Abstract
Epigenetic regulation of gene enhancer elements is important for establishing and maintaining the identity of cells. Gene enhancer elements are thought to exist in either active or poised states distinguishable by chromatin features, but a complete understanding of the regulation of enhancers is lacking. Here, by using mouse embryonic stem cells and their differentiated derivatives, as well as terminally differentiated cells, we report the coexistence of multiple, defined classes of enhancers that serve distinct cellular functions. Specifically, we found that active enhancers can be subclassified based on varying levels of H3K4me1, H3K27ac, and H3K36me3 and the pSer2/5 forms of RNA polymerase II. The abundance of these histone modifications positively correlates with the expression of associated genes and cellular functions consistent with the identity of the cell type. Poised enhancers can also be subclassified based on presence or absence of H3K27me3 and H3K9me3, conservation, genomic location, expression levels of associated genes, and predicted function of associated genes. These findings not only refine the repertoire of histone modifications at both active and poised gene enhancer elements but also raise the possibility that enhancers associated with distinct cellular functions are partitioned based on specific combinations of histone modifications.
Enhancers are distal-acting elements that orchestrate the regulation of genes critical for cell lineage specification (Bulger and Groudine 2010). Genomic studies have identified several key features of gene enhancer elements. Specifically, enhancer elements correlate with the location of mono-methylated lysine 4 of histone H3 (H3K4me1), are located in open chromatin that is hypersensitive to DNase digestion, and often exhibit cell type–specific localization patterns (Barski et al. 2007; Heintzman et al. 2007, 2009; Koch et al. 2007; Xi et al. 2007; Lupien et al. 2008; Schnetz et al. 2010). Although a large number of regions in the genome display these characteristics, only a fraction of the H3K4me1-marked elements are actively engaged in modulating transcription in a given cell type (Heintzman et al. 2009; Creyghton et al. 2010; Rada-Iglesias et al. 2011). These elements are referred to as active enhancers. Other H3K4me1-marked enhancers modulate transcription in response to differentiation cues or other cellular stimuli and are thus considered poised (Creyghton et al. 2010; Rada-Iglesias et al. 2011). Consistent with this model, active enhancers associated with pluripotency factor genes (i.e., POU5F1 and NANOG), are inactivated upon human embryonic stem cell (hESC) differentiation, while poised enhancers associated with genes involved in early developmental processes are activated (Rada-Iglesias et al. 2011).
Although recent studies support the existence of both active and poised classes of enhancer elements in hESCs and mESCs, the histone marks associated with poised enhancers were recently reported to differ between hESCs and mouse embryonic stem cells (mESCs). Specifically, in hESCs, poised enhancers contain H3K4me1 but are also trimethylated at K27 of histone H3 (H3K4me1+, H3K27me3+) (Rada-Iglesias et al. 2011). In mESCs, poised enhancers contain H3K4me1 but are not reported to show covalent modification of H3K27 (H3K4me1+, H3K27−) (Creyghton et al. 2010). In this study, we integrated ChIP-seq data sets for enhancer binding proteins and histone modifications to better understand the chromatin-level regulation of enhancer elements. The results support the co-existence of multiple epigenetically and functionally distinct active and poised enhancer classes in mESCs. These findings refine and expand the chromatin signatures associated with enhancer elements and suggest that combinations of histone modifications at enhancers can be used to compartmentalize genes based on their expression levels and cellular functions.
Results
Identification of three major enhancer classes in mESCs
We previously mapped the distribution of two enhancer binding proteins, CHD7 and P300 (also known as EP300), on mESC chromatin by combining chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) (Schnetz et al. 2010). Here, we integrated these data sets with DNase-seq and publically available ChIP-seq data for H3K4me1, H3K27ac, and H3K27me3. Consistent with previous studies, we identified both active (H3K4me1+, H3K27ac+) and poised (H3K4me1+, H3K27−) classes of enhancers in mESCs (Creyghton et al. 2010). In addition, we identified a third class of enhancers that, similar to those reported in hESCs (Rada-Iglesias et al. 2011), contained significant levels of H3K27me3 (Fig. 1A,B; Supplemental Fig. 1A,B). These three classes were also observed when enhancers were defined solely on the basis of H3K4me1 and/or H3K27ac enrichment (Supplemental Fig. 2). All three classes were depleted of H3K4me3 (Supplemental Fig. 3). We note that both the H3K4me1+, H3K27− and H3K4me1+, H3K27me3+ enhancer classes contain lower levels of H3K4me1 than does the H3K4me1+, H3K27ac+ class. This finding is consistent with previous reports showing decreased H3K4me1 at previously described poised mESC and hESC enhancers (Creyghton et al. 2010; Rada-Iglesias et al. 2011).
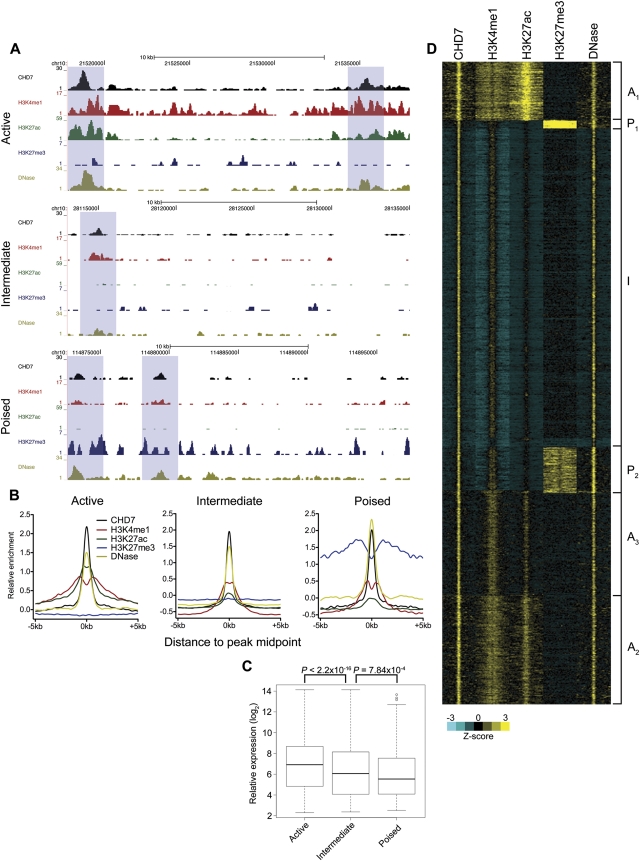
Figure 1.
