Abstract
The characterization of transcriptional networks (TNs) is essential for understanding complex biological phenomena such as development, disease, and evolution. In this study, we have designed and implemented a procedure that combines in silico target screens with zebrafish and mouse validation, in order to identify cis-elements and genes directly regulated by Pax6. We chose Pax6 as the paradigm because of its crucial roles in organogenesis and human disease. We identified over 600 putative Pax6 binding sites and more than 200 predicted direct target genes, conserved in evolution from zebrafish to human and to mouse. This was accomplished using hidden Markov models (HMMs) generated from experimentally validated Pax6 binding sites. A small sample of genes, expressed in the neural lineage, was chosen from the predictions for RNA in situ validation using zebrafish and mouse models. Validation of DNA binding to some predicted cis-elements was also carried out using chromatin immunoprecipitation (ChIP) and zebrafish reporter transgenic studies. The results show that this combined procedure is a highly efficient tool to investigate the architecture of TNs and constitutes a useful complementary resource to ChIP and expression data sets because of its inherent spatiotemporal independence. We have identified several novel direct targets, including some putative disease genes, among them Foxp2; these will allow further dissection of Pax6 function in development and disease.
Complex biological processes such as organogenesis require finely tuned spatiotemporal regulation of multiple developmental pathways. This is achieved at a number of levels including transcriptional regulation, translation control, and protein modification. Recently, the deciphering of transcriptional networks (TNs) has become feasible with the availability of multiple vertebrate genome sequences, the ever-increasing information on transcription factor (TF) binding sites, and large biological data sets, such as tissue-specific expression profiles. The combination of these types of data with the concept of evolutionary conservation has led to some successful studies identifying direct targets of selector gene transcription (Mann and Carroll 2002), including those for eyeless, the Drosophila Pax6 gene (Ostrin et al. 2006), and, in vertebrates, targets of Atoh7 (Ath5) and Pax2/5/8 were identified and validated in medaka (Del Bene et al. 2007; Ramialison et al. 2008).
We have developed a process that combines an in silico approach based on evolutionary conservation and the use of available experimentally tested binding sites to predict similar TF binding sites genome-wide, followed by validation of predictions in zebrafish and mouse. The known binding sites were extracted from the literature and used to generate Pax6 binding site hidden Markov models (HMMs) that allow genome-wide prediction of a new set of putative Pax6 binding sites that are expected to function as enhancers for genes regulated directly by Pax6. The choice of Pax6 as a paradigm for this study was based on its important roles in development and in human disease. During development, Pax6 is required for correct patterning of the nervous system (Stoykova et al. 1996; Ericson et al. 1997; Engelkamp et al. 1999; Holm et al. 2007; Brill et al. 2008; Simpson et al. 2009), eyes (Ashery-Padan et al. 2000; Marquardt et al. 2001), and pancreas (St-Onge et al. 1997; Ashery-Padan et al. 2004).
PAX6 haploinsufficiency in humans results predominantly in eye anomalies such as aniridia, lenticular-corneal adhesions (van Heyningen and Williamson 2002), and, rarely, microphthalmia (V van Heyningen, pers. comm.). In some cases, cognitive impairment (Heyman et al. 1999; Ticho et al. 2006), mental retardation, and cerebellar ataxia (Graziano et al. 2007) were reported. Structural and functional brain anomalies have also been observed (Sisodiya et al. 2001; Mitchell et al. 2003; Bamiou et al. 2007). Similar phenotypes are seen in animal models, such as the mouse Pax6 mutant Smalleye (Hill et al. 1991; Estivill-Torrus et al. 2001; Davis et al. 2003), In the mouse, homozygous loss of function was shown to lead to anophthalmia, severe brain malformation, and absence of olfactory system and endocrine pancreas function, all leading to neonatal lethality. Additionally, mouse conditional inactivation models provide further information on late functions when early lethality of the full knockout precludes late studies (Davis-Silberman et al. 2005; Tuoc et al. 2009). A pax6b missense mutant sunrise (sri) has also been described in zebrafish, showing a relatively mild eye phenotype seen only in homozygotes which are viable and fertile (Kleinjan et al. 2008). However, morpholino-induced knockdown of one or both zebrafish pax6 genes, pax6a and pax6b, allows us to explore the role of these genes in early embryonic development. Zebrafish is a particularly good model to investigate eye development and disease because of the similarity between zebrafish and human eyes, both at a molecular and morphological level (Goldsmith and Harris 2003; Glass and Dahm 2004; Fadool and Dowling 2008). In contrast to mice with eyes specialized for nocturnal life, both zebrafish and humans have evolved eyes for diurnal life including cone-dense retinas (Goldsmith and Harris 2003), which are also biochemically more similar than mouse to human retinas. For example, guanylate cyclase activator 1a is expressed in zebrafish and human retinas but not in the mouse (Imanishi et al. 2002). Such species-specific distinction is significant, given the clinical importance of cone degeneration syndromes.
Several studies, aimed at better understanding the architecture and diverse functions of Pax6 networks, have investigated differential expression of potential target genes in specific tissues and under various experimental conditions, including the use of mutant versus wild type comparisons (Chauhan et al. 2002; Holm et al. 2007; Visel et al. 2007; Sansom et al. 2009; Wolf et al. 2009). PAX6 binding sites have been identified in mouse embryonic cortex, using ChIP (Sansom et al. 2009). Comparison of the data sets from these studies shows that there is not one fixed set of genes regulated by Pax6 but several, depending on the spatiotemporal environment. Although there is overlap, the majority of genes in each target set are not present in other sets. Furthermore, binding of Pax6 to a cis-enhancer does not always correlate with function; for example, only 22% of the genes associated with adjacent Pax6 binding sites show expression level differences in the cortex when Pax6 levels are altered (Sansom et al. 2009).
There is a need for Pax6 target screens that are not spatiotemporally restricted, as these can complement and drive “wet” lab-based studies and be used as a theoretical framework to predict the Pax6 transcription networks, identifying genes that may transmit and modulate specific Pax6 functions in different tissues or cell types at different stages of the life cycle.
Results
In silico identification and analysis of Pax6 targets
To achieve better insight into the role of Pax6 in neuronal development, particularly eye development, we began by manually annotating experimentally validated Pax6 binding sites (BSs) from research articles in PubMed. From these, we identified 29 binding sites. Close inspection of the BSs showed that there is only a low degree of similarity between some of these (Supplemental Fig. S1). This led us to develop a semiautomated procedure to identify a reasonable number of BSs with a relatively high level of similarity (Supplemental Fig. S2A,B). These were used to make a HMM to identify the best matches among the remaining ones. This process was iterated until the best new matches were deemed too deviant, by visual inspection.
This selection procedure retrieved 16 similar Pax6 BSs (Supplemental Fig. S2B) which were used to compare HMM- and position weight matrices (PWM)-based strategies. PWM and HMM were generated and run, using parallel procedures (as described in Fig. 1A for HMMs). The results show that both strategies are largely equivalent for high stringency levels, but the use of HMMs combined with thresholds based on experimentally validated BSs is, at least computationally, more stringent (Supplemental Fig. S3A–D).
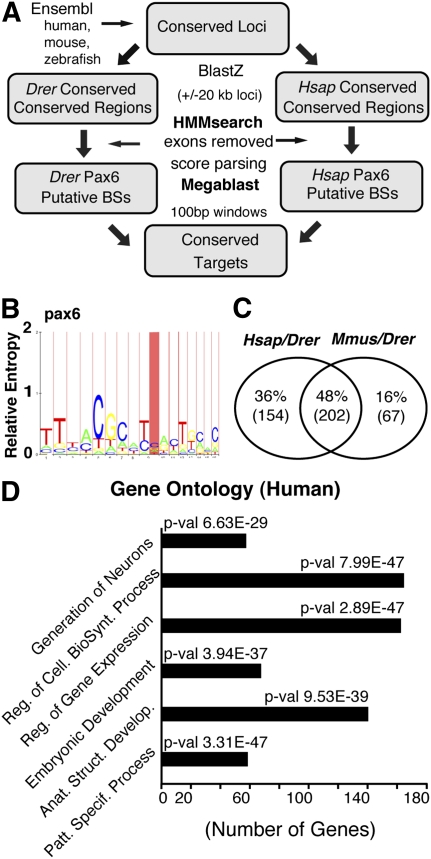
Figure 1.
