Abstract
The olfactory receptor (OR) genes are the largest mammalian gene family and are expressed in a monogenic and monoallelic fashion in olfactory neurons. Using a high-throughput approach, we mapped the transcription start sites of 1085 of the 1400 murine OR genes and performed computational analysis that revealed potential transcription factor binding sites shared by the majority of these promoters. Our analysis produced a hierarchical model for OR promoter recognition in which unusually high AT content, a unique epigenetic signature, and a stereotypically positioned O/E site distinguish OR promoters from the rest of the murine promoters. Our computations revealed an intriguing correlation between promoter AT content and evolutionary plasticity, as the most AT-rich promoters regulate rapidly evolving gene families. Within the AT-rich promoter category the position of the TATA-box does not correlate with the transcription start site. Instead, a spike in GC composition might define the exact location of the TSS, introducing the concept of “genomic contrast” in transcriptional regulation. Finally, our experiments show that genomic neighborhood rather than promoter sequence correlates with the probability of different OR genes to be expressed in the same olfactory cell.
The mammalian nervous system is composed of diverse cell types produced by distinct transcription programs. It is well established that positional information, which allows alternative interpretation of signaling cues, plays a dominant role in establishing neuronal fates. In the spinal cord, for example, signaling gradients dictate distinct gene expression programs responsible for different columnar and segmental identities (Dasen et al. 2005). In most of the cases examined, the final interpreters of the differentiation signals are the promoters of the responding genes, which together with distant enhancer sequences are targeted by particular combinations of transcription factors.
The olfactory system constitutes an extreme example of neuronal diversity, as each olfactory sensory neuron monoallelically expresses one out of approximately 1400 OR genes (Buck and Axel 1991; Chess et al. 1994). Sensory neurons expressing a particular receptor are scattered in a pattern that seems stochastic but are restricted to a subregion of the olfactory epithelium, called a zone (Ressler et al. 1993; Vassar et al. 1993). Two main models describe molecular mechanisms that might give rise to the observed expression characteristics (Shykind 2005; Fuss and Ray 2009). At one extreme, a combinatorial model states that 1400 distinct combinations of transcription factors act on the 1400 different OR promoters to drive expression of only one OR gene per neuron. On the other hand, a completely stochastic model proposes that the cell produces the observed monogenic and monoallelic expression pattern by choosing one among 2800 equivalent OR promoters. Elements of both models could work in concert; a combinatorial process sets zonal boundaries, and a stochastic choice within the zonal repertoire of OR genes results in the expression of only one OR allele. In this scenario, one would predict that OR genes that are expressed in the same zone share similar promoter information that allows expression within the zone and prevents expression in other areas of the olfactory epithelium. Alternatively, OR promoters may not contain information that restricts their expression to specific domains of the olfactory epithelium. In this case, zonal expression might be regulated by genomic location, the local chromatin environment, or by the action of long-range enhancers. A comprehensive OR promoter analysis could provide insight into the regulatory logic of OR choice by pinpointing regulatory sequence similarities and differences across the OR family.
Progress has been made in characterizing regulatory elements in OR promoters and the transcription factors that bind them. The major study of transcription factor binding sites in 200 OR promoters identified two classes of motifs—homeodomain and O/E (Olf1/Early B-cell factor)–like sites (Michaloski et al. 2006). Genetic experiments showed that, directly or not, OR expression requires two homeodomain proteins, LHX2 and EMX2. Loss of LHX2 prevents the maturation of most olfactory neurons, resulting in perturbation of type II (mammal-specific) OR expression and maintenance of some but not all type I (fish-like) ORs; EMX2 deletion causes 75% of ORs to lose expression and up-regulates expression of others (Levi et al. 2003; Hirota and Mombaerts 2004; Hirota et al. 2007; McIntyre et al. 2008). While homeodomain and O/E factors are certainly important for regulation of OR expression, additional factors likely contribute either to expression in the olfactory neurons or to restriction to specific zones. However, a more comprehensive promoter analysis that would correlate promoter properties with expression differences is not possible due to the low proportion of mapped OR promoters. For this reason and to obtain a better understanding of the contribution of proximal regulatory sequences to OR expression, we sought a high-throughput mapping of OR transcriptional start sites (TSSs).
To accomplish this, we designed a custom, high-density tiling microarray that covers the olfactory genome at 4-bp resolution and hybridized to it capped OR transcripts prepared by RLM-RACE (Liu and Gorovsky 1993; Michaloski et al. 2006). With this method, we mapped 1085 odorant receptor transcriptional start sites, which expands the mapped OR TSSs fivefold. Largely agreeing with the previous study, our computational analysis reveals potential transcription factor binding motifs that might be involved in OR expression and demonstrates a stereotypic positioning of O/E sites upstream of OR TSSs. In addition, we find that OR promoters are extremely AT-rich, a genomic property restricted to and shared with other genes with extreme evolutionary plasticity. Using these features, together with the epigenetic properties of OR genes, we can predict OR promoters among murine promoters with ∼80% specificity. However, different computational approaches failed to identify strong correlations between promoter motifs and zonal expression. On the contrary, our analysis suggests that the genomic location of an OR gene correlates better than promoter similarity with OR expression in particular cell types.
Results
Using RNA ligation-mediated rapid amplification of cDNA ends (RLM-RACE) on total RNA from the main olfactory epithelium, we generated libraries of capped odorant receptor 5′ ends that were reverse-transcribed and amplified using degenerate primers against conserved transmembrane domains III, V, VI, and VII (Buck and Axel 1991; Malnic et al. 1999; Michaloski et al. 2006). We reverse-transcribed and amplified capped RNAs in two ways (Supplemental Fig. S1). To generate a high-stringency library, we reverse-transcribed using degenerate primers in transmembrane domains V, VI, and VII and amplified using nested PCR with degenerate primers in TMIII (GEO accession number GSM647450) (Fig. 1). After analyzing this data set, we sought to increase our coverage by decreasing our stringency (GEO accession numbers GSM647451, GSM647452). For the high-coverage data set, we reverse-transcribed and amplified using the same degenerate primer sets. We did this individually for each of six sets listed in Supplemental Methods and pooled the RACE products for hybridization. By hybridizing these libraries to a 4-bp-resolution olfactory tiling array, we have mapped the 5′ non-coding exons of 1085 OR genes (Cheng et al. 2005).
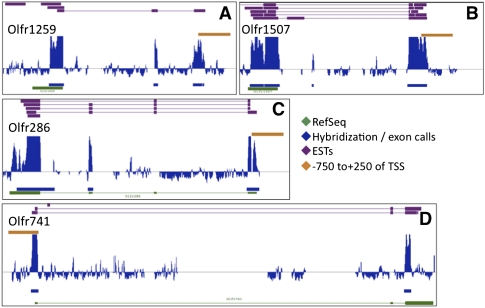
Figure 1.
One thousand eighty-five olfactory receptor 5′ structures mapped by high-throughput RLM-RACE. For selected ORs (A–D), RefSeq records (green), ESTs (purple), summary hybridization patterns and computed exons (blue), and promoter calls (orange) are displayed in IGB. Our hybridization patterns match RefSeq and EST records well. As shown for Olfr286, the tiling array cannot detect exons that occur in RepeatMasked (and therefore untiled) areas of the genome or across probes that do not map uniquely. For scale, the orange bar is 1000 bp in each panel.