Identification of multiple enhancer classes. (A) Example ChIP-seq profiles of each enhancer class in mESCs. Data were visualized using the UCSC Genome Browser. Putative active, intermediate, and poised enhancers are highlighted in blue boxes. (B) Aggregate plots of CHD7, H3K4me1, H3K27ac, H3K27me3, and DNase hypersensitivity signal centered on the CHD7 peak midpoint. (C) Boxplot of expression levels of genes associated with each enhancer class. P-values were calculated by the Wilcoxon rank-sum test. (D) Heatmap of CHD7-bound enhancers generated by k-means cluster analysis. Each window represents signal ±5 kb of the CHD7 peak midpoint. Active clusters are designated A1-3, the intermediate cluster is designated I, and the poised clusters are designated P1-2.
Expression analyses indicated that H3K4me1+, H3K27ac+ associated genes were expressed at significantly higher levels than H3K4me1+, H3K27ac− associated genes (CHD7 and P300 P < 2.2 × 10−16), which in turn were expressed at higher levels than H3K4me1+, H3K27me3+ associated genes (CHD7, P = 7.84 × 10−4; P300, P = 0.085) (Fig. 1C; Supplemental Fig. 1C). In keeping with the published designations, we classified H3K4me1+, H3K27ac+ enhancers as active and H3K4me1+, H3K27me3+ enhancers as poised. However, because genes associated with the H3K4me1+, H3K27ac− class of enhancers were expressed at an overall level ranking between the active and poised classes, we hypothesized that this class represents a unique, “intermediate” class of enhancers, and designated it as such. Of 12,332 CHD7-bound enhancers, 5181 (42%) were classified as active, 6115 (50%) were intermediate, and 986 (8%) were poised. Of 4239 P300-bound enhancers, 1929 (45.5%) were active, 1892 (44.6%) were intermediate, and 417 (9.8%) were poised.
We previously assayed a large set of putative enhancer sites in mESCs using luciferase reporter assays (Schnetz et al. 2010). Reanalysis of this data set revealed that, of 30 enhancers that we classified here as active based on chromatin signatures (H3K4me1+, H3K27ac+), 20 were positive in the luciferase reporter assay (67%) (Supplemental Fig. 4). Of 15 classified here as either intermediate or poised, only three were positive (20%), which is a significantly smaller proportion than that observed for active enhancers (P = 0.0084 by Z-test). These results verify that the chromatin signatures we associate with each enhancer class are predictive of their activity in vivo. We also verified that enhancers that are inactive in mESCs become active later in development using available data from the VISTA enhancer browser (Visel et al. 2006). Specifically, we found that 75 of 95 intermediate and 22 of 39 poised mESC enhancers were active at mid-gestation in the developing mouse embryo.
DNase-seq (Crawford et al. 2006), used to measure the accessibility of chromatin, revealed that all three classes of enhancers were located in open regions of chromatin, consistent with previous reports of nucleosome depletion at enhancers (Giresi et al. 2007; Mito et al. 2007; He et al. 2010). Poised enhancers had a slightly more open chromatin conformation than either active or intermediate enhancers (Fig. 1B; Supplemental Fig. 1B), potentially indicating the presence of the Polycomb proteins necessary to maintain the H3K27me3 mark. Consistent with this hypothesis, substantial binding of the Polycomb group proteins EZH2, SUZ12, and RNF2 (also known as RING1B) was detected at poised, but not active or intermediate, enhancers (Supplemental Fig. 5).
Subclassification of enhancer classes
Through cluster analysis, we found that both active and poised enhancers segregated into multiple subclasses distinguishable by the levels of H3K4me1, H3K27ac, and/or H3K27me3 (Fig. 1D; Supplemental Fig. 1D). Most notably, the active class of enhancers could be divided into three subclasses, which we designated as A1, A2, and A3. As previous reports have demonstrated transcription of ncRNA from enhancers (Guttman et al. 2009; De Santa et al. 2010; Kim et al. 2010; Ørom et al. 2010), we investigated potential transcriptional chromatin signatures at active enhancers. Specifically, we examined the levels of H3K36me3, associated with transcriptional elongation, and the levels of serine 2 and 5-phosphorylated RNA polymerase II (pSer2 and pSer5 pol II) within each active enhancer subclass. We observed marked enrichment of H3K36me3 and both phosphorylated forms of pol II, especially pSer5, within clusters A1 and A2 and to a lesser extent within cluster A3 (Fig. 2A). Little or no enrichment of these factors was seen at intermediate or poised enhancers (Fig. 2B). The abundance of H3K36me3 and pSer2/5 RNA pol II positively correlated with expression of associated genes (see Fig. 8, rightmost column). We next analyzed RNA-seq data from mESCs and detected RNA transcripts at the active A1 and A2 enhancer clusters (Fig. 2B). Similar results were obtained for P300-defined enhancers (data not shown). Our results support previous studies demonstrating that some enhancers are transcriptionally active (Guttman et al. 2009; De Santa et al. 2010; Kim et al. 2010; Ørom et al. 2010), although we currently do not know whether the transcripts we observe are eRNA or another class of enhancer-associated RNA. Alternatively, because these RNAs do not appear to correspond to any known class of noncoding RNA, the signal we detect may simply reflect genomic background noise. Regardless, these findings suggest that a subset of active enhancers acquire a chromatin signature resembling that of transcriptionally active genes. The results also suggest that H3K36me3 and pSer2/5 pol II, in addition to H3K27ac, can distinguish active from intermediate and poised enhancers.
