Abstract
Amphibian genomes differ greatly in DNA content and chromosome size, morphology, and number. Investigations of this diversity are needed to identify mechanisms that have shaped the evolution of vertebrate genomes. We used comparative mapping to investigate the organization of genes in the Mexican axolotl (Ambystoma mexicanum), a species that presents relatively few chromosomes (n = 14) and a gigantic genome (>20 pg/N). We show extensive conservation of synteny between Ambystoma, chicken, and human, and a positive correlation between the length of conserved segments and genome size. Ambystoma segments are estimated to be four to 51 times longer than homologous human and chicken segments. Strikingly, genes demarking the structures of 28 chicken chromosomes are ordered among linkage groups defining the Ambystoma genome, and we show that these same chromosomal segments are also conserved in a distantly related anuran amphibian (Xenopus tropicalis). Using linkage relationships from the amphibian maps, we predict that three chicken chromosomes originated by fusion, nine to 14 originated by fission, and 12–17 evolved directly from ancestral tetrapod chromosomes. We further show that some ancestral segments were fused prior to the divergence of salamanders and anurans, while others fused independently and randomly as chromosome numbers were reduced in lineages leading to Ambystoma and Xenopus. The maintenance of gene order relationships between chromosomal segments that have greatly expanded and contracted in salamander and chicken genomes, respectively, suggests selection to maintain synteny relationships and/or extremely low rates of chromosomal rearrangement. Overall, the results demonstrate the value of data from diverse, amphibian genomes in studies of vertebrate genome evolution.
When the genomes of living species are compared, patterns of variation emerge that implicate a diversity of mechanisms—whole-genome duplications (Postlethwait et al. 2000), convergent patterns of chromosome evolution (Bellott et al. 2010), dynamic chromosomal rearrangements (Nakatani et al. 2007), and lineages-specific changes in genome size and gene content (Hughes and Friedman 2008), just to name a few. The ability to identify mechanisms that have shaped the evolution of vertebrate genomes is directly related to the breadth of the comparisons that are made. If species that present unique and variable genome structures are not included in comparative studies, perspective will be limited and inferences that are made about genome evolution will be incomplete.
Very few studies have attempted to generate and compare genomic information from amphibians, and yet, amphibians present some of the largest and most diverse genome structures of all vertebrates (Morescalchi 1973; Duellman and Trueb 1986; Green and Sessions 1991; Vinogradov 1998). In particular, salamanders are famous for the range of variation they show in genome size and karyotype. This variation is partly explained by phylogeny—ancestral species present the largest genome sizes and more complex karyotypes, with relatively high numbers of macro- and microchromosomes (Sessions 2008). For example, the primitive hellbender (Cryptobranchus alleganiensis) has 112 pg/haploid nucleus (N) and 30 chromosome pairs consisting of both micro- and macrochromosome types. In contrast, the more recently derived Mexican axolotl (Ambystoma mexicanum) has >20 pg/N, with few macrochromosomes (n = 14) and no microchromosomes. Similar phylogenetic patterns of karyotypic variation are also observed among anuran amphibians (Duellman and Trueb 1986). The more recently derived western clawed frog (Xenopus tropicalis) has only 10 pairs of macrochromosomes, while primitive tailed frogs (Ascaphus trueii) have 42 pairs of chromosomes, including microchromosomes.
Modern reptiles, and especially birds, present much smaller genomes than amphibians, but the karyotypes of many species resemble those of basal amphibians. For example, the chicken genome (1.25 pg/N) consists of 36 chromosome pairs, with micro- and macrochromosome types. Indeed, most reptiles present micro- and macrochromosome types (Olmo 2008), and the majority of these are highly conserved among species, as determined by chromosome painting and reciprocal mapping of orthologous genes (Shetty et al. 1999; Guttenbach et al. 2003; Matsuda et al. 2005; Kayang et al. 2006; Stapley et al. 2008; Dalloul et al. 2010; Nanda et al. 2011) (but, see Kawai et al. 2007; Hansmann et al. 2009; Nie et al. 2009). The observation of microchromosomes between basal amphibians and reptiles suggests that karyotype evolution was also conservative prior to the divergence of reptile lineages. In support of this idea, Burt (2002) proposed that approximately half of the chromosomes in the avian genome arose from fission of ancestral chromosomes, but many others (including microchromosomes) correspond to ancient chromosomes that were conservatively retained from the tetrapod ancestor. Nakatani et al. (2007) also showed conservation of gene orders between reconstructed proto-vertebrate chromosomes and chicken microchromosomes. Presumably, these ancestral chromosomes have been maintained in vertebrate genomes for hundreds of millions of years and were present in the amphibian ancestor. Indeed, the most proximate outgroup to the tetrapod lineage also has a karyotype that is similar to primitive amphibians (Bogart et al. 1994). Given this presumed ancestral structure, more recently evolved amphibian lineages would be expected to show differences in genome structure. This is because salamanders and frogs are deeply diverged (∼250 MYA) (Zhang and Wake 2009) and the more recently evolved lineages within both groups present lower chromosome numbers and smaller genome sizes (Duellman and Trueb 1986; Sessions 2008).