Bioinformatics approaches used to identify the PAX6 target sites and the initial analysis of results. (A) Flow diagram of the HMM approach used to identify evolutionarily conserved direct targets of pax6. The procedure to identify pax6 target enhancers and associated genes is based on the screen of evolutionarily conserved noncoding genomic sequences from orthologous loci in zebrafish and human (or mouse). (B) The screens were performed using models based on pax6 binding sites or their respective reverse complement sequences. (C) 48% (202) of the putative target genes were found in both pairwise comparisons, while 16% (67) are mouse/zebrafish and 36% (154) human/zebrafish specific. (D) The full set of human conserved target genes was analyzed using gene ontology. The identified target genes are enriched for transcription factor activity and developmental regulation as well as for neuron generation functions. The x-axis represents the number of genes for each of the highest over-represented molecular function GO term which are named on the y-axis, and the associated P-values are shown.
Using the selected BSs, we then generated species-specific HMM models and used these (Fig. 1A) in combination with the HMMER software package (Fig. 1B ; Eddy 1998) to identify predicted BSs genome-wide within transcribed regions, excluding exonic regions but covering 20 kb on either side of each transcription unit. This thresholded process revealed two lists of putative BSs at sites showing evolutionary conservation from zebrafish to human and zebrafish to mouse. We identified 874 human and 1032 mouse putative BSs, corresponding to 903 and 746 zebrafish BSs, respectively. Applying the stringent criterion that BSs have to be at least 40 bp distant from other predicted BSs, reduced these numbers to 654 human and 819 mouse BSs, and 813 and 640 in zebrafish, respectively (Supplemental Table S4). The predicted BSs can be assigned to 327 human genes and 272 mouse genes, corresponding to 356 and 269 zebrafish genes, respectively (Supplemental Table S4). Inspection of the list of zebrafish putative target genes shows that 48% (202 genes) are found in both the human/zebrafish and mouse/zebrafish conserved sets. Of the remainder, 16% (67 genes) are mouse/zebrafish specific and 36% (154 genes) are human/zebrafish specific (Fig. 1C). The full annotated lists of BSs within human and mouse gene homologues, with chromosomal nucleotide positions, are shown in Supplemental Tables S5–S8. Gene targets that coincide between the two mammals are highlighted in yellow. Multiple binding sites associated with a single gene are clearly observed, reinforcing the concept of homotypic clustering (Gotea et al. 2010).
To characterize the identified sets of mouse and human genes at a functional level, we investigated gene ontology and expression data. Gene ontology analysis shows enrichment, in both sets of predicted mammalian target genes, for the GO terms related to embryonic development, patterning, and transcriptional regulation (Fig. 1D; Supplemental Fig. S4). In addition, the set of human genes was also highly enriched for genes involved in the generation of neurons (Fig. 2). Pax6 is known to be a key regulator of central nervous system and eye development. To determine whether the target sets were enriched for brain and eye specific genes, we combined UniGene expression data analysis with a bootstrap method to determine the statistical significance of the findings. In our list of human genes, we found 192 eye- (P = 0.0456), 229 brain- (P = 0.0022), and 130 heart- (P = 0.7614) associated genes. Similarly, among the mouse genes, we found 133 eye (P = 0.8254), 154 brain (P = 0.1218), and 74 heart (P = 0.9758). The P-values associated with the number of genes in each category were computed relative to the full sets of mouse or human UniGene genes. Differences in the relative number of eye genes between the two sets are discussed later. The overall results show enrichment in eye and brain genes, while, for the control organ, the heart, there was no significant enrichment. Therefore, our in silico procedure is able to identify classes of genes, particularly in human data, that play a role in the development of organs where Pax6 function is crucial.
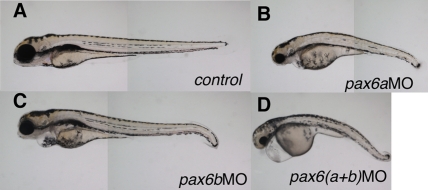
Figure 2.
pax6 knockdown in zebrafish results in reduced size, particularly of nervous system structures. (A) Representative control sibling. Knockdown with morpholino oligonucleotides at 1–2-cell stages of (B) pax6a, (C) pax6b, and (D) both pax6a and pax6b genes leading to a phenotype with smaller eyes and reduced central nervous system structures, e.g., neural tube, in comparison to control siblings The abnormal phenotype is present from early stages (24 hpf), and persists long after 48 hpf (B–D). Treated embryos die before day 4.
Characterization of pax6 morpholino models
While disruption of Pax6 function is known to be detrimental for normal development in mouse and human, little is known about the role of pax6 in zebrafish development. In zebrafish, there are two pax6 paralogues (or co-orthologs), pax6a and pax6b. They have partially overlapping expression patterns during early development. Previously, we characterized a pax6b missense mutation, sunrise (sri), which is homozygous viable and fertile (Kleinjan et al. 2008). The pax6bsri/sri embryos, as well as adults, have a mild phenotype with reduced lens size as the only obvious phenotype (Kleinjan et al. 2008). To improve the characterization of pax6 function during zebrafish development and to establish a suitable model for the validation of the in silico target gene results, we used microinjection of morpholino oligonucleotide(s) at the 1–2-cell stage (Nasevicius and Ekker 2000). The morpholinos were designed to target the translation start sites of pax6a (pax6aMO) and pax6b (pax6bMO), respectively (see Methods and Supplemental Table S2).
Specificity of the morpholinos was addressed by designing three distinct morpholinos against pax6a and 2 morpholinos against pax6b. For both genes, the phenotypes that resulted from the morpholino microinjections were highly similar, regardless of which gene-specific morpholino was used. In addition, we confirmed, by western-blot analysis, that a reduction of Pax6 protein level is observed in morpholino-injected embryos (Supplemental Fig. S5). Unfortunately, microinjection of even very low doses of pax6 mRNA induces abnormalities (Kleinjan et al. 2008); therefore, it was not possible to rescue the morpholino-induced phenotypes.
Embryos injected with pax6aMO or pax6bMO or both display small body size, reduced neural tube girth, morphologically abnormal brain, and small eyes. All aspects of these phenotypes were very robust, with >95% of the embryos exhibiting them (>>500 embryos were examined). The phenotype for pax6a knockdown is slightly more severe than for pax6b, and the simultaneous knockdown of both genes gives rise to a similar but even more severe phenotype (Fig. 2).
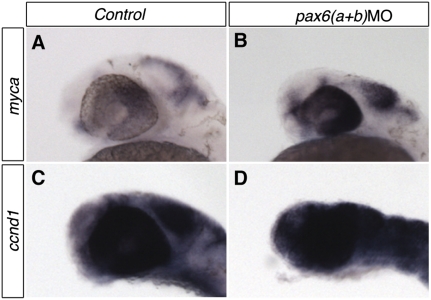
Since decreases in body-, brain-, and eye-size, were obvious phenotypes in the morpholino-injected embryos, we investigated the levels of apoptosis and cell proliferation within the affected tissues. No alteration in apoptosis levels (data not shown) was observed, using Tunel assays, but we found abnormally high maintenance of expression for two proliferation markers, myca and ccnd1, within the eye and brain (Fig. 3), suggesting that injection of the pax6 morpholinos gives rise to a proliferation defect that may secondarily result in aberrant differentiation.