Using the high-stringency data set, we mapped 650 mouse OR 5′ UTRs. The high-coverage set added about 450 OR maps, some de novo and some by increasing significance of weak signals also present in the high-stringency data. We identified exonic intervals computationally by thresholding array peaks and called most distal exonic signal relative to particular odorant receptor genes transcription start sites. We curated these designations and assigned gene names to intervals (using GIN) to generate a final set of transcriptional start sites and putative promoters (Cesaroni et al. 2008). Bar files summarizing the data and TSS calls are available in GEO; representative hybridization patterns are shown in Figure 1. Occasionally, UTRs were different in 5′ extent across the data sets, probably representing differential amplification of alternative splice products. In cases of conflict, we chose the map from the stringent data set.
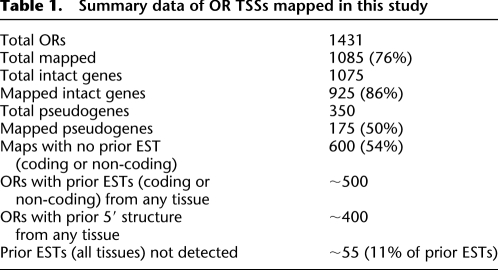
Our 1085 5′-UTR maps comprise 76% of the 1431 known OR genes and pseudogenes. We visually compared a subset of 300 maps from our data to published OR EST and RefSeq records or lack thereof and used these comparisons to estimate frequencies for the whole data set (Table 1; Pruitt et al. 2007). More than half of our data set represent ORs with no previous coding or non-coding ESTs deposited in GenBank (Benson et al. 2004; Feldmesser et al. 2006). We detected 86% of intact genes and 50% of pseudogenes. When our data could be compared to ESTs in the database, our data matched the published data with accuracy higher than 95% (Fig. 1A). Only 5% of our 5′ UTRs have extra or alternative UTR exons relative to ORs that are already well-covered by ESTs (Fig. 1B) (Young et al. 2003). However, our structures often contained larger or extra exons when compared to the few ORs with 5′ non-coding RefSeq records (Fig. 1D), suggesting that our analysis provides a better coverage of the OR transcriptome. Therefore, our data provide high throughput and accurate description of the transcription start sites of OR genes. Since genetic analysis suggests that (for most OR genes tested) proximal promoter sequences located <500 bp upstream of the TSS are sufficient to recapitulate OR-like expression in the MOE, we committed our analysis to the region 1 kb upstream of the TSS.
Table 1.
Summary data of OR TSSs mapped in this study
Promoter motif analysis
According to a hierarchical model of OR transcriptional control, we expected to find two types of regulatory circuits in OR promoters. One circuit would be responsible for OR transcription in the main olfactory epithelium (MOE) and would be common to most OR promoters; the other would impose zonal restrictions to OR expression and would vary across promoters of genes expressed in different zones. To identify regulatory modules that are responsible for OR expression in the MOE, we looked for common motifs in the whole set of 1085 OR promoters. We searched for known transcription factor binding sites (TFBS) using the Genomatix transcription factor binding site database, and we used the Weeder, Gibbs Recursive Sampler, and MEME algorithms to identify novel motifs (Bailey and Elkan 1994; Thompson et al. 2003; Pavesi et al. 2004; Cartharius et al. 2005).
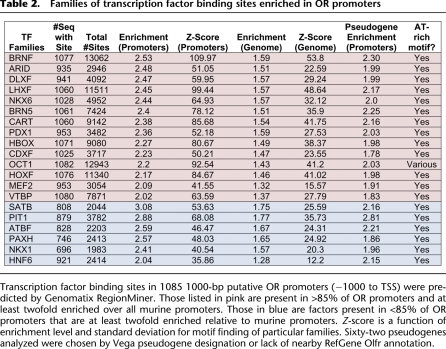
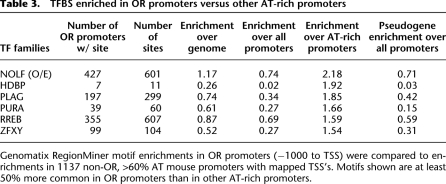
Using the Genomatix TFBS library, we searched for families of sites enriched in the OR promoter data set relative to all murine promoters (Cartharius et al. 2005). The analysis identified many expected classes of motifs—homeodomains and homeoboxes, in particular LHX and DLX sites—along with many other families that have not been correlated with OR expression in previous reports (Table 2). To find transcription factor families that may act across all OR genes, we selected those that were present in at least 85% of promoters and were enriched at least twofold relative to all murine promoters in the Genomatix database. This yielded 14 families (Table 2, pink). As expected, the group included many variants of homeodomain sites, in agreement with genetic observations that suggest a role for LHX2 and EMX2 in OR regulation (Hirota et al. 2007; McIntyre et al. 2008). We found no qualitative differences in TFBS enrichment between intact and transcribed, pseudogenized OR promoters, as the same types of motifs appear enriched in both types of promoters. This is not surprising given that the frequency of OR choice is not affected by the ability of the selected OR to produce receptor protein; the fact that the transcription of pseudogenous transcripts is not stable does not stem from promoter differences, as suggested by our computational analysis and verified by genetic experiments (Shykind et al. 2004). However, it is worth mentioning that we do observe a slightly reduced representation for most of these motifs in the promoters of OR pseudogenes, which might represent an evolutionary drift that could ultimately result in the degeneration of these promoters and the transcriptional incapacitation of OR pseudogenes.
Table 2.
Families of transcription factor binding sites enriched in OR promoters
Based on our RNA-seq analysis in the MOE (Magklara et al. 2011) and published microarray expression data, members of each transcription factor family predicted to bind enriched sites are expressed in the olfactory epithelium (Sammeta et al. 2007). For example, the CART and MEF2 families of transcription factors are represented by very specific expression of Uncx and Mef2b, respectively (Saito et al. 1996; Su et al. 2002; Sammeta et al. 2007). Finally, several classes of POU domain transcription-factor binding sites were predicted in the OR promoters, including OCT1, BRNF, and BRN5 types. These probably represent another major requirement for OR expression that could be explored by future genetic experiments.
