Abstract
Whole-genome sequencing (WGS) with new short-read sequencing technologies has recently been applied for genome-wide identification of mutations. Genomic rearrangements have, however, often remained undetected by WGS, and additional analyses are required for their detection. Here, we have applied a combination of WGS and genome copy number analysis, for the identification of mutations that suppress the growth deficiency imposed by excessive initiations from the Escherichia coli origin of replication, oriC. The E. coli chromosome, like the majority of bacterial chromosomes, is circular, and DNA replication is initiated by assembling two replication complexes at the origin, oriC. These complexes then replicate the chromosome bidirectionally toward the terminus, ter. In a population of growing cells, this results in a copy number gradient, so that origin-proximal sequences are more frequent than origin-distal sequences. Major rearrangements in the chromosome are, therefore, readily identified by changes in copy number, i.e., certain sequences become over- or under-represented. Of the eight mutations analyzed in detail here, six were found to affect a single gene only, one was a large chromosomal inversion, and one was a large chromosomal duplication. The latter two mutations could not be detected solely by WGS, validating the present approach for identification of genomic rearrangements. We further suggest the use of copy number analysis in combination with WGS for validation of newly assembled bacterial chromosomes.
Most bacteria, including E. coli, have one circular chromosome. Replication of this chromosome is initiated by assembling a complex of DnaA-ATP at the origin of chromosome replication, oriC. This subsequently leads to assembly of two complete replisomes in a highly controlled process (Mott and Berger 2007). Replication proceeds bidirectionally from oriC to a termination region, ter, at the opposite side of the circular chromosome. Because the replication time may exceed the generation time in fast-growing bacteria, such cells will display overlapping rounds of replication (Cooper and Helmstetter 1968).
The DnaA protein is the master regulator of initiation of chromosomal DNA replication in Escherichia coli. DnaA binds ATP and ADP with similar affinities, but only the former is active in the initiation process (Kaguni 2006; Mott and Berger 2007; Katayama et al. 2010). In E. coli, several mechanisms regulate the amount and activity of the DnaA protein (Nielsen and Lobner-Olesen 2008). One such mechanism is the RIDA (regulatory inactivation of DnaA) process (Katayama et al. 1998), which reduces the activity of the DnaA protein by hydrolysis of the ATP to ADP. The Hda protein, encoded by the hda gene, forms a complex with DNA-bound β-subunits of the DNA polymerase III holoenzyme to stimulate the intrinsic ATPase activity of DnaA. Hda is therefore instrumental for the RIDA process (Kato and Katayama 2001). Inactivation of hda results in an increase in the ATP bound form of the DnaA protein, which, in turn, leads to overinitiation of chromosome replication from oriC and compromised cell growth (Kato and Katayama 2001). DnaA-ATP also serves as a repressor of the nrdAB genes encoding ribonucleotide reductase (Gon et al. 2006; Olliver et al. 2010). Loss of Hda is, therefore, also expected to result in deprivation of dNTPs in the cell, which could further contribute to the growth defect of hda mutant cells.
Once formed, Hda-deficient cells rapidly accumulate suppressor mutations, termed hsm (hda suppressor mutation) (Riber et al. 2006). Previously, eight hsm mutant strains were isolated, but the suppressor mutation was only identified in one of these (a point mutation in the dnaA gene) (Riber et al. 2006). Identification and characterization of hsm mutations is expected to advance our understanding of how chromosome replication in E. coli is controlled and prompted the present study to identify the mutations of the remaining hsm strains.
Traditional genetic approaches to map unmarked mutations in E. coli require a readily identified phenotype of the mutation in question. These approaches include Hfr mating and co-transduction frequencies with known markers that identify a candidate region for sequencing. The main disadvantages of these classical mapping techniques are that they are time-consuming and inefficient (not all mutations can be mapped this way), and the genetic tools needed are only available for a few organisms. Large chromosomal rearrangements are particularly difficult to map with genetic tools and have only been identified in a few cases, including the rrnD–rrnE inversion in E. coli strain W3110, which was mapped with DNA–RNA hybridization (Hill and Harnish 1981). The emergence of complete genomic sequences allows comparison of genomes of related bacteria. Such comparisons indicate that fixed inversions have a preference to pivot around oriC (Eisen et al. 2000). Optical mapping has also been applied to identify genomic rearrangements and linking these to sequence information (Zhou et al. 2004).
