Abstract
Genetic reference populations in model organisms are critical resources for systems genetic analysis of disease related phenotypes. The breeding history of these inbred panels may influence detectable allelic and phenotypic diversity. The existing panel of common inbred strains reflects historical selection biases, and existing recombinant inbred panels have low allelic diversity. All such populations may be subject to consequences of inbreeding depression. The Collaborative Cross (CC) is a mouse reference population with high allelic diversity that is being constructed using a randomized breeding design that systematically outcrosses eight founder strains, followed by inbreeding to obtain new recombinant inbred strains. Five of the eight founders are common laboratory strains, and three are wild-derived. Since its inception, the partially inbred CC has been characterized for physiological, morphological, and behavioral traits. The construction of this population provided a unique opportunity to observe phenotypic variation as new allelic combinations arose through intercrossing and inbreeding to create new stable genetic combinations. Processes including inbreeding depression and its impact on allelic and phenotypic diversity were assessed. Phenotypic variation in the CC breeding population exceeds that of existing mouse genetic reference populations due to both high founder genetic diversity and novel epistatic combinations. However, some focal evidence of allele purging was detected including a suggestive QTL for litter size in a location of changing allele frequency. Despite these inescapable pressures, high diversity and precision for genetic mapping remain. These results demonstrate the potential of the CC population once completed and highlight implications for development of related populations.
Genetic reference populations in model organisms provide a powerful system in which to study complex phenotypes including disease-related traits. These strain panels provide a framework for integrating data across multiple phenotype domains spanning molecular, morphological, physiological, and behavioral traits. However, their breeding history can greatly influence the resulting population, the breadth and extent of phenotypic diversity, and the mapping precision of the resulting cross.
There has been heightened interest in these populations with the advent of systems genetics (Chesler et al. 2003; Kadarmideen et al. 2006; Churchill 2007). Several such panels have been or are being developed for a variety of species, including yeast, Drosophila (Ayroles et al. 2009), Arabidopsis (Kover et al. 2009), maize (Buckler et al. 2009), Caenorhabditis elegans (Johnson and Wood 1982; Li et al. 2006; Rockman and Kruglyak 2009), and mice (Chesler et al. 2008; Iraqi et al. 2008; Morahan et al. 2008). Efforts are also being made to expand (Peirce et al. 2004) and maximize the potential of existing populations (Bennett et al. 2010). In the construction of these populations, there is a unique opportunity to observe the impact of selection and inbreeding depression on the range of phenotypic diversity and genetic structure (Ayroles et al. 2009; Rockman and Kruglyak 2009).
The Collaborative Cross (CC) is an emerging mouse recombinant inbred (RI) panel that was designed to extend the available mouse genetic reference populations (Churchill et al. 2004) by combining the genomes of eight genetically and phenotypically diverse founder strains—A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ—that capture almost 45 million SNPs present in laboratory mice (Roberts et al. 2007; Yang et al. 2007). Three sites have been involved in the production of the CC (Chesler et al. 2008; Iraqi et al. 2008; Morahan et al. 2008). Here we report on the genetic analysis of phenotypes collected at Oak Ridge National Laboratory (ORNL) from the first cross of progenitors through the eighth generation of inbreeding (Chesler et al. 2008).
Randomized matings and fully traceable lineages in the ORNL population allowed us to observe the dynamics of viability and drift on allelic frequencies, phenotypic diversity, and the nature of allelic effects underlying QTLs. Our analyses address phenotypic diversity, heritability, and genetic mapping and reveal the impact of novel allelic combinations in this population.
Results
Variation in breeding productivity and fecundity in the Collaborative Cross
A total of 650 CC lines were initiated at ORNL (Chesler et al. 2008) from eight inbred progenitor strains consisting of five common inbreds (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/H1LtJ) and three wild-derived inbreds (CAST/EiJ, PWK/PhJ, WSB/EiJ). The G1 generation consists of two-way cross progeny; the G2 generation consists of four-way cross progeny, and the G2:F1 generation consists of the eight-way cross progeny. The G2:F1s were brother–sister-mated to make the first inbreeding G2:F2 generation. Successive sib matings created the G2:Fn generations. Individuals in the G2:F1 generation contain the maximum genetic diversity with genetic material from all eight progenitors and are at the theoretical maximum level of heterozygosity of alleles identical by descent.
Among the G1 crosses, mean litter size was lower for crosses involving one or two wild-derived lines (Fcross type (2,51) = 6.07; p < 0.0043). The pattern of mean differences reveals outcrossing depression in litter size for some crosses. As previously reported, NZO/HlLtJ × PWK/PhJ and NZO/HlLtJ × CAST/EiJ matings were avoided due to exceedingly poor mating performance, and PWK/PhJ × 129/SvImJ gave rise to infertile males (Chesler et al. 2008). The mean litter size in the crosses was lower than both parental means for the 129/SvImJ × NZO/HlLtJ, but not for the reciprocal NZO/HlLtJ × 129/SvImJ cross. The mean litter size was between that of the two progenitors for C57BL6/J × WSB/EiJ and its reciprocal, 129/SvImJ × PWK/PhJ and its reciprocal, CAST/EiJ × NOD/HlLtJ, WSB/EiJ × A/J, and WSB/EiJ × 129/SvImJ. For all other crosses, the majority of which were crosses between common laboratory strains, an increase in litter size was observed.
Among the G2 crosses, patterns of both depression and heterosis were again observed. G2 mean litter sizes were compared to G1 litter sizes for each of 576 four-way crosses (G1 × G1). Among these, 19 exhibited decreased litter size indicative of depression, 330 exhibited heterosis, and the remainder fell between their G1 parental means. All four-way crosses in the G2 generation were classified into 15 configurations based on whether the parents were offspring of common × common, common × wild-derived, or wild-derived × wild-derived strains. Analysis of change in litter sizes (δLS) from G1 to G2 reveal significant effects of configuration (F(15,561) = 3.00; p < 0.0002) and number of wild-derived lines in the four-way cross (F(3,572) = 7.39; p < 0.0001). With increasing numbers of wild-derived lines among the four grandparents, δLS is increasingly positive (δLS0 < δLS1 < δLS2 < δLS3).
Effects of genetic composition are dependent on the direction of the cross. When the common × common derived parent is female, the change in litter size from the previous generation is lower than for all other configurations (F(1,561) = 6.29; p < 0.012), and the litter size itself is higher. As a further test of the directionality of mating effects, we directly compared each cross of common × common derived males or females to other configurations (e.g., {[common × common] × [wild × common]} vs. {[common × wild] × [common × common]}). We detected significant effects only for the difference between CC × WW and WW × CC crosses, such that wild-derived males resulted in an increase in litter size when crossed to common females than do wild-derived females (F(1,561) = 6.29; p < 0.012) crossed to common males. This was tested again by comparing the effect of crossing wild-derived males to that of common-derived males into common-derived females, which again reveals a greater increase in litter size when the male is wild-derived, CC × CC versus CC × WW (F(1,561) = 5.04 p < 0.025).
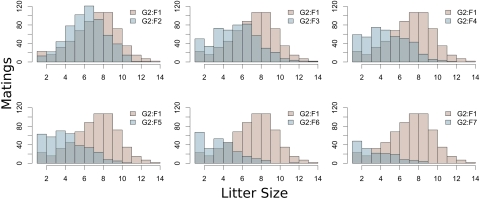
Fecundity and reproductive fitness among the advanced inbreeding lines typically varied outside the range of progenitors. Dispersion of CC fertility-related phenotypes, obtained from G2:F5 and subsequent generations, exceeds that of the inbred progenitors for measures including mean litter size, parental age at first litter, and interval from mating to first litter (Kolmogorov-Smirnov, p < 0.05) (Supplemental Table 1). However, litter sizes in these lines cover a smaller range than the G2:F1 population (Fig. 1). The shrinking range was unilateral such that the maximum litter size obtained among the lines in inbreeding generations was successively lower than that observed in the G2:F1, a phenomenon considered consistent with overdominance-related inbreeding depression.
Figure 1.
Distribution of litter size across generations. The inbreeding generations lose the extreme litter size values observed in the G2:F1s, indicating that no recessive allele produces this value. When such a phenomenon is observed in inbreeding lines relative to a natural population, the result is attributable to overdominance as a mechanism of inbreeding depression.
However, purging of deleterious allele combinations, manifest in the loss of lines during inbreeding, is also observed. Among the inbreeding generations, there is an inverted-U-shaped pattern of the proportion of lines lost in each generation relative to prior generations, peaking at G2:F6, in which 20% of lines were lost relative to the preceding generation. The G2:F1 generation lost <1%, G2:F2 lost 0%, G2:F3 lost 4%, G2:F4 lost 10%, G2:F5 lost 16%, G2:F7 lost 14%, and G2:F8 lost 8%, resulting in 296 lines at the G2:F9 generation.