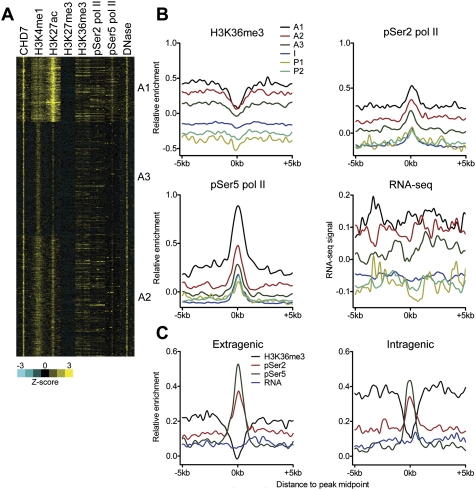
Figure 2.
Sub-classification of active enhancers. (A) Heatmap showing levels of CHD7, histone modifications, pSer2/5 pol II, and DNase-seq signals for each active enhancer cluster. Each window represents signal ±5 kb of the CHD7 peak midpoint. (B) Aggregate plots of H3K36me3, pSer2 pol II, pSer5 pol II, and RNA-seq signal at each enhancer subclass centered on the CHD7 peak midpoint. (C) Aggregate plots showing H3K36me3, pSer2 pol II, pSer5 pol II, and RNA-seq signal at CHD7-centered active enhancers located in extragenic and intragenic regions.
Figure 8.
Heatmap summarizing histone modifications, DNase hypersensitivity, pSer2/5 RNA pol II, RNA expression, and expression of associated genes at each enhancer subclass in mESCs.
The substantial levels of H3K36me3, pSer2/5 pol II, and RNA-seq signal observed at active enhancers led us to test whether enhancers located within actively transcribed genes bias the above findings. We therefore divided our active enhancers into intragenic and extragenic classes and determined the H3K36me3, pSer2/5 pol II, and RNA-seq signals for each class. Both extra- and intragenic active enhancers contain substantial levels of H3K36me3, phosphorylated pol II, and RNA-seq signal (Fig. 2C), suggesting that our results are not biased by ongoing transcription through intragenic enhancers. We also examined H3K4me1, H3K27ac, and H3K27me3 at enhancers in the active, intermediate, and poised classes and found no substantial differences in these modifications between extra- and intragenic enhancers (Supplemental Fig. 6).
H3K9me3 distinguishes poised enhancers from active and intermediate enhancers
We next tested whether repressive modifications other than H3K27me3 could distinguish poised from other enhancer classes. We therefore reclustered the CHD7-bound enhancers in mESCs using H3K9me3, instead of H3K27me3. We observed clusters with features similar to those observed when the data were clustered with H3K27me3 (Fig. 3A,B). We then determined the overlap between each class of H3K9me3-clustered enhancers and H3K27me3-clustered enhancers. Strikingly, the enhancers we designated as poised based on H3K9me3 levels were identical to those defined as poised based on H3K27me3 levels. Other enhancer classes also displayed a high degree of overlap between the two analyses (Fig. 3C). These findings indicate that H3K9me3, independent of H3K27me3, can be used to identify poised enhancers. Moreover, these findings suggest that Polycomb silencing is not necessarily the sole mechanism involved in enhancer poising.
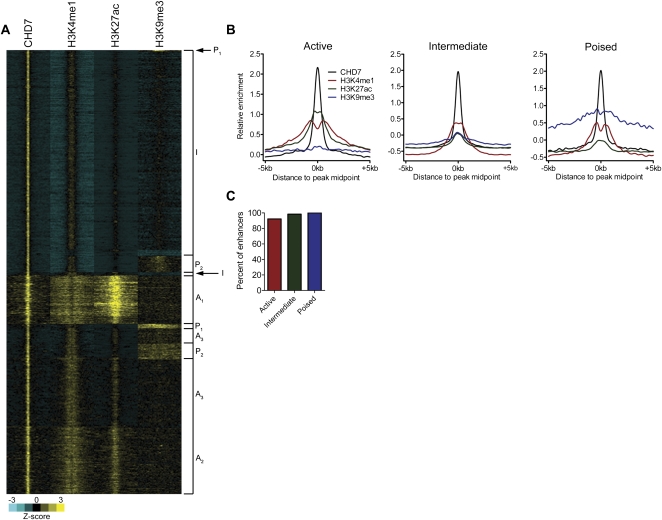
Figure 3.
H3K9me3 can distinguish poised from active and intermediate enhancers. (A) Heatmap of CHD7-bound enhancers showing cluster analysis with H3K9me3. Subclass designations from Figure 1D were used to classify each H3K9me3-clustered enhancer class. (B) Aggregate plots of CHD7, H3K4me1, H3K27ac, and H3K9me3 at each enhancer class following H3K9me3 clustering. (C) Percentage of enhancers in each class after H3K9me3 clustering that overlapped with enhancers in the corresponding class after H3K27me3 clustering.