An alternative to the classical genetic mapping of mutations is based on whole-genome sequencing (WGS) by “next-generation DNA sequencing technologies” (NGS) (Shendure and Ji 2008), in which massive parallel sequencing of millions of short reads of DNA sequence can provide many-fold coverage of the entire genome. The many short reads can be aligned to a reference sequence similar to the experimental sequence in resequencing experiments and the differences enumerated.
There are several examples of the use of short read WGS for mutation detection. Srivatsan et al. (2008) sequenced a selection of Bacillus subtilis laboratory strains, using Illumina WGS, and identified a variety of mutations including two synthetic relA-suppressing mutations, each residing in a separate relA homolog and each having only a partial suppressing effect. Davis and Waldor (2009) sequenced rnaE mutants of Vibrio cholera, using Illumina WGS, in a search for rnaE suppressors and report single-nucleotide substitutions and single-nucleotide indels compared to the reference sequence.
Traditional WGS may not solve the difficulty with detection of large chromosomal rearrangements. This was demonstrated by Herring and Palsson (2007), who resequenced E. coli W3110 and used E. coli MG1655 as a reference sequence with the Comparative Genome Sequencing (CGS) service provided by Nimblegen Systems Inc. They reported that CGS was efficient in detection of SNPs (single-nucleotide polymorphisms), small indels, IS element insertions, and deletions compared to a reference sequence, but they failed to detect the known rrnD–rrnE inversion in W3110 (Hill and Harnish 1981). In a long-term E. coli adaptation experiment, SNPs, small indels, three larger deletions, and IS element insertions were readily detected. However, a major inversion between citC and gatZ (Schneider et al. 2000) was not detected (Barrick et al. 2009). These publications collectively indicate that point mutations and small insertions or deletions (indels) can be identified by short-read-based WGS, whereas larger chromosomal rearrangements are difficult to identify due to the limited ability of these methods to span the repeated sequences surrounding the chromosomal rearrangements.
Here we report the Illumina WGS sequencing of eight hsm strains. Apart from detecting point mutations and small insertions or deletions, the additional use of copy number analysis of template DNA isolated from fast-growing bacterial cultures allowed for easy detection of large chromosomal rearrangements. These rearrangements would have been very difficult to identify with classical genetic methods. Our results suggest that chromosomes are frequently rearranged, but only a few rearrangements are fixed.
Results and Discussion
Detection of point mutations and small indels
In previous work (Riber et al. 2006), we isolated eight independent E. coli strains with hda-suppressing mutations and named the mutations hsm-1 to hsm-8 (hda suppressor mutation). We sequenced the origin of replication, oriC, from all hsm strains, and we further sequenced the dnaA gene and the ygfZ gene in four of the hsm strains (Riber et al. 2006), since mutations in ygfZ were reported to suppress hda deficiency (Ote et al. 2006). We found that the hsm-2 strain had a point mutation in the dnaA gene that substituted phenylalanine with valine at position 349 in the protein and that this mutation was likely to cause hda suppression (Riber et al. 2006). The seven other hsm mutations remained unidentified.
Since hsm mutations arise spontaneously with a high frequency, it would be very difficult to map and eventually sequence the hsm mutations using any genetic approach. We have therefore investigated the use of the Illumina sequencing platform to detect mutations genome-wide without any prior mapping.
We obtained 5 to 13 million reads per mutant, each with a length of 35 nt, to cover the 4.62-Mb genome for each sample (Supplemental Table S1). The short sequences were aligned to the published sequence of E. coli MG1655 using a set of Perl scripts that we tentatively call “From Reads to Results, R2R” (Supplemental Table S1).
We found seven mutations to be common to some or all strains, and we found one unique mutation in each of hsm-1–hsm-6 (Supplemental Table S2).
In the two strains (hsm-7 and hsm-8) in which we failed to detect any point mutations or small indels, we identified larger rearrangements by copy number analysis of exponentially growing cultures (see below). We subsequently analyzed read frequencies from exponentially growing cultures of the hsm-1 to hsm-4 mutants and verified that they did not carry any large chromosomal rearrangement (data not shown).