Physiological, morphological, and behavioral variation in the Collaborative Cross
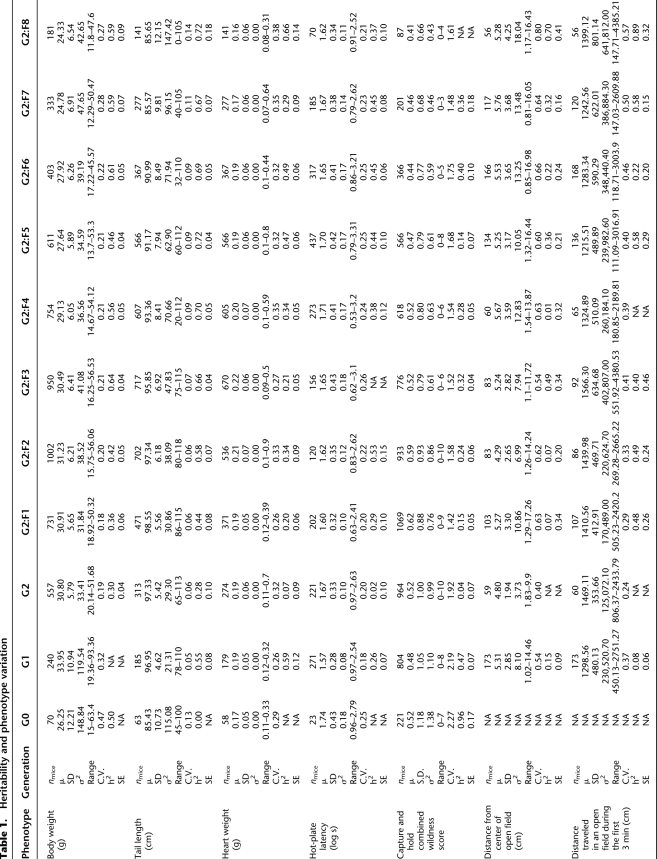
Phenotypic variance for many traits increased in the G1 (first outcrossing) generation (Table 1) relative to progenitor lines. However, phenotypic variation decreased somewhat in the G2 (second generation of outcrossing) and beyond. For all morphometric measures, the distribution of individual scores showed greater dispersion in the CC lines relative to the progenitors, (Kolmogorov-Smirnov, p < 0.05). As expected from the wide range of body sizes among the progenitor strains, indicators of overall growth vary widely among the CC lines. For example, among progenitors, body weight ranged from 25.56 ± 0.85 g (WSB/EiJ) to 29.66 ± 2.16 g (NZO/HlLtJ), whereas in the inbreeding generations (G2:F5–G2:F8), this measure ranged from 23.65 g to 32.00 g. Tail length, an index of linear growth, ranged from 67.92 ± 7.98 mm (PWK/PhJ) to 94 ± 7.94 mm (NOD/LtJ) among the progenitors, whereas it ranged from 59.04 mm to 112.00 mm among the inbreeding generations. Heart weight and adiposity index also showed considerable variation among the progenitors and in the CC offspring, but the CC range was within that of the progenitors.
Table 1.
Heritability and phenotype variation
Phenotypic variation in the CC relative to a two-progenitor genetic reference population
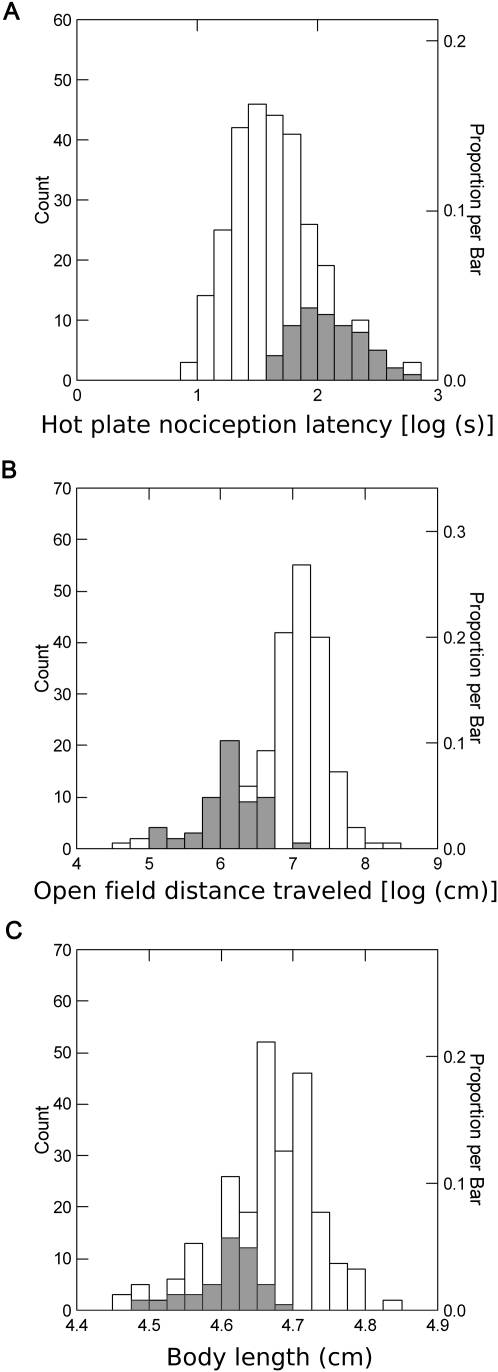
While it is interesting to note the spread of phenotypic variation relative to progenitor lines, increased phenotypic variation is expected due to transgressive segregation, the random assortment of increasor and decreasor alleles that were once in a fixed assortment in the progenitor lines. Therefore, we assessed phenotypic variation in the CC in contrast to a relatively large and widely used genetic reference population, the BXD recombinant inbred lines, for which we have previously reported phenotypic values for diverse behavioral measures (Philip et al. 2010) obtained under the same conditions as our Collaborative Cross phenotypic data. Phenotypic values observed in generations G2:F5 and beyond for thermal nociception, distance traveled in the open field, and body length spanned a far greater range than those obtained in the BXD RI strain panel (Fig. 2). Interestingly, the ranges of BXD phenotypes were not centered within the CC phenotypic range (Fig. 2).
Figure 2.
Comparison of phenotypic distributions among the CC and BXD mice. Shaded bars represent the phenotypic distribution of the BXD population. (A) Distribution of thermal nociception across the CC and BXD populations. (B) Distribution of distance traveled in the open field across the CC (during the first 3 min) and BXD populations (during the first 5 min). (C) Distribution of body lengths across the CC and BXD populations. Note that in all the three examples the phenotypic range of the BXD population is contained within one side of the distribution of CC phenotypes.
Restoration of continuous behavioral variation in the Collaborative Cross
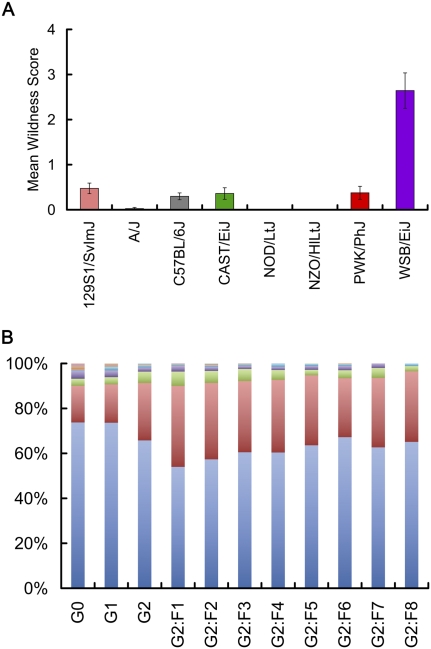
It has long been suspected that the development of inbred mouse strains has selected for docility. Behavioral wildness is expected to exhibit a wide range in the CC due to the inclusion of wild-derived progenitor strains. Behavioral wildness scores in the CC progenitors show high within-strain consistency (intraclass correlation = 0.95). The inbred strains differ categorically, with WSB/EiJ having an extreme high value and the other strains having relatively low scores (Fig. 3), a phenomenon that has been related to possible selection for easy handling in derivation of inbreds. The CC population at ORNL was derived using software-assisted mating to randomize the choice of individuals who continued on to the next generation. Twenty-six percent of individuals in the G0 population had wildness scores above 0, and of these, 37% were WSB/EiJ mice. The proportion of extreme high scores dropped in the G1 generation, indicating that WSB/EiJ alleles are being diluted, while the proportion of mice with wildness scores >0 increases in the G2 and G2:F1 generations, reflecting possible segregation of wildness alleles at multiple loci and greater diversity in combinations of alleles. Wildness scores stabilize through inbreeding to a mean value of ∼0.43, slightly higher than that of inbred progenitors other than WSB/EiJ. Importantly, there is a discontinuity in the score distribution in G0 mice, but not in the CC lines. The continuous distribution of wildness scores in the CC lines indicates segregation of multiple loci within the population.
Figure 3.
Wildness variation among CC progenitors, outcrossed (G1 and G2) and inbreeding generations (G2:F1 to G2:F8). (A) Mean wildness scores among CC progenitors. Among the progenitors, behavioral wildness resembles a discrete trait with high wildness in WSB/EiJ and low wildness in the other lines. (B) Proportion of mice in CC generations with wildness scores of 1 (blue), 2 (red), and >2 (other colors). Among the outcrossed generations, wildness scores increase, and among the inbreeding generations, a greater proportion of intermediate values is observed. In general, while more mice have scores >1 in the CC lines, fewer extreme high scores were observed, suggesting a restoration of continuous variation of this phenotype.