Figure 3.
pax6a and pax6b knockdown causes abnormal maintenance of proliferation markers in the eye and CNS. (A,C) Normal expression of the proliferation markers myca and ccnd1 is down-regulated in the eye and CNS by 32 hpf. (B,D) In embryos co-injected with pax6aMO and pax6bMO, abnormal maintenance of expression of both markers is observed.
Validation of Pax6 target genes by whole-mount in situ analysis
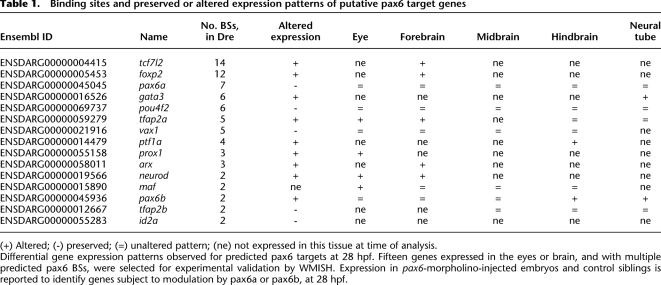
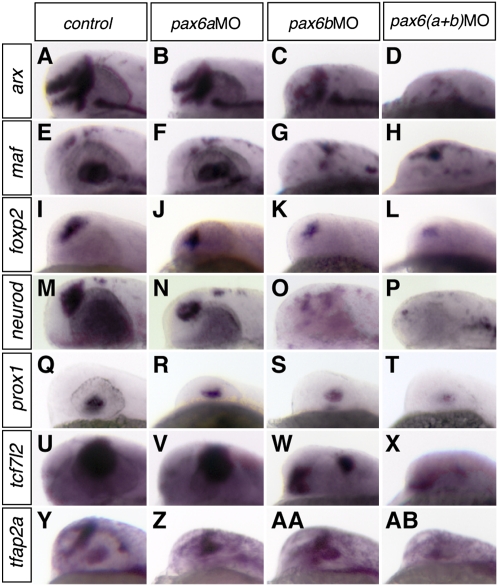
Using morpholino knockdown of pax6a, pax6b, or both in embryos, we set up a screen to validate experimentally some of the computationally predicted targets, using whole-mount in situ hybridization (WMISH) to assess gene expression. We used two criteria to choose 15 putative target genes for testing: (1) known expression in eye or brain, and (2) ≥2 predicted pax6 BSs at the gene locus (Table 1). As Pax6 is known to play an important role in early proliferation and differentiation, we chose to perform the screen at 28 hpf (hours post fertilization). Many neuronal cells, including those in the retina, are still proliferating at this stage. Retinal ganglion cells, the first retinal cell type to differentiate (Glass and Dahm 2004), have not yet begun to be generated. Not all predicted binding sites are expected to be demonstrable at this, or any other, single stage. Moreover, WMISH may not reveal subtle changes in gene expression in a subset of target cells within strongly expressing tissues. Of the 15 genes tested, 10 (66.6%) show altered expression in the morpholino-injected embryos compared to sibling controls at 28 hpf. Eye and forebrain are the structures most frequently affected, with seven genes showing disrupted expression (arx, maf, foxp2, neurod, prox1, tcf7l2, and tfap2a) (Fig. 4). Three predicted target genes showed altered neural tube expression—gata3, pax6b, and maf (Supplemental Fig. S6)—and two were midbrain/hindbrain region genes: ptf1a and pax6b (Supplemental Fig. S7). Among the set of genes with disturbed eye or forebrain expression, Arx and Neurod1 have been previously documented as direct or indirect PAX6 targets in the mouse embryonic cortex (Visel et al. 2007). Expression of maf in the lens disappears in the morphant embryos because the structure is not present anymore. Tfap2a and Pax6 have been reported to co-regulate lens development (Makhani et al. 2007) and, with Prox1, they were shown to modulate Sox2 expression (Lengler et al. 2005). Pax6 misexpression is known to induce Tcf7l2 in the diencephalon (Matsunaga et al. 2000), and conversely, Tcf7l2 (previously named Tcf4) diencephalon expression was found to be lost in Pax6-null mice at E11.5 (Cho and Dressler 1998). These earlier data support the validity of our method, but the majority of our direct target predictions (Supplemental Tables S5–S8) is novel. For example, the prediction of multiple BSs in the region of the foxp2 gene and the observed alteration of expression in the tectum of morpholino-injected embryos provide new evidence that pax6 plays a key role in regulating critical neural function genes. In zebrafish, early foxp2 expression is observed predominantly in the telencephalon, but it is also seen at 36 hpf in the anterior commissure (Bonkowsky and Chien 2005), an interhemispheric connection that is absent in a high proportion of aniridia patients with known heterozygous PAX6 mutations (Sisodiya et al. 2001).
Table 1.
Binding sites and preserved or altered expression patterns of putative pax6 target genes
Figure 4.
pax6a and pax6b knockdown disrupts the expression of putative target genes in the eye and brain. Of the 15 putative targets analyzed by WMISH, seven display altered expression within the eye or brain, at 28 hpf: (A–D) arx, (E–H) maf, (I–L) foxp2, (M–P) neurod, (Q–T) prox1, (U–X) tcf7l2, (Y–AB) tfap2a. The loss of expression of maf in the eye, B, illustrates that loss of tissue (the lens) can lead to observed loss of gene expression.
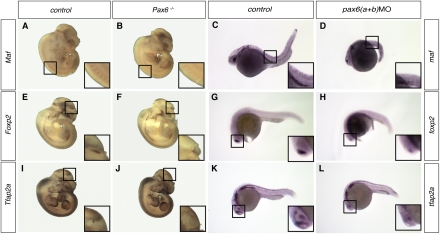
The design of the selection process, used to define the Pax6 targets predicted here, ensures that the BSs lie in evolutionarily conserved elements. Therefore, we set out to study some of the target genes that we had tested in zebrafish in the mouse, where null mutants are available. Having noted the loss of Tcf7l2 diencephalon expression in homozygous Smalleye (Pax6−/−) mice (Cho and Dressler 1998), we set out to investigate the expression pattern of three other genes, Maf, Foxp2, and Tcfap2a, at E11.5, the mouse developmental stage equivalent to 28 hpf in zebrafish. The severity of the abnormalities in Pax6-null mice is generally greater than seen in the double morpholino-treated zebrafish. For example, the eye is absent in mutant mice. We found the Maf expression level reduced in the neural tube of Pax6−/− mice, just as it is in pax6abMO zebrafish (Fig. 5A–D). Absence of Foxp2 expression from the dorsolateral telencephalon (Fig. 5E–H) and of Tcfap2a from the lateral diencephalic prosomere 1 was seen in Pax6−/− embryos (Fig. 5I–L), mirroring the severe reduction of foxp2 and tfap2 expression in the forebrain of pax6MO fish embryos.
Figure 5.
pax6a and pax6b disruption causes similar expression pattern changes in mouse and zebrafish. pax6a and pax6b morpholino knockdown in zebrafish and Pax6 null mutation in the mouse lead to similar disturbance in expression pattern. (A–D) Maf (maf) expression in the neural tube is reduced in both species when Pax6 (pax6a and pax6b) function is disrupted. (E–H) The prominent lateral telencephalon expression domain of Foxp2 (foxp2) is severely reduced. (I–L) Tfap2a (tfpa2) expression in prosomere 1 is abolished.
Altered expression of a predicted target gene in a knockdown or null mutant embryo does not demonstrate that the gene is a direct target. The specificity of the regulatory alteration is, however, emphasized by the fact that only some expression sites are affected. For example, pax6MO zebrafish neurod expression is turned off in the forebrain and eye but remains unaltered in the lateral line placodes and in the pancreas bud (Supplemental in situ data).