Copy number analysis can detect chromosomal rearrangements
The replication time in fast-growing E. coli exceeds the doubling time, and the oriC proximal regions will, therefore, have more than a twofold higher copy number than ter proximal regions. The frequency of a sequence on the circular chromosome in a balanced culture with doubling time τ and replication time C will depend on the fractional distance m from the origin to the terminus (moriC = 0, mter = 1) of this sequence. Assuming that the replication fork velocity is constant from initiation at oriC to termination at ter and is the same for each of the two replication forks, the frequency Nx of a sequence X at position mx is given by (Bremer and Churchward 1977):
which can be rewritten as:
The slope of log2 Nx plotted as a function of m thus becomes:
A plot of log2 Nx as a function of m gives straight lines with maximum at oriC and minimum at ter. Any deviation from the straight line indicates either that the replication speed C varies with the position on the chromosome or that the position m is wrong, as it will be if part of the chromosome is missing, relocated, inverted, or duplicated. Variations in replication speed will show as bends on a continuous curve, whereas chromosomal rearrangements will show as a discontinuous curve.
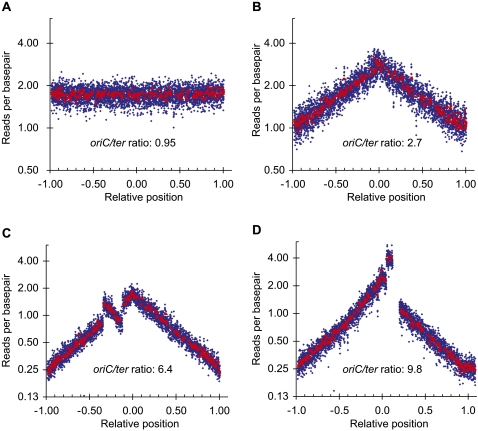
We prepared chromosomal DNA from an exponentially growing culture and a stationary-phase culture of the wild-type strain MG1655 and used the frequency of reads per base pair to analyze copy number variations. The read frequency of the stationary-phase culture where replication does not take place (Fig. 1A) was uniform for all parts of the chromosome, whereas the read frequency of the exponentially grown culture (Fig. 1B) showed a different distribution with the origin proximal regions being 2.7-fold more frequent compared to the ter proximal regions. The “tent-shape” of the curve reflects the bidirectionality of the DNA replication.
Figure 1.
Chromosomal rearrangements detected by read frequency analysis. The number of reads starting at each base is plotted as a function of the relative position on the reference chromosome; oriC is set to 0, and ter is set to ±1 to reflect the bidirectional DNA replication. (Blue rhombus) The average of 1000-bp windows; (red squares) the average of 10.000-bp windows. Any window containing repeat sequences is omitted. The oriC/ter ratio is the ratio of reads per base pair at oriC to reads per base pair at ter. Sequencing template was isolated from: (A) an overnight culture of MG1655; (B) an exponentially growing culture of MG1655; (C) an exponentially growing culture of hsm-7; and (D) an exponentially growing culture of hsm-8. (C,D) Clear deviations from the pattern of the host strain MG1655 (B) indicating an inversion and a duplication, respectively.
