Abstract
Eukaryotic chromatin is a combination of DNA and histone proteins. It is established fact that epigenetic mechanisms are associated with DNA and histones. Initial studies emphasize on core histones association with DNA, however later studies prove the importance of linker histone H1 epigenetic. There are many types of linker histone H1 found in mammals. These subtypes are cell specific and their amount in different types of cells varies as the cell functions. Many types of post-translational modifications which occur on different residues in each subtype of linker histone H1 induce conformational changes and allow the different subtypes of linker histone H1 to interact with chromatin at different stages during cell cycle which results in the regulation of transcription and gene expression. Proposed O-glycosylation of linker histone H1 promotes condensation of chromatin while phosphorylation of linker histone H1 is known to activate transcription and gene regulation by decondensation of chromatin. Interplay between phosphorylation and O-β-GlcNAc modification on Ser and Thr residues in each subtype of linker histone H1 in Homo sapiens during cell cycle may result in diverse functional regulation of proteins. This in silico study describes the potential phosphorylation, o-glycosylation and their possible interplay sites on conserved Ser/Thr residues in various subtypes of linker histone H1 in Homo sapiens.
Introduction
Eukaryotic genome is packaged into a structure known as chromatin. The basic structural unit of chromatin called as nucleosome is composed of DNA and proteins [1]. The major proteins involved in chromatin structure are histone proteins. Histone proteins are of five types: H1, H2A, H2B, H3 and H4 [2-4]. Histone H1 is known as linker histone while the other four histone proteins are collectively known as core histones. This DNA-protein complex is the tempelate for a number of essential cell processes including transcription recombination, repair and replication. Histone H1 is located on the linker DNA that goes between the nucleosomes in chromatin structure [5]. Linker DNA which is associated with linker histone H1 interconnects core particles, varies in length, depending on species and tissue [6]. Organization of DNA into nucleosomes by histone proteins and folding of nucleosomes into higher-order chromatin structure is generally believed to compact DNA and make it inaccessible to transcription factors [7]. Linker histones H1 are necessary for modulating chromatin structure and function at multiple levels [8].
Organisms contain a variety of subtypes of linker histone which exhibit significant sequence divergence and distinct patterns of expression differentiation and development [9]. The H1 linker histones are the most divergent group. Usually nine subtypes of linker histone H1 are present in mammals including H1.1, H1.2, H1.3, H1.4, H1.5, H1o, H1Foo, H1.t [10] and H1.x [11]. Linker histone sub-types are classified according to their tightly regulated expression pattern during embronyal development and cell differentiation [12]. All known sub-types of linker histone contain a common domain structure. Linker histones consist of a short N-terminal, a highly conserved central globular domain and a long C-terminal domain [13]. Somatic cells contain almost all sub-types of linker histone H1 [12]. In vitro, H1-containing chromatin shows strong inhibition of transcription [14] and transcriptionally active chromatin typically depleted in H1 compared with inactive chromatin [15]. Wisniewski et al. showed that many of the mapped modification sites which are thought to be involved in binding to nucleosomal DNA are located within the globular domain region of the different subtypes of the linker histone H1 [16]. H1 depletion results in a dramatic lengthening of chromosomes, which suggests an important role in mitotic chromosome condensation [17]. The presence of these large number of various H1 histone subtypes and their possible post-translational modifications, make it very clear that H1 histones play numerous structural and functional roles in chromatin [18]. Until now, no specific role for the various variants has been established but it is known that the mouse histone H1.2 binds preferentially to a regulatory sequence within a mouse H3.2 replication-dependent histone gene [19].