In the absence of amphibian comparative gene maps, it has not been possible to test for the presence of ancestral tetrapod chromosomes in amphibian genomes. It also has not been possible to exploit amphibians as outgroups to reconstruct the evolutionary origin of amniote chromosomes. Here we address these issues using a newly constructed gene map of the Mexican axolotl—Tiger salamander genome. Using data from this map and comparative data from human, chicken, and X. tropicalis, we present four primary results. First, we show extensive conservation of synteny among vertebrate orthologs and a positive correlation between the length of conserved segments and genome size. The results suggest that synteny relationships were maintained as genomes expanded and contracted in size during vertebrate evolution. Second, we show that orthologs defining chicken chromosomes are discretely ordered in the Ambystoma and X. tropicalis genomes. Thus, ancestral chromosome segments were present in the amphibian ancestor, and their structures were subsequently conserved during amphibian phylogenesis. Third, we show that some ancestral chromosome segments are ordered in the same manner along Ambystoma and X. tropicalis linkage groups, while other ancestral segments are independently distributed among salamander and frog chromosomes. These patterns identify chromosome fusion as an important mechanism of vertebrate genome evolution—ancestral segments were fused before and after the divergence of salamanders and frogs. Fourth, we use the outgroup perspective provided by Ambystoma and X. tropicalis data to reconstruct the origin of chicken chromosomes by fission, fusion, and retention of ancestral chromosomes.
Results
Construction of an Ambystoma gene map
Single nucleotide polymorphisms were identified from orthologous A. mexicanum and A. t. tigrinum EST contigs that are available from Sal-Site (Putta et al. 2004; Smith et al. 2005a). We used SNPs from 917 EST contigs and individuals of the AxTg mapping panel (Voss and Shaffer 1997) to build a linkage map consisting of 17 groups (Supplemental Fig. 1). This number of linkage groups approaches the Ambystoma haploid chromosome number (n = 14) and is comprised mostly of gene-based markers that allow orthologous loci and conserved chromosomal segments to be identified and compared among vertebrate genomes. The three largest linkage groups are >400 cM and the overall map size is ∼4200 cM. Assuming that 1 cM = 7 Mb (4200 cM/30 Gb), Ambystoma LG1 is predicted to contain more DNA than is present in the entire human genome.
Comparative gene mapping
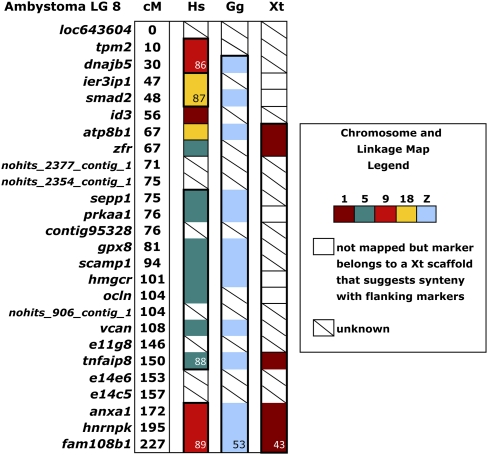
The order of genes on Ambystoma linkage groups was compared with the positions of presumptive orthologs in the human, chicken, and X. tropicalis genomes. Orthologs of contiguously linked genes from the Ambystoma map were sometimes rearranged locally along human and chicken chromosomes. Local change in the position of genes is expected when comparing taxa that diverged several hundred million years ago. Accordingly, we identified conserved segments as regions where two or more contiguous genes from the Ambystoma map located to the same region of a chromosome or linkage group in another species (Supplemental Fig. 2; Supplemental Table 1). Overall, comparative mapping revealed the well-documented pattern of increased interchromosomal rearrangement in the mammalian lineage leading to human, and greater conservation of gene order between the chicken and amphibian genomes (Smith and Voss 2006; Helsten et al. 2010). For example, Figure 1 shows that orthologs of genes on Ambystoma linkage group (LG) 8 map to a single chicken chromosome (Z) and a distinct region of X. tropicalis LG1, but these orthologs are distributed among three different chromosomes (5, 9, and 18) in the human genome.
Figure 1.
Organization of orthologs among human and chicken chromosomes and Ambystoma mexicanum/A. tigrinum and Xenopus linkage maps. This diagram shows chromosomal rearrangements in the human genome (chromosomes 5, 9, 18) and conservation of genes demarking Ambystoma LG8, chicken Z, and X. tropicalis LG1.