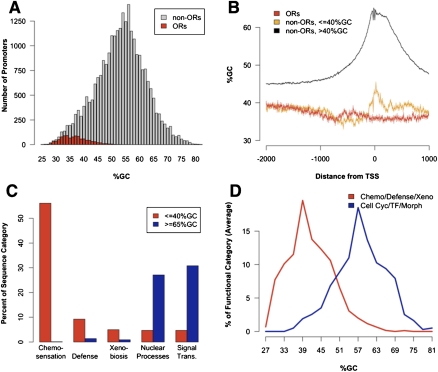
We repeated the transcription factor binding site enrichment analysis using other portions of the OR genomic locus—1 kb downstream from the TSS or 1 kb upstream of the CDS. Surprisingly, enrichment characteristics in these data sets were almost identical to those in the promoters (data not shown); moreover, RepeatMasking did not alter the TFBS distribution. Thus, regulatory potential may simply be diffuse in OR genomic loci, as has been suggested by genetic experiments (Rothman et al. 2005). Another possibility is that the enrichment of these families in OR sequences is a consequence of the high AT content of OR genomic loci. The mouse genome has an average 58% AT content but is GC-rich near promoter sequences as shown in Figures 2A and 2B (Waterston et al. 2002; Akan and Deloukas 2008). In contrast, OR promoters average 63% AT (regardless of RepeatMasking) and, unlike average murine promoters, do not become more GC-rich toward the TSS (Fig. 2B). Therefore, the TFBS enrichment we identified could be a consequence of this extreme nucleotide composition, since the identified motifs are AT-rich sequences. To test this, we performed the same TFBS prediction analysis on four randomly generated 1000-bp sequences with 63% AT content and observed that 4/4 sequences contain 8/10 of the most common motifs found in Table 2. While this sequence bias is probably important for OR regulation, it skews the analysis toward AT-rich binding sites and convolutes the interpretation of our data. To establish an AT-rich baseline for comparison, but also to understand the evolutionary or regulatory significance of the use of promoter sequences with extreme AT content, we constructed a secondary promoter data set consisting of all the non-OR mouse promoters that have similar AT content to OR promoters: those >60% AT.
Figure 2.
Promoters of ORs and other plastic gene families are extremely AT-rich. (A) Olfactory receptor TSS's were mapped, and the %GC distribution of OR (red) and non-OR (gray) promoters (−750 bp to +250 bp relative to TSS) was compared. (B) %GC around the TSS was plotted for ORs (red) and non-ORs sorted by average GC content (orange, black). (C) Promoters ≤40% GC (red) or ≥65% GC (blue) were categorized by gene function (skewing of each category into AT or GC significant, p < 10−10, χ2) (see Methods). (D) Full GC content distribution of averaged biased functional groups (for statistics, see Supplemental Fig. S2; Supplemental Table 1).
As shown in Figure 2C and Supplemental Table 1, annotation analysis of the approximately 1360 non-OR and 846 OR genes driven by AT-rich promoters reveals common themes even though they perform various biological functions and do not share recent common ancestry. Even when Olfactory Receptor genes are excluded from this list, the majority of these genes are organized in genomic clusters characterized by significant copy number variation and polymorphism (Supplemental Table S2; Wong et al. 2007; Liu et al. 2009, 2010; Nicholas et al. 2009; Young et al. 2008; Waszak et al. 2010). Most of these genes encode transmembrane or secreted proteins expressed in barrier epithelia that selectively communicate with and protect from the external world (Fig. 2C; Supplemental Fig. S1; Supplemental Table S1). Finally, a significant portion of these genes have variegated, mutually exclusive, monogenic, or even monoallelic expression patterns (Sampsell and Held 1985; Held et al. 1995; Hollander et al. 1998; Singh et al. 2003; Chess 2005). Thus, AT-rich sequences regulate a diverse group of genes that expanded fast to accommodate adaptation and remain highly plastic, like the OR genes, in contrast with GC-rich promoters that regulate evolutionarily stable gene families organizing development and morphogenesis (Fig. 2D; Supplemental Fig. S1; Davidson and Erwin 2006; Bernardi 2007).
Distinguishing OR promoters from other AT-rich promoters
Seeking distinction between OR and non-OR AT-rich promoters, we identified a few binding sites enriched in OR promoters over the rest of the AT-rich promoters in the mouse genome (Table 3). Of these, the O/E (“Nolf”) site is by far the most common and most enriched. Gibbs and MEME analysis in this larger ensemble of OR promoters revealed O/E consensuses that appear in subsets of OR promoters (Fig. 3E). Since the GC-rich O/E sites appear frequently on mouse promoters with average GC content and are not enriched in OR promoters relative to the murine promoterome as a whole, this motif might provide a significant distinction between OR and non-OR AT-rich promoters. This analysis is most likely saturated since additional search for common, novel motifs that would distinguish ORs from the other AT-rich promoters, using Weeder or Gibbs, failed to reveal any other differences.
Table 3.
TFBS enriched in OR promoters versus other AT-rich promoters
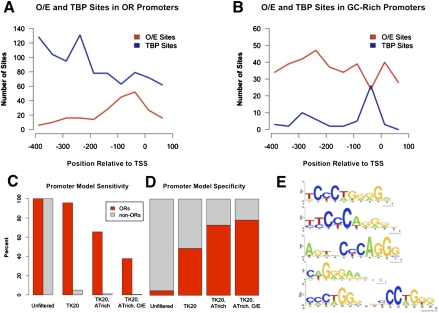
Figure 3.
O/E sites define the OR TSS. (A) OR promoters (n = 1085 for O/E, 243 for TBP) and (B) GC-rich promoters (≥65% GC in −750 to +250 region; n = 1098) were searched for O/E and TBP sites using strings. (O/E) TCCCTGGGG, up to one mismatch; (TBP) TATAWW. Sites were plotted by position relative to TSS. (C,D) Murine promoters were filtered by H4K20-Me3 (“TK20”) coverage of at least 50% of the gene body, then by AT content of at least 60%, and then by presence of summary O/E site (TCCC or CCCT string) within 100 bp upstream of the TSS. Of 501 murine promoters meeting the filters, 383 drive ORs (specificity: 78%; sensitivity: 38%). The numbers of genes passing each filter and statistical significance can be found in Supplemental Table S3. (E) Gibbs Recursive Sampler and MEME identify new O/E sites in OR promoters.
To obtain a better understanding of the possible role of O/E sites in OR transcription, we plotted the position of this motif in relationship to the TSS, both in OR promoters and in GC-rich promoters. As seen in Figure 3B, the distribution of O/E sites is random in GC-rich promoters, except at the region defined by the TATA-box, where we observe decreased O/E frequency, as expected due to locally higher AT content. In contrast, the distribution of O/E sites in OR promoters clumps ∼50 bp upstream of the TSS, in agreement with previous observations in a smaller sample of OR promoters (Fig. 3A; Michaloski et al. 2006). This distribution is strikingly reminiscent of TATA-boxes in GC-rich mouse promoters (Fig. 3B). Conversely, the distribution of TATA-box sequences in OR promoters (and the rest of the AT-rich promoters for that matter) (data not shown) appears diffuse, suggesting that this is not the determining sequence of the transcription start site of OR genes.
The analysis presented above implies that the high AT content of OR promoters, together with stereotypic presence of an O/E site near the TSS, distinguishes OR promoters from the rest of the mouse promoters. Moreover, we recently showed that in the olfactory epithelium, OR loci are marked by the hallmarks of constitutive heterochromatin, H3K9me3 and H4K20me3 (Magklara et al. 2011). Since these trimethyl marks are deposited with very high specificity on OR loci, we reasoned that they also contribute to the distinction of the OR genome from the rest of the euchromatic genome. To test the hypothesis that OR promoters are defined by three layers of specificity, namely, genomic, genetic, and epigenetic signature, we applied successive filters to the mouse promoterome and asked whether we can enrich for OR promoters with sufficient specificity and sensitivity. As seen in Figure 3, ∼80% of promoters with AT content >40% that are marked by H4K20me3 (which overlaps completely with H3K9me3) and have an O/E site within 100 bp upstream of the TSS are OR promoters, an impressive 17-fold enrichment over their total prevalence among mouse promoters (Fig. 3D; Supplemental Table S3). Importantly, these filters provide such high specificity while retaining significant specificity, as these three parameters identify ∼38% of the mapped OR promoters (Fig. 3C; Supplemental Table S3).