Enhancers classes in mESCs are distinguishable by conservation, genomic location, and predicted function of associated genes
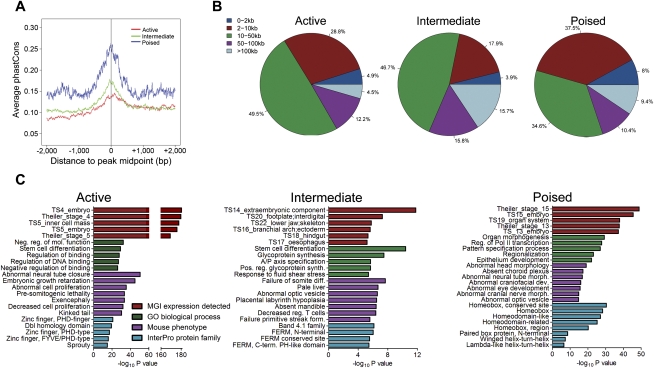
We next investigated whether the active, intermediate, and poised enhancer classes are distinguishable by features other than chromatin signatures. Indeed, poised enhancers were more conserved among vertebrates than were either active or intermediate enhancers (Fig. 4A; Supplemental Fig. 1E). Interestingly, the conservation of active and intermediate enhancers appears to be distributed asymmetrically about the center of the CHD7 peak. This unexpected observation may suggest that these enhancers are not direction independent, or indicate a flaw in the methods used for alignment or conservation analysis. Additionally, intermediate enhancers were located farther (>50 kb) from transcription start sites than active or poised enhancers, while poised enhancers were located closer to TSSs (<10 kb) than active or intermediate enhancers (Fig. 4B; Supplemental Fig. 1F). We then computationally predicted the functions of genes regulated by the enhancers in each class using GREAT (Genomic Regions Enrichment of Annotations Tool) (McLean et al. 2010). GREAT associates cis-regulatory elements (in this case enhancers) with a putative target gene(s), and then uses annotations from numerous ontologies to associate the target gene(s) with function. GREAT then calculates the statistical enrichments for the associations between the enhancers and the annotations, so that statistically significant associations can be easily identified and further investigated. We first utilized GREAT to associate genes in each class with mouse genome informatics (MGI) expression data, which provides information on spatiotemporal gene expression during mouse development. Genes associated with active enhancers were among those expressed at Theiler Stage (TS) 4–5, the time at which the ES cells were derived from the ICM blastocyst. In comparison, genes associated with intermediate and poised enhancers were expressed later in embryonic development (TS14–22). The results of the GREAT analysis also revealed differences in mouse phenotypes associated with genes in each enhancer class. Specifically, defects in cell proliferation were related to genes associated with active enhancers, while a broad array of embryonic phenotypes were linked to genes associated with the intermediate enhancer class. Poised enhancer-associated genes were primarily associated with defects in neural development (Fig. 4C; Supplemental Fig. 1G). Notably, poised enhancers were also significantly associated with genes encoding homeobox transcription factors, including members of the developmentally important SOX, FOX, and TBX families. In addition, poised enhancers were associated with GO biological functions such as “transcription factor activity” and “sequence-specific DNA binding” as well as the “transcription factor complex” term of the GO cellular component ontology (Supplementary Data 3). In contrast, intermediate enhancers displayed no significant associations with GO biological function terms but were associated with a broad range of Pathway Commons terms such as cell junction organization, biological oxidation, and insulin receptor signaling (Supplementary Data 3). These observations suggest that a major function of poised enhancers is to regulate the expression of developmentally important transcription factors, while the intermediate enhancers regulate a broad range of cellular processes.
Figure 4.
Distinguishing features of enhancer subclasses. (A) Average phastCons plot for each enhancer class in a 4-kb window centered on the CHD7 peak midpoint. (B) Distribution of enhancers in each class relative to known transcription start sites. (C) Results of functional annotation of each enhancer class using GREAT. The −log10 of the binomial test P-value is reported.
Fate of mESC enhancer classes upon differentiation into neural precursor cells
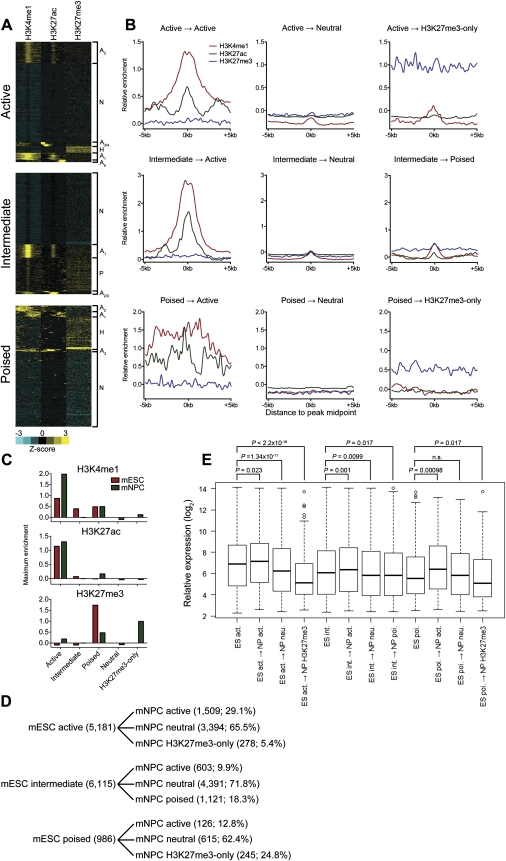
We used available H3K4me1, H3K27ac, H3K27me3, and H3K9me3 ChIP-seq data from neural precursor cells (mNPCs) derived from cultured mESCs to investigate the status of ES cell enhancers upon neural differentiation. Upon neural differentiation, enhancers that were designated as active in the mESCs either retained the active chromatin signature (H3K4me1+, H3K27ac+), lost both H3K4me1 and H3K27ac and became “neutral” (H3K4me1−, H3K27−), or lost H3K4me1 and acquired H3K27me3 (H3K4me1−, H3K27me3+, or “H3K27me3-only”). The H3K27me3-only class of enhancers did not contain H3K9me3. Poised mESC enhancers showed a similar fate; i.e., they either lost both H3K27me3 and H3K9me3 and acquired H3K27ac to become active (H3K4me1+, H3K27ac+), lost all marks to become neutral, or lost H3K4me1 and H3K9me3 and retained H3K27me3 (H3K4me1−, H3K27me3+). Intermediate mESC enhancers were also found to transition to active and neutral states but, in contrast to active and poised enhancers, failed to transition to the H3K4me1−, H3K27me3+ state. We also found that, of enhancers in all three classes, only those designated as intermediate in the mESCs assumed the poised (H3K4me1+, H3K27me3+, H3K9me3+) state in the mNPCs (Fig. 5A–C; Supplemental Fig. 7). The percentage of each mESC enhancer class that transitioned to each state in mNPCs is indicated in Figure 5D. These observations suggest that the intermediate class of enhancers is uniquely versatile, able to assume the active and poised states upon differentiation. Moreover, the findings indicate that removal of both H3K27me3 and H3K9me3 repressive marks occurs upon enhancer activation, which in turn suggests that enhancer activation could involve mechanisms besides those associated with reversal of Polycomb silencing.