Validation of Pax6 binding sites by chromatin immunoprecipitation and reporter transgenesis
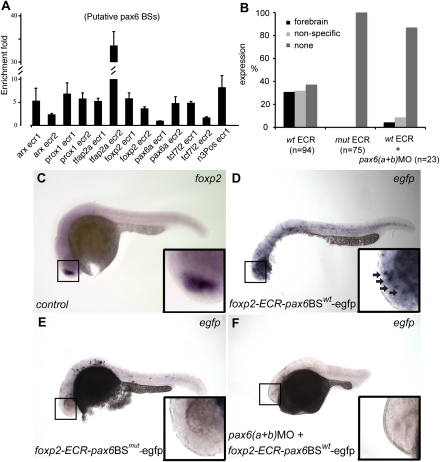
The expression pattern changes resulting from pax6 disruption, together with the presence of evolutionarily conserved Pax6 BSs, provide strong circumstantial evidence for direct regulation of target genes by Pax6. To confirm that the regulation is direct, we performed ChIP, using pax6-specific antibodies with chromatin from 28-hpf zebrafish embryos. We selected two putative pax6 BSs for each of five genes that show differential expression in pax6MO and control embryos, plus two novel predicted BSs within the pax6a locus. As a positive control, we used the pax6 BS in the sox2 enhancer N3 (Inoue et al. 2007) and, as negative control, a similar-sized sequence, 1 kb upstream of N3, that we call N3neg. We also chose to study two predicted pax6a enhancers, because Pax6 is known to auto-regulate (Kleinjan et al. 2004; Manuel et al. 2007), and we wanted to show that genes can be direct targets even if they do not show a qualitative difference in our limited WMISH screen. Of the 12 BSs tested, one of the pax6a predicted BSs did not show any enrichment. All the others showed enrichment that ranged from almost twofold to 32-fold. These results demonstrate that the combined procedure is highly accurate (Fig. 6A).
Figure 6.
pax6 directly regulates the novel predicted enhancers. (A) Immunoprecipitation of chromatin from 28-hpf zebrafish embryos, using anti-Pax6 antibodies and control rabbit IgG, was followed by real time PCR to confirm enrichment for predicted BSs. The Pax6 in vivo occupancy is site-dependent, leading to enrichment values from two- to 32-fold (y-axis). The x-axis shows each tested Pax6 BS (ECR1 and ECR2 for six different target genes). (B) Summary expression data for the reporter transgenic zebrafish embryos for foxp2 ECR1: wild type (wt), mutant (mut), and wild type with pax6a and pax6b knockdown (wt+pax6(a+b)MO), showing specific forebrain, nonspecific, or no expression. (C) foxp2 WMISH showing prominent expression in forebrain at 28 hpf. (D) foxp2 ECR1 wt reporter study employing EGFP WMISH to reveal signal in subset of cells showing full foxp2 expression in C. (E) Absence of forebrain EGFP expression when reporter driven by foxp2 ECR1 mutated at the pax6 binding site. (F) Absence of EGFP reporter expression when pax6(a+b) expression is down-regulated by double morpholino co-injection.
Finally, we examined functionally the role of the most interesting and novel Pax6 target predicted, foxp2, using transient reporter transgenics, in zebrafish. foxp2 ECR1 gave a robust pattern of reporter expression at 28 hpf (Fig. 6B,D) when EGFP expression was examined by WMISH. The pattern was consistent with the site of expression of foxp2 in the brain (Fig. 6C,D) and was disrupted either by mutation of the predicted pax6 BS (Fig. 6E) or by co-injection of a mixture of pax6aMO with pax6bMO (Fig. 6F).
Discussion
We have developed a novel integrated procedure to characterize the core architecture of transcriptional networks. While previous studies have relied on the use of position weight matrices (Stormo 2000) to identify BSs, we decided to use profile HMM instead. HMMs allow abstraction of the probability distribution of the nucleotides at each position, while PWMs are based on the nucleotide distribution at each BSs site separately, which incorrectly assumes that these are independent. In addition, HMMs are also better than PWMs at identifying matches with small indels or partial matches (Marinescu et al. 2005), and this capability has the potential to be useful for the identification of noncanonical BSs. Moreover, we have shown that for Pax6, the HMM approach that we have taken gives more stringent results than a similar PWM-based approach, using patser (Hertz and Stormo 1999). This high level of stringency is the most adequate for use in transcriptional studies focused on evolutionary transcriptional conservation across relatively large evolutionary distances, such as from zebrafish to mammals. This is based on the knowledge that transcriptional conservation is evolutionarily relatively fast. For these reasons, studies such as ours are very useful for highlighting the core of these networks, because they are specifically known to be under higher positive selection pressures (Odom et al. 2007; Schmidt et al. 2010).
The possibility of identifying transcription factor BSs quickly and defining the enhancers in which they reside and the genes that are directly regulated by a given DNA binding transcription factor, in combination with other types of data, such as gene expression profiles, allows us to reconstruct molecular networks and identify new key modulators of development and disease. The procedure is based on evolutionary conservation, and it is spatiotemporally independent and can be used to define the genetic framework in a wide range of experimental studies. Our aim was to identify enhancers linked to genes directly regulated by Pax6, a protein whose function is essential for development and which is disrupted in distinct forms of human disease. The identification of a full spectrum of target genes will provide a better understanding of the diversity of developmental processes in which Pax6 functions and also directly pinpoint enhancers regulating target genes whose disruption could potentially give rise to human disease or disease susceptibility.
The evolutionarily conserved genomic loci analyzed in this Pax6 target screen were restricted to sequence windows including genes and +/− 20 kb flanking each gene. These distances were selected for efficient scanning that covers ∼80% of all intergenic regions (Supplemental Fig. S8). It is also well established that a high proportion of regulatory elements for one gene lie within one or more introns of neighboring genes (Becker et al. 2007). It has also been estimated in genome-wide association studies, carried out by measuring allelic expression in lymphoblastoid cell lines, that <5% of eQTLs marked by SNPs lie >20 kb upstream of transcription start sites (Veyrieras et al. 2008).
The screen has identified hundreds of novel putative Pax6 BSs and the associated target genes. The gene sets are enriched for transcription factor activity and the human/zebrafish conserved set for neurogenesis. In addition, there is enrichment for genes expressed in eye and brain, particularly between zebrafish and human. The differences between the human and mouse sets in the relative enrichment for eye-expressed genes are due to human eye genes that may have mouse orthologues but are not present in the mouse set of targets. This is probably due to a combinatorial effect of evolutionary divergence of tissue-specific transcriptional regulation (Odom et al. 2007; Schmidt et al. 2010) and specific evolutionary pressures on the core Pax6 network resulting from the closer similarity in visual function between human and zebrafish than with the mouse (Fadool and Dowling 2008). In spite of these species differences, the results are in agreement with known Pax6 functions, both as a selector gene and as a gene essential for both proliferation and differentiation during development, particularly for the neuronal lineage.
Owing to the lack of available null mutations for pax6a and pax6b in zebrafish, we used morpholino oligonucleotides to induce pax6 knockdown. Injection into early fertilized oocytes results in embryos with smaller eyes and reduced brain- and neural tube-phenotypes that are similar to the mouse Smalleye model and also fit in with observations in human PAX6 haploinsufficiency syndromes and very rare compound heterozygotes (van Heyningen and Williamson 2002). In zebrafish, these outcomes may be the consequence of a proliferation defect that might also result in abnormal differentiation, since cells acquire distinct fates, depending on how long they are exposed to morphogens (Dessaud et al. 2007). The prolonged maintenance of proliferation may result in increased exposure to morphogenetic cues, which can impact on the cell fate distribution (Duparc et al. 2007). This might be a factor in the etiology of the human PAX6 mutant phenotype as well.