Post-translational modifications (PTMs) of linker histone H1 play very important role in regulation of chromatin structure, transcriptional regulation, gene activity [17] and controlling the accessibility of transcription factors to chromatin structure [20]. A working model of the cell cycle has slowly been constructed from the discovery of cyclins 22 years ago. This model is composed of protein phosphorylation, acetylation timed expression of cyclins, and well orchestrated cell division. Nevertheless, a detailed mechanism of the cell cycle is still incomplete [21-23]. Transcriptional activation of genes starts with the dissociation of linker histone H1 from linker DNA [24]. Phosphorylation of linker histone is required for efficient cell cycle progression by enzyme CDK2 [25]. These kinases requires a consensus sequence (S/T)PXZ or (S/T)PXK for phosphorylation (where X is any amino acid and Z is a basic amino acid) and this consensus sequence is found in many linker histone H1 variants which become phosphorylated [26]. It is found that PKC is also involved in phosphorylation of linker histone variants during regulation of gene expression in cell cycle [27]. Phosphorylation of linker histone regulates transcription and gene expression by reducing the electrostatic binding of linker histone to DNA in chromatin [28]. In vivo phosphorylation of the linker histone tails influence both the binding to mononucleosomes and the aggregation of polynucleosomes [29]. The phosphorylation of linker histones at their N and C-terminal tails during the cell cycle influence its functions for enhancing decondensation which in turn regulate transcription and gene expression. This phosphorylation and dephosphorylation is a common regulatory mechanism for protein functions [30].
O-Glycosylation is also very important PTM of proteins. During O-Glycosylation one molecule of N-acetylglucosamine (O-β-GlcNAc) is introduced on Ser or Thr residue by enzyme O-GlcNAc transferases (OGT). Addition of O-β-GlcNAc can inhibit phosphorylation on Ser or Thr residue and is reciprocal with phosphorylation on some well studied proteins, such as RNA polymerase II, estrogen receptor, and the c-Myc proto-oncogene product [31-34]. These studies suggest that O-GlcNAc may function as a global regulator of cell growth and division. Deletion of OGT in mouse embryonic fibroblasts is associated with delayed growth, increased expression of the cyclin inhibitor p27, and death. A reduction in O-GlcNAc levels results in cell growth defects, by the lowering UDP-GlcNAc levels to 5% of normal [35,36]. Studies in Xenopus demonstrated maturation defects in oocytes when microinjected with galactosyltransferase which prevents O-GlcNAc removal. Meanwhile incubation of Xenopus oocytes with the O-GlcNAcase inhibitor PUGNAc altered progression of oocytes through progesterone-mediated maturation [37-40].
In 1994, Kim et al. first time observed the o-GlcNAc modification in mouse linker histone H1. They also observed same PTM on core histones [41]. In 2005, Slawson et al. showed that increased O-GlcNAc resulted in growth defects linked to delay in G2/M progression, altered mitotic phosphorylation, and cyclin expression. Over expression of O-GlcNAcase, the enzyme that removes O-GlcNAc, induces amitotic exit phenotype accompanied by a delay in mitotic phosphorylation, altered cyclin expression, and pronounced disruption in nuclear organization. Overexpression of the O-GlcNAc transferase, the enzyme that adds O-GlcNAc, results in a polyploid phenotype with faulty cytokinesis. Notably, O-GlcNAc transferase is concentrated at the mitotic spindle and mid body at M phase. These data suggest that dynamic O-GlcNAc processing is a pivotal regulatory component of the cell cycle, controlling cell cycle progression by regulating mitotic phosphorylation, cyclin expression, and cell division [42]. On the basis of above observations, Kaleem et al (2008) used bioinformatics tools to predict o-glycosylation on human core histone H3, even though there was no experimental proof of that PTM on histone H3 [43].
Interplay between O-β-GlcNAc modification and phosphorylation on the same amino acid residues has been observed in several nuclear and cytoplasmic proteins [44]. These PTMs are dynamic and result in temporary conformational changes and regulate many functions of the proteins. The alternation of these two modifications on the same or neighboring residue may modulate the specific function of the proteins either by enhancing or inhibiting the functional capacity. Residues where O-β-GlcNAc and phosphorylation compete for each other are known as Yin Yang sites [45]. These Yin Yang sites can be predicted and analyzed using various computer-assisted neural network-based programs, which can help us to determine proteins regulatory functions by accessing their modification potentials.