Figure 5.
Fate of mESC enhancer classes upon differentiation into neural precursor cells. (A) Heatmaps of enhancer-associated histone modifications in mNPCs defined by active (top), intermediate (middle), and poised (bottom) classes in mESCs. Each window represents ±5 kb of the CHD7 peak midpoint in mESCs. H3K9me3 was also present at poised enhancers derived from the intermediate class (data not shown). (B) Aggregate plots of enhancer-associated histone modifications for each mNPC enhancer class derived from each mESC class. (C) Bar plot of the average maximum signal for each histone modification in each mESC and mNPC enhancer class. The plot indicates that poised enhancers derived from intermediate-class enhancers contain significant levels of both H3K4me1 and H3K27me3, which is less apparent in the aggregate plot in B. (D) Summary of chromatin states achieved upon neural differentiation. The number and percentage of each enhancer state achieved are indicated. (E) Boxplot of expression levels of genes associated with each mNPC enhancer class compared to the average expression of genes in the mESC class from which they were derived. P-values were calculated by the Wilcoxon rank-sum test.
We next defined enhancer clusters in mNPCs based on the levels of H3K4me1, H3K27ac, and H3K27me3, i.e., not considering CHD7 or P300. As expected, all three enhancer classes were detected (Supplemental Fig. 8). However, only 19.7% of all enhancers in mNPCs was derived from any of the three mESC enhancer classes, consistent with previous studies demonstrating a high degree of cell type specificity in the distribution of H3K4me1 (Heintzman et al. 2007, 2009; Koch et al. 2007; Lupien et al. 2008).
We compared the expression of genes associated with each enhancer class in mNPCs to the expression of genes in the mESC class from which they were derived. Genes associated with enhancers that transitioned from poised to active showed a significant increase in expression in mNPCs versus mESCs. Genes associated with active enhancers that transitioned to the neutral or H3K27me3-only state showed significantly lower expression than did the genes in the originating class (Fig. 5E, cf. box 1 [leftmost] to boxes 3,4). Likewise the expression of intermediate-associated genes was reduced upon transition to the neutral or poised states (Fig. 5E, cf. box 5 to boxes 7,8). Lastly, poised-associated genes showed a significant decrease in expression upon transition to the H3K27me3-only state (Fig. 5E, cf. box 9 to box 11). These results verify that the enhancer states assumed upon neural differentiation affect gene expression in a manner consistent with the chromatin signature of each enhancer class. Lastly, consistent with the ectodermal identity of mNPCs, mNPC active enhancers derived from mESC intermediate enhancers were primarily associated with ectodermal expression terms.
Expression and phenotypic analysis of enhancer-associated genes correlated with germ layer
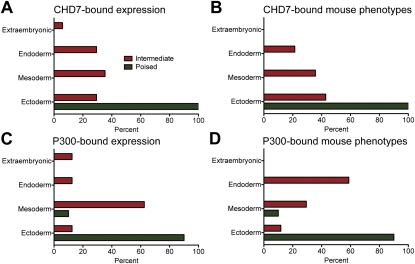
Using GREAT, we examined the spatiotemporal expression patterns of genes associated with each class and then determined whether the anatomical structure associated with each annotation was derived from extra-embryonic tissue, ectoderm, mesoderm, or endoderm. For example, if we detected a significant association between enhancers and genes expressed in an annotation such as TS17_brain, we designated the expression term as “ectodermal.” By using this approach, we found that genes associated with intermediate enhancers were expressed in cell lineages derived from extra-embryonic tissue, endoderm, ectoderm, and mesoderm (Fig. 6A,C). In contrast, genes associated with poised enhancers were expressed in lineages predominantly derived from ectoderm. Consistent with these findings, intermediate enhancer-associated genes were linked to mouse phenotypes involving malformations of organs derived from all three germ layers, while poised enhancer-associated genes were primarily linked to ectoderm-related phenotypes (Fig. 6B,D). These results are not biased for enhancers bound by CHD7 or P300, as similar results were observed when enhancer classes were defined purely by their epigenetic signatures and reannotated with GREAT (Supplemental Fig. 9). These results suggest that poised enhancers preferentially regulate genes associated with ectodermal development, although functional studies are clearly necessary to test the biological relevance of these findings.
Figure 6.
Expression and phenotypic analysis of enhancer-associated genes, correlated with germ layer. The spatiotemporal expression patterns of genes associated with each enhancer class in the developing mouse embryo were determined using GREAT. The germ layer origin of the tissue linked to each annotation was then determined (see Methods). Shown is the percentage of expression annotation terms corresponding to each germ layer, as well as extra-embryonic tissue, for CHD7-bound (A) and P300-bound (C) enhancers. Mouse phenotypes resulting from the mutation of genes associated with each enhancer class were also determined using GREAT. Shown is the percentage of mouse phenotypes, classified by the germ layer of origin of the affected tissue, for CHD7-bound (B) and P300-bound (D) enhancers.