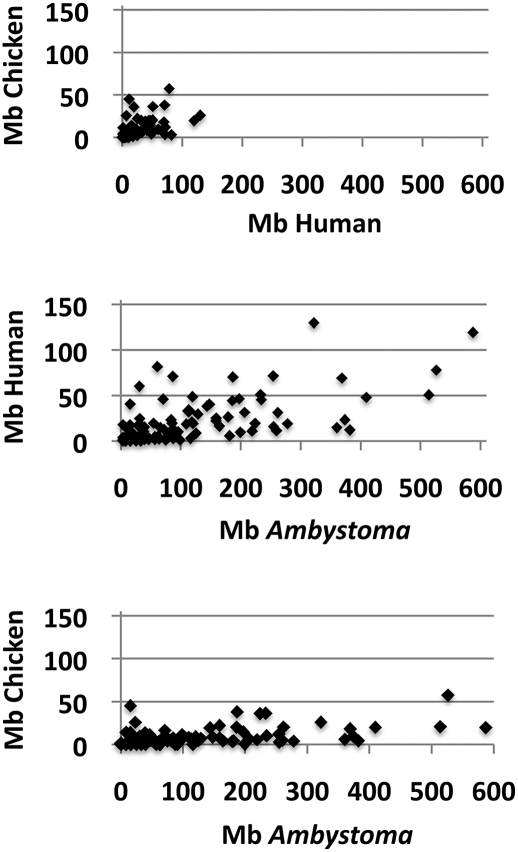
A total of 134, 78, and 58 conserved segments were identified between the Ambystoma map and the human genome, chicken genome, and X. tropicalis linkage map, respectively. More conserved segments were identified between Ambystoma and human, in part because the latter has more gene annotations than any other vertebrate. An analysis of 94 conserved segments among human, chicken, and Ambystoma was then performed to compare the sizes of homologous segments deriving from vertebrates with very different genome architectures (Fig. 2). The ratio of human to chicken physical distance was >1 for 89% (84/94) of comparisons, and with the exception of a region where genes are densely organized in the chicken genome (chromosome 25), these values were relatively constant across segments (n = 84; Avg = 4.8; SD = 4.8). Thus, on average, human conserved segments are approximately four times longer in physical distance than homologous chicken segments. The ratio of Ambystoma linkage distance to human and chicken was also >1 for 93% (173/186) of comparisons. Assuming again that 1 cM = 7 Mb in the Ambystoma genome, conserved Ambystoma segments are estimated to be 14 and 51 times longer on average than homologous human (n = 86; Avg = 14.1 Mb; SD = 18.2) and chicken (n = 91, Avg = 51.4 Mb; SD = 101.8) segments, respectively. These results suggest that gene orders have been maintained across homologous vertebrate chromosome segments that differ by >500 Mb in size (Fig. 2).
Figure 2.
Analysis of the physical size of 94 conserved segments among human, chicken, and Ambystoma. To obtain estimates of physical size for Ambystoma, 1 cM was assumed to equal 7 Mb.
Along alignments of linkage groups and chromosomes, conserved segments were occasionally disrupted by markers of unknown orthology, single nonsyntenic markers, or incomplete mapping data. It is possible that some of these nonsyntenic markers correspond to lineage-specific paralogs. Ignoring these cases, we extended the definition of conserved synteny to encompass consecutive conserved segments of the same type. For example, chicken conserved segments 66, 67, and 68 contain genes that map to chromosome 4, but they are separated on the Ambystoma map by genes (rad23b and chdh) that map to different chicken chromosomes (Fig. 3). Ignoring these markers unites conserved segments 66–68 into a single, synteny group with markers spanning 75% of chicken chromosome 2 and nearly the entire length of Ambystoma LG 12. Similarly, we found that gene orders spanning the lengths of other chicken chromosomes are arranged discretely as units along Ambystoma linkage groups (Supplemental Fig. 2). Nine Ambystoma LGs (5, 7, 8, 9, 11, 12, 13, 14, 17) showed a 1: 1 correspondence to either a whole chicken chromosome or X. tropicalis LG, or to a large, discrete region within a chicken chromosome or X. tropicalis LG. These patterns of long-range segment conservation are consistent with the idea that some chicken chromosomes correspond to ancestral chromosome segments.
Figure 3.
Organization of orthologs among human and chicken chromosomes and Ambystoma mexicanum/A. tigrinum and X. tropicalis linkage maps. The diagram shows that chicken conserved segments 66, 67, and 68 likely form a large synteny group, excepting the location of two genes (RAD23B and CHDH) that map to different chicken chromosomes.