Although a yin/yang relationship between phosphorylation and O-GlcNAcylation on histone H3 has been proposed; the direct evidence for O-glycosylation of histones was never been described. Recent studies by Sakabe et al (2010-2011) first time proved the O-GlcNAc modification on histones and also mapped glycosylation sites with specific immunological, enzymatic and mass spectrometric techniques. They also insist to include O-GlcNAc modification as part of histone code. They showed that histone O-GlcNAcylation increases with heat shock and this increase is concomitant with DNA condensation [46,47]. The present work describe potential phosphorylation, O-Glycosylation and their possible interplay sites which influence condensation, decondensation and transcriptional and gene regulation during cell cycle in various subtypes of linker histone H1.
Materials and Methods
The sequences of different types of linker histone H1 of many species mostly mammals have been described by many workers [10,11,16]. The sequence data used for predicting phosphorylation and glycosylation sites for different subtypes of linker histone H1 of human was retrieved from the SWISS-PROT [48] sequence database. The primary accession numbers for each subtype of linker histone in human are Q02539 (H1.1), P16403 (H1.2), P16402 (H1.3), P10412 (H1.4), P16401 (H1.5), Q81ZA3 (H1oo), P22492 (H1.T), P07305 (H1.0) and Q 92522 (H1.X). BLAST search was made using the NCBI database of non-redundant sequences [49]. The search was done for all organisms' sequences with expect value set to 10 using blosum 62 matrix and low complexity filter selecting nr database. Hits with highest bits score and zero expect value were selected. The four to five sequences of each subtype of linker histone H1 from different selected species were selected to find out conserved residues in Homo sapiens linker histone H1. All selected sequences were multiple aligned using CLUSTALW [50]. All the sequences of subtypes of linker histone H1 present in Homo sapiens were aligned to get the conservation status of subtypes.
Post-translational modifications prediction methods
Phosphorylation sites on Ser, Thr and Tyr residues were predicted by using NetPhos 2.0 (http://cbs.dtu.dk/services/NetPhos/) server [51]. NetPhos 2.0 is a neural network-based method for the prediction of potential phosphorylation sites.
NetPhosK 1.0 server (http://cbs.dtu.dk/services/NetPhosK) [52] was used to predict kinase specific phosphorylation sites in human histone H1.
Phospho.ELM database (http://phospho.elm.eu.org/) was used for the determination of the experimentally verified phosphorylation sites [53] present on various linker histone H1 subtypes in different species. The Phospho.ELM database contains a collection of experimentally verified Ser, Thr and Tyr sites in eukaryotic proteins.
To predict potential O-β-GlcNAc modification sites, YinOYang 1.2 (http://www.cbs.dtu.dk/services/YinOYang/) was used. This method is also capable of predicting the potential phosphorylation sites as well and hence predicting the Yin Yang sites [54-56].
Neural networks-based prediction methods
Artificial neural networks based methods have been extensively used in biological sequence analysis and predicting the potentials for modifications [57]. The methods developed using machine learning approach includes memorizing the neural networks with the sequence environment windows of phosphorylated/glycosylated and non-phosphorylated/non-glycosylated sites. During this learning process the input data of phosphorylated/glycosylated and non-phosphorylated/non-glycosylated sites is presented to neural networks in the form of binary codes of 21 digits. A threshold value in form of bits is set for positive hit and zero for negative hits. The learning process and performance is checked with the data reserved for cross validation using statistical equations. During learning, the error is computed and weights given to each neuron are set to get the maximum correct predictions.
Results
Allignment of sequences for determination of conserved status of Ser/Thr residues within different linker histone subtypes
Each human linker histone subtype was aligned with other species. Conserved and conserved substituted Ser and Thr residues within each subtype were determined (Data not shown). These nine subtypes were also aligned with each other to find conserved residues within subtypes (Figure 1).
Figure 1.