Intermediate and poised enhancer classes are not exclusive to embryonic cell types
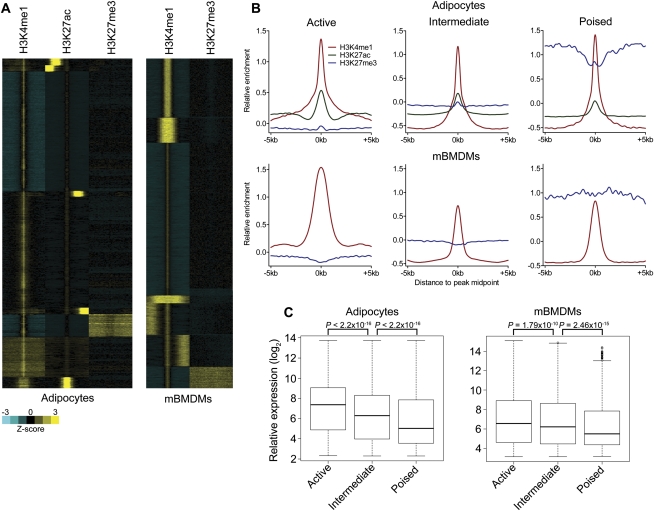
We next determined if multiple enhancer classes existed in terminally differentiated cell types, using available ChIP-seq data from 3T3L1 fibroblast-derived adipocytes and bone marrow–derived macrophages (mBMDMs). The results indicate the presence of active, intermediate, and poised enhancers in both cell types (Fig. 7A,B). Similar to the results in mESCs, genes associated with active enhancers in both cell types were expressed significantly higher than were genes associated with intermediate enhancers. Genes associated with poised enhancers were expressed at a significantly lower overall level than were intermediate enhancer-associated genes (Fig. 7C).
Figure 7.
Identification of multiple enhancer classes in terminally differentiated cells. (A) Heatmaps demonstrating the presence of active, intermediate, and poised enhancer classes in adipocytes and mBMDMs. Each window represents ±5 kb of the H3K4me1/H3K27ac peak midpoint in adipocytes or the H3K4me1 peak midpoint in mBMDMs. (B) Aggregate plots of enhancer-associated histone modifications for each adipocyte and mBMDM enhancer class. (C) Average expression of genes associated with each enhancer class in adipocytes and mBMDMs. P-values were calculated by the Wilcoxon rank-sum test.
We next functionally annotated each enhancer class using GREAT. Active enhancer-associated genes in both cell types were associated with GO biological processes that were consistent with the known functions of each cell type (i.e., lipid biosynthetic process and regulation of lipid storage in adipocytes; regulation of cytokine production and regulation of immune response in mBMDMs). Mouse phenotypes resulting from mutation of active-associated genes were also consistent with the known functions of each cell type (i.e., abnormal triglyceride level and abnormal adipose tissue physiology in adipocytes; abnormal immune cell physiology and abnormal bone marrow morphology/development in mBMDMs) (Supplemental Table 4). Genes linked to intermediate enhancers, as in mESCs, were associated with a diverse array of biological processes and mouse phenotypes (Supplemental Table 4). Poised-associated genes were associated with metabolic processes related to lipid storage and synthesis in adipocytes. Poised enhancers were also significantly associated with genes encoding lipases, the dysregulation of which leads to abnormal triglyceride levels, increased circulating cholesterol, and increased circulating lipid levels in mice (Supplemental Table 4). In mBMDMs, poised-associated genes were associated with transcriptional functions as well as some immunological phenotypes (Supplemental Table 4). Additionally, and similar to the poised class in mESCs, the poised class in mBMDMs was significantly associated with homeobox transcription factors.
Discussion
In this report, we integrated multiple ChIP-seq data sets to identify epigenetic characteristics of enhancer elements in mESCs. We report several novel findings. First, it is clear that multiple subclasses of active enhancers can be distinguished in undifferentiated and terminally differentiated cell types. In mESCs, active enhancer subclasses show varying levels of H3K4me1 and H3K27ac and may also contain H3K36me3 and the pSer2/5 forms of RNA pol II, which are generally associated with transcriptionally active chromatin. The presence of H3K36me3 and phosphorylated RNA pol II as well as RNA transcripts detected by RNA-seq at these enhancers suggests that these marks can distinguish highly active enhancers from less active enhancers. Correlations with gene expression support this notion. Second, it is clear that at least two classes of poised enhancers co-exist in multiple cell types: one class marked with H3K4me1 and not modified at H3K27, and the second marked with both H3K4me1 and H3K27me3. These two poised enhancer classes are distinguishable by not only H3K27me3 but also H3K9me3. The chromatin signatures of each enhancer class are summarized in Figure 8. Upon neural differentiation, both types of poised enhancers can transition to active states defined by H3K4me1 and H3K27ac, and both types of enhancers can lose marks and enter a chromatin “neutral” state. However, in contrast to poised enhancers containing H3K27me3, poised enhancers devoid of H3K27me3 can acquire H3K27me3 upon differentiation. In addition, genes associated with the H3K4me1+, H3K27− class are expressed at higher levels than are H3K4me1+, H3K27me3+ associated genes. Based on the chromatin features and the expression level of associated genes, we propose that the H3K4me1+, H3K27− elements represent an “intermediate” class of enhancers. The intermediate and poised classes are further distinguishable by conservation, genomic location, and predicted function.