Reconstructing the origin of avian chromosomes
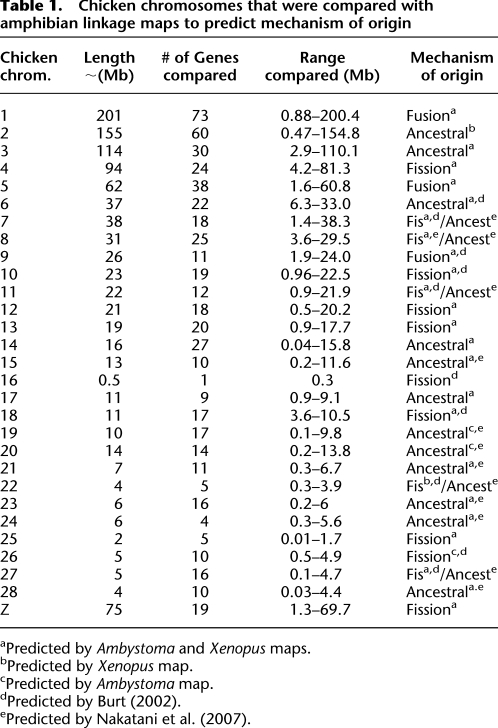
Marker coverage was sufficiently dense between the amphibian maps and chicken genome to resolve the ordered structures of chicken chromosomes and infer mechanisms associated with their origin (Table 1). For example, chicken chromosomes 8, 11, and 25 are ordered on Ambystoma LG1 (Supplemental Fig. 2). As orthologous genes for these chromosomes are also linked on X. tropicalis LG7, chicken chromosomes 8, 11, and 25 likely were linked in the amphibian ancestor. If chicken 8, 11, and 25 were linked in the tetrapod ancestor, then these chromosomes must have arisen during evolution by chromosome fission; alternatively, if these chromosomes were unlinked in the tetrapod ancestor, the inference would be chromosome fusion in the stem lineage of extant amphibians and ancestral chromosome retention in the lineage leading to chicken. Using such logic and data from the literature, we predict that three chicken chromosomes originated by fusion, 9–14 originated by fission, and 12–17 evolved directly from ancestral tetrapod chromosomes (Table 1). We confirmed by comparative mapping of chicken and zebra finch genomic sequences that these inferences are likely to apply generally to avian species (Supplemental Table 3). We only found evidence for two fusion/fission events between these taxa: (1) Fusion of a microchromosome (zebra finch 4A) to a large chromosome likely occurred within the chicken lineage, giving rise to modern chicken chromosome 4. These chromosomes presumably correspond to ancestral avian chromosomes 4 and 10, which are uncharacteristically labile within the avian karyotype (Guttenbach et al. 2003; Griffin et al. 2007; Völker et al. 2010). (2) Fission of a chromosome (chicken chromosome 1) within the zebra finch lineage (Itoh and Arnold 2005; Völker et al. 2010). This fission event was documented previously by chromosomal painting of finch, chaffinch, blackbird, and redwing (Derjusheva et al. 2004; Itoh and Arnold 2005), and more recently by comparative mapping (Völker et al. 2010).
Table 1.
Chicken chromosomes that were compared with amphibian linkage maps to predict mechanism of origin
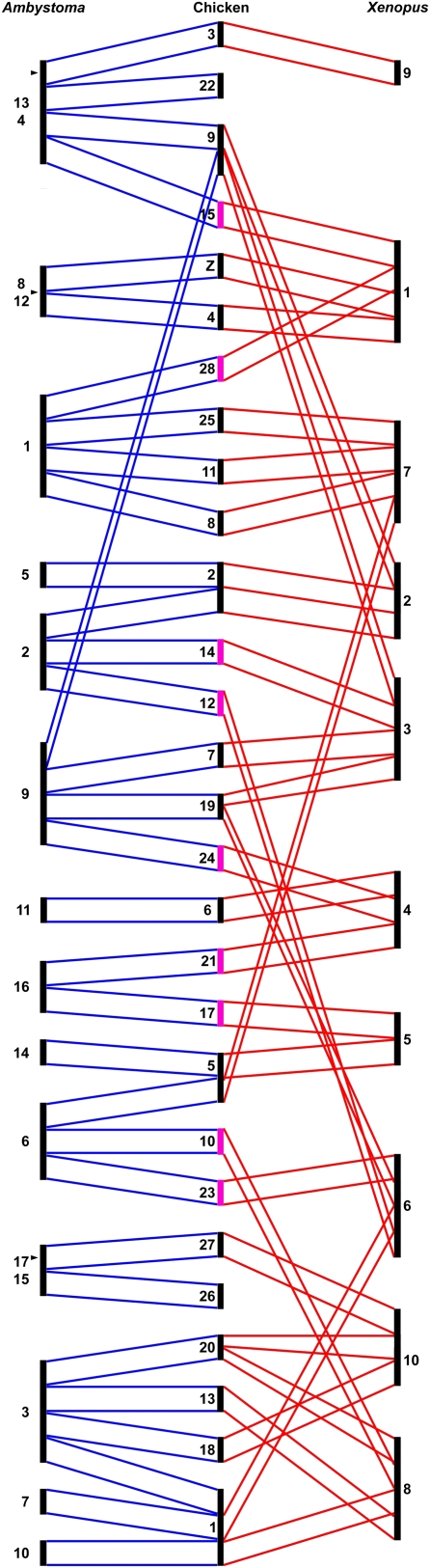
The evidence for retention of ancestral tetrapod chromosomes is indirect, but striking. Ten segments corresponding to chicken chromosomes are arranged differently between Ambystoma and Xenopus linkage groups (chicken 10, 12, 14, 15, 17, 21, 23, 24, 27, and 28) (Fig. 4). Such a pattern is expected if modern chicken chromosomes correspond to ancestral chromosome segments that were fused into different combinations as anuran and salamander lineages diverged independently. As an alternative explanation, the pattern could be explained by a high rate of interchromosomal rearrangement within one of the amphibian lineages. However, only three segments corresponding to chicken chromosomes were rearranged uniquely among Ambystoma or Xenopus linkage groups (chicken 2, 19, 20). Moreover, random chromosomal rearrangements are not expected to result in conservation of synteny between markers defining whole chicken chromosomes and large tracts of salamander and frog linkage groups.