Parent–offspring phenotypic correlations
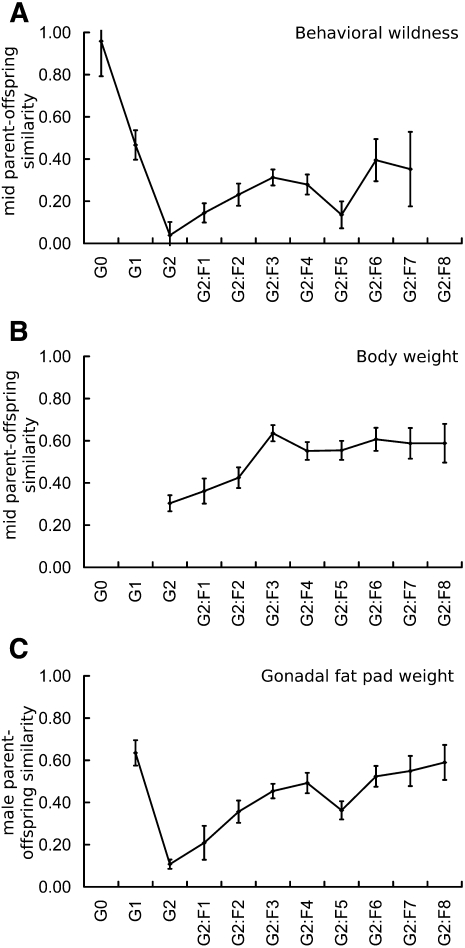
For most of the traits, parent–offspring similarity increases for several generations during inbreeding, followed by a single generation drop after which the heritabilities resume their increase. This is likely due to the effects of attrition of lines and the consequent restriction of phenotypic range resulting from lost allele combinations. For example, the similarity of parent–offspring behavioral wildness scores increases during the first three generations of inbreeding (G2:F1–G2:F3), starts to decline in generations G2:F4 and G2:F5, and resumes its increase at the G2:F6 generation (Fig. 4A). Parent–offspring regression coefficients for each morphological phenotype increase progressively with inbreeding through generation G2:F6, as allele fixation increases. By G2:F6, body weight (Fig. 4B), tail length, heart weight, kidney weights, fat-pad mass (Fig. 4C), and adiposity regression coefficients range from 0.67 to 0.75 (Supplemental Table 1), values greater than those reported for mapping crosses in other model systems (Kunes et al. 1990; Kramer et al. 1998; Gaya et al. 2006). Similarity of body weight between parents and offspring in the CC is estimated at 0.73, exceeding values typically reported for F2 crosses and estimates obtained by strain intraclass correlations in other mouse RI panels (e.g., 0.48 in LXS RI strains) (Bennett et al. 2005). This is likely a manifestation of both the expanded range of phenotypic variation in the Collaborative Cross and an increase in the similarity of individuals with inbreeding.
Figure 4.
Parent–offspring similarity estimates for three phenotypes. Parent–offspring regression coefficients typically increase during inbreeding with a single generation drop in correlation. For behavioral wildness, parent–offspring similarity increases until the G2:F5 generation followed by a single generation of drop (A). Similar trends exist for body weight (B) and gonadal fat-pad weight (C).
Parent–offspring regression coefficients for reproductive traits are lower than those for morphometric parameters (Rocha et al. 2004) but are nonetheless statistically different from 0 beginning in generation G2:F2 (p < 0.0001) for litter size, G2:F3 (p < 0.0001) for maternal age at birth of the first litter, and G2:F2 (p < 0.02) for time from mating to first litter. By G2:F6, similarity estimates for litter size and time to first litter reach 0.22 and 0.09, respectively. These values compare favorably with other published reports for heritability of litter size (0.10–0.17) and time to first litter (0.05) (Eisen 1978; Peripato et al. 2004; Canario et al. 2006). Although relatively low in early generations, parent–offspring similarity estimates for wildness reach 0.40 by G2:F6 (Fig. 4A).
Stability of allele frequency through the inbreeding process
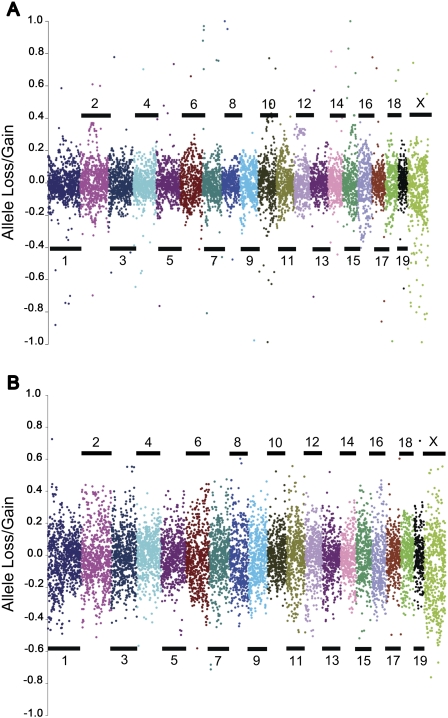
Lines from G2:F1 and G2:F7 were genotyped to track changes in allele frequencies across generations. Progenitor alleles entered the population at a minimum minor allele frequency (MAF) of 0.125 for all autosomal locations, and it is expected that allele frequencies remain stable throughout each generation and genome location. Efforts were made to balance alleles on the sex chromosomes during initial breeding (Chesler et al. 2008). Expected X-chromosome allele frequencies are dependent on the position of the progenitor strains in each line's mating scheme and were adjusted accordingly. Distribution of strain-specific alleles (MAF = 0.125) grouped by progenitor strain of origin indicates that there is no statistically significant deviation from expectation for the MAF at the majority of these loci using a χ2 test for goodness of fit with a family-wise false discovery rate of 0.05. Only 95 strain-specific genotyped SNPs showed significant deviations when comparing progenitor allele frequencies to those of G2:F1 allele frequencies. Sixty-two were from PWK/PhJ, 15 of these were located on Chr16, and 22 were on ChrX. Another 21 of these 95 loci were from CAST/EiJ, and 18 were on ChrX. The remaining loci originated from WSB/EiJ (eight loci), A/J (one locus), and C57BL/6J (three loci). Genotyped loci from the three other progenitor strains, namely, 129S1/SvImJ, NZO/HlLtJ, and NOD/LtJ, showed no significant deviation from expected segregation patterns. Comparison of G2:F1 strain-specific alleles relative to those of G2:F7 resulted in only 13 loci with significant deviations from expected frequencies. Nine of these loci were from PWK/PhJ, while the remaining four loci were from CAST/EiJ. Twelve loci show declining allele frequencies throughout both outcrossing and inbreeding. Of these, eight loci were from PWK/PhJ and four were from CAST/EiJ. The strain-specific allele distributions spread from G2:F1 to G2:F7, indicating that overall alleles at some loci increase in frequency while other alleles decrease in frequency through the process of inbreeding (Fig. 5). A comparison of observed local allele frequencies relative to expected frequencies in the G0 to G2:F1 and G2:F1 to G2:F7 generations reveals that this increase in variance occurs on a genome-wide level (Fig. 6). In general, observed changes in allele frequencies relative to expected frequencies were less pronounced following outcrossing (Fig. 6A) than during inbreeding (Fig. 6B).
Figure 5.
Strain-specific Minor Allele Frequencies (MAF) at the G2:F1 and G2:F7 generations. Strain-specific MAF in the G2:F1 (A) generation depicts less variation than the G2:F7 (B) generation, with some spread of allele frequencies evident in the G2:F7 generation. Allele frequency distributions become asymmetrical for some strains. The distribution becomes right skewed for PWK/PhJ, indicating that more loss than gain has occurred, and left skewed for WSB/EiJ.
Figure 6.
Assessment of allele loss during inbreeding. Comparison of the percent allele loss between (A) progenitors (G0) and final outcross generation (G2:F1). (B) G2:F1 and the seventh inbreeding generation (G2:F7). Positive values indicate SNPs with an increase in minor allele frequency, while negative values indicate a decrease (allele loss) in minor allele frequency. The y-axis indicates the percent change from G0 allele frequency.
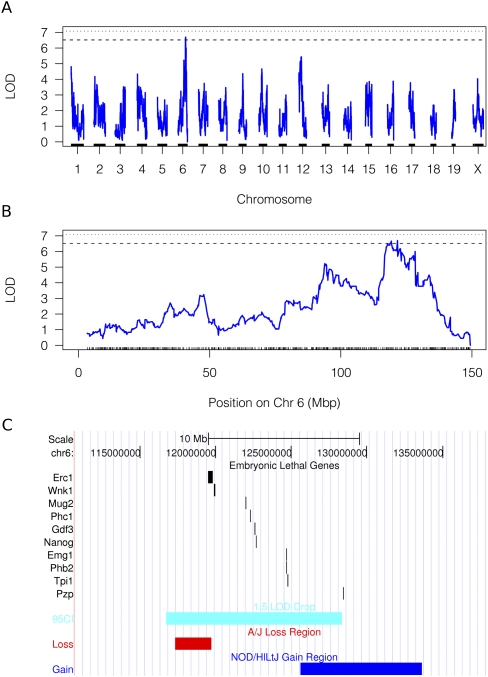
Figure 7.