To validate the results of our Pax6 target identification procedure experimentally, we chose 15 genes expressed in the eye or brain, which are associated with multiple predicted pax6 BSs. Ten of these genes (66%) showed altered expression in pax6MO embryos compared with controls at 28 hpf, a stage that precedes retinal differentiation. Three of these genes were also assessed in E11.5 Pax6−/− mouse embryos where homozygous null mice survive to birth. For all three genes, we found equivalent expression pattern changes in the zebrafish and mouse models upon Pax6 disruption. Together with the published Pax6-dependent regulation of predicted target gene Tcf7l2 (Cho and Dressler 1998), the collected data allow us to conclude that our target selection procedure is highly efficient in finding Pax6 regulated genes and simultaneously delivers (part of) the evolutionarily conserved regulatory network that centers around this important selector gene.
To demonstrate direct regulation, we were able to show, using ChIP with chromatin from 28-hpf zebrafish embryos, that pax6 binds to selected targets, namely nine predicted enhancers (corresponding to five validated genes) and to a novel predicted enhancer from the pax6a locus. We found that with the exception of one of the pax6a predicted enhancers, all of the others tested bound Pax6, with up to 32-fold enrichment. The pax6a results allow us to highlight that our validation screen is a qualitative screen, and the experimental validation is stage-dependent. Thus, although Pax6 binds one tested pax6a enhancer, no alteration in the overall pax6a expression pattern was detected in pax6MO embryos. Furthermore, the genes that do not show altered expression patterns at 28 hpf may be active targets of pax6 at different stages, including in adult tissues.
For Foxp2, the major novel Pax6 target, we complemented the ChIP experiments by generating and analyzing transient reporter transgenics in zebrafish with the fopx2 ECR1 enhancer. This element drives expression in the forebrain, in a region topologically equivalent to where Foxp2 is expressed (Fig. 5E) and overlaps with Pax6 expression (Theil et al. 1999). This reporter transgenic expression pattern can be disrupted by either morpholino-driven down-regulation of pax6 or by mutation of the putative ECR1 pax6 BS. Thus, the HMM approach developed here for target binding site identification is very successful in identifying cis-elements in core vertebrate transcriptional networks.
Uncovering direct interactions between regulatory genes and their downstream targets is a vital step toward the discovery of the gene regulatory networks that control development, patterning, and maintenance of organ function. Here, we have used a combined strategy of bioinformatic target prediction and both in vivo and in vitro validation, using two different animal models, zebrafish and mouse, to reveal new players in the core networks involving the developmental regulator Pax6. A number of genes that are found on the target lists (Supplemental Tables S5–S8) are well-established interactors or documented targets of Pax6. These include Pax2 (Schwarz et al. 2000), Dach1, Six3/Six6, Eya1 (Purcell et al. 2005), and Nr2e1 (Schuurmans et al. 2004). In addition, while this work was in progress, Hoxd4-driven antero-posterior patterning in mouse and zebrafish was shown to be directly regulated by Pax6/pax6 (Nolte et al. 2006). Similarly, a regulatory region of Six6 was shown to be directly bound by Pax6 (Tetreault et al. 2009). Regulation of Ctnnd2 by Pax6 was also reported (Zhang et al. 2010). Moreover, in some instances, predicted targets have been described as upstream regulators of Pax6; for example, binding of homeodomain proteins Meis1/2 is required for Pax6 expression in lens development (Zhang et al. 2002), while Pbx1 binding is additionally needed in murine (Zhang et al. 2006) and zebrafish (Delporte et al. 2008) pancreas. It is intriguing and instructive to find that these genes are also predicted targets for Pax6 binding (Supplemental Tables S5–S8), since there are many instances emerging where reciprocal regulation is observed in transcription factor networks (Kondoh and Kamachi 2010). One other important aspect to be taken in consideration is that, although our screen is restricted to a +/− 20-kb region (that is, a 40-kb distance between genes), in some cases we have identified enhancers that are further away than that within neighboring loci, e.g., ELP4 and WWOX harbor enhancers for PAX6 and MAF, respectively (Jamieson et al. 2002; Kleinjan et al. 2008). Knowing this, we have complemented our human target list with the two proximal and distal neighboring genes (Supplemental Table S5).
The reliability of the list of predicted targets is also emphasized by the significant frequency with which the genes appearing in Supplemental Tables S5–S8 are found in reports of altered gene expression studies describing comparison of the transcriptomes in Pax6-null and wild type mice (Holm et al. 2007; Visel et al. 2007). Although such lists include indirect targets, clearly the coincidence of their appearance on these lists increases our confidence in accepting them as real binding targets for Pax6. Another plausible subset of target genes found in Supplemental Tables S5–S8 includes genes with a known role in human eye or neurological disease (e.g., choroideremia: CHM, retinoschisis: RS1, mental retardation: ARX, glaucoma with associated anomalies: LMX1B) (OMIM—http://www.ncbi.nlm.nih.gov/omim), or with other relevant phenotypes in model systems (e.g., Arx, Barhl2, Emx2, Gbx2, Gli3, Lmx1b, Mab21l2, Neurod1, Nr2e1, Nkx2-2, Pou3f2, Pou4f2, Six6, Vax1, Zic3) (MGI— http://www.informatics.jax.org/). Disease and mouse phenotype reference numbers in OMIM and MGI are shown in Supplemental Table S9.
Finally, it is important to remember that this HMM approach only identifies a subset of predicted transcription factor binding sites. As discussed, some of the 29 identified known Pax6 binding sites were excluded by the strategy of selecting similar enough sites to allow the generation of the HMM. Furthermore, there are some additional known sites, such as the three well-defined Pax6-Sox2 co-binding sites (Kondoh and Kamachi 2010), each of which contains a specified distinct Pax6-binding sequence. These are so divergent from each other that they could not produce an HMM.
These results will form the basis for further exploration of the many essential roles of PAX6 in development and human disease. For example, one of the novel targets, FOXP2, is a protein known to play a key role in language and speech functions (Fisher and Scharff 2009). Interestingly, PAX6 haploinsufficiency has been associated with structural and functional brain anomalies and learning disability (Heyman et al. 1999; Sisodiya et al. 2001; Bamiou et al. 2007; Graziano et al. 2007; Maekawa et al. 2009) and with behavioral and functional brain changes in rodents, too (Tuoc et al. 2009; Umeda et al. 2010). Another of the targets, TCF7L2, is associated with Type 2 diabetes (Grant et al. 2006; Helgason et al. 2007; Scott et al. 2007). In addition to the previously discussed diencephalon co-expression, both TCF7L2 and PAX6 are also expressed in the endocrine pancreas and involved in the regulation of insulin expression and maintenance of glucose levels. Using the recent information about open chromatin sites in human islet tissue (Gaulton et al. 2010), we were able to show that three of the 12 sites identified in the TCF7L2 region were among our Pax6 target predictions for this gene (data not shown). These observations illustrate the power of transcription factor target screens for the discovery and extension of regulatory networks.
Methods
Computational predictions
Manipulation of tools and data
The manipulation of data and automation of bioinformatic tools was performed using Perl scripts, developed in-house and available in the Supplemental Material.
Sequence retrieval and orthology definition
The gene orthology data between H. sapiens, M. musculus, and D. rerio, with description and annotation of respective genomic locus coordinates, were retrieved from Ensembl, using the Biomart tool (http://www.ensembl.org/biomart/). Extended gene locus sequence, spanning from 20 kb upstream of to 20 kb downstream from each gene, was retrieved from Ensembl, using Perl scripts and the Ensembl Perl API (http://www.ensembl.org/info/data/api.html). The Ensembl assemblies used were the human NCBI36, the mouse NCBIM37, and the zebrafish Zv7danRer5.
Identification and retrieval of evolutionarily conserved regions
To identify sequences that are evolutionarily conserved between pairs of orthologous gene loci, interspersed repeats and low complexity DNA sequences were first masked, using RepeatMasker (Smit et al. 1996–2004). The masked sequences were then blasted against each other with BLASTZ (Schwartz et al. 2003) using the following conditions: H=2200, T=0, W=6, K=2200. This allowed us to define the coordinates for conserved regions that were then used locally to extract the respective sequences.