Sequence alignment of different subtypes of linker histone H1 present in Homo sapiens. The residues highlighted in red show conserved and conserved substitution regions in Ser and Thr residues, while the regions highlighted in yellow show that Ser and Thr residues which are conserved in maximum subtypes but not present in all of the subtypes in linker histone H1. The consensus sequences (motifs) for phosphorylation are shown in square lines.
Prediction of phosphorylated S/T residues with motif
(S/T)PXZ and (S/T)PXK motifs were searched for each linker histone H1 subtypes. Sequences within boxes showed the specific motifs (Figure 1). These residues are given in Table 1.
Table 1.
Phosphorylation and O-β-GlcNAc site map of Homo sapiens
| Substrate | Phosphorylation Sites by NetPhos | Experimentally known | Predicted by Motif | Yin Yang sites | Conserved | Conserved sub | |
|---|---|---|---|---|---|---|---|
| H1.1 | SER | 33, 41, 51, 52, 53, 91, 106, 114, 115, 123, 135, 145, 148, 164, 165 | 1, 35, 103, 183 | 183 | 33, 52, 53, 114, 164, 165 | 41, 43, 51, 53, 60, 106, 183 | 1, 48, 52, 91, 103 |
| THR | 94, 151, 161, 173, 199, 203 | 151 | 151 | 161, 173, 199, 203 | 94 | 101, 151, 11, 164, 203 | |
| H1.2 | SER | 35, 50, 54, 104, 112, 149, 172, 187 | 1, 172 | 172 | 30, 50, 187 | 1, 40, 58, 77, 102, 104, 172, 187 | 35, 85, 88, 112 |
| THR | 30, 91, 145, 153, 166 | 30 | 30, 145, 153 | 145, 166 | 44, 91, 95, 98 | 3, 153 | |
| H1.3 | SER | 36,51, 55, 104, 113, 150, 173, 188, 204 | 188 | 173, 188 | 35, 51, 188, 204 | 36, 41, 51, 58, 79, 89, 102, 104, 188 | 1, 86 |
| THR | 18, 92, 146, 154, 167, 179 | 18 | 18, 146, 154 | 146 | 3, 45, 92, 96, 99 | 154 | |
| H1.4 | SER | 26, 35, 50, 54, 103, 112, 150, 171, 186 | 35, 171, 186 | 171, 186 | 35, 50, 186 | 1, 35, 40, 50, 54, 57, 78, 85, 88, 101, 103, 112, 150, 171, 186 | 172, 188 |
| THR | 17, 91, 145, 153, 202 | 17 | 17, 145, 153 | 17, 145, 202 | 3, 17, 91, 95, 98, 145 | 141, 153, 202 | |
| H1.5 | SER | 17, 43, 53, 106, 115, 172, 188 | 17, 172, 188 | 17, 172 | 17, 43, 53 | 1, 43, 60, 80, 104, 106, 115 | 17, 53, 88, 91, 172 |
| THR | 10, 24, 38, 94, 137, 154 | 137,154 | 10, 137, 154 | 10, 38 | 38 | 3, 8, 47, 98, 101, 154 | |
| H1.0 | SER | 6, 18, 21, 44, 48, 65, 70, 97, 103, 123, 130, 185 | 123 | 6, 21, 44, 97, 103, 123, 130 | 4, 6, 21, 28, 44, 45, 55, 65, 70, 89, 91, 103, 130, 170, 184, 185 | 18, 97, 115 | |
| THR | 109, 118, 134, 140, 152, 161 | 118, 140, 152 | 134, 161 | 1, 5, 22, 76, 77, 83, 109, 118, 123, 134, 140, 152 | 161 | ||