Why do mouse cells contain three classes of enhancers? With respect to active enhancers, we propose a hypothetical model wherein the variable levels of the active histone modifications H3K4me1, H3K27ac, and H3K36me3 function to dictate the expression level of the associated genes. We speculate that the presence of these marks in high abundance is likely to provide a more permissive chromatin environment for recruitment of regulatory factors than the presence of these marks in lower abundance, which in turn would allow for fine-tuning of gene expression. With respect to the intermediate and poised enhancers, the intermediate class is associated with genes implicated in a diverse array of biological processes that is not necessarily specific to any particular cell type. In contrast, the poised class is associated with genes with generally more specialized functions (i.e., lipases in adipocytes, homeobox transcription factors in mESCs and mBMDMs). In addition, misexpression of many poised associated genes would be particularly deleterious. For example, poised enhancers are often associated with homeobox transcription factors in mESC, the misexpression of which can have severe developmental consequences (Schulte et al. 1999; Mathers and Jamrich 2000; Wu et al. 2003; Sunmonu et al. 2009). This notion is supported by mouse phenotypes associated with mutation of genes linked to each class. Mutation of mESC poised-associated genes causes abnormal neural tube development, abnormal cranial nerve morphology, and abnormal forebrain, midbrain, and hindbrain development (i.e., Gbx2, Hes1, Otx2, Pax2), which are generally considered very severe. In contrast, the majority of phenotypes linked to intermediate-associated genes are less severe, including “pale liver,” polycystic kidney, and uremia. Thus, it stands to reason that transcription factors that are particularly dosage sensitive have evolved to incorporate an additional layer of regulation, presumably mediated by H3K27 and H3K9 methyltransferases, at highly conserved enhancer elements.
We propose a hypothetical model wherein the combination of histone modifications at gene enhancer elements provides a mechanism for the cell to partition genes according to their level of expression and function in a given cell type. We hypothesize that our findings, in which only a fraction of all possible histone modifications were investigated, represent the “tip of the iceberg” with respect to functional refinement of gene enhancer elements. We speculate that additional modifications will serve to further refine enhancer classes as they are analyzed and as the functions of protein-coding genes are further delineated.
Fundamental questions regarding the epigenetic state of enhancers remain. First, it is not known if the histone modifications associated with enhancers are a cause or consequence of an enhancer adopting a particular state. For example, it is not clear whether the presence of H3K27ac on an H3K4me1-marked enhancer is responsible for the establishment of the active state, or is a result of enhancer activation. Second, given that enhancers are proposed to interact with their target promoters via chromatin looping (Bulger and Groudine 2010), the chromatin state of a given enhancer could be influenced by association with its target promoters, or vice versa. Studies integrating ChIP-seq and genome-wide maps of chromatin interactions generated using the Hi-C (Lieberman-Aiden et al. 2009) or ChIA-PET (Li et al. 2010) methods could help shed light on this matter.
Methods
Sequencing data alignment and analysis
The following publically available ChIP-seq data sets were obtained as SRA-lite files from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/): mESC CHD7 (SRX022492) (Schnetz et al. 2010), mESC P300 (SRX022493) (Schnetz et al. 2010), mESC H3K4me1 (SRX027330) (Creyghton et al. 2010), mNPC H3K4me1 (SRX000581) (Meissner et al. 2008), 3T3L1-derived adipocyte H3K4me1 (SRX019386) (Mikkelsen et al. 2010), mBMDM H3K4me1 (SRX019782) (Heinz et al. 2010), mESC H3K4me3 (SRX023508), mESC H3K9me3 (SRX014428) (Bilodeau et al. 2009), mNPC H3K9me3 (SRX001939) (Mikkelsen et al. 2007), mESC H3K27ac (SRX027331) (Creyghton et al. 2010), mNPC H3K27ac (SRX027338) (Creyghton et al. 2010), 3T3L1-derived adipocyte H3K27ac (SRX019387) (Mikkelsen et al. 2010), mESC H3K27me3 (SRX001921) (Mikkelsen et al. 2007), mNPC H3K27me3 (SRX001936) (Mikkelsen et al. 2007), 3T3L1-derived adipocyte H3K27me3 (SRX019388) (Mikkelsen et al. 2010), mBMDM H3K27me3 (SRX025081) (Heinz et al. 2010), mESC H3K36me3 (SRX001922) (Mikkelsen et al. 2007), mESC RNA pol II pSer2 (SRX017057) (Rahl et al. 2010), mESC RNA pol II pSer5 (SRX017056) (Rahl et al. 2010), mESC Ezh2 (SRX003847) (Ku et al. 2008), mESC Suz12 (SRX003849) (Ku et al. 2008), mESC Ring1b (SRX003848) (Ku et al. 2008), mESC input (SRX027352) (Creyghton et al. 2010), mNPC input (SRX001940) (Mikkelsen et al. 2007), 3T3L1-derived adipocyte input (SRX019362) (Mikkelsen et al. 2010), and mBMDM input (SRX016346) (Heinz et al. 2010). mESC RNA-seq data were obtained from the SRA (SRX019275) (Guttman et al. 2010). mESC DNase-seq data were previously described (Schnetz et al. 2010). SRA-lite files were converted to FASTQ using the fastq-dump utility of the SRA toolkit, and FASTQ files were aligned to the mm8 genome assembly with Bowtie (Langmead et al. 2009), allowing two mismatches per read and discarding reads with more than one reportable alignment. Peaks were detected with MACS (Zhang et al. 2008) using an aligned input DNA sample as control. Wiggle tracks stepped at 100-bp intervals were generated and visualized on the UCSC Genome Browser. We then determined the median signal in fifty 200-bp windows ±5 kb of each peak midpoint, Z-score transformed the data to standardize samples with different normal distributions, clustered the data with Gene Cluster 3.0 (de Hoon et al. 2004), and visualized clustered data with Java TreeView (Saldanha 2004).