Figure 4.
Conserved syntenies among Ambystoma mexicanum/A. tigrinum, chicken, and X. tropicalis genomes. Proposed ancestral chromosomal blocks are delimited by blue lines drawn between Ambystoma and chicken chromosomes. Red lines indicate conserved synteny between chicken chromosomes and X. tropicalis linkage groups. The length of each chromosome corresponds to the number of conserved blocks that are contained within that chromosome. Chicken chromosomes that correspond to blocks that were independently fused in Ambystoma and X. tropicalis are highlighted in pink. Arrowheads mark positions where Ambystoma linkage groups 4–13, 8–12, and 15–17 likely join, based upon previous mapping data (Smith et al. 2005b) and conserved synteny.
Discussion
The structure of vertebrate genomes has been shaped by many different mechanisms over several hundred million years of evolution. This has yielded considerable diversity in genome size and karyotype among the various vertebrate groups, and in particular, amphibians (Sessions 2008). Unfortunately, the amphibian perspective has rarely been incorporated into comparative studies of vertebrate genome evolution (Voss et al. 2001; Smith and Voss 2006, 2007; Hellsten et al. 2010). This is explained by the large and complex genomes that amphibians present, and the nature of science to focus on organisms with small and simple genome structures. However, large salamander genomes are being prioritized for sequencing (Haussler et al. 2009), and insights that will come from these efforts are prefaced in the results of this study. Although amphibians, and especially salamanders, present extremely large genomes, we show that the structure of amphibian genomes is highly conserved relative to other vertebrates. We show that chromosome segment sizes have been conserved under very different scenarios of genome size evolution—gene loss and genome size reduction in the case of chicken (Hughes and Friedman 2008) and moderate and extreme increases in genome size in humans and salamanders, respectively. The basis of this conservation is largely explained by the retention of ancestral vertebrate chromosome segments in both amphibian and amniote lineages. Our study clarifies the role of fission versus retention of ancestral chromosomes in the origin of avian chromosomes, and predicts a greater role for chromosome fusion in vertebrate evolution (Nakatani et al. 2007). In amphibians, ancestral chromosome segments were fused to yield mosaic arrangements while retaining the integrity of these segments over hundreds of millions of years of evolution.
The discrete ordering of ancestral chromosomal segments in two modern amphibian genomes provides a validation of Burt (2002) hypothesis concerning the origin of avian chromosomes. Based on a comparison of relatively few chicken and fish conserved syntenies, Burt (2002) predicted that many chicken microchromosomes might correspond to ancestral tetrapod chromosomes. Our finding of these chromosome segments in extant amphibian genomes, coupled with the observation of microchromosomes and high chromosome numbers in basal amphibian lineages (Morescalchi 1973; Duellman and Trueb 1986; Sessions 2008; Vinogradov 1998) and coelacanth (Bogart et al. 1994) supports a model whereby ancestral tetrapod chromosomes were fused during amphibian phylogenesis (Fig. 5). Some of the fusion events preceded the divergence of salamanders and anurans. This inference is supported by the following observation: Some chicken chromosomes are ordered the same way on Ambystoma and X. tropicalis linkage groups. For example, consider the linked arrangement of genes defining chicken chromosomes 8 and 11 on Ambystoma LG1 and X. tropicalis LG7. Nakatani et al. (2007) showed that chickens 8 and 11 likely trace their ancestry to independent chromosomes in the ancestral jawed vertebrate. Because genes defining these chromosomes are not linked in other vertebrates, the most parsimonious explanation is fusion in the lineage of extant amphibians and subsequent retention of this linkage relationship in salamanders and anurans.