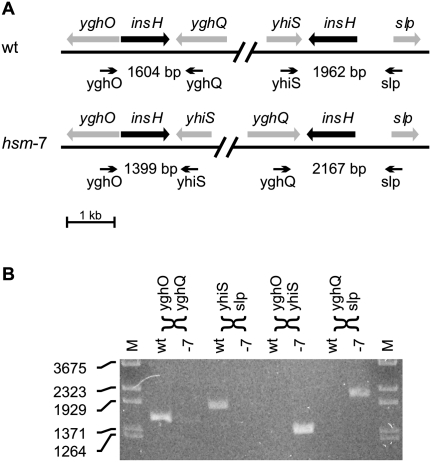
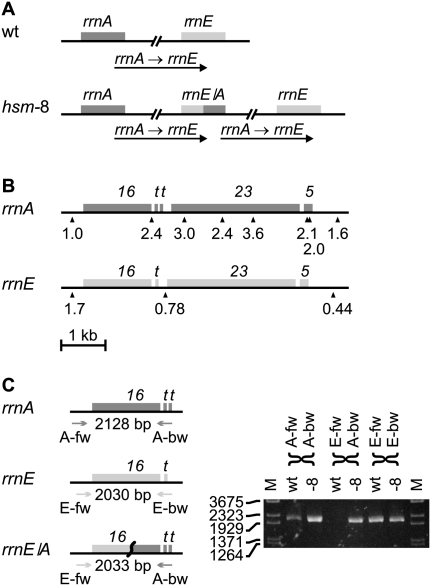
We also grew hsm-7 and hsm-8 strains exponentially before preparing the chromosomal DNA. The read frequency distribution generated from these two strains indicated a ratio of oriC to ter that was increased in hsm-7 (6.4) and hsm-8 (9.8) compared to wild type (2.7) (Fig. 1). Previous publications have noted similar patterns in strains in which the activity of DnaA protein was increased by DnaA overproduction (Atlung et al. 1987; Simmons et al. 2004), the dnaAcos mutation (Simmons and Kaguni 2003) or the seqA mutation (von Freiesleben et al. 1994; Riber et al. 2006). The increased oriC-to-ter ratio in the hsm-7 and hsm-8 strains suggests that the activity of the DnaA protein is still elevated and that the rearrangements only partially relieve the defects of Hda deficiency. The shape of the read frequency distribution of hsm-7 and hsm-8 deviated clearly from the “tent-shape.” A region of hsm-7 covering 521,000 bp showed an inverted distribution compared to wild type (Fig. 1C). This region is flanked by two IS5 insertion sequences in opposite orientations that provide a 1203-bp inverted repeat sequence ideal for generating an inversion by recombination (Fig. 2A). Indeed, we verified the inversion by PCR analysis (Fig. 2B). For the hsm-8 strain, a region from rrnA to rrnE ribosomal operons had twice the expected read depth compared to wild type (Fig. 1D), indicating that this region is duplicated. The rrnA and rrnE operons are oriented in the same direction, and they provide long, near perfect repeat sequences ideal for recombination (Fig. 3A). A detailed analysis of the read frequencies of the few unique sequences within each operon indicated that recombination had taken place between the two genes for 16S RNA rrsA and rrsE (Fig. 3B). We verified this chromosomal rearrangement with PCR analysis across the rrnE–rrnA junction (Fig. 3C). The absence of other detectable mutations in hsm-7 and hsm-8 suggests that the observed chromosomal rearrangements cause the hsm phenotype. We propose that the hsm phenotype in hsm-7 and hsm-8 is caused by altered expression of one or more gene(s) within the inverted and duplicated regions, respectively, resulting from their altered copy number(s).
Figure 2.
The chromosome of hsm-7 is inverted between two IS5 elements. The read frequency analysis of hsm-7 (Fig. 1C) indicates an inversion between two inverted copies of the insH gene encoding the IS5 transposase and trans-activator. (A) Genetic map of the two copies of insH and their neighboring genes in the wild-type (wt) and in the suggested hsm-7 configuration. The heads of the arrows indicate the approximate locations of PCR primers designed to differentiate between the wt and the hsm-7 configuration by amplifying the insH genes with neighboring sequences. The expected size of each PCR product is indicated. (B) PCR analysis of MG1655 DNA (wt) and hsm-7 DNA (-7) with the primers shown in A. MG1655 shows the expected wild-type fragments of 1604 bp and 1962 bp. hsm-7 shows the 1399-bp and the 2167-bp fragment expected from the inversion.
Figure 3.