Generation of hidden Markov models (HMMs) for the Pax6 binding site
Pax6 bibliography was mined to extract experimentally validated binding sites for the Pax6(-5a) isoform (references are available in the Supplemental Material) We found a total of 29, of which 16 could be used to make species-specific forward and reverse complemented HMMs, using the HMMER software package (Eddy 1998). The models were adjusted for species specificity to incorporate any differences in nucleotide frequency between species. This was accomplished by generating null models that contain such information for each one of the three species.
The selection of the 16 binding sites for generation of the models was achieved by an iterative procedure that used HMMs produced from the initial set of similar binding sites and identifying others by similarity. The iterative procedure stopped when none of the remaining sequences had significant sequence similarity to the ones already selected, as assessed by manual inspection.
Generation of position weight matrices (PWMs) for the Pax6 binding site
We used the sequence information from the 16 experimentally validated pax6 binding sites to define the PWM according to the nucleotide occurrence at each position. A Perl script was used to do this (see Supplemental Material).
Comparison of PWM- and HMM-based procedures
The genomic regions that are conserved between the species' pairs that were analyzed were screened using hmmsearch (HMMER) and patser, using the pax6 HMM and pax6 PWM, respectively. For the HMM data, we split the data according to the threshold that can be determined by screening known BSs with the model. We then calculated percentile for all the results data sets and compared them using Perl scripts to identify co-identification of the same putative binding sites by both methods. The heatmap was generated using the heatmap.2 function of R (http://www.r-project.org/).
Identification of putative pax6 binding sites and of putative direct gene targets
The conserved sequences were screened using the pax6 HMM models in order to identify putative binding sites. The score threshold used was species-specific and defined for each model as the minimum score within the sequences that were used to generate the models. To select the putative binding sites further, we removed those that overlapped exons, and used megablast (Zhang et al. 2000) on the remaining ones to identify those at the center of a 100-bp highly conserved sequence interval (with at least 70% sequence similarity to an equivalent interval in its respective orthologous locus in the other species).
The identification of the gene targets was based on the association of the predicted binding site with the extended locus in which it is found.
Gene ontology over-representation
The gene ontology characterization of the target sets was performed using the default parameters from g:Profiler (Reimand et al. 2007), as implemented in the web site, http://biit.cs.ut.ee/gprofiler/, providing as input the sets of mouse or human genes predicted to be directly regulated by PAX6.
Gene expression characterization
The computational characterization of gene expression was performed using the UniGene expression data (available from files in ftp://ftp.ncbi.nih.gov/repository/UniGene/). The Ensembl gene IDs were converted into UniGene IDs using the Biomart tool from Ensembl (http://www.ensembl.org/), and these were used to parse the expression data. We used a Monte-Carlo approach, with 5000 test sets for each species, to determine the statistical significance of the results.
Analysis of the coverage of intergenic regions
For any of the genes that are conserved between either zebrafish and human, or zebrafish and mouse, or for all the genes in any of the three species, we identified the distance between them and the nearest proximal or distal gene. This was performed using gene data downloaded from Ensembl, using the Biomart tool and Perl scripts developed in-house.
Zebrafish embryo collection
General maintenance, collection, and staging of zebrafish were carried out according to The Zebrafish Book (Westerfield 2000). The approximate stages are given in hours post fertilization (hpf) at 28°C and are determined according to morphological criteria.
Zebrafish whole mount in situ hybridization
Whole-mount in situ hybridization (WMISH) reactions were carried out according to published protocols (Thisse et al. 1993), at 28 hpf. The riboprobes for the putative target genes were transcribed from PCR templates, amplified from genomic DNA using the oligonucleotides shown in Supplemental Table S1.
Mouse expression pattern analysis
Whole-mount in situ hybridization reactions were carried out according to published protocols (Wilkinson 1992), using fetuses at 11.5-d post coitum (E11.5). The riboprobes for the putative target genes were transcribed from DNA amplicons defined for Eurexpress II (http://www.eurexpress.org/). These were: Foxp2–T9351, Tcfap2a–T6939, and Maf –T40542.
Antisense morpholino oligonucleotide injections
The pax6a and pax6b antisense morpholino oligonucleotides (MO) (Gene Tools) were directed against the 5′ sequence near the start of translation (Supplemental Table S2). The embryos were injected through the chorion of 1 to 2-cell stage embryos with a volume of 1.4 nl to deliver a mass of 3.5 ng. For each MO, at least 100 embryos were injected, and we ascertained that the phenotype was fully penetrant and of consistent severity. Co-injection of pax6a and pax6b morpholinos was also performed, using the same final masses. Standard control morpholino-injected embryos did not differ from wild type uninjected embryos.
Chromatin immunoprecipitation and QPCR
Pax6 ChIP was performed using the protocol described by Sansom and coworkers (Sansom et al. 2009), using 28-hpf zebrafish embryos for the preparation of chromatin. The antibodies used were: Pax6 antibody (B5790; Abcam) and rabbit control IgG - ChIP Grade (ab46540; Abcam). The only modification was to dissociate embryonic cells mechanically before fixing them for 30 min.
The relative occupancy values for both Pax6 and IgG ChIPs were calculated by determining the apparent IP efficiency (ratios of the amount of ChIP-enriched DNA over that of the input sample) and normalized to the level observed at a control region (N3 neg—1 kb upstream of the sox2 enhancer N3), which was defined as 1.0. The fold-enrichment was calculated by normalizing the relative occupancy for Pax6 with the relative occupancy for IgG. The primers used are shown in Supplemental Table S3.
Zebrafish transient reporter transgenesis
The zebrafish transient reporter transgenics were generated as described in Kleinjan et al. (2008), using the Tol2-2way system with a reporter cassette containing the zebrafish gata2 minimal promoter, GFP and a poly A, adapted from a similar construct from the Becker laboratory (Navratilova et al. 2009). A putative 385-bp enhancer fragment was PCR-amplified from zebrafish genomic DNA and cloned into the Gateway P4P1r entry vector using the following primers:
foxp2_attB4: 5′-aggggacaactttgtatagaaaagttggcgcgccatgacttttacagttgcagc-3′
foxp2_attB1r: 5′-aggggactgcttttttgtacaaacttgtgtctctggtcacccaagca-3′
The predicted Pax6 binding site in the foxp2 enhancer (5′-atggaatttacagtgcgcctccagtagtaactccattcgg-3′) was mutated using the Quickchange II site-directed mutagenesis kit (Stratagene) with the following mismatch oligonucleotides: 5′-atggaatttacagtgctacaccagtagtaactccatttcccatcgg-3′ and 5′-ggaaatggagttactactggtgtagcactgtaaattccatacagcca-3′.
Western blot analysis
Ice-cold Ringer's solution with “complete protease inhibitor cocktail” tablets (Roche) was added to the zebrafish embryos. The embryos were homogenized, using a Polytron PT 3100 system (Thermo Fisher Scientific; setting 27,000 rpm, 30 sec), and the homogenates were centrifuged at 13,000 rpm for 1 min. The supernatants were collected and concentrated using Microcon YM-3 columns (Millipore).
Protein concentration was measured using the Bradford assay (Bio-Rad) and NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific).
The proteins were resolved using a 4%–12% Nu-PAGE gel and electroblotted onto Hybond-P PVDF membrane (Amersham/ GE Healthcare). The PVDF membrane was blocked in 5% nonfat milk in PBST (phosphate-buffered saline-Tween 20; 3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, 0.05% Tween 20, pH 7.4) overnight at 4°C. The membrane was probed with anti-PAX6 antibody (AB5790; Abcam) for 1 h (1:5000 dilution in 1× TBS), followed by donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase (GE Healthcare) (1:5000 dilution in 1× TBS). Immunoreactive protein bands were detected with enhanced chemiluminescence reagent (ECL) according to the manufacturer's instructions (Amersham Pharmacia Biotech). Loading was checked by stripping the blot with Restore Stripping Buffer (Pierce 21059) and reprobing using anti-beta actin (Sigma A5441), followed by HRP goat anti-mouse (SC2064; Santa Cruz Biotechnology) secondary antibody.