Genetic mapping of litter size. A genome-wide scan (A) reveals a suggestive QTL on chromosome 6 (B) for litter size. Within the confidence interval are several alleles that have decreasing frequency in the inbreeding CC lines and several genes associated with embryonic lethality (C).
Quantitative trait loci mapping in the CC
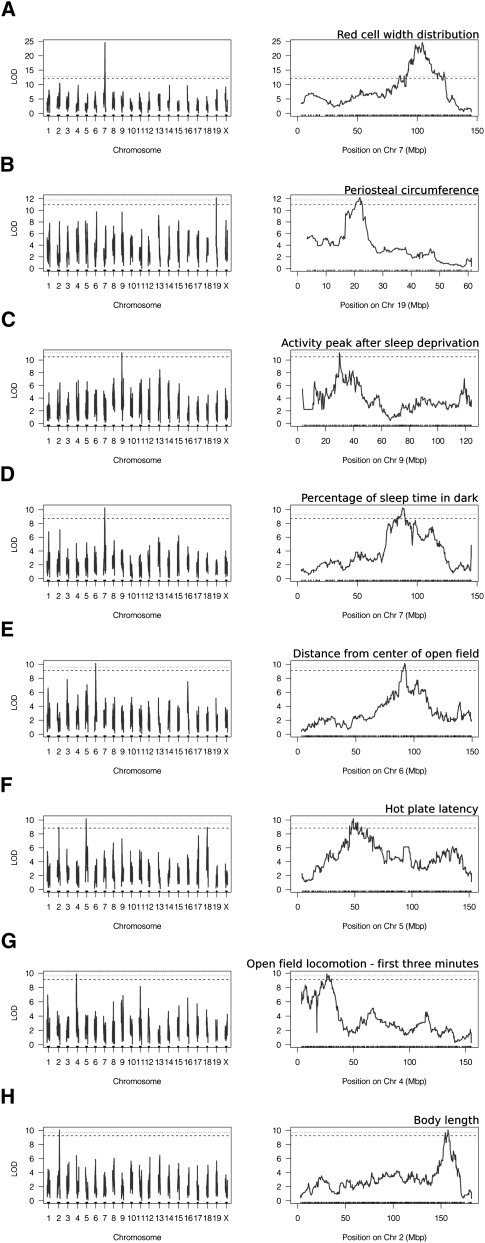
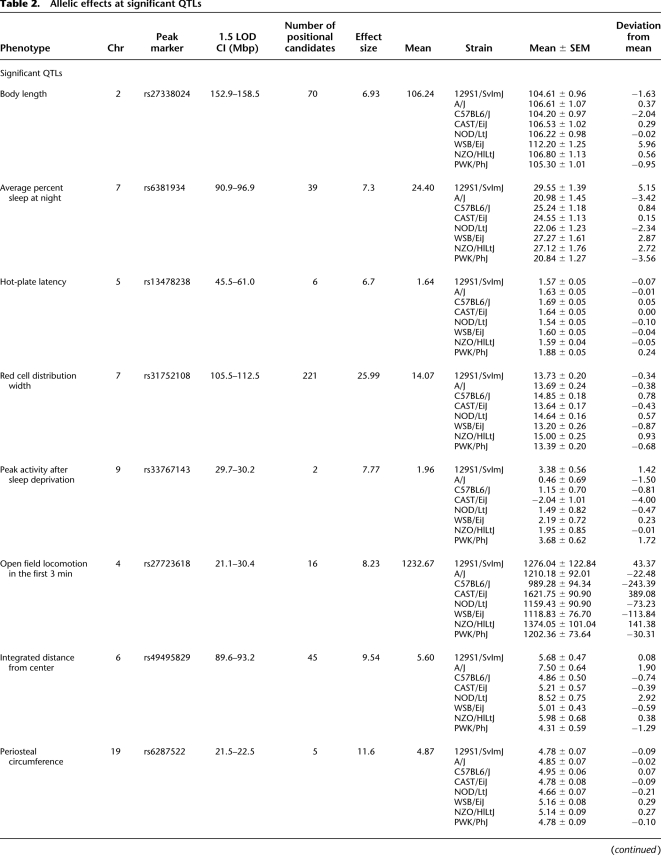
A total of 102 reproductive, behavioral, physiological, and morphological traits were subject to QTL mapping, although several of the measures were highly correlated, including 50 sleep-related parameters and several similar blood measures. Approximately 72 unique though not statistically independent phenotypes were considered. Among these, eight statistically significant (p < 0.05) main-effect QTLs were detected: red cell distribution width, hot-plate latency, average percent of sleep time in the dark, distance traveled in open field during the first 3 min, periosteal circumference of the femur, peak activity onset after sleep deprivation, body length, and cumulative distance from the center of the open field (Fig. 8). An additional nine suggestive QTL were detected (p < 0.10) (Table 2). Confidence intervals around the significant QTL peaks, estimated using 1.5 LOD drop-off, had an average width of 3.98 Mb and contained an average of 50 genes each, with fewer than 10 candidate coding genes for three of the traits. The narrowest 1.5 LOD drop interval detected is 530 kb for peak activity time in hours from activity onset after sleep deprivation (Chr9: 29.70–30.03 Mb; Build 36 between RefSNPs rs33767143 and rs6264816), which harbors coding sequence for a microRNA and two genes: Ntm (neurotrimin) and Snx19. The largest interval detected for a statistically significant locus was 6.96 Mb for red blood cell distribution width, containing 232 positional candidate genes.
Figure 8.
Significant genome-wide QTLs. (A) Red blood cell width distribution. (B) Periosteal circumference. (C) Peak activity time in hours from dark onset after sleep deprivation. (D) Average percentage of sleep time over dark cycles for all baseline days. (E) Average minimum distance of the center of the mouse from the absolute center of the open field (cm). (F) Thermal nociception. (G) Open field locomotion in the first 3 min. (H) Body length. Horizontal lines indicate genome-wide significance thresholds based on 1000 permutations. Dotted lines are genome-wide significant thresholds at p ≤ 0.05; dashed lines indicate genome-wide suggestive thresholds at p ≤ 0.10.
Table 2.
Allelic effects at significant QTLs
Periosteal circumference was mapped to a 1.5 LOD interval from 21.55 to 22.51 Mb on Chr19 (Build 36), containing five candidate genes, including Tmem2 and Trpm3. For thermal nociception, the 1.5 LOD drop interval (Chr5: 45.44–60.99 Mb) around the peak marker contains just six genes: Slit2, Pacrgl, Gpr125, Dhx15, Sod3, and Kcnip4. The 1.5 LOD drop interval for average minimum distance from the center of the open field (Chr6: 89.59–93.20 Mb), average percentage of sleep time over dark cycles for all baseline days (Chr7: 90.92–96.94 Mb), and red cell distribution width (Chr7: 105.55–112.52 Mb), respectively, map onto gene-rich genomic regions with 45, 39, and 221 genes, the latter containing a cluster of 130 Olfr family members in addition to three compelling functional candidates, hemoglobins Hbb-b1, Hbb-bh1, and Hbb-y.
A suggestive QTL was found on Chr6 for the regulation of litter size. This locus harbors several genes for which alleles have been annotated to embryonic lethality (Fig. 7). At this locus, A/J alleles are associated with higher litter sizes and NZO with lower litter sizes. Interestingly, these alleles have a significant decrease in frequency (rs13478994, p < 0.003; rs29829339, p < 0.02), suggesting that for any surviving line with an A/J allele at this location, litter size is elevated.
Significant QTLs for some traits are the effect of alleles from single strains, while for other traits, the effect is due to alleles present among multiple strains (Table 2). For body length, individuals with alleles originating from WSB/EiJ at the QTL were significantly different from individuals with alleles derived from other strains. The QTL for hot-plate thermal nociception was driven by significant differences among individuals with alleles derived from PWK/PhJ and relative to those with other alleles except for C57BL6/J. For gonadal fat-pad mass, alleles specific to PWK/PhJ were significantly different from other alleles except for CAST/EiJ and 12S1/SvImJ. For peak activity onset after sleep deprivation, wild-derived strains PWK/PhJ and CAST/EiJ differ significantly from WSB/EiJ, CAST/EiJ is significantly different from 129S1/SvImJ, PWK/PhJ is significantly different from A/J, and A/J is significantly different from 129SvImJ. For other measures, a more complex pattern of strain-specific effects emerges (Table 2). A composite interval mapping approach enabled the detection of additional loci for several phenotypes (Supplemental Table 3). For example, a search for additional QTLs for open field locomotor behavior, conditioned on the Chr4 locus, reveals an additional locus on Chr9 at rs33324954, from 108 to 113 Mb, consistent with previously reported QTL for this phenotype (Hitzemann et al. 2002).