The rrnA–rrnE region of the chromosome of hsm-8 is duplicated. The read frequency analysis of hsm-8 (Fig. 1D) indicates a duplication of the rrnA–rrnE region. This duplication can be caused by homologous recombination between sequences repeated in the rrnA and rrnE ribosomal RNA operons creating a rrnE/A chimeric operon and duplicating the entire sequence between rrnA and rrnE. (A) The duplication in hsm-8 compared to wild type (wt). (B) A detailed read frequency analysis was made to narrow down the recombination point. (16) rrs genes for 16S RNA; (t) genes for tRNA; (23) rrl genes for 23S RNA; (5) rrf genes for 5S RNA. The read frequency of a 500-bp window on each side of the rrnA and rrnE operons and at positions with unique sequences (shown by triangles) within the rrn operons in hsm-8 was normalized to the read frequencies of the same positions in wt. The relative read frequency of the 500-bp window to the left of rrnA was set to 1. The read frequency shifts from 1.0 to higher than 2 on the opposite side of the 16S RNA gene. This indicates that the entire part of the rrnA operon downstream from the 16S RNA gene is duplicated. In the rrnE operon the read frequency drops fourfold from the left side to the right side of the operon. This fourfold drop is in agreement with Figure 1D and indicates a long DNA replication time for the rrnE operon. The read frequency at the only measurable point in the rrnE operon is lower than half the read frequency of the left side and higher than the read frequency of the right side and is less conclusive. The combined analysis of rrnA and rrnE indicates that recombination took place within the 16S RNA genes. (C, left) PCR primers were designed to amplify the rrsA, rrsE, and the rrsE/A chimeric genes. (Heads of the arrows) The approximate locations of the primers and the expected PCR product sizes are indicated. (C, right) PCR products obtained from MG1655 (wt) and from hsm-8 (-8). hsm-8 shows the native rrnA and rrnE operons as well as the rrnE/A chimeric operon, whereas MG1655 only shows the native rrnA and rrnE operons.
Large chromosomal rearrangements may be frequent, but few are fixed
A number of clinical E. coli isolates have now been completely sequenced. Rasko et al. (2008) compared 10 completely sequenced E. coli genomes from four different pathovars. They observed only one large inversion in the EHEC strain EDL933 compared to the other E. coli strains. Similarly, Touchon et al. (2009) compared 12 clinical E. coli isolates and six clinical Shigella isolates representing four different species and found only a limited number of rearrangements in the E. coli isolates, whereas the Shigella isolates had 16 to 64 rearrangements compared to E. coli. The Shigella isolates also had a much higher number of IS-like elements in their genome (from 549 to 1165) compared to most E. coli isolates (from 42 to 150). One exceptional E. coli isolate, IAI39, had 224 IS-like elements and 10 rearrangements compared to other E. coli.
The low frequency of large chromosomal rearrangements found in these studies contrasts with our observation of both a large inversion and a large segmental duplication in only eight samples. Tandem duplications of regions between rRNA operons have been known for long to occur in a small percentage of cells in a population of Salmonella enterica cells (Anderson and Roth 1981), and this is likely true for E. coli as well. Tandem duplications may form in a RecA-independent way, but duplication loss is RecA-dependent and the steady state of duplication frequency is a balance between formation and loss due to a fitness cost of duplication (Reams et al. 2010). This fitness cost may explain why duplications are rarely fixed in populations. The duplications may meanwhile provide the substrate for more stable mutations before they are lost, leading to, for instance, short junction mutations (Kugelberg et al. 2006) or point mutations (Pranting and Andersson 2011). We suggest that also inversions may be formed frequently and then lost again because of a fitness cost. In the case of our hsm mutants, the chromosomal rearrangements provide the host an immediate advantage, i.e., the ability to stay alive, and hence there is a selection for keeping the suppressing rearrangement. The ease of formation and removal of chromosomal rearrangements may explain why they are rare in wild isolates compared to laboratory strains.
In general, large chromosomal rearrangements in closely related species/strains may easily be overlooked in resequencing experiments with short read sequencing technologies. In this case, the inversion was between 1203 bp perfectly inverted repeats, and the duplication was between ∼5 kb nearly perfectly repeats. The paired-end technology and the recent mate pair technology from Illumina may span 200–500 bp and 2–5 kb repeats, respectively. Paired-end sequencing will not span any of these repeat sequences. Mate pair sequencing is most efficient in combination with single-end or paired-end sequencing and will thus require additional sequencing.