| H1.T | SER | 8, 42, 52, 54, 86, 107, 111, 118, 126, 128, 137, 140, 142, 165, 180, 187, 204 | 177 | 142, 180 | 8, 54, 118, 180, 204 | 1,42, 44, 52, 54,61,81, 105,107,140, 142,165, 180 | 8, 35, 126, 128, 137, 187, 189, 204 |
| THR | 131, 148, 158, 159, 162, 203 | 158, 159 | 148, 158, 159, 162, 203 | 3, 21, 99, 102, 148, 158 | 10, 48, 131, 145, 203 | ||
| H1oo | SER | 8, 11, 13, 14, 16, 20, 21, 23, 26, 32, 42, 73, 161, 211, 229, 230, 235, 243, 245, 246, 260, 262, 263, 276, 336, 337, 340, 341 | 276 | 8, 13, 14, 16, 26, 73, 229, 262, 336, 337, 340, 341 | 5, 8, 12, 13, 20, 67, 110, 118, 221, 236 | 7, 122, 219, 231, 241, 249 | |
| THR | 72, 194, 256, 278, 319 | 256, 319 | 66, 81, 97, 116, 231 | 19, 209 | |||
| H1.X | SER | 31, 33, 39, 92, 113, 154, 171 | 31, 33 | 33 | 49, 65, 66, 92, 113, | 27, 31, 133 | |
| THR | 55 | 101 | 12, 13, 55, 87 | ||||
Acquiring of experimentally verified S/T/Y residues
Data for experimentally confirmed S/T/Y residues was obtained from Phospho ELM and UniprotKB (http://www.uniprot.org) is given in Table 1. All histone H1 subtypes phosphorylated during cell cycle except H1oo.
Prediction of Phosphorylation Sites
NetPhos 2.0 server was used for the prediction potential for phosphorylation of possible Ser and Thr residues among all known subtypes of linker histone H1. All the subtypes of linker histone H1 showed high potential for phosphorylation as shown in Figure 2. The predicted Ser and Thr residues are shown in Table 1.
Figure 2.
Graphical presentation of potential for phosphate modification at Ser, Thr and Tyr residues in different subtypes of linker histone H1 in Homo sapiens. Here blue vertical line show the phosphorylation potential of Ser, green vertical lines show the phosphorylation potential of Thr residues, redlines show phosphorylation potential of Tyr residues, and gray horizontal lines show threshold for modification potential in each subtype of linker histone H1.
Prediction of Kinases involved in Phosphorylation
Different kinases are involved in phosphorylation of Ser and Thr residues of linker histone H1 subtypes. Almost each kinase predicted is involved in phosphorylation of two or more residues. The predicted kinases involved in phosphorylation by NetPhos K 1.0 are shown in Table 2.
Table 2.
Protein kinases invoved in phosphorylation of different subtypes of linker histone H1 in Homo sapiens
| Histone H1 Sub-types | Enzymes for Phosphorylation HUMAN | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PKC | PKA | CDC2 | CDK5 | GSK3 | P38 MAPK | RSK | PKG | ||
| H1.1 | SER | 33, 52, 104, 106, 115, 123, 145, 148, 164 | 41,123, | 51, 52, 53 | 182 | 182 | 164, 165 | 11, 165 | |