Discussion
Given the powerful resources for mouse genetics and genomics, there is a tremendous value to increasing the genetic diversity of existing populations through the intercrossing of mice from diverse origins. The Collaborative Cross is one such effort to accomplish this goal, and as implemented at ORNL, was designed to minimize systematic effects of selection (Chesler et al. 2008). Phenotype analysis of the CC breeding population provided a unique opportunity to monitor complex traits as the underlying genetic material is recombined, heterozygosity is gained and lost, and novel allele combinations are created. This enabled us to evaluate the consequences of diversity and selection on the resulting population and its application to complex trait analysis. These data demonstrate the trans-generational phenotypic impact of randomizing allele combinations produced from a pool of extremely high genetic diversity intended to approximate what is found in a natural population (Roberts et al. 2007).
Although the population has not yet reached the homozygosity required for a genetic reference population, the majority of loci are already fixed with ∼95% being identical by state (IBS) and ∼75% estimated to be identical by descent from founders (IBD) at the G2:F7 generation (Broman 2005; Chesler et al. 2008). In general, allele frequencies did not deviate from expectation, but for a few loci, evidence of significant change in allele frequency was detected. We also observe that asymmetric drift was observed with trends toward greater loss of PWK/EiJ, CAST/EiJ, and WSB/EiJ alleles (Fig. 5), and that this occurred primarily during the inbreeding rather than the outcrossing phase of the breeding project, suggesting that allele purging may have occurred through inbreeding. We do not observe region-specific excess of heterozygotes.
Our phenotypic analyses highlight the relative contributions of novel allelic combinations and heterozygosity in determining the extent of phenotypic variation. In general, traits exhibit the broadest phenotypic range in generation G1, the generation produced from pairwise mating of the eight progenitor strains. In this generation each wild-derived allele is crossed with a common allele or different wild-derived allele. This introduction of rare haplotypes results in an increase in phenotypic variation consistent with heterosis. Recombination during the second and third generations of breeding the CC has the potential to create new allele combinations that could potentially drive phenotypic ranges beyond the preceding generation. However, we observed that the second generation of outcrossing (four-way crosses), often results in a decreased phenotypic variance. This may result from the reestablishment of homozygosity at common haplotypes, or it may reflect outcrossing depression due to incompatible heterozygous states. Only genotypic analysis of these generations can discriminate among these possibilities. The fact that trait ranges begin to compress in generations G2 and G2:F1 suggests that this pressure toward decreased variance has a greater impact on trait distributions than increased haplotype recombination, also occurring during these generations, which would increase variance.
Phenotypic variance decreases as a function of inbreeding, suggesting either loss of rare alleles, loss of rare allele combinations between loci, or simple loss of heterozygosity. Parent–offspring regression coefficients are determined by two factors—range and covariance. In early generations, heritability is low due to a lack of covariance; in the higher generations, range restriction occurs as extreme lines are lost or disadvantageous alleles are purged. Changes in parent–offspring regression coefficients through inbreeding generations indicate that during the generations of greatest loss of lines, there is also a retraction of phenotypic range. This suggests that embryos entered one of two states, a non-viable state in which the fixation of alleles or allele combinations results in lethality, or a viable state in which allele fixation results in a reduced range of allelic variants and phenotypic diversity due to purging of deleterious or disadvantageous alleles.
The range of trait values in late inbreeding generations provides an indication of the breadth of phenotypic variation that will exist in the finished CC strains. For the traits described herein, and despite manifestation of inbreeding depression as shown for litter size (Fig. 1) and wildness (Fig. 3), the ranges of values typically exceed those obtained across the eight progenitor strains. Metabolism and adiposity are expected to exhibit extreme variation across the finished CC strains due to the inclusion of obesity-prone (NZO/H1LtJ) and lean wild-derived strains in the progenitors. Consistent with this expectation, the adiposity index varied 80-fold among G2:Fn lines. Kidney and heart weight, when adjusted for body weight, provide an indication of physiological function. These measures varied approximately threefold among offspring in generation G2:F5 and higher.
Phenotypic variation greatly exceeds that of the largest existing mouse genetic reference populations and typically exceeds that of the progenitor strains. This “transgressive segregation” is not due entirely to heterosis, as it persists well into the inbreeding process, and is therefore more likely reflective of the combinatorial effects of allele configurations at multiple loci. Comparisons to the BXD recombinant inbred strain panel reveal that phenotypic variation is greater in the CC, with almost double the range for several traits. The observation that BXD phenotypes are clustered on either the high or low end of the range of CC phenotypes indicates that the differences are likely due to major effects of alleles from wild-derived progenitor strains (Yang et al. 2007). Analysis of allelic effects at regulatory QTLs confirms that many of the effects are explained by alleles found only in one or more of the wild-derived strains (Table 2). However, effects of common alleles are also readily detectable, particularly when conditioning on these major effect loci (Supplemental Table 3).
Following traits in each parent–offspring transition across generations enabled the resemblance of relatives to be monitored as inbreeding progressed. Parent–offspring similarity is an important predictor of heritable variance that can be explained in genetic mapping analysis. In the CC generations, these measures also allow estimation of whether most of the trait-determining loci have been fixed and to what extent the added genetic variability results in added environmental sensitivity. Overall, parent–offspring similarity is consistent with previous published studies, for example, body weight, h2G2:F8 = 0.59, at the high end of published estimates ranging from 0.40 to 0.60 (Eisen 1978; Jones et al. 1992; Kramer et al. 1998; Valdar et al. 2006). For total distance traveled in open field, h2G2:F8 = 0.61, greater than previous estimates of 0.20–0.50 (DeFries et al. 1978), although below that which is observed in some genetic reference populations. By six generations of inbreeding (G2:F6), parent–offspring similarity estimates for morphometric traits approach 75%. Interestingly, each of these traits exhibits a decline in heritability in an earlier generation (typically G2:F4), followed by a progressive increase through G2:F8. A similar decline is observed for reproductive and behavioral traits, although for some (e.g., time to first litter) it is not clearly followed by an increase in heritability in the subsequent generation. Based on the breeding interval (G2:F1–G2:F8) represented in Figure 3, we cannot determine if heritability estimates will continue to rise in subsequent generations or if a plateau has been reached.
The process of creating the Collaborative Cross exposed a pool of segregated alleles to the selection pressures of outcrossing and inbreeding depression. We observed inbreeding depression in several phenotypes but focused our analyses of this phenomenon on litter size because it is the one most likely to alter allele composition and, thus, the resulting diversity and precision of the cross. While most experimental evidence thus far has indicated partial dominance as a mechanism of inbreeding depression, our analyses of these processes revealed some experimental evidence for both partial dominance and overdominance mechanisms of inbreeding depression. This is expected, as these effects are integrated over multiple loci with multiple mechanisms of action. Overdominance models predict that the maximum phenotypic value can only be obtained in heterozygotes, and, therefore, no individual line in the inbreeding population would have a phenotypic value equal to the maximum value observed in the heterozygous base population (Lynch and Walsh 1996). Our analysis of litter size (Fig. 1) reveals that this is, indeed, the case, although it is conceivable that in a much larger population size, such an individual line could be found. Other evidence of overdominance or heterozygous advantage, in the form of regionally specific excess of heterozygotes, was not detected. In partial dominance, purging of deleterious recessive alleles results in decreased phenotypic range. This is the more commonly observed mechanism of inbreeding depression, and we have found evidence for this in the inbreeding CC lines. The detection of a locus that influences litter size (Fig. 8) coinciding with a region of allele loss is consistent with partial dominance and the purging of deleterious alleles. It is important to note that each locus is alone viable as a homozygous allele, given a compatible genetic background. The creation of the G2:F1 population segregates the background genetic material and can introduce incompatibilities. Those mice carrying the Chr6 alleles with decreasing frequencies have higher average litter sizes than those that do not, consistent with a complementary allele in the background that has rescued this effect.
A randomly mating base population is essential to the study of mechanisms of inbreeding depression. The G2:F1 generation differs from a natural base population in that deleterious recessive alleles have been purged in the process of creating the inbred founder strains. Comparison of the phenotypic values of the CC lines to their progenitors enables investigation of combinatorial variation across loci but does not allow for evaluation of effects of heterozygosity and allele purging. Ayroles et al. (2009) approximated a naturally occurring population in Drosophila by simultaneously performing randomized outcrossing and inbreeding of a panel of lines. Such an analysis will soon be possible through the comparison of heterogeneous stocks derived from the Collaborative Cross including the Diversity Outcross (J:DO; The Jackson Laboratory, Bar Harbor ME). Another important test of mechanisms of inbreeding depression involves the analysis of a single generation outcross of the finished CC strains.