In summary, we mapped the transcription start sites of 1085 OR promoters. Computational analysis revealed that 14 TFBS are enriched in OR promoters versus the rest of the mouse promoters. However, the same modules can be found in random computer-generated AT-rich sequences and are shared by 1360 non-OR, AT-rich promoters that also regulate rapidly evolving genes. By using the other AT-rich promoters as a baseline for our analysis, we discovered that O/E sites provide some distinction between OR and non-OR AT-rich promoters and mark the position of the OR TSS, in a similar fashion to TATA-box definition of the TSS in regular promoters. Together these two features, combined with the epigenetic signature of OR loci, yield a 17-fold computational enrichment of OR promoters among murine promoters.
Zonal promoter analysis
A combinatorial model of OR gene regulation predicts that ORs expressed in a particular zone share common TF binding motifs. Unfortunately, zone information has been ascribed to only 98 type II and about 100 type I ORs (Miyamichi et al. 2005), and we could not successfully detect significant differences in TFBS enrichment among the four zones using such a small sample. Analysis of these limited zonal subgroups by enrichment of known TFBS and by the wordCount method (described in Methods) are presented in Supplemental Figure 2. To obtain information from an OR subgroup with an expression pattern that is defined with more strict criteria, we compared the promoters of the approximately 100 type I ORs, which are almost exclusively expressed dorsally, to type II ORs that are expressed in the rest of the MOE (Hirota et al. 2007). Apart from their phylogenetic differences (type I ORs are more ancient fish-like ORs) (Niimura and Nei 2005, 2006), genetic experiments suggest that type I ORs are expressed in neurons of a different lineage than type II neurons (Hirota et al. 2007; Bozza et al. 2009). When analyzed separately for TFBS enrichment, type I and type II promoters were extremely similar. Ranked by enrichment relative to all murine promoters, the top 30 matrix families were the same for both groups, although the order was slightly different. Below this cutoff, three matrix families were enriched in one type but not the other—PAX1 and NBRE sites in type I promoters and GABF sites in type II promoters (Supplemental Table S4). Fold change for PAX1 was greater than two standard deviations of the mean fold difference between promoter types; NBRE and GABF were outside one standard deviation of this mean. Future genetic experiments could address whether these represent meaningful differences in the regulation of the two OR types. Notably, in our epigenetic analysis of OR loci (Magklara et al. 2011), we found that type I ORs have significantly lower levels of H3K9me3 and H4K20me3 in chromatin preparations from the total olfactory epithelium, providing the only distinction between the two types of ORs. Thus, the genomic location and epigenetic profile of these genes correlate with their expression zone, while promoter sequence does not. Unfortunately, we do not have the tools to test the seductive speculation that type I ORs have high levels of these trimethyl marks only in zone I, where they can be expressed. In other words, and based on the filtering analysis in Figure 3, it is possible that these repressive histone marks have dual functions: to repress but also to label loci for later activation.
As existing modular methods for parsing promoters failed to produce models of zonal regulation, we sought to determine whether zonal expression correlates with bulk similarity of the associated promoters. Therefore, we built a simple tool to cluster distantly related or unrelated sequences by counting the presence or absence of k-mers irrespective of their position in the larger sequence. Instead of looking for evolutionary homology or shared single motifs, we compare the total complement of “words” in each promoter to those in each other promoter (described in Supplemental Fig. 3).
We ran the k-mer clustering method using a variety of parameters and checked whether clustering could sort promoters of known zone. Altered parameters included word size (8-mers were most informative) distance metric, RepeatMasking, reverse complement collapse, and number of total word occurrences for inclusion. Under all test parameters, promoters of a particular expression zone or OR class are distributed across k-mer clusters. We were unable to identify parameters that produced coherent zonal clusters among the approximately 200 OR promoters with mapped zone, not even for the approximately 100 type I ORs that are expressed exclusively in zone I. Supplemental Figure S2D depicts one representative analysis.
Finally, we obtained highly quantitative RNA-seq data from FACS-sorted mature olfactory neurons (Magklara et al. 2011). Although OR genes are among the most highly expressed genes, our RNA-seq analysis reveals a wide range of expression differences at the tissue level. This variability mostly reflects differences in the frequency of choice for each OR and to a lesser degree differences in the transcription rate of each chosen OR, at least at the sensitivity level of RNA in situ hybridization experiments (data not shown). We asked whether these quantitative differences in OR expression levels could be attributed to promoter characteristics. We compared the promoters of the most highly expressed ORs (top 5%) with the promoters of ORs that have the lowest expression levels (bottom 5%), with an average approximately 200-fold difference in mRNA levels. Again, this analysis did not reveal qualitative or major quantitative (different abundance of certain motifs) differences among the strongest and weakest OR promoters (Supplemental Table S5). Notably, however, as in the case of the pseudogene promoters, there is a trend: Promoters of the most highly expressed ORs have on average slightly more copies of the 14 most enriched TFBS. This is in agreement with recent experiments suggesting that the number of homeobox binding sites on transgenic OR promoters could influence their transcription frequencies (Vassalli et al. 2011).
In summary, promoters of ORs expressed in the same zone did not cluster, nor did they clump when graphed by their final distance order in the data set (Supplemental Fig. S2D). Nor did zonal expression associate with enrichment for known or novel TFBSs (Supplemental Fig. S2A–C). Thus, based on several distinct computational approaches, we were able to identify only one weak promoter model, for zone 2. We were unable to model other zones using linear promoter sequence. This suggests that linear promoter sequence is insufficient to give zonal identity to OR expression. However, our analysis is challenged by the lack of a clear understanding of zonal identity of OR expression and the small proportion of ORs with mapped expression zone. To overcome this, we sought a more defined system for restrictive OR expression, provided by two olfactory placode-derived cell lines (Illing et al. 2002; Pathak et al. 2009).
Contextual expression
Seeking to examine whether promoters of ORs expressed in similar contexts resemble one another, we profiled the OR transcriptome of the OP6 and OP27 olfactory placode cell lines (R Lane, in prep.). OR expression in these lines was recently demonstrated to be monogenic in each cell but various across a cultured population. The two cell populations expressed about 80–100 ORs, and, surprisingly, ORs from each zone were represented in the transcribed OR populations of each OP cell type. Interestingly, however, type I ORs are not expressed in either cell type or differentiation state, supporting the notion of their being expressed in a different lineage under a different regulatory logic (Bozza et al. 2009).