To generate lists of putative enhancers in mESCs, mNPCs, 3T3L1-derived adipocytes, and mBMDM, we combined the lists of H3K4me1 and H3K27ac MACS peaks into a single file (H3K4me1 peaks alone were used for mBMDM, as H3K27ac data were not available), merging peaks <10 bp apart into a single peak. Transcription start sites were removed by intersecting this file with a list of coordinates representing ±1 kb upstream of and downstream from all TSSs in the mm8 genome assembly using the UCSC Tables Browser (http://genome.ucsc.edu/cgi-bin/hgTables?command=start). This yielded lists of 76,001 (mESC), 51,329 (mNPC), 48,841 (3T3L1-derived adipocytes), and 49,475 (mBMDM) putative distal enhancers. To generate lists of CHD7- and P300-bound mESC enhancers, the lists of CHD7 and P300 MACS peaks, with TSSs removed as above, were intersected with the list of H3K4me1/H3K27ac-defined mESC enhancers, yielding 12,332 CHD7-bound and 4329 P300-bound enhancers. Lists of 1-kb regions centered on the midpoint of active, intermediate, and poised enhancer sites defined by CHD7 and P300 in mESCs are listed in Supplementary Data 1, and those defined by H3K4me1/H3K27ac in mESCs, mNPCs, 3T3L1-derived adipocytes, and mBMDMs are given in Supplementary Data 2.
Annotation of enhancers
For the following analyses, we converted lists of mm8 coordinates representing 1-kb windows centered on each peak midpoint to mm9 using the UCSC LiftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Location analysis was performed with the Location Analysis feature of the ChIP-seq tool set (http://havoc.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi). For analysis of extragenic versus intragenic enhancers, location analysis sites with a region ID of “gene” were considered intragenic, while all others were considered extragenic. Conservation analysis was performed with the Conservation/Aggregate Datapoints feature of the Cistrome analysis pipeline (http://cistrome.dfci.harvard.edu/ap/), using a 4-kb window and the average vertebrate phastCons metric. Functional annotation was performed with GREAT (McLean et al. 2010) using the default basal plus extension parameters and the whole-mouse genome as background. The −log10 of the raw binomial P-value was reported, and all ontologies were also significant by FDR Q-value. In cases where the hypergeometric test was saturated due to the large number of associated genes, significance was assessed solely by the region-based binomial test. Complete GREAT results are provided as Supplementary Data 3 and 4.
Correlation of enhancer classes with expression
Microarray data sets for mESCs and mNPCs (GSE8024), 3T3L1-derived adipocytes (GSE20752), and mBMDM (GSE22935) were obtained from GEO (http://www.ncbi.nlm.nih.gov/geo/). Replicates from each cell type were RMA-normalized using the affy R package (Irizarry et al. 2003) and averaged. For genes represented by multiple probes, the probe with the highest average expression value was retained for analysis. Lists of genes in each category of enhancers were obtained by downloading the region-gene association file for each class from GREAT. Duplicate gene names in each region–gene association file were discarded, and files were merged to the normalized expression data. Statistical significance between groups was assessed by the Wilcoxon rank-sum test.
Analysis of germ layer specificity of enhancer classes
The top 20 ontologies in the MGI expression and mouse phenotype categories for each enhancer class were determined with GREAT, using lists of intermediate and poised enhancers defined by CHD7, P300, or the combined H3K4me1/H3K27ac peak list described above. Terms without a defined associated germ layer (i.e., “TS13_embryo”, “abnormal cell proliferation”) were not considered. The number of terms representing the germ layers (ectoderm, mesoderm, endoderm) and the extra-embryonic component were compared against the total number of terms obtained for all germ layers. The relative contribution of each germ layer to the total number of expression or phenotype terms was represented as a percentage of the total terms tested.
Acknowledgments
We thank Peter Harte for helpful discussions during the course of this work. This work was supported by grants from the National Institute of General Medical Sciences (5T32GM008614-15 to G.E.Z.), the National Institute of Child Health and Development (R01HD056369 to P.C.S.), and the National Human Genome Research Institute (5R01HG004722 to P.C.S.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.122382.111.
References
- Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23: 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2010. Enhancers: The abundance and function of regulatory sequences beyond promoters. Dev Biol 339: 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. 2006. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 16: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C-L, Natoli G 2010. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384 doi: 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15 doi: 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. 2007. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17: 691–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fullwood M, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi H-S, Tennakoon C, et al. 2010. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11: R22 doi: 10.1186/gb-2010-11-2-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers P, Jamrich M 2000. Regulation of eye formation by the Rx and pax6 homeobox genes. Cell Mol Life Sci 57: 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143: 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S 2007. Histone replacement marks the boundaries of cis-regulatory domains. Science 315: 1408–1411 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A 2004. Java Treeview: extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, LaFramboise T, et al. 2010. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet 6: e1001023 doi: 10.1371/journal.pgen.1001023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Furukawa T, Peters MA, Kozak CA, Cepko CL 1999. Misexpression of the Emx-related homeobox genes cVax and mVax2 ventralizes the retina and perturbs the retinotectal map. Neuron 24: 541–553 [DOI] [PubMed] [Google Scholar]
- Sunmonu NA, Chen L, Li JYH 2009. Misexpression of Gbx2 throughout the mesencephalon by a conditional gain-of-function transgene leads to deletion of the midbrain and cerebellum in mice. Genesis 47: 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA 2006. VISTA Enhancer Browser, a database of tissue-specific human enhancers. Nucleic Acids Res 35Suppl 1: D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L-Y, Li M, Hinton DR, Guo L, Jiang S, Wang JT, Zeng A, Xie JB, Snead M, Shuler C, et al. 2003. Microphthalmia resulting from Msx2-induced apoptosis in the optic vesicle. IOVS 44: 2404–2412 [DOI] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RDG, Chenoweth JG, Tesar PJ, Furey TS, et al. 2007. Identification and characterization of cell type–specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3: e136 doi: 10.1371/journal.pgen.0030136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nusbaum C, Myers R, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]