Unlike the progenitor strains or any other panel of inbred mouse strains, the phenotypic diversity of the CC is the result of independent, randomized combinations of alleles that support accurate and precise mapping of complex traits. QTL analysis in the emerging CC reveals that with a sample size of about 250 lines with only one or two mice per line, it is possible to map genetic loci with high precision. In several instances, the CC had sufficient mapping resolution to identify one or a few candidate genes. Mapping of “activity after sleep deprivation” revealed only three positional candidates, a microRNA and two genes—Ntm (neurotrimin), which encodes a neural cell-adhesion molecule that plays a role in brain development (Chen et al. 2001), and Snx19. Allelic variation of neurotrimin could affect sleep, wake, and activity by influencing developmental patterns in central nervous system structure or through dynamic effects on adult neuronal functions. Sleep deprivation maximizes the magnitude of slow-wave activity (SWA), which is directly proportional to the duration of prior periods of wakefulness (Achermann and Borbely 2003). Increasing evidence suggests a functional role for sleep and high SWA in synaptic plasticity (Krueger et al. 2008; Hanlon et al. 2009). Periosteal circumference at the midshaft of the femur mapped to five candidates—Gda, 1110059E24Rik, Tmem2, Fam108b, and Trpm3, a transient receptor potential channel gene. Periosteal circumference is one of many measures for skeletal size and is correlated with total body size and total skeletal size; therefore, the gene underlying this QTL could be responsible for global effects on body size. Mechanistic analysis of the causative locus is the best method to interpret the specificity of these effects. Neither 1110059E24Rik nor Fam108b appears to be expressed in osteoblasts or osteoclasts. The Tmem2 gene is highly expressed in primary osteoblasts, but the function of this gene in bone biology is unknown. The Gda gene appears to be moderately expressed in osteoclasts, but like Tmem2, the function of this gene in bone biology has not been previously determined. Expression of Trpm3 has been demonstrated in human osteoblast-like cell lines, and other genes in this family have a known role in osteoblast differentiation (Abed et al. 2009).
Our QTL mapping analyses found more QTLs than expected by chance, although our single-locus modeling results are expected to account for a fraction of total genetic variance. More comprehensive modeling of multiple sources of trait variation reveals additional loci. Using a composite interval mapping approach, with each main effect locus as a cofactor in the search for additional loci, revealed additional detected loci (Supplemental Table 3). It is particularly important to note that our QTL effects are typically of large size and rare allele origin. These effects may be obscuring the detection of the common allelic variation typically observed in conventional crosses of closely related inbred strains. The high diversity inflates total phenotypic variance, and therefore, without accounting for these large effects, we do not expect to see smaller-effect QTLs.
Our studies conducted in the production colony of the Collaborative Cross reveal that, despite the observed and unavoidable consequences of inbreeding, tremendous behavioral, morphological, and physiological variation remains, sufficient to allow precise QTL mapping using conventional sample sizes. The CC strains, their F1 hybrid progeny, and their outcrossed sister population together provide a resource for further exploration of fundamental genetic questions in a tractable but sufficiently large and diverse mammalian population. In our mapping analyses we found large-effect wild-derived alleles at loci regulating disease-relevant phenotypes. Systematic characterization of wild-derived lines in structured breeding schemes will provide many new alleles of interest to biomedical research. The effective sizes of the alleles segregating in the CC appear large relative to our prior experience with mice, suggesting that the potential to detect QTLs for disease-related phenotypes is greater in the CC than in conventional mapping populations.
Methods
All procedures, including husbandry and animal euthanasia described below, were approved by the Institutional Animal Care and Use Committee of Oak Ridge National Laboratory and were conducted in compliance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Housing
Mice were housed in the William L. and Liane B. Russell Vivarium at the Oak Ridge National Laboratory. Mice at ORNL received irradiated Purina Diet #5053 and Harlan Softcob bedding, with one nestlet enrichment device in each cage. Most mice were maintained in ventilated racks that provided 99.997% HEPA filtered air to each cage. Mice from wild-derived progenitor strains were housed in a quiet room within static micro-isolator cages to facilitate breeding. Room temperatures and humidity were maintained at 70°F and 30%–70%, respectively. Animals were kept under a 14:10 light:dark cycle, with light intensities maintained at 3 lux at a distance of 30 inches from the floor surface. Water was delivered via an automatic watering system chlorinated to 3–5 ppm.
Breeding
The breeding strategy for the Collaborative Cross at ORNL has been described elsewhere (Churchill et al. 2004; Chesler et al. 2008). Briefly, mice from each of the eight inbred progenitor strains consisting of five common inbreds (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/H1LtJ) and three wild-derived inbreds (CAST/EiJ, PWK/PhJ, WSB/EiJ) were randomly assigned to one of a roughly balanced set of breeding schemes or lines, which dictate the order in which strains are crossed. The strains were crossed pairwise to create a G1 generation, and these were crossed pairwise to make the four-way G2 generation, then crossed again to make the G2:F1 generation. The G2:F1s were crossed and the progeny randomly assigned to one of three mating pairs, of which one was randomly chosen as the priority pair to contribute to the next generation. If this mating was non-productive, the offspring of the next ranked pair were used. To prevent die-out at advanced generations, back-crossing was also used in rare instances. A line was considered lost if no progeny were born after several breeding attempts using these strategies.
Subjects
A total of 650 lines were initiated according to the Complex Trait Consortium Protocol (Churchill et al. 2004). Of these, 414 lines with at least one male or one female survived to the G2:F5 generation, while the remaining lines were lost during the inbreeding process. Litters were weaned at ∼3 wk of age into breeding pairs determined by the CCWORKS software-assisted mating system developed by K.F.M. (Chesler et al. 2008). Litter size, parental age at birth of first litter, and length of the interval from mating to the first litter were recorded as measures of fecundity. Mice were maintained in breeding pairs until they entered the phenotyping protocol. Retired breeders from the cross population were phenotyped on a variety of behavioral, physiological, and morphological measures. Phenotyping was performed across generations to evaluate trait heritability. Phenotyping and genotyping were performed in at least one breeding pair per line from generations G2:F5–G2:F8 for QTL analysis. Most of the genotyped mice came from the G2:F5 generation. From this generation, 235 lines were tested. At this generation the population is estimated to be ∼75% inbred at the genotyped loci (Chesler et al. 2008), based on allelic identity by descent calculations performed using the R/ricalc package (Broman 2005). Because assays were added to the phenotyping pipeline throughout the course of the breeding project, not all mice were evaluated on all measures. Approximately 7500 mice of both sexes (except for gonadal fat-pad mass and testes weight) from up to 626 lines were phenotyped for body weight, tail length, fasting plasma glucose level, weights of kidney and heart, and behavioral and physiological traits; of these, about 3000 have been phenotyped for adiposity based on perigonadal fat-pad weight. Sample sizes by generation along with phenotypic means, variances, and ranges are provided in Supplemental Table 1.
Mice did not enter the phenotyping queue until the birth of their grand-progeny to ensure that lines could be propagated. Average age of testing was 38.5 wk. More than 95% of the mice were phenotyped between the ages of 20 and 100 wk. Of the total number of mice phenotyped, 52% were females and 48% were males.
General phenotyping methods
The phenotyping strategy was designed to capture a panel of complex traits amenable to high-throughput analyses while broadly reflecting behavior, morphology, and physiology. The panel of phenotypes is summarized in Supplemental Table 1. Upon birth of grand-progeny, mice were separated into individual pens for phenotyping. The individuals were then moved to a holding room, where they were housed for 3 wk prior to testing, when females were checked to ensure that they were not pregnant at the time of testing. Mice were housed in this room throughout testing. At the start of the week prior to the first test, an elbow-shaped PVC pipe was added to each cage as a standardized enrichment device. At this point in the queue, mice intended to begin testing the following week remained in place on one side of one rack.
Wildness
During separation for phenotyping, mice were scored on the behavioral wildness scale (Wahlsten et al. 2003), for which scores were obtained for the mouse's response to capture and holding by the technician. The scores were combined to form a total wildness score. In the original method for this test (Wahlsten et al. 2003), the maximum score over multiple trials is recorded. In this study a single measure was obtained for each mouse.
Activity monitoring
All activity monitoring tests (open field, light/dark, and modified visual cliff) were administered on separate days. The testing was carried out in a temperature-, noise-, and light-controlled room. Mice were acclimated to the room for 1 h before testing, and the light intensity for each arena's four corners and center point was adjusted to 300 ± 10 lux. Each mouse was picked up by the tail and then lowered gently into the center of the arena with its nose pointed east. All activity was recorded by a videocamera mounted above the open field, and all activities were scored in real time by body point tracking using a Noldus Ethovision XT tracking system (Noldus Information Technology).
Open field
The open field apparatus is a white, square, opaque Plexiglas box (39 × 39 × 39 cm) with a red floor, illuminated evenly at 300 ± 10 lux in a 9-foot × 15-foot room. Each mouse was placed in the center of the box for a 10-min trial, during which the following parameters were recorded: number of crossings into the center of the field, distance traveled, the time spent in the periphery, the time spent immobile, defecation, and urination.
Light/dark box
To test for anxiety-related behavior, a dark insert divided the open field apparatus into light/dark compartments. The compartments are separated with a guillotine door that is closed during placement of mice in the chamber. Mice were placed in the light compartment, and the latency to enter the dark was obtained, along with the percent time spent in light, total light–dark transitions, distance traveled in the light side, fecal boli, and urinations observed over a 10-min trial. Behavioral phenotypes on the open field and light/dark arena tests were acquired at ORNL and analyzed at the University of Chicago by G.S. and A.A.P.