| THR | 3, 94, 101, 118, 127, 132, 142, 151, 161, 173, 199, 203 | 151 | 151 | ||||||
| H1.2 | SER | 50, 57, 85, 101 103, 112, 149, 172, 187 | 35 | 50 | 37, 149 | ||||
| THR | 30, 91, 98, 125 153, 164, 166, | 145, 153, | 30 | ||||||
| H1.3 | SER | 51, 58, 86, 102 104, 113, 150, 173, 188, 204 | 36, | 51, | 188, | 36, | |||
| THR | 29, 92, 99, 154 167, 210 | 9, | 17,146, 154, | 146 | 17, 146, 179, | 210 | |||
| H1.4 | SER | 26, 50, 57, 85 101, 103, 112, 149, 171 | 26, 35, | 50, | 187 | 187 | 187 | 171 | 26, 35, 149, |
| THR | 91, 98, 141, 153, 202 | 17, 145, 153, | 17, | ||||||
| H1.5 | SER | 53, 88, 104, 106, 115, 172, 188, | 60, | 17, 172, 188 | 172 | 188 | |||
| THR | 24, 38, 94, 101 137, 154, 186, | 38, | 137, 154, | 10, 137, | 8, 38, 154, | ||||
| H1.O | SER | 18, 44, 45, 55, 70, 91, 103, 123, 130, 170, 184, 185 | 18, 28, 44, | 4, 6, 21, | 18, 185 | ||||
| THR | 22, 76, 109, 118, 134, 152, 161 | 89, | 140 | 22, 109 | |||||
| H1.F | SER | 20, 73, 124, 211, 235, 243, 256, 260, 263, 268, 276, 306, 335, 336, 337, 341 | 42, | 13, 14, 16, 21, 45, | 11, | 23, 276 | 73, | 207, 243, 256 | |
| THR | 17, 103, 194, 266, 278, 297 | 72, | 266, | ||||||
| H1.T | SER | 35, 86, 89,105, 107, 111, 118, 121, 126, 128 137, 165, 187, 189, 204 | 42, 61, 86, 187 | 1, 33, 35, 44, 54, 111, 180 | 180 | 180 | 187 | ||
| THR | 102, 119, 131, 144, 148, 158, 162, 203 | 159 | 203 | ||||||
| H1.X | SER | 27, 33, 39, 92, 113, 154, 204 | 39, 49, 66, | 65, | 31 | 31 | 39 | 39, 204 | |
| THR | 87, 135, 140 | 135 | 189 | ||||||
Prediction of O-Linked Glycosylation Sites
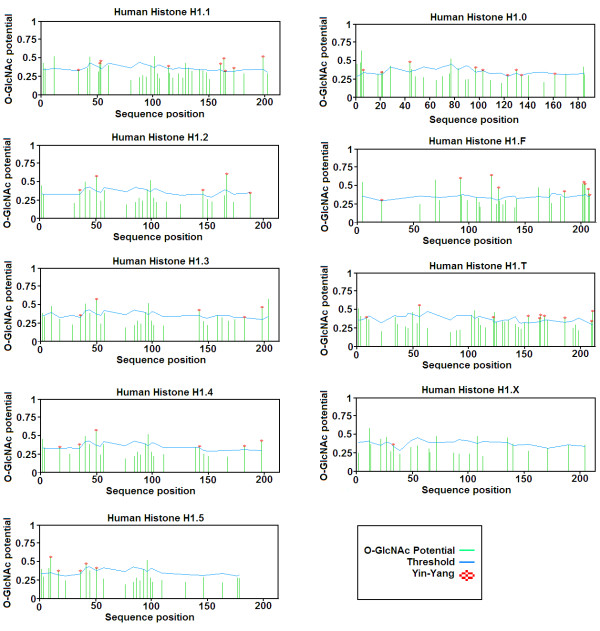
Prediction results for O-linked glycosylation sites showed that all subtypes of linker histone H1 have very high potential for O-β-GlcNAc modification Table 3. There are many predicted Yin Yang sites in each subtype of linker histone which are shown by an asterisk as shown in Figure 3.
Table 3.
Proposed Ser/Thr residues for interplay of phosphorylation and O-β-GlcNAc modification in different subtypes of linker histone H1 in Homo sapiens
| SUBSTRATE | Proposed Yin Yang sites | Proposed Fn-Yin Yang sites | |
|---|---|---|---|
| H1.1 | SER | 103, 183 | 41, 51, 91, 104, 106, 182 |
| THR | 203 | 94, 203 | |
| H1.2 | SER | 187 | - |
| THR | - | - | |
| H1.3 | SER | 188 | 104 |
| THR | 146 | 92, 154 | |
| H1.4 | SER | 35, 186 | 54, 103, 112, 171 |
| THR | 17, 45, 202 | 91, 153 | |
| H1.5 | SER | 17 | 106, 115, 172 |
| THR | - | - | |
| H1.0 | SER | 21, 44, 97, 103, 123, 130 | - |
| THR | 134, 161 | - | |
| H1.T | SER | 54, 180, 204 | 42, 52, 107, 126, 