Since the cultured cells are exposed to a homogeneous extracellular environment and were expanded from a single cell, we reasoned that they share a common transcription factor milieu under the culturing conditions. Therefore, if OR promoters contain instructive (rather than simply permissive) information for OR expression, promoters of the expressed genes should share common characteristics that distinguish them from the non-expressed ORs. We repeated known TFBS enrichment analyses on promoters of the four expressed subsets (OP6 and OP27 differentiated and undifferentiated) and searched for motifs with significant enrichment differences over the rest of the 1085 mapped OR promoters. Again, this analysis failed to produce a predictive model that could pick out promoters active in this cell line (R Lane, in prep.; data not shown). Novel motifs found in expressed groups by the Gibbs algorithm or Genomatix CoreSearch were no more common in the expressed groups than in the whole promoter group and were not information rich (e.g., WWWWWW).
For a more detailed analysis, we zoomed in on a large, OP-expressed cluster on chromosome 2 and asked what united the expressed subset and divided it from the non-expressed ORs—promoter elements or genomic locus. We ordered the promoters of all ORs on chromosome 2 using our k-mer method. This produced two ordering indices for ORs—linear order on the chromosome (where, for example, the OR closest to 0 bp in the chromosome is assigned index 1) and promoter word composition distance order. We show the positions of OP27-expressed ORs within each of these indices in Figure 4. The expressed set clumps according to linear order in the genome (Fig. 4B) but is smoothly distributed when indexed by promoter similarity (Fig. 4A).
Figure 4.
ORs on chromosome 2 expressed in olfactory placode cells are similar in genomic locus but not in promoter sequence. ORs on chromosome 2 were indexed by (A) promoter sequence homology order (determined by the bottom level of the Hopach fuzzy clustering tree of pairwise distance measures generated by the wordcount method outlined in Figure S3) or (B) linear order along the chromosome. Index numbers of ORs expressed in undifferentiated OP27 cells were graphed, although expression patterns in the other three conditions were similar. After RepeatMasking, only 74 chromosome 2 promoters were long enough to be analyzed. The linear index contains 288 ORs.
Discussion
The molecular mechanisms regulating the monogenic and monoallelic expression of OR genes remain poorly understood. A major obstacle in the understanding of OR gene regulation was the lack of knowledge of the exact locations of the promoters of most of these genes. Here we used a novel, high-throughput method to map the transcription start site of most OR promoters and subsequently performed a comprehensive computational analysis in an attempt to understand the logic behind OR regulation.
Putative transcription factor binding sites on OR promoters
While the putative OR promoters identified through 5′-exon mapping are not conserved in their overall sequence, there are 14 transcription factor binding sites that are over-represented in OR promoters over the rest of the murine promoters (Table 1). Although any combination of 10 of these motifs can accurately predict OR promoters, AT-rich motif enrichment does not distinguish OR promoters from genomic regions flanking the OR promoters. It is possible that many of these TFBSs are not biologically relevant and their computed abundance is a tautological consequence of the AT sequence bias.
Alternatively, these elements may function through a diffuse arrangement along several kilobases upstream of and downstream from the TSS. Given the extremely high levels of OR transcription within each neuron, it is not unlikely that there might be an additive effect of multiple transcription factors bound across each OR locus. In support of this, knockin genetic manipulation of homeobox and O/E sites of olfactory receptor gene M71 suggested the existence of redundant transcription factor binding sites within that locus (Rothman et al. 2005). Given our recent discovery that OR clusters are packed in extremely compacted and repressive chromatin structure prior to OR expression (Magklara et al. 2011), then the repetitive assembly of the same combination of transcription factors for kilobases upstream of and downstream from the TSS could be crucial for OR activation; if the machinery responsible for OR activation “opens” the trimethyl-marked chromatin locally without knowledge of the underlying genomic sequence, the chances of finding an OR promoter within a cluster would be very low. In contrast, with the diffuse and repetitive arrangement of transcription factors that we describe here, any local chromatin opening would allow for the proper combination of transcription factors to bind to the DNA and to further propagate histone demethylation and chromatin remodeling until this reaches an OR promoter. In this case, the promoter could be defined by the stereotypic presence of an O/E motif (Fig. 3A), or a local spike of GC content that generates “genomic contrast” recognized by the transcription machinery (Affolter et al. 2008). The well-established existence of a feedback signal generated by the synthesis of OR protein could be responsible for locking the selected promoter in this transcriptional state and preventing chromatin opening from expanding to other OR loci (Lewcock and Reed 2004).
AT-rich promoters regulate rapidly expanding gene families
The realization that OR promoters have a distinct AT content, so different from the majority of the mouse promoters, provoked the search for other promoters with similar characteristics. Interestingly, there are an additional 1360 murine promoters with nucleotide composition similar to ORs (Fig. 2A,B). These promoters retain a high AT content throughout diverse mammalian species, although there is no conservation at the sequence level. Even in evolutionarily older vertebrates, such as fish and frogs, the genomic regions surrounding these genes have a high AT content, although the results in these organisms are harder to interpret due to an overall higher AT content in these genomes. These observations raise important questions regarding the regulatory significance of the preservation of nucleotide composition over large genomic regions (isochors) without simultaneous conservation of sequence information. Recent studies have shown that DNA shape is more tightly conserved than linear sequence (Parker et al. 2009). In addition, many DNA-binding proteins contact the minor groove, especially those with preference for AT-rich motifs, instead of or in addition to the major groove (Joshi et al. 2007; Rohs et al. 2009, 2010). AT content is a major contributor to the shape of the DNA molecule, particularly the minor groove, and it is possible that factors such as AT content affect DNA shape over a large region and determine the context in which transcriptional machinery understands short, modular sites, such as the O/E site we find near the OR TSS.
However, the significance of high AT content might extend beyond regulatory functions. As we mentioned, genes regulated by these promoters play crucial roles in vertebrate evolutionary adaptation: Their products sense the environment, protect from pathogens, and detoxify and metabolize new compounds that would appear in novel ecological niches. Under such selective pressure, these genes evolved extremely fast; the explosion of olfactory receptor gene family members from about 150 in fish to 1500 in frog and other tetrapods is a prime example. However, evolutionary plasticity comes with a price: Genes that use AT-rich promoters demonstrate significant intra-species copy number variation and sequence polymorphisms, as well as large oscillations in number of family members among related species. Furthermore, most of these rapidly expanding genes have sporadic expression, ranging from the monogenic regulation of chemoreceptors and protocadherins to the variegated expression of NK cell receptors (Held et al. 1995; Singh et al. 2003; Chess 2005). For the majority of these families, mutually exclusive transcription provides functional specificity to the expressing cell; for others, it might simply afford the expression of one family member at a very high level (Sampsell and Held 1985). Did the stochastic expression patterns of these genes arise because a deterministic regulatory system did not evolve as quickly as the gene families expanded? We propose that AT-rich promoters provide a genomic platform for rapid evolution and compatibility with non-deterministic gene regulation. Unusual DNA or chromatin architecture, the ability to form long-range interactions, or propensity to segregate in distinct nuclear territories could promote diversification and allow mutually exclusive expression of multigenic families, providing a solution to the problem of evolutionary adaptation (Tajbakhsh et al. 2000; Ribich et al. 2006; Savarese and Grosschedl 2006; Dekker 2008; Segal and Widom 2009). In effect, the partitioning of genes with terminally differentiated functions into the AT genome may allow rapid change in these cassettes to the benefit of the animal while the core cellular and developmental machinery encoded in the GC genome remains static. One can imagine how mutations in the AT genome might cause only minor harm, while alterations to the GC genome lead to developmental Armageddon, congenital disease, and carcinogenesis.