Sleep
The morning after the Light–Dark test, each mouse was then placed in its own chamber atop a piezoelectric grid and chamber system for a 5-d sleep analysis (Flores et al. 2007; Donohue et al. 2008). The mice had access to food and water ad lib while in the chamber. The room was maintained on a 12:12-h light:dark cycle. Mice were placed in the chambers between 9 and 10 a.m. on Day 1 and were removed on Day 5 at the same time. The data acquisition computer, food, and water were checked daily; otherwise, the mice remained undisturbed, except during the sleep deprivation test on Day 3. On this day, the experimenter disturbed the mice by changing bedding, taking away nestlets, and placing the mice in brown paper bags. Measures recorded and analyzed consist of activity onset, time of peak activity, sleep bout length, total sleep time, sleep bout length after sleep deprivation, peak activity after sleep deprivation, and activity onset after sleep deprivation. Data were acquired at ORNL and analyzed at The University of Kentucky by M.S., K.D.D., and B.F.O.
Hot plate
After 30 min of habituation to the testing room, mice were placed on a metal surface (IITC Inc. Hotplate Analgesia Meter Model 39) maintained at 54°C (±0.2°C) (HP54) within a transparent Plexiglas cylinder (15 cm D; 22.5 cm H) with a Plexiglas lid. The latency to respond with a jump or hindpaw lick or shake/flutter was measured to the nearest 0.1 sec with a stopwatch. Two latencies were recorded per mouse with intra-trial separation of 30 sec and maximum trial duration of 30 sec. If no response occurred within 30 sec, the mouse was removed from the hot plate. The apparatus was thoroughly cleansed with MB-10 (QuipLabs) between each mouse tested.
Tail-clip
Two days after hot plate testing, the tail-clip mechanical nociception test was performed. As in the hot plate, mice were allowed 30 min of habituation. The enclosure is a Plexiglas-bound arena measuring 13.5 in L × 16 in W × 15 in H, open at the front. Each mouse was lightly restrained in a denim pocket. An alligator clip with a rubber cuff around each jaw, exerting ∼600 g of force, was applied to the tail 1 cm from the base and vertically oriented with respect to the table. The mouse was immediately removed from the holder, and the latency to lick, bite, or grab the clip or bring the head within 1 cm of the clip was measured with a stopwatch to the nearest 0.1 sec, after which the clip was immediately removed. Each mouse was tested only once with maximum trial duration of 60 sec. If no response occurred by 60 sec, the tail clip was removed. The enclosure and clip were thoroughly cleansed with MB-10 (QuipLabs) between mice, and a clean denim pocket was used for each mouse.
Blood counts
Blood was drawn for phenotypic assays following completion of behavioral testing. Samples for counts of major peripheral cell types and analysis of blood chemistry were collected by retro-orbital sinus puncture into tubes containing EDTA as an anti-coagulant for blood counts and lithium heparin for blood chemistry. Blood count phenotypes were obtained using Scil-Vet ABC blood analyzer (Scil) and include lymphocyte counts and percentages (LYM, LYM %), monocyte counts and percentages (MONO, MONO %), glucocorticoid-remediable aldosteronism count and percentages (GRA, GRA %), eosinophil count and percentages (EOS, EOS %), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), mean corpuscular volume (MCV), mean platelet volume (MPV), neutrophils, hematocrit (HCT), hemoglobin (HGB), platelet (PLT), red blood cell (RBC), and white blood cell counts (WBC).
Blood chemistry
All measurements were obtained using the Abbot i-STAT Chem8 panel (Abbot). This is a cartridge-based system that obtains measures of sodium (Na), potassium (K), chloride (Cl), ionized calcium (iCa), TCO2, glucose (Glu), urea nitrogen (BUN)/urea, creatinine (Crea), hematocrit (Hct), hemoglobin (Hgb), and anion gap.
Fasting plasma glucose
Approximately 1 wk after blood collection for these phenotypes, mice were fasted overnight, and blood was drawn again for assay of fasting plasma glucose level measured using a handheld glucometer (Bayer Glucometer Elite XL; Bayer AG).
Dissection methods
One week after blood collection, mice were fasted overnight, then euthanized and dissected for measurements of morphometric phenotypes and collection of tissue samples. Body weight, tail length, and weights of heart, kidney, perigonadal fat pads, spleen, and testes were measured and recorded into the MouseTrack database (Baker et al. 2004). Weights were regression-adjusted to an average age of 39 wk. The adiposity index was calculated as (gonadal fat-pad weight/body weight) × 100. The presence of any obvious pathologies (e.g., spontaneous neoplasms) was recorded in the MouseTrack database. A tail biopsy was collected from each mouse as a source of DNA.
Morphometry
Morphological traits included body weight (overall growth), tail length (linear growth), and the weights of gonadal fat pad (leanness/obesity), kidney, and heart (organs in which weight reflects functional demand). These measures were scaled to body weight to reflect overall differences in organ function (e.g., hypertrophy due to increased function demand) (Hamet et al. 1998). Perigonadal fat-pad weight is highly correlated with independent measures of carcass fatness, and the weight of this depot relative to body weight is widely used as a standard measure of adiposity (West et al. 1994a,b, 1995; York et al. 1997). Periosteal circumference was measured as previously described (Ackert-Bicknell et al. 2009). In short, the hind axial skeleton (hind limbs, pelvis, lumbar spine, and attached muscular) was placed in 95% ethanol for a minimum period of 14 d. The femurs were then isolated from the musculature, and periosteal circumference was measured using the SA Plus densitometer (Orthometrics, Stratec SA Plus Research Unit). The bones were scanned using thresholds of 710 and 570 mg/cm3 such that cortical bone areas and surfaces could be accurately determined. Periosteal circumference measures were made at the exact midshaft of the femur.
Genotyping and SNP selection
A custom array using the Illumina iSelect platform for the Infinium system was developed for SNP genotyping. A subset of 11,969 SNPs were chosen from the NIEHS-Perlegen combined SNP panel (Yang et al. 2007). A sliding window was used to search for the minimum-sized sets of SNPs that could discern all eight progenitor strains. Once a set of SNPs was identified that together could discriminate all eight founder haplotypes within the window or the window length reached 1.86 Mb, the search was terminated and a new window was started. The resulting array enabled genotyping at more than 1200 windows within which the haplotypes from the progenitor strains could be identified. This algorithm ensured that for a given number of SNPs on the array, the maximum density of informative markers could be used, while still ensuring that at any locus, ancestry can be traced back to one of eight founders.
Data analysis
Statistical analyses were performed within the MouseTrack database environment (http://mouse.ornl.gov) (Baker et al. 2004), which includes SAS v. 9.1.3 tools for univariate statistics, univariate and multivariate outlier detection, modeling of litter size, and calculation of parent–offspring regression coefficients (Chesler et al. 2008). For modeling of cross type and other effects on litter size and inbreeding depression, we used mixed models with random effects of specific crosses within cross types. Heritability for the progenitor generation was estimated using strain intraclass correlations, and for subsequent generations, it was estimated using parent–offspring regression by calculating the slope of the regression between the midparent (mean of the two parents) and offspring trait values. We note that these estimates cannot be extrapolated to heritability defined as transmission of phenotypic values in wild randomly mating populations, for which it is customary to adjust regression coefficients by relatedness of the two samples, nor is that our objective. We use unadjusted parent–offspring relations for the monitoring of resemblance among relatives throughout the inbreeding process. For the majority of traits, untransformed data were used to estimate resemblance of relatives. To more closely reflect genetic contribution to tissue function rather than allometric relationship to body weight, organ weights were first adjusted to body weight using a regression model in which individual trait values were regressed to the population mean. Only data from extant lines in the terminal generation studied were used to calculate parent–offspring similarity estimates.
QTL mapping
QTL mapping was undertaken using the R/HAPPY package developed based on multipoint HAPPY dynamic-programming algorithm and regression models (Mott et al. 2000). Trait data from G2:F5 generations and greater were used in the mapping analysis. Sample sizes ranged from 160 to 293 lines with at most one male and/or one female per line. Each trait was assessed for normality prior to its mapping. Appropriate data transformations were applied to trait data that deviated significantly from normality. For each trait an additive model genome scan was performed. For each marker, R/HAPPY (Mott et al. 2000) returns a −logP value. Additionally, for each marker using the residual sums of square under the null hypothesis of no segregating QTL (RSS0) and alternative hypothesis (RSS1, additive or full model) LOD scores were calculated by
This statistic can be used to determine significant and/or suggestive QTLs. Genome-wide significance thresholds were obtained using a modified permutation algorithm rather than the one available in the R/HAPPY package. The R/HAPPY package treats each observation as an independent observation and permutes the phenotype between subjects while keeping the genotypes fixed. In the mapping data used in the Collaborative Cross, we use two subjects per line (one male and one female). This introduces a dependent relationship among subjects (sib pairs). Therefore, lines are permuted rather than subjects, and sex is randomized within lines, while keeping the genotypes fixed. One thousand permutations were performed, and permutation-based genome-wide thresholds were used to identify significant (5%, 10%) and suggestive (63%) QTLs. Confidence intervals around significant and/or suggestive QTL peaks were defined using a one-LOD drop. Genes within this interval are considered candidate genes.