Acknowledgments
Some of this work was supported by a Marie Curie award to P.C. (FP6-MEIF-CT-2005-025389). K.J.C. was funded by the EVI-GENORET European consortium. We wish to thank Fernando Casares (CABD [Centro Andaluz de Biologia del Desarrollo]) for the ccnd1 and myca RNA probes, Paul Perry (MRC-HGU) for imaging support, and Susan Lynas (MRC-HGU) and Keith Erskine (MRC-HGU) for technical help. We appreciated the support and the opportunity for stimulating discussions with Nick Hastie and Richard Baldock.
Authors' contributions: P.C., S.P., S.B., D.A.K., and V.v.H. conceived and designed experiments and analyzed the data. All the bioinformatics work was designed and carried out by P.C. K.J.C. carried out the western blot work and helped optimize the ChIP protocol. S.P. analyzed the cell cycle and apoptosis, and P.C. performed the morpholino experiments, the in silico and “wet” screens, the phenotypic characterization, and the Pax6 ChIP and real time PCR. P.C. and V.v.H. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124115.111.
References
- Ashery-Padan R, Marquardt T, Zhou XL, Gruss P 2000. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev 14: 2701–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Zhou XL, Marquardt T, Herrera P, Toube L, Berry A, Gruss P 2004. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol 269: 479–488 [DOI] [PubMed] [Google Scholar]
- Bamiou DE, Free SL, Sisodiya SM, Chong WK, Musick F, Williamson KA, van Heyningen V, Moore AT, Gadian D, Luxon LM 2007. Auditory interhemispheric transfer deficits, hearing difficulties, and brain magnetic resonance imaging abnormalities in children with congenital aniridia due to PAX6 mutations. Arch Pediatr Adolesc Med 161: 463–469 [DOI] [PubMed] [Google Scholar]
- Becker TS, Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, et al. 2007. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 17: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowsky JL, Chien CB 2005. Molecular cloning and developmental expression of foxP2 in zebrafish. Dev Dyn 234: 740–746 [DOI] [PubMed] [Google Scholar]
- Brill MS, Snapyan M, Wohlfrom H, Ninkovic J, Jawerka M, Mastick GS, Ashery-Padan R, Saghatelyan A, Berninger B, Gotz M 2008. A Dlx2-and Pax6-dependent transcriptional code for periglomerular neuron specification in the adult olfactory bulb. J Neurosci 28: 6439–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BK, Zhang WY, Cveklova K, Kantorow M, Cvekl A 2002. Identification of differentially expressed genes in mouse Pax6 heterozygous lenses. Invest Ophthalmol Vis Sci 43: 1884–1890 [PMC free article] [PubMed] [Google Scholar]
- Cho EA, Dressler GR 1998. TCF-4 binds beta-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech Dev 77: 9–18 [DOI] [PubMed] [Google Scholar]
- Davis J, Duncan MK, Robison WG, Piatigorsky J 2003. Requirement for Pax6 in corneal morphogenesis: A role in adhesion. J Cell Sci 116: 2157–2167 [DOI] [PubMed] [Google Scholar]
- Davis-Silberman N, Kalich T, Oron-Karni V, Marquardt T, Kroeber M, Tamm ER, Ashery-Padan R 2005. Genetic dissection of Pax6 dosage requirements in the developing mouse eye. Hum Mol Genet 14: 2265–2276 [DOI] [PubMed] [Google Scholar]
- Del Bene F, Ettwiller L, Skowronska-Krawczyk D, Baier H, Matter JM, Birney E, Wittbrodt J 2007. In vivo validation of a computationally predicted conserved Ath5 target gene set. PLoS Genet 3: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delporte FM, Pasque V, Devos N, Manfroid I, Voz ML, Motte P, Biemar F, Martial JA, Peers B 2008. Expression of zebrafish pax6b in pancreas is regulated by two enhancers containing highly conserved cis-elements bound by PDX1, PBX, and PREP factors. BMC Dev Biol 8: 53 doi: 10.1186/1471-213X-8-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J 2007. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature 450: 717–720 [DOI] [PubMed] [Google Scholar]
- Duparc RH, Abdouh M, David J, Lepine M, Tetreault N, Bernier G 2007. Pax6 controls the proliferation rate of neuroepithelial progenitors from the mouse optic vesicle. Dev Biol 301: 374–387 [DOI] [PubMed] [Google Scholar]
- Eddy SR 1998. Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V 1999. Role of Pax6 in development of the cerebellar system. Development 126: 3585–3596 [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J 1997. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90: 169–180 [DOI] [PubMed] [Google Scholar]
- Estivill-Torrus G, Vitalis T, Fernandez-Llebrez P, Price DJ 2001. The transcription factor Pax6 is required for development of the diencephalic dorsal midline secretory radial glia that form the subcommissural organ. Mech Dev 109: 215–224 [DOI] [PubMed] [Google Scholar]
- Fadool JM, Dowling JE 2008. Zebrafish: A model system for the study of eye genetics. Prog Retin Eye Res 27: 89–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Scharff C 2009. FOXP2 as a molecular window into speech and language. Trends Genet 25: 166–177 [DOI] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. 2010. A map of open chromatin in human pancreatic islets. Nat Genet 42: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass AS, Dahm R 2004. The zebrafish as a model organism for eye development. Ophthalmic Res 36: 4–24 [DOI] [PubMed] [Google Scholar]
- Goldsmith P, Harris WA 2003. The zebrafish as a tool for understanding the biology of visual disorders. Semin Cell Dev Biol 14: 11–18 [DOI] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I 2010. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res 20: 565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SFA, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. 2006. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38: 320–323 [DOI] [PubMed] [Google Scholar]
- Graziano C, D'Elia AV, Mazzanti L, Moscano F, Guidi SG, Scarano E, Turchetti D, Franzoni E, Romeo G, Damante G, et al. 2007. A de novo nonsense mutation of PAX6 gene in a patient with aniridia, ataxia, and mental retardation. Am J Med Genet A 143A: 1802–1805 [DOI] [PubMed] [Google Scholar]
- Helgason A, Palsson S, Thorleifsson G, Grant SFA, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen YX, Chen GJ, Reynisdottir I, et al. 2007. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 39: 218–225 [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15: 563–577 [DOI] [PubMed] [Google Scholar]
- Heyman I, Frampton I, van Heyningen V, Hanson I, Teague P, Taylor A, Simonoff E 1999. Psychiatric disorder and cognitive function in a family with an inherited novel mutation of the developmental control gene PAX6. Psychiatr Genet 9: 85–90 [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BLM, Ton CCT, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V 1991. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354: 522–525 [DOI] [PubMed] [Google Scholar]
- Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M, Gotz M 2007. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci 34: 99–119 [DOI] [PubMed] [Google Scholar]
- Imanishi Y, Li G, Sokal I, Sowa ME, Lichtarge O, Wensel TG, Saperstein DA, Baehr W, Palczewski K 2002. Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur J Neurosci 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Kamachi Y, Matsunami H, Imada K, Uchikawa M, Kondoh H 2007. PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development. Genes Cells 12: 1049–1061 [DOI] [PubMed] [Google Scholar]
- Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, et al. 2002. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis, and coloboma. Hum Mol Genet 11: 33–42 [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Childs AJ, van Heyningen V 2004. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev Biol 265: 462–477 [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, Seawright A, Hever AM, Yeyati PL, van Heyningen V, et al. 2008. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet 4: e29 doi: 10.1371/journal.pgen.0040029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H, Kamachi Y 2010. SOX-partner code for cell specification: Regulatory target and underlying molecular mechanisms. Int J Biochem Cell Biol 42: 391–399 [DOI] [PubMed] [Google Scholar]
