cytoplasmic domain is essential for basolateral sorting in vivo and has roles in spermatogenesis and hematopoiesis. Dev Biol 337: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinchuk JE, Henderson NL, Burn TC, Huber R, Ho SP, Link J, O'Neil KT, Focht RJ, Scully MS, Hollis JM, et al. 2000. Aspartyl beta-hydroxylase (Asph) and an evolutionarily conserved isoform of Asph missing the catalytic domain share exons with junctin. J Biol Chem 275: 39543–39554 [DOI] [PubMed] [Google Scholar]
- Dupuis J, Siegmund D 1999. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics 151: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant C, Mott R 2010. Bayesian quantitative trait locus mapping using inferred haplotypes. Genetics 184: 839–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Runyan C, Shoemaker A, Surani A, Wylie C 2009. Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development 136: 1295–1303 [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. 2009. Ensembl 2009. Nucleic Acids Res 37: D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Morris KH, Ishikawa A, Falconer VM, Oliver F 1998. Test of candidate gene–quantitative trait locus association applied to fatness in mice. Heredity 81: 630–637 [DOI] [PubMed] [Google Scholar]
- Kong A, Wright F 1994. Asymptotic theory for gene mapping. Proc Natl Acad Sci 91: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551 doi: 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman E 1975. Nonparametrics: Statistical Methods Based on Ranks. Holden-Day, San Francisco [Google Scholar]
- Liu EY, Zhang Q, McMillan L, de Villena FP, Wang W, 2010. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics 26: i199–i207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SJ, Long AD 2007. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176: 1261–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Flint J 2008. Prospects for complex trait analysis in the mouse. Mamm Genome 19: 306–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell-Huynh DA, Weinreb A, Castellani LW, Mehrabian M, Doolittle MH, Lusis AJ 1995. Genetic factors in lipoprotein metabolism. Analysis of a genetic cross between inbred mouse strains NZB/BINJ and SM/J using a complete linkage map approach. J Clin Invest 96: 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim H, Dunn KJ, Aronzon A, Mac S, Cheng M, Lamoreux ML, Tilghman SM, Pavan WJ 2000. Spatially restricted hypopigmentation associated with an Ednrbs-modifying locus on mouse chromosome 10. Genome Res 10: 17–29 [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: Implications for QTL discovery and systems genetics. Mamm Genome 18: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Larsson O 2007. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics 8: 48 doi: 10.1186/1471-2105-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvella P 1956. Steel, a new dominant gene in the house mouse. J Hered 47: 123–128 [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA 2008. An imputed genotype resource for the laboratory mouse. Mamm Genome 19: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW 2002. Genetic dissection of complex and quantitative traits: From fantasy to reality via a community effort. Mamm Genome 13: 175–178 [DOI] [PubMed] [Google Scholar]
- Valdar W, Flint J, Mott R 2006a. Simulating the collaborative cross: Power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172: 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J 2006b. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38: 879–887 [DOI] [PubMed] [Google Scholar]
- Valdar W, Holmes CC, Mott R, Flint J 2009. Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F 2007. On the subspecific origin of the laboratory mouse. Nat Genet 39: 1100–1107 [DOI] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, de Villena FP, Churchill GA 2009. A customized and versatile high-density genotyping array for the mouse. Nat Methods 6: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Fan GC, Dong M, Altschafl B, Diwan A, Ren X, Hahn HH, Zhao W, Waggoner JR, Jones LR, et al. 2007. Sarcoplasmic reticulum calcium overloading in junctin deficiency enhances cardiac contractility but increases ventricular automaticity. Circulation 115: 300–309 [DOI] [PubMed] [Google Scholar]