Acknowledgments
The production and characterization of the Collaborative Cross was funded by the Office of Biological and Experimental Research, Office of Science, US Department of Energy to E.J.C., with initial support for production of the Collaborative Cross from the Ellison Medical Foundation to E.J.C., David W. Threadgill (UNC), and Dabney K. Johnson (ORNL). Support for design and production of the genotyping array came from the Center of Genomics and Bioinformatics, University of Tennessee Health Science Center to K.F.M., NIGMS to G.A.C., and ORNL Laboratory Directors Research and Development Fund to E.J.C. and B.H.V. Genotyping was performed through Oak Ridge National Laboratory TTRF to E.J.C. Dr. Roumyana Yordanova performed early heritability analyses. Dr. Arnold M. Saxton, University of Tennessee, performed early mapping analyses using alternate methods and provided helpful feedback on this manuscript. Additional phenotyping was supported by DA021198 to W.R.L., FA9550-05-1-0464 to B.F.O., MH079103 to A.A.P., and the Jackson Laboratory, Nathan Shock Center of Excellence in the Basic Biology of Aging, AG025707 to C.L.A.B.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113886.110. Freely available online through the Genome Research Open Access option.
References
- Abed YY, Beltrami G, Campanacci DA, Innocenti M, Scoccianti G, Capanna R 2009. Biological reconstruction after resection of bone tumours around the knee: Long-term follow-up. J Bone Joint Surg Br 91: 1366–1372 [DOI] [PubMed] [Google Scholar]
- Achermann P, Borbely AA 2003. Mathematical models of sleep regulation. Front Biosci 8: s683–s693 [DOI] [PubMed] [Google Scholar]
- Ackert-Bicknell CL, Shockley KR, Horton LG, Lecka-Czernik B, Churchill GA, Rosen CJ 2009. Strain-specific effects of rosiglitazone on bone mass, body composition, and serum insulin-like growth factor-I. Endocrinology 150: 1330–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RR, et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Galloway L, Jackson B, Schmoyer D, Snoddy J 2004. MuTrack: a genome analysis system for large-scale mutagenesis in the mouse. BMC Bioinformatics 5: 11 doi: 10.1186/1471-2105-5-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link PJ, Lu L, Chesler EJ, Johnson TE 2005. Genetics of body weight in the LXS recombinant inbred mouse strains. Mamm Genome 16: 764–774 [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, et al. 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 20: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. 2009. The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Canario L, Roy N, Gruand J, Bidanel JP 2006. Genetic variation of farrowing kinetics traits and their relationships with litter size and perinatal mortality in French Large White sows. J Anim Sci 84: 1053–1058 [DOI] [PubMed] [Google Scholar]
- Chen S, Gil O, Ren YQ, Zanazzi G, Salzer JL, Hillman DE 2001. Neurotrimin expression during cerebellar development suggests roles in axon fasciculation and synaptogenesis. J Neurocytol 30: 927–937 [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Wang J, Lu L, Qu Y, Manly KF, Williams RW 2003. Genetic correlates of gene expression in recombinant inbred strains: A relational model system to explore neurobehavioral phenotypes. Neuroinformatics 1: 343–357 [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, et al. 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome 19: 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA 2007. Recombinant inbred strain panels: a tool for systems genetics. Physiol Genomics 31: 174–175 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- DeFries JC, Gervais MC, Thomas EA 1978. Response to 30 generations of selection for open-field activity in laboratory mice. Behav Genet 8: 3–13 [DOI] [PubMed] [Google Scholar]
- Donohue KD, Medonza DC, Crane ER, O'Hara BF 2008. Assessment of a non-invasive high-throughput classifier for behaviours associated with sleep and wake in mice. Biomed Eng Online 7: 14 doi: 10.1186/1475-925X-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen EJ 1978. Single-trait and antagonistic index selection for litter size and body weight in mice. Genetics 88: 781–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A, Flores JE, Deshpande H, Picazo J, Xie XM, Franken P, Heller HC, Grahn DA, O'Hara BF 2007. Pattern recognition of sleep in rodents using piezoelectric signals generated by gross body movements. IEEE Trans Biomed Eng 54: 225–233 [DOI] [PubMed] [Google Scholar]
- Gaya LG, Ferraz JB, Rezende FM, Mourao GB, Mattos EC, Eler JP, Michelan Filho T 2006. Heritability and genetic correlation estimates for performance and carcass and body composition traits in a male broiler line. Poult Sci 85: 837–843 [DOI] [PubMed] [Google Scholar]
- Hamet P, Pausova Z, Dumas P, Sun YL, Tremblay J, Pravenec M, Kunes J, Krenova D, Kren V 1998. Newborn and adult recombinant inbred strains: A tool to search for genetic determinants of target organ damage in hypertension. Kidney Int 53: 1488–1492 [DOI] [PubMed] [Google Scholar]
- Hanlon EC, Faraguna U, Vyazovskiy VV, Tononi G, Cirelli C 2009. Effects of skilled training on sleep slow wave activity and cortical gene expression in the rat. Sleep 32: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Malmanger B, Cooper S, Coulombe S, Reed C, Demarest K, Koyner J, Cipp L, Flint J, Talbot C, et al. 2002. Multiple Cross Mapping (MCM) markedly improves the localization of a QTL for ethanol-induced activation. Genes Brain Behav 1: 214–222 [DOI] [PubMed] [Google Scholar]
- Iraqi FA, Churchill G, Mott R 2008. The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort. Mamm Genome 19: 379–381 [DOI] [PubMed] [Google Scholar]
- Johnson TE, Wood WB 1982. Genetic analysis of life-span in Caenorhabditis elegans. Proc Natl Acad Sci 79: 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LD, Nielsen MK, Britton RA 1992. Genetic variation in liver mass, body mass, and liver:body mass in mice. J Anim Sci 70: 2999–3006 [DOI] [PubMed] [Google Scholar]
- Kadarmideen HN, von Rohr P, Janss LL 2006. From genetical genomics to systems genetics: potential applications in quantitative genomics and animal breeding. Mamm Genome 17: 548–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R 2009. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551 doi: 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M, Vaughn T, Pletscher L, King-Ellison K, Adams E, Erickson C, Cheverud J 1998. Genetic variation in body weight growth and composition in the intercross of large (LG/J) and small (SM/J) inbred strains of mice. Genet Mol Biol 21: 211–218 [Google Scholar]
- Krueger JM, Rector DM, Roy S, Van Dongen HPA, Belenky G, Panksepp J 2008. Sleep as a fundamental property of neuronal assemblies. Nat Rev Neurosci 9: 910–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes J, Kren V, Klir P, Zicha J, Pravenec M 1990. Genetic determination of heart and kidney weights studied using a set of recombinant inbred strains: the relationship to blood pressure. J Hypertens 8: 1091–1095 [DOI] [PubMed] [Google Scholar]
- Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JA, Hazendonk E, Prins P, Plasterk RH, Jansen RC, et al. 2006. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet 2: e222 doi: 10.1371/journal.pgen.0020222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B 1996. Genetics and analysis of quantitative traits. Sinauer, Sunderland, MA [Google Scholar]
- Morahan G, Balmer L, Monley D 2008. Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mamm Genome 19: 390–393 [DOI] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW 2004. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet 5: 7 doi: 10.1186/1471-2156-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peripato AC, De Brito RA, Matioli SR, Pletscher LS, Vaughn TT, Cheverud JM 2004. Epistasis affecting litter size in mice. J Evol Biol 17: 593–602 [DOI] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, et al. 2010. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav 9: 129–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome 18: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha JL, Eisen EJ, Siewerdt F, Van Vleck LD, Pomp D 2004. A large-sample QTL study in mice: III. Reproduction. Mamm Genome 15: 878–886 [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet 5: e1000419 doi: 10.1371/journal.pgen.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Cookson WO, Rawlins JN, Mott R, Flint J 2006. Genetic and environmental effects on complex traits in mice. Genetics 174: 959–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlsten D, Metten P, Crabbe JC 2003. A rating scale for wildness and ease of handling laboratory mice: results for 21 inbred strains tested in two laboratories. Genes Brain Behav 2: 71–79 [DOI] [PubMed] [Google Scholar]
- West DB, Goudey-Lefevre J, York B, Truett GE 1994a. Dietary obesity linked to genetic loci on chromosomes 9 and 15 in a polygenic mouse model. J Clin Invest 94: 1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DB, Waguespack J, York B, Goudey-Lefevre J, Price RA 1994b. Genetics of dietary obesity in AKR/J × SWR/J mice: segregation of the trait and identification of a linked locus on chromosome 4. Mamm Genome 5: 546–552 [DOI] [PubMed] [Google Scholar]
- West DB, Waguespack J, McCollister S 1995. Dietary obesity in the mouse: interaction of strain with diet composition. Am J Physiol 268: R658–R665 [DOI] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F 2007. On the subspecific origin of the laboratory mouse. Nat Genet 39: 1100–1107 [DOI] [PubMed] [Google Scholar]
- York B, Lei K, West DB 1997. Inherited non-autosomal effects on body fat in F2 mice derived from an AKR/J × SWR/J cross. Mamm Genome 8: 726–730 [DOI] [PubMed] [Google Scholar]