Application of copy number analysis for assembly of de novo sequencing
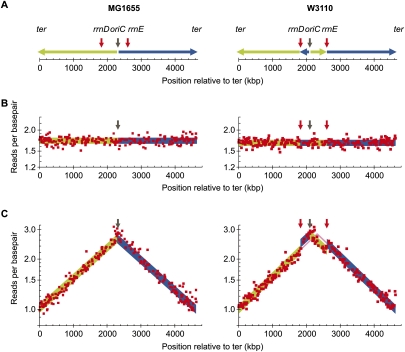
We suggest that an additional copy-number analysis derived from using sequencing templates from fast-growing cultures is a cost- and labor-effective strategy for detection of assembly errors in de novo sequencing projects. To test this hypothesis, we mapped the sequence reads obtained from MG1655 with the sequence of W3110 as a reference sequence (Fig. 4). W3110 is almost identical to MG1665 except for the inversion between rrnD–rrnE (Fig. 4A; Hayashi et al. 2006). Mapping reads obtained from a stationary-phase culture, where all sequences are present in the same copy number, did not reveal the inversion in W3110 compared to MG1655 (Fig. 4B). In contrast, a clear discontinuity in the curve was observed when the reads obtained from the fast-growing culture were mapped to W3110 compared to MG1655 (Fig. 4C, cf. right and left part of panel) revealing the inversion (Fig. 4C). This demonstrates that genomic copy number analysis can greatly contribute to detection of assembly errors in de novo sequencing of bacterial genomes and to the detection of large chromosomal rearrangements in resequencing bacterial genomes.
Figure 4.
Simulation of detection of errors in de novo assembled bacterial sequences. (A) Chromosomal DNA is replicated bidirectionally from oriC (black arrow) indicated by the green line (left arm) and blue line (right arm). Strain W3110 (right) carries an inversion between the ribosomal operons rrnD and rrnE (red arrows) but is otherwise very similar to MG1665 (left). This inversion includes the origin of replication, oriC, and displaces oriC 215 kb to the left compared to MG1655. (Green and blue) The inverted chromosomal arms. Reads generated from DNA template of a stationary phase (B) and an exponentially growing culture of MG1655 (C) were mapped to the genomic sequence of MG1655 (left) and W3110 (right). The number of reads starting at each base pair was calculated for a 10-kb-wide sliding window (red squares), and windows covering repeat sequence were removed. The ideal read frequencies were calculated using an oriC/ter ratio of 2.7 and shown with the same color code as in A.
A specific variation in the copy number of different positions in a genome is required to benefit from copy number analysis. The extent of such variations depends on the ratio of the time required to replicate the genome, C, to the doubling time of the organism, τ. This analysis can therefore be applied directly to all fast-growing organisms with one or few origins for DNA replication. Also, slow-growing organisms may be susceptible to copy number analysis. For instance, Lundgren et al. (2004) demonstrated that the hyperthermophilic archaeons Sulfolobus acidocaldarius and Sulfolobus solfataricus show significant variations in copy number along the chromosome even with doubling times of 3–4 h. This led us to suggest that most cultivable prokaryotes can be subjected to copy number analysis.
Conclusion
We have detected one specific mutation in each of eight isolated hsm mutant strains, as well as seven other mutations common to most or all the strains solely by analyzing the short read sequences. This yield is superior to classical genetic mapping techniques, and our analysis required only a fraction of the workload of classical techniques. The detected mutations comprise point mutations, single and dinucleotide indels, insertion and deletion of transposable IS elements, insertions and deletions between short tandem repeats, and major genomic rearrangements. The chromosomal rearrangements identified in this study were only detected because we used DNA template isolated from a fast-growing bacterial culture and combined sequence determination and copy number variation analysis. These rearrangements would not have been detectable using the standard paired-end sequencing technique since the paired sequences usually span <500 bp, and in these cases, the repeated sequences enclosing the rearrangements exceed 1000 bp. The frequency with which we observed large chromosomal rearrangements led us to speculate that they may normally constitute a significant fraction of mutations.