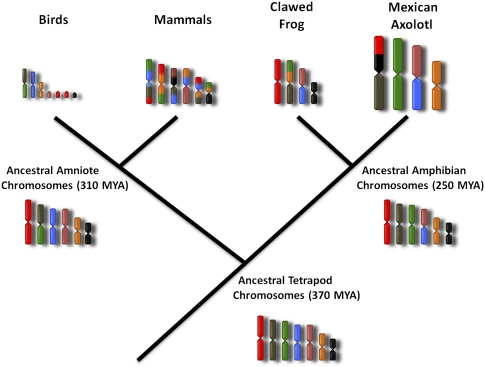
Figure 5.
A generalized model of vertebrate chromosomal evolution. The ancestral vertebrate karyotype has remained relatively stable over the last ∼370 million years, and large segments of ancestral chromosomes are still retained among lineages. Different combinations of ancestral chromosomes fused together as chromosome numbers decreased in parallel within anuran and salamander lineages. Gene orders defining ancestral chromosomal segments have been rearranged locally during evolution, but synteny has been maintained in the face of dramatic increases and decreases of interstitial genomic content and genome size. Some avian chromosomes are predicted to correspond to entire ancestral chromosomes, although fission and fusion played a role in shaping the generalized avian karyotype. Divergence dates were taken from the literature as follows: tetrapod ancestor (Ruta et al. 2003), amniote ancestor (Hedges and Kumar 2004; Riesz and Muller 2004), amphibian (salamander-frog) ancestor (Zhang and Wake 2009). (MYA) Million years ago.
We also propose that some ancestral chromosomes were fused after the divergence of salamander and anuran lineages. Among frogs and amphibians, some of the more derived lineages present fewer chromosomes, a convergent pattern indicating selective pressure to reduce chromosome number. Our results suggest that during the process of chromosome number reduction in Ambystoma and Xenopus, ancestral chromosome segments corresponding to chicken microchromosomes underwent independent fusions, yielding reduced chromosome numbers in both taxa, but with a different distribution of ancestral segments among chromosomes. Given the deep divergence time of frog and salamander lineages (∼250 MYA) (Zhang and Wake 2009), there was considerable time for ancestral chromosomes to fuse independently in lineages leading to X. tropicalis and Ambystoma. It is interesting that the fundamental structure of ancestral chromosomes has been maintained in amphibian genomes for hundreds of millions of years, despite extensive changes in karyotype and genome size among amphibian lineages. For example, Ambystoma LG8 has undergone a >20-fold expansion relative to the size of the chicken Z chromosome (∼75 Mb). While there are a few examples of chromosome fusion and fission among avian taxa (Guttenbach et al. 2003; Derjusheva et al. 2004; Itoh and Arnold 2005; Griffin et al. 2007; Hansmann et al. 2009; Nie et al. 2009), reptilian karyotypes are generally conservative (Olmo 2008). These patterns suggest a role for selection in maintaining the integrity of ancestral chromosomal segments. One possibility is that selection has acted to maintain the spatial organization of regulatory elements and genes during vertebrate evolution (Larkin et al. 2009; Nie et al. 2010). These patterns may also reflect relatively low rates of interchromosomal rearrangement in fishes, amphibians, and birds, while still allowing for intrachromosomal rearrangement of loci (Smith and Voss 2006; Völker et al. 2010).
The results from this study lend a critical amphibian perspective to ongoing debates concerning the ancestral tetrapod karyotype and origin of amniote chromosomes. It was originally proposed that chromosomes within amniote genomes originated primarily by chromosomal fission of an ancestral genome with ∼12 chromosome pairs (Postlethwait et al. 2000). Ambystoma and X. tropicalis do present relatively few chromosomes, and these approximate the number of the presumptive vertebrate ancestor. As we show here, some chromosomes of these amphibians are actually the product of independent fusion events and, therefore, chromosome counts do not provide direct insight into tetrapod ancestral chromosome number. Rather, this information is gained by understanding the distribution of gene homologies in evolutionarily informative lineages and mechanisms of karyotype evolution (Griffin et al. 2007). Our results support an evolutionary model that predicts at least twice as many chromosomes for the tetrapod ancestor (Nakatani et al. 2007; Putnam et al. 2008), approximating the numbers of primitive amphibians, coelacanth, and some nonmammalian amniotes (e.g., birds).