- Lengler J, Bittner T, Munster D, Gawad AEA, Graw J 2005. Agonistic and antagonistic action of AP2, Msx2, Pax6, Prox1, and Six3 in the regulation of Sox2 expression. Ophthalmic Res 37: 301–309 [DOI] [PubMed] [Google Scholar]
- Maekawa M, Iwayama Y, Nakamura K, Sato M, Toyota T, Ohnishi T, Yamada K, Miyachi T, Tsujii M, Hattori E, et al. 2009. A novel missense mutation (Leu46Val) of PAX6 found in an autistic patient. Neurosci Lett 462: 267–271 [DOI] [PubMed] [Google Scholar]
- Makhani LF, Williams T, West-Mays JA 2007. Genetic analysis indicates that transcription factors AP-2 alpha and Pax6 cooperate in the normal patterning and morphogenesis of the lens. Mol Vis 13: 1215–1225 [PubMed] [Google Scholar]
- Mann RS, Carroll SB 2002. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev 12: 592–600 [DOI] [PubMed] [Google Scholar]
- Manuel M, Georgala PA, Carr CB, Chanas S, Kleinjan DA, Martynoga B, Mason JO, Molinek M, Pinson J, Pratt T, et al. 2007. Controlled overexpression of Pax6 in vivo negatively auto-regulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development 134: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu VD, Kohane IS, Riva A 2005. MAPPER: A search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics 6: 79 doi: 10.1186/1471-2105-6-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P 2001. Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105: 43–55 [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Araki I, Nakamura H 2000. Pax6 defines the di-mesencephalic boundary by repressing En1 and Pax2. Development 127: 2357–2365 [DOI] [PubMed] [Google Scholar]
- Mitchell TN, Free SL, Williamson KA, Stevens JM, Churchill AJ, Hanson IM, Shorvon SD, Moore AT, van Heyningen V, Sisodiya SM 2003. Polymicrogyria and absence of pineal gland due to PAX6 mutation. Ann Neurol 53: 658–663 [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC 2000. Effective targeted gene “knockdown” in zebrafish. Nat Genet 26: 216–220 [DOI] [PubMed] [Google Scholar]
- Navratilova P, Fredman D, Hawkins TA, Turner K, Lenhard B, Becker TS 2009. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev Biol 327: 526–540 [DOI] [PubMed] [Google Scholar]
- Nolte C, Rastegar M, Amores A, Bouchard M, Grote D, Maas R, Kovacs EN, Postlethwait J, Rambaldi I, Rowan S, et al. 2006. Stereo specificity and PAX6 function direct Hoxd4 neural enhancer activity along the antero-posterior axis. Dev Biol 299: 582–593 [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrin EJ, Li YM, Hoffman K, Liu J, Wang KQ, Zhang L, Mardon G, Chen R 2006. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res 16: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell P, Oliver G, Mardon G, Donner AL, Maas RL 2005. Pax6-dependence of Six3, Eya1, and Dach1 expression during lens and nasal placode induction. Gene Expr Patterns 6: 110–118 [DOI] [PubMed] [Google Scholar]
- Ramialison M, Bajoghli B, Aghaallaei N, Ettwiller L, Gaudan S, Wittbrodt B, Czerny T, Wittbrodt J 2008. Rapid identification of PAX2/5/8 direct downstream targets in the otic vesicle by combinatorial use of bioinformatics tools. Genome Biol 9: R145 doi: 10.1186/gb-2008-9-10-r145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J 2007. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 35: W193–W200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan YL, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ 2009. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet 5: e1000511 doi: 10.1371/journal.pgen.1000511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, et al. 2004. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J 23: 2892–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W 2003. Human-mouse alignments with BLASTZ. Genome Res 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Cecconi F, Bernier G, Andrejewski N, Kammandel B, Wagner M, Gruss P 2000. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development 127: 4325–4334 [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TI, Pratt T, Mason JO, Price DJ 2009. Normal ventral telencephalic expression of Pax6 is required for normal development of thalamocortical axons in embryonic mice. Neural Dev 4: 19 doi: 10.1186/1749-8104-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, Stevens JM, Kendall BE, Shorvon SD, Hanson IM, Moore AT, et al. 2001. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet 28: 214–216 [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P 1996–2004. RepeatMasker Open-3.0. <http://repeatmasker.org>
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P 1997. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 387: 406–409 [DOI] [PubMed] [Google Scholar]
- Stormo GD 2000. DNA binding sites: Representation and discovery. Bioinformatics 16: 16–23 [DOI] [PubMed] [Google Scholar]
- Stoykova A, Fritsch R, Walther C, Gruss P 1996. Forebrain patterning defects in small eye mutant mice. Development 122: 3453–3465 [DOI] [PubMed] [Google Scholar]
- Tetreault N, Champagne MP, Bernier G 2009. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol 327: 541–550 [DOI] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Ruther U 1999. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development 126: 3561–3571 [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH 1993. Structure of the zebrafish Snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119: 1203–1215 [DOI] [PubMed] [Google Scholar]
- Ticho BH, Hilchie-Schmidt C, Egel RT, Traboulsi EI, Howarth RJ, Robinson D 2006. Ocular findings in Gillespie-like syndrome: Association with a new PAX6 mutation. Ophthalmic Genet 27: 145–149 [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Radyushkin K, Tonchev AB, Pinon MC, Ashery-Padan R, Molnar Z, Davidoff MS, Stoykova A 2009. Selective cortical layering abnormalities and behavioral deficits in cortex-specific Pax6 knock-out mice. J Neurosci 29: 8335–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda T, Takashima N, Nakagawa R, Maekawa M, Ikegami S, Yoshikawa T, Kobayashi K, Okanoya K, Inokuchi K, Osumi N 2010. Evaluation of Pax6 mutant rat as a model for autism. PLoS ONE 5: e15500 doi: 10.1371/journal.pone.0015500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heyningen V, Williamson KA 2002. PAX6 in sensory development. Hum Mol Genet 11: 1161–1167 [DOI] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4: e1000214 doi: 10.1371/journal.pgen.1000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Carson J, Oldekamp J, Warnecke M, Jakubcakuva V, Zhou X, Shaw CA, Alvarez-Bolado G, Eichele G 2007. Regulatory pathways analysis by high-throughput in situ hybridization. PLoS Genet 3: 1867–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 4th ed University of Oregon Press, Eugene, OR [Google Scholar]
- Wilkinson D. 1992. Whole mount in situ hybridization of vertebrate embryos. In In situ hybridization: A practical approach (ed. D Wilkinson), pp.75–83. IRL Press at Oxford University Press, Oxford, UK. [Google Scholar]
- Wolf LV, Yang Y, Wang J, Xie Q, Braunger B, Tamm ER, Zavadil J, Cvekl A 2009. Identification of Pax6-dependent gene regulatory networks in the mouse lens. PLoS ONE 4: e4159 doi: 10.1371/journal.pone.0004159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214 [DOI] [PubMed] [Google Scholar]
- Zhang X, Friedman A, Heaney S, Purcell P, Maas RL 2002. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev 16: 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rowan S, Yue YZ, Heaney S, Pan Y, Brendolan A, Selleri L, Maas RL 2006. Pax6 is regulated by Meis and Pbx homeoproteins during pancreatic development. Dev Biol 300: 748–757 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lu JP, Suter DM, Krause KH, Fini ME, Chen B, Lu Q 2010. Isoform- and dose-sensitive feedback interactions between paired box 6 gene and delta-catenin in cell differentitation and death. Exp Cell Res 316: 1070–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]