Distinguishing OR promoters from other promoters
The unusually high AT content of OR promoters and the overall organization of OR genes in AT-rich isochores provide an obvious distinction, even at the genomic level, between OR promoters and the majority of the mouse promoters. However, this genomic signature would not be sufficient to distinguish OR promoters from the additional 1360 AT-rich promoters discussed earlier, the majority of which are not transcribed in olfactory sensory neurons based on our RNA-seq analysis. This distinction might be accomplished by the unique combination of a cell-type specific epigenetic signature, such as the trimethylation of H3K9 and H4K20, that is extremely specific for OR loci and the stereotypic placement of different variants of the O/E consensus near the TSS. Indeed, if we ask computationally how many promoters combine high enrichment for H3K9me3/H4K20me3 in the olfactory epithelium, AT content ≥60%, and presence of O/E sites within 100 bp of the TSS, we achieve 38% sensitivity (38% of OR promoters fall in this group) and 78% specificity (78% of these promoters regulate ORs).
Lack of zonal information in OR promoters
Although our analysis revealed a signature for the OR promoterome that likely instructs expression in the MOE, it did not succeed in identifying motifs correlated with zonal identity. Complementary approaches failed to identify strong correlations between modular promoter sequence signatures and zonal expression. The same holds true for the comparison of type I and type II OR promoters, the comparison of promoters of highly and lowly expressed ORs, or a comparison between expressed and non-expressed ORs in olfactory placode cell lines. This was unexpected given that OR transgenes with minimal promoter information can recapitulate the sporadic expression pattern of endogenous OR genes (Rothman et al. 2005; Vassalli et al. 2011). One explanation for this is that regulatory information might also be encoded in transcribed parts of the gene or even in the coding sequence, as has already been shown regarding the silencing of OR transgenes (Nguyen et al. 2007). It is also possible that minigenes do not abide by the epigenetic regulation of the endogenous ORs and therefore have different regulatory requirements from the endogenous OR genes. Alternatively, it is possible that a “zone” is a complicated region consisting of multiple mini-zones in each of which only a handful of OR genes can be expressed (Miyamichi et al. 2005). In this scenario, looking for motifs shared by many OR promoters is pointless, and, instead, a search should be focused on a very limited number of very similar OR promoters that drive expression in neighboring cells. While such an approach could reveal TFBSs shared by very homologous promoters, it would not explain genetic data suggesting that approximately 40 ORs have the potential to be expressed in the same neurons (Serizawa et al. 2003). Consistent with this is our observation that a heterologous promoter with the potential to be expressed in every olfactory neuron, the promoter of the olfactory marker protein gene (OMP), is expressed in a monogenic, monoallelic, and zonal fashion, when subjected to the epigenetic regulation of a neighboring OR gene (Magklara et al. 2011). This observation is in complete agreement with our computational prediction that zonal information is not encoded in the promoter sequences of OR genes.
Genomic coordinates as a potential source of specificity in OR expression
If any of the genetic and epigenetic parameters examined above failed to reveal the logic behind sub-tissue-level OR expression, then the question remains: What organizes OR expression in distinct zones? Our analysis revealed an intriguing correlation between genomic location and expression properties. The subset of OR genes that we found to be expressed in an olfactory placode cell line is concentrated in specific genomic locations rather than being evenly distributed in the OR genome. Similarly, ORs that reside only in specific clusters are expressed during the early development of the olfactory epithelium (Rodriguez-Gil et al. 2010). The most parsimonious explanation for these observations is that sensitivity to the positional information for each sensory neuron stems from distant enhancer elements that reside outside of the inaccessible OR heterochromatin and contain sequence information sufficient to interpret transcription factor gradients. The existence of such regulatory elements near specific OR clusters could provide an explanation for the concentration of highly expressed ORs at specific genomic coordinates (data not shown). Indeed, the H element, which is located 75 kb upstream of an OR cluster in chromosome 14, interacts most frequently with the most proximal OR gene, Olfr1507 (MOR28), making it the second most transcribed OR gene in our RNA-seq data set (Serizawa et al. 2003; Lomvardas et al. 2006). Future studies will test whether similar H-like elements are located proximal to the other highly transcribed ORs and whether these enhancers are responsive to spatiotemporal cues, providing some specificity in a stochastic regulatory process.
Methods
RLM-RACE
We isolated total RNA from the olfactory epithelium using TRIzol (Invitrogen), selected for capped transcripts using RLM-RACE (GeneRacer; Invitrogen), and reverse-transcribed 5′ ends using degenerate primers against conserved OR transmembrane domains III, V, VI, or VII (Buck and Axel 1991; Malnic et al. 1999). Amplification was initially performed using nested primers against TMIII, and this data set was confirmed and expanded by two further biological isolations and unnested amplification. Primer sequences and the method schematic can be found in Supplemental Methods.
Mice
Experiments were performed on adult (6–8 wk old) C57/Bl6 mice. Each RNA isolation pooled five mice.
Array design and hybridization
The 49 murine odorant receptor clusters and 100-kb flanking sequence were RepeatMasked and tiled at 4-bp resolution on an Affymetrix custom tiling array (GeneChip CustomExpress format 49-7875) (Smit 1996-2007). Non-OR GPCRs (e.g., Vomeronasal Receptors, Opsin) and genes expressed specifically in non-MOE tissue (e.g., protamine) were included as negative controls; many genes expressed in all cells of the olfactory epithelium were included as positive controls (e.g., CNGa2, OMP). Following amplification in the presence of dUTP, 2–7-μg dsDNA samples were fragmented with UDG/APE and labeled with terminal transferase (GeneChip WT Double Stranded DNA Terminal Labeling Kit; Affymetrix); OP cell dsDNA was fragmented with DNase I (NEB) and labeled with terminal transferase (Roche) and biotin-11-ddUTP (Perkin-Elmer).
Computational exon assignment
We processed hybridization signals with both MAT and TAS, yielding similar results (Affymetrix 2005-2007; Johnson et al. 2006). Intervals were generated in IGB (Affymetrix) by thresholding in reference to existing OR gene structures. Galaxy was used to join nearby intervals, and these were assigned computationally to the nearest OR gene and given a strand identity (Zhang and Firestein 2002; Giardine et al. 2005). The most 5′ base pair of the interval on the coding strand was called the transcriptional start site (TSS). TSS and gene name assignments were curated to remove duplicates. Ambiguous assignments were resolved by comparing across hybridizations and 5′-non-coding exons lacking corresponding coding region signal were discarded. While cross-hybridization was of some concern in coding sequences, the high tiling density and masking of ambiguous probes in array design mitigate these concerns. Moreover, while OR coding sequences are conserved, 5′ UTRs are not and thus did not cause cross-hybridization. When possible, the more stringent nested preparation was used for mapping. The less stringent data set, amplified with the same primer used for reverse transcription, was used to identify transcripts that could not be mapped under nested conditions and to confirm the more stringent data. Approximately 90% of these promoter assignments are unambiguous; for the rest, alternative splicing or alternative promoter usage might contribute to the difficulty of assigning the TSS. For the analysis presented here, we chose the most 5′ potential TSSs; more conservative promoter selection did not affect the overall AT-content distribution of OR promoters.