Methods
Genetic linkage analysis
This study used 570 previously genotyped molecular markers from Smith et al. (2005b) and 337 new markers that were developed using expressed sequence tag (EST) contigs from Sal-Site (Putta et al. 2004; Smith et al. 2005a). The EST markers were scored for species-specific polymorphisms using a previously described AxTg mapping family and genotyping methods (Voss and Shaffer 1997). Primer sequences, diagnostic polymorphisms, and polymorphism detection assays are summarized in Supplemental Table 2. Linkage groups were built using MultiPoint 2.2 (Korol et al. 2003) and visualized using MapChart 2.1 (Voorips 2002). Linkage groups were assembled by first identifying sets of markers that formed linear linkage groups supported at a recombination frequency value of 0.025. Then, markers were added step-wise at higher recombination frequency values (0.05, 0.075, 0.10…0.4) and linkage group distances were estimated using the Kosambi (1944) mapping function.
Comparative mapping
EST contigs were used as query sequences in BLASTx searches of the human and nrRefSeq protein databases at NCBI. The official NCBI Gene ID associated with the best matching sequence was assigned to the marker associated with the EST contig. The positions of orthologous loci in the human and chicken genomes were obtained from the NCBI Gene database. The location of X. tropicalis genes was inferred from data presented in Hellsten et al. (2010). Conserved syntenic blocks of loci were identified among Ambystoma and X. tropicalis linkage groups and chicken and human genome assemblies. A conserved syntenic block was defined when orthologs of two or more syntenic loci in one species was also observed to be syntenic in another species. To further confirm inferences of karyotype evolution in a second avian species, a chicken/zebra finch map was generated and compared by downloading the whole-genome sequence alignment net from the UCSC Genome Browser. The alignment net was filtered to retain all conserved segments with >25 kb in gap-free alignment.
Acknowledgments
This research was supported by grant R24-RR016344 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). The project also used resources developed under Multidisciplinary University Research Initiative grant (W911NF-09-1-0305) from the Army Research Office, and resources from the Ambystoma Genetic Stock Center, which is funded by grant DBI-0951484 from the National Science Foundation (S.R.V.). The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of NCRR, NIH, ARO, or NSF.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116491.110.
References
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, et al. 2010. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogart JP, Balon EK, Bruton MN 1994. The chromosomes of the living coelacanth and their remarkable similarity to those of one of the most ancient frogs. J Hered 85: 322–325 [DOI] [PubMed] [Google Scholar]
- Burt DW 2002. Origin of avian microchromosomes. Cytogenet Genome Res 96: 97–112 [DOI] [PubMed] [Google Scholar]
- Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Ann Blomberg L, Bouffard P, Burt DW, Crasta O, Crooijmans RP, et al. 2010. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol 8: 9 doi: 10.1371/journal.pbio.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derjusheva S, Kurganova A, Habermann F, Gaginskaya E 2004. High chromosome conservation detected by comparative chromosome painting in chicken, pigeon and passerine birds. Chromosome Res 12: 715–723 [DOI] [PubMed] [Google Scholar]
- Duellman WE, Trueb L 1986. Biology of amphibians. McGraw-Hill, San Francisco, CA [Google Scholar]
- Green DM, Sessions SK, ed. 1991. Amphibian cytogenetics and evolution. Academic, San Diego, CA [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Skinner BM 2007. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res 117: 64–77 [DOI] [PubMed] [Google Scholar]
- Guttenbach M, Nanda I, Feichtinger W, Masabanda JS, Griffin DK, Schmid M 2003. Comparative chromosome painting of chicken autosomal paints 1–9 in nine different bird species. Cytogenet Genome Res 103: 173–184 [DOI] [PubMed] [Google Scholar]
- Hansmann T, Nanda I, Volobouev V, Yang F, Schartl M, Haaf T, Schmid M 2009. Cross-species chromosome painting corroborates microchromosome fusion during karyotype evolution of birds. Cytogenet Genome Res 126: 281–304 [DOI] [PubMed] [Google Scholar]
- Haussler D, O'Brien SJ, Ryder OA, Barker FK, Clamp M, Crawford A, Hanner R, Hanotte O, Johnson WE, McGuire JA, et al. 2009. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered 100: 659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Kumar S 2004. Precision of molecular time estimates. Trends Genet 20: 242–247 [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R 2008. Genome size reduction in the chicken involved massive loss of ancestral protein coding genes. Mol Biol Evol 25: 2681–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Arnold AP 2005. Chromosomal polymorphism and comparative painting analysis in the zebra finch. Chromosome Res 13: 47–56 [DOI] [PubMed] [Google Scholar]
- Kawai A, Nishida-Umehara C, Ishijima J, Tsuda Y, Ota H, Matsuda Y 2007. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet Genome Res 117: 92–102 [DOI] [PubMed] [Google Scholar]