Methods
Strains and growth conditions
Our isolate of the E. coli K-12 strain MG1655 was originally obtained as CGSC6300 from the Coli Genetic Stock Center and have traveled through the laboratories of Donald Biek and Martin G. Marinus; the isolate was stored as a frozen glycerol stock in these laboratories and in our laboratory (MG Marinus, pers. comm.). Several variants of MG1655 are currently in circulation in various laboratories, some of which contain a major chromosomal deletion in the fnr region (Soupene et al. 2003). Overall, the MG1655 sequenced here is quite similar to the strain for which the sequence is published as U00096 (Blattner et al. 1997).
Construction of the hda:cat insertion and isolation of the MG1655 derived strains ALO1917 (hsm-1), ALO2515 (hsm-2), ALO2516 (hsm-3), ALO2517 (hsm-4), ALO2518 (hsm-5), ALO2519 (hsm-6), ALO2520 (hsm-7), and ALO2521 (hsm-8) were described previously (Riber et al. 2006).
Strains were grown in AB minimal medium (Clark and Maaløe 1967) supplemented with 10 μg/mL thiamine, 0.2% glucose, and 0.5% acid-hydrolyzed casamino acids (BD Difco Bacto Casamino Acids, technical grade). Chromosomal DNA was extracted (Grimberg et al. 1989) for sequencing either from stationary-phase cultures or sampled at OD450 ∼0.2 from exponentially growing cultures as indicated.
Sequencing, sequence analysis R2R, and read frequency analysis
DNA was prepared for sequencing following the Illumina recommended procedure. Briefly, genomic DNA was randomly sheared to 200–800 bp using a Nebulizer. DNA fragments were blunt ended, and A-overhangs were added to the 3′ end of the fragments. Sequencing adapters were ligated to the fragments, and the library was subjected to 18 cycles of PCR amplification. The final library was sequenced using the GA II (Illumina) instrument following the protocol of the manufacturer. Base calling of the 35-bp short read sequences was done using Illumina's analysis GOAT Pipeline.
We mapped the short reads to the published sequence of strain MG1655 (or W3110, AC_000091) as a resequencing analysis using our own set of Perl scripts, that we call R2R. The key steps of R2R are (1) indexing of the reference sequence and call of any sequence with length equal to or longer than the read length and present two or more times in the reference sequence as a repeat sequence; (2) mapping and quality monitoring of the short reads with the reference sequence; and (3) presentation, call of mutations, and statistics of read frequencies and sequence coverage, etc. The mapping of reads was done in the following order: First, all matching reads were mapped and subtracted; the remaining reads were quality-filtered; all reads with one misfit were mapped and subtracted; all reads with two misfits were mapped and subtracted; and the remaining reads were analyzed for having at least 15 exactly matching nucleotides from either end. We call this latter class of reads “indel-reads” since they potentially mark sites for insertions or deletions. The filter for quality was set so the first 30 nt should be called with an average quality of at least 35 using the Illumina raw quality, which is a phred-like value with a maximum value of 40 for one nucleotide.
Mapped reads are presented together with reference sequence and annotations, and in the same run tables of called mutations are generated. A nucleotide is set “confirmed” if a matching read overlaps this nucleotide, and the read extends for at least n nucleotides before and after. For SNPs, simple indels, and read statistics, n was set to 3; for calling duplications between short tandem repeats, n was set to 7. SNPs and simple indels were called with the criteria that at least two reads should indicate a mutation, and the number of matching reads should be <50% of the nonmatching reads. Duplications between short tandem repeats were called with the criteria that at least 10 indel reads should mark the position, and the number of matching reads should be less than twice the indel reads.
Read frequency analysis was visualized with Microsoft Excel, using the R2R-generated frequency data as input.
R2R is available at http://milne.ruc.dk/R2R/; details of the R2R scripts will be reported elsewhere.
Acknowledgments
The Wilhelm Johannsen Centre for Functional Genome Research was established by the Danish National Research Foundation. This study was supported by the Lundbeck Foundation (N.T.), the Danish Natural Science Research Council, and the Novo Nordisk Foundation (to A.L.-O.). We thank Martin G. Marinus for supplying E. coli strain MG1655 and for critical reading of the manuscript, and we thank Christa Persson for technical assistance.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRP003789.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.117416.110.