Promoter analysis
We called 1000 bp 5′ of transcriptional start sites “promoters.” These 1085 1000-mer sequences were searched for known transcription factor binding sites using Genomatix RegionMiner and MatInspector; sequence was mined for novel motifs using Weeder, Gibbs Recursive Sampler, MEME, and Genomatix CoreSearch. We obtained similar results in these analyses with and without RepeatMasking.
To examine AT/GC content, we extracted the region from −750 to +250 relative to the TSS for every non-Olfr protein-coding gene in RefGene and for our OR TSSs and calculated GC content for each using the EMBOSS GeeCee tool (Rice et al. 2000; Karolchik et al. 2004; Taylor et al. 2007). Non-coding RNAs were removed because TSSs are often poorly annotated. A small group of RefGene maps in the current annotation is not RACE-based; we consider our findings robust in the face of this noise as distributions were similar using wider and shifted windows. The EMBOSS Freak tool was used to plot GC content as a function of distance from TSS with window 10 bp and step 10 bp (Rice et al. 2000). In Figure 2C, promoters ≤40% GC or ≥65% GC were selected. Forty percent was chosen as the lower cutoff to include 75% of OR genes; 65% was chosen for the upper cutoff so as to include similar numbers of genes (about 2000) in each category. Duplicates were removed from each list, and all annotations for each gene were collected from DAVID, UCSC, MGI, and Weitzmann GeneCards (Kent et al. 2002; Safran et al. 2002; Huang et al. 2009; Bult et al. 2010). Each gene was assigned exclusively to one category among those listed below and in Supplemental Methods; assignment was hierarchical such that proteins fitting more than one category were assigned preferentially. Percentages of the AT- or GC-rich promoters with functions of interest were then plotted. A full description of the categories can be found in Supplemental Methods. To assess statistical significance, we added the AT and GC categories together, counted total occurrences of each function, calculated an expected number in AT and GC based on hypothetical random distribution around the murine AT/GC composition midpoint, and compared these values to the actual findings.
For Figure 2D and Supplemental Figure S2, existing KEGG (Xenobiotic Metabolism by Cytochrome p450) and GO (Transcription Factor Activity, Cell Cycle) categories were used when possible (Ashburner et al. 2000; Kanehisa and Goto 2000). When no existing category captured a function of interest (Chemosensation, Morphogen, Innate Defense and Barriers), a category was constructed by collecting all RefGene names with applicable prefixes, listed in Supplemental Methods (DeFranco et al. 2007). GC-content distribution was plotted as percent of promoters of each functional category in 3% GC bins (26%–28%, 29%–31%, etc.). For Figure 2D, Chemosensory/Defense/Xenobiotic and Cell Cycle/Transcription Factor/Morphogen categories were combined by averaging the three percentages at each GC content to give equal weight to categories with varying numbers of constituent genes. Supplemental Figure S2 shows each functional distribution individually.
The GC-rich promoter group in Figure 3 contains a randomly selected group of 1098 promoters of at least 65% GC. O/E and TBP position plots used IUPAC string searches through Genomatix and Nolf site searches using Genomatix MatBase.
wordCount
The wordCount method of sequence clustering is described in the text. Code was written in Perl and R and is available as Supplemental Material. In general, RepeatMasked sequences were clustered and records were discarded if the RepeatMasked sequence was shorter than 500 bp.
RepeatMasking
Analysis 2A was repeated after RepeatMasking, and the distribution of enriched functional groups shown in Supplemental Figure S2 was examined (Smit et al. 1996). RepeatMasking slightly narrowed the overall %GC distribution of murine promoters but did not affect skewed distribution of functional categories toward one pole or the other. RepeatMasking did not affect TFBS distribution. Promoter clustering and ordering were performed using Hopach (Magalhaes et al. 2007).
OP cell culture
RNA isolation and RLM-RACE were performed as for the tissue samples. Olfactory placode cell culture, data analysis, and extensive qPCR validation of “on” and “off” genes will appear in a forthcoming paper (R Lane, in prep.).
Data access
Hybridization data can be accessed at GEO series accession GSE26373; sample accessions GSM647450, GSM647451, and GSM647452.
Acknowledgments
We are thankful to Nirao Shah, Nadav Ahituv, Hiten Madhani, Keith Yamamoto, and members of our laboratory for critical reading of the manuscript. Mice used for our experiments were treated in accordance with IACUC and Federal guidelines under the protocol AN078602-03A. These experiments were supported by grants from the NIH Director's New Innovator Award Program (DP2OD006667), the NIDCD (R03DC010273), and the Sandler Program in Basic Sciences. E.J.C. and B.M.C. are supported by NSF graduate fellowships (NSF GRFP).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.120162.110.