- Kayang BB, Fillon V, Inoue-Murayama M, Miwa M, Leroux S, Fève K, Monvoisin JL, Pitel F, Vignoles M, Mouilhayrat C, et al. 2006. Integrated maps in quail (Coturnix japonica) confirm the high degree of synteny conservation with chicken (Gallus gallus) despite 35 million years of divergence. BMC Genomics 7: 101 doi: 10.1186/1471-2164-7-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korol A, Ronin Y, Minkov D, Brivin E, Mester D, Korostishevsky M, Malkin I, Frenkel Z, Orion O, Cohen L, et al. 2003. MultiPoint 2.2. http://www.multiqtl.com
- Kosambi D 1944. The estimation of map distances from recombination values. Ann Eugen 12: 172–175 [Google Scholar]
- Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA 2009. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res 19: 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Nishida-Umehara C, Tarui H, Kuroiwa A, Yamada K, Isobe T, Ando J, Fujiwara A, Hirao Y, Nishimura O, et al. 2005. Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res 13: 601–615 [DOI] [PubMed] [Google Scholar]
- Morescalchi A 1973. Amphibia. In Cytotaxonomy and vertebrate evolution (ed. Chiarelli AB, Campanna E), pp. 233–348 Academic, London, UK [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res 17: 1254–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, Benisch P, Fetting D, Haaf T, Schmid M 2011. Synteny conservation of chicken macrochromosomes 1-10 in different avian lineages revealed by cross-species chromosome painting. Cytogenet Genome Res 132: 165–181 [DOI] [PubMed] [Google Scholar]
- Nie W, O'Brien PC, Ng BL, Fu B, Volobouev V, Carter NP, Ferguson-Smith MA, Yang F 2009. Avian comparative genomics: reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes)–an atypical species with low diploid number. Chromosome Res 17: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H, Crooijmans RP, Lammers A, van Schothorst EM, Keijer J, Neerincx PB, Leunissen JA, Megens HJ, Groenen MA 2010. Gene expression in chicken reveals correlation with structural genomic features and conserved patterns of transcription in the terrestrial vertebrates. PLoS ONE 5: e11990 doi: 10.1371/journal.pone.001990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo E 2008. Trends in the evolution of reptilian chromosomes. Integr Comp Biol 48: 486–493 [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res 10: 1890–1902 [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071 [DOI] [PubMed] [Google Scholar]
- Putta S, Smith JJ, Walker JA, Rondet M, Weisrock DW, Monaghan J, Samuels AK, Kump DK, King DC, Maness NJ, et al. 2004. From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genomics 5: 54 doi: 10.1186/1471-2164-5-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesz RR, Muller J 2004. Molecular timescales and the fossil record: a paleontological perspective. Trends Genet 20: 237–241 [DOI] [PubMed] [Google Scholar]
- Ruta M, Coates MI, Quicke DJ 2003. Early tetrapod relationships revisited. Biol Rev Camb Philos Soc 78: 251–345 [DOI] [PubMed] [Google Scholar]
- Sessions SK 2008. Evolutionary cytogenetics in salamanders. Chromosome Res 16: 183–201 [DOI] [PubMed] [Google Scholar]
- Shetty S, Griffin DK, Graves JA 1999. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res 7: 289–295 [DOI] [PubMed] [Google Scholar]
- Smith JJ, Voss SR 2006. Gene order data from a model amphibian: New perspectives on vertebrate genome structure and evolution. BMC Genomics 7: 219 doi: 10.1186/1471-2164-7-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Voss SR 2007. Chromosomal regions from bird and mammalian sex chromosomes were linked ancestrally. Genetics 177: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Putta S, Walker JA, Kump DC, Samuels AK, Monaghan JR, Weisrock DW, Staben C, Voss SR 2005a. Sal-Site: Integrating new and existing ambystomatid research and information resources. BMC Genomics 6: 181.doi: 10.1186/1471-2164-6-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kump K, Walker JA, Parichy DM, Voss SR 2005b. A genetic linkage map for tiger salamander and mexican axolotl (Ambystoma): Enabling gene mapping, comparative genomics, and PCR marker development in Ambystoma. Genetics 171: 1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Birkhead TR, Burke T, Slate J 2008. A linkage map of the zebra finch Taeniopygia guttata provides new insights into avian genome evolution. Genetics 179: 651–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE 1998. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry 31: 100–109 [DOI] [PubMed] [Google Scholar]
- Völker M, Backström N, Skinner BM, Langley EJ, Bunzey SK, Ellegren H, Griffin DK 2010. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res 20: 503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorips RE. 2002. MapChart 2.1. http://www.plant.wageningenur.nl/ products/mapping/MapChart.
- Voss SR, Shaffer HB 1997. Adaptive evolution via a major gene effect: paedomorphosis in the Mexican axolotl. Proc Natl Acad Sci 94: 14185–14189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss SR, Smith JJ, Gardiner DM, Parichy DM 2001. Conserved vertebrate chromosomal segments in the large salamander genome. Genetics 158: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wake DB 2009. Higher-level salamander relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol 53: 492–508 [DOI] [PubMed] [Google Scholar]