References
- Anderson P, Roth J 1981. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci 78: 3113–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T, Løbner-Olesen A, Hansen FG 1987. Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in E. coli. Mol Gen Genet 206: 51–59 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247 [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- Bremer H, Churchward G 1977. An examination of the Cooper-Helmstetter theory of DNA replication in bacteria and its underlying assumptions. J Theor Biol 69: 645–654 [DOI] [PubMed] [Google Scholar]
- Clark DJ, Maaløe O 1967. DNA replication and the division cycle in Escherichia coli. J Mol Biol 23: 99–112 [Google Scholar]
- Cooper S, Helmstetter CE 1968. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol 31: 519–540 [DOI] [PubMed] [Google Scholar]
- Davis BM, Waldor MK 2009. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: one fewer porin is enough. Nucleic Acids Res 37: 5757–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Heidelberg JF, White O, Salzberg SL 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol 1: RESEARCH0011 doi: 10.1186/gb-2000-1-6-research0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gon S, Camara JE, Klungsoyr HK, Crooke E, Skarstad K, Beckwith J 2006. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J 25: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg J, Maguire S, Belluscio L 1989. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res 17: 8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, et al. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2: 2006.0007 doi: 10.1038/msb4100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CD, Palsson BO 2007. An evaluation of Comparative Genome Sequencing (CGS) by comparing two previously-sequenced bacterial genomes. BMC Genomics 8: 274 doi: 10.1186/1471-2164-8-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CW, Harnish BW 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci 78: 7069–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni JM 2006. DnaA: Controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol 60: 351–375 [DOI] [PubMed] [Google Scholar]
- Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94: 61–71 [DOI] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8: 163–170 [DOI] [PubMed] [Google Scholar]
- Kato J, Katayama T 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J 20: 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc Natl Acad Sci 103: 17319–17324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R 2004. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci 101: 7046–7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott ML, Berger JM 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5: 343–354 [DOI] [PubMed] [Google Scholar]
- Nielsen O, Lobner-Olesen A 2008. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep 9: 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olliver A, Saggioro C, Herrick J, Sclavi B 2010. DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli. Mol Microbiol 76: 1555–1571 [DOI] [PubMed] [Google Scholar]
- Ote T, Hashimoto M, Ikeuchi Y, Su'etsugu M, Suzuki T, Katayama T, Kato J 2006. Involvement of the Escherichia coli folate-binding protein YgfZ in RNA modification and regulation of chromosomal replication initiation. Mol Microbiol 59: 265–275 [DOI] [PubMed] [Google Scholar]
- Pranting M, Andersson DI 2011. Escape from growth restriction in small colony variants of Salmonella typhimurium by gene amplification and mutation. Mol Microbiol 79: 305–315 [DOI] [PubMed] [Google Scholar]
- Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190: 6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reams AB, Kofoid E, Savageau M, Roth JR 2010. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184: 1077–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riber L, Olsson JA, Jensen RB, Skovgaard O, Dasgupta S, Marinus MG, Løbner-Olesen A 2006. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev 20: 2121–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156: 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H 2008. Next-generation DNA sequencing. Nat Biotechnol 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Simmons LA, Kaguni JM 2003. The dnaAcos allele of Escherichia coli: hyperactive initiation is caused by substitution of A184V and Y271H, resulting in defective ATP binding and aberrant DNA replication control. Mol Microbiol 47: 755–765 [DOI] [PubMed] [Google Scholar]
- Simmons LA, Breier AM, Cozzarelli NR, Kaguni JM 2004. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol Microbiol 51: 349–358 [DOI] [PubMed] [Google Scholar]
- Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, Prasad G, Paliy O, Charernnoppakul P, Kustu S 2003. Physiological studies of Escherichia coli strain MG1655: Growth defects and apparent cross-regulation of gene expression. J Bacteriol 185: 5611–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet 4: e1000139 doi: 10.1371/journal.pgen.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344 doi: 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV, Schaechter M 1994. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol 14: 763–772 [DOI] [PubMed] [Google Scholar]
- Zhou S, Kile A, Bechner M, Place M, Kvikstad E, Deng W, Wei J, Severin J, Runnheim R, Churas C, et al. 2004. Single-molecule approach to bacterial genomic comparisons via optical mapping. J Bacteriol 186: 7773–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]