References
- Affolter M, Slattery M, Mann RS 2008. A lexicon for homeodomain–DNA recognition. Cell 133: 1133–1135 [DOI] [PubMed] [Google Scholar]
- Affymetrix 2005-2007. Tiling array tools. http://www.affymetrix.com/partners_programs/programs/developer/TilingArrayTools/index.affx
- Akan P, Deloukas P 2008. DNA sequence and structural properties as predictors of human and mouse promoters. Gene 410: 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36 AAAI Press, Menlo Park, CA [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL 2004. GenBank: update. Nucleic Acids Res 32: D23–D26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G 2007. The neoselectionist theory of genome evolution. Proc Natl Acad Sci 104: 8385–8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, Vassalli A, Fuss S, Zhang JJ, Weiland B, Pacifico R, Feinstein P, Mombaerts P 2009. Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron 61: 220–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R 1991. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65: 175–187 [DOI] [PubMed] [Google Scholar]
- Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT 2010. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res 38: D586–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 [DOI] [PubMed] [Google Scholar]
- Cesaroni M, Cittaro D, Brozzi A, Pelicci PG, Luzi L 2008. CARPET: a web-based package for the analysis of ChIP-chip and expression tiling data. Bioinformatics 24: 2918–2920 [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chess A 2005. Monoallelic expression of protocadherin genes. Nat Genet 37: 120–121 [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell 78: 823–834 [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM 2005. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123: 477–491 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH 2006. Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800 [DOI] [PubMed] [Google Scholar]
- DeFranco AL, Locksley RM, Robertson M 2007. Immunity: the immune response in infectious and inflammatory disease. New Science Press, UK. [Google Scholar]
- Dekker J 2008. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J Biol Chem 283: 34532–34540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser E, Olender T, Khen M, Yanai I, Ophir R, Lancet D 2006. Widespread ectopic expression of olfactory receptor genes. BMC Genomics 7: 121 doi: 10.1186/1471-2164-7-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss SH, Ray A 2009. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol Cell Neurosci 41: 101–112 [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: A platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W, Roland J, Raulet DH 1995. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature 376: 355–358 [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P 2004. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci 101: 8751–8755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Omura M, Mombaerts P 2007. Differential impact of Lhx2 deficiency on expression of class I and class II odorant receptor genes in mouse. Mol Cell Neurosci 34: 679–688 [DOI] [PubMed] [Google Scholar]
- Hollander GA, Zuklys S, Morel C, Mizoguchi E, Mobisson K, Simpson S, Terhorst C, Wishart C, Golan DE, Bhan AK, et al. 1998. Monoallelic expression of the interleukin-2 locus. Science 279: 2118–2121 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Illing N, Boolay S, Siwoski JS, Casper D, Lucero MT, Roskams AJ 2002. Conditionally immortalized clonal cell lines from the mouse olfactory placode differentiate into olfactory receptor neurons. Mol Cell Neurosci 20: 225–243 [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS 2006. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS 2007. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi G, Puche AC, Mantero S, Barbieri O, Trombino S, Paleari L, Egeo A, Merlo GR 2003. The Dlx5 homeodomain gene is essential for olfactory development and connectivity in the mouse. Mol Cell Neurosci 22: 530–543 [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR 2004. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci 101: 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gorovsky MA 1993. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res 21: 4954–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Ventura M, Cellamare A, Chen L, Cheng Z, Zhu B, Li C, Song J, Eichler EE 2009. Analysis of recent segmental duplications in the bovine genome. BMC Genomics 10: 571 doi: 10.1186/1471-2164-10-571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Hou Y, Zhu B, Cardone MF, Jiang L, Cellamare A, Mitra A, Alexander LJ, Coutinho LL, Dell'Aquila ME, et al. 2010. Analysis of copy number variations among diverse cattle breeds. Genome Res 20: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413 [DOI] [PubMed] [Google Scholar]
- Magalhaes TR, Palmer J, Tomancak P, Pollard KS 2007. Transcriptional control in embryonic Drosophila midline guidance assessed through a whole genome approach. BMC Neurosci 8: 59 doi: 10.1186/1471-2202-8-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour K, Moutoufaris F, Carey C, et al. 2011. An epigenetic signature for monoallelic olfactory receptor expression. Cell 135: 555–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB 1999. Combinatorial receptor codes for odors. Cell 96: 713–723 [DOI] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS 2008. Emx2 stimulates odorant receptor gene expression. Chem Senses 33: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski JS, Galante PA, Malnic B 2006. Identification of potential regulatory motifs in odorant receptor genes by analysis of promoter sequences. Genome Res 16: 1091–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H 2005. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci 25: 3586–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L 2007. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell 131: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas TJ, Cheng Z, Ventura M, Mealey K, Eichler EE, Akey JM 2009. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res 19: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M 2005. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene 346: 13–21 [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M 2006. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet 51: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Hansen L, Abaan HO, Tullius TD, Margulies EH 2009. Local DNA topography correlates with functional noncoding regions of the human genome. Science 324: 389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N, Johnson P, Getman M, Lane RP 2009. Odorant receptor (OR) gene choice is biased and non-clonal in two olfactory placode cell lines, and OR RNA is nuclear prior to differentiation of these lines. J Neurochem 108: 486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G 2004. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res 32: W199–W203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB 1993. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73: 597–609 [DOI] [PubMed] [Google Scholar]
- Ribich S, Tasic B, Maniatis T 2006. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci 103: 19719–19724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gil DJ, Treloar HB, Zhang X, Miller AM, Two A, Iwema C, Firestein SJ, Greer CA 2010. Chromosomal location-dependent nonstochastic onset of odor receptor expression. J Neurosci 30: 10067–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B 2009. The role of DNA shape in protein–DNA recognition. Nature 461: 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS 2010. Origins of specificity in protein–DNA recognition. Annu Rev Biochem 79: 233–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P 2005. The promoter of the mouse odorant receptor gene M71. Mol Cell Neurosci 28: 535–546 [DOI] [PubMed] [Google Scholar]
- Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, Ben-Dor U, Esterman N, Rosen N, Peter I, et al. 2002. GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics 18: 1542–1543 [DOI] [PubMed] [Google Scholar]
- Saito T, Lo L, Anderson DJ, Mikoshiba K 1996. Identification of novel paired homeodomain protein related to C. elegans unc-4 as a potential downstream target of MASH1. Dev Biol 180: 143–155 [DOI] [PubMed] [Google Scholar]
- Sammeta N, Yu TT, Bose SC, McClintock TS 2007. Mouse olfactory sensory neurons express 10,000 genes. J Comp Neurol 502: 1138–1156 [DOI] [PubMed] [Google Scholar]
- Sampsell BM, Held WA 1985. Variation in the major urinary protein multigene family in wild-derived mice. Genetics 109: 549–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F, Grosschedl R 2006. Blurrind cis and trans in gene regulation. Cell 126: 248–250 [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H 2003. Negative feedback regulation ensures the one receptor–one olfactory neuron rule in mouse. Science 302: 2088–2094 [DOI] [PubMed] [Google Scholar]
- Shykind BM 2005. Regulation of odorant receptors: one allele at a time. Hum Mol Genet 14: R33–R39 [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G 2004. Gene switching and the stability of odorant receptor gene choice. Cell 117: 801–815 [DOI] [PubMed] [Google Scholar]
- Singh N, Ebrahimi FA, Gimelbrant AA, Ensminger AW, Tackett MR, Qi P, Gribnau J, Chess A 2003. Coordination of the random asynchronous replication of autosomal loci. Nat Genet 33: 339–341 [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P 1996-2007. RepeatMasker Open-3.0. http://www.repeatmasker.org
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci 99: 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh J, Luz H, Bornfleth H, Lampel S, Cremer C, Lichter P 2000. Spatial distribution of GC- and AT-rich DNA sequences within human chromosome territories. Exp Cell Res 255: 229–237 [DOI] [PubMed] [Google Scholar]
- Taylor J, Schenck I, Blankenberg D, Nekrutenko A 2007. Using Galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics 19: 10.5.1–10.5.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W, Rouchka EC, Lawrence CE 2003. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res 31: 3580–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Feinstein P, Mombaerts P 2011. Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Mol Cell Neurosci 46: 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R 1993. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74: 309–318 [DOI] [PubMed] [Google Scholar]
- Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, Khen M, Stütz AM, Schlattl A, Lancet D, Korbel JO 2010. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol 6: e1000988 doi: 10.1371/journal.pcbi.1000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, et al. 2007. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet 80: 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Shykind BM, Lane RP, Tonnes-Priddy L, Ross JA, Walker M, Williams EM, Trask BJ 2003. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol 4: R71 doi: 10.1186/gb-2003-4-11-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Endicott RM, Parghi SS, Walker M, Kidd JM, Trask BJ 2008. Extensive copy-number variation of the human olfactory receptor gene family. Am J Hum Genet 83: 228–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S 2002. The olfactory receptor gene superfamily of the mouse. Nat Neurosci 5: 124–133 [DOI] [PubMed] [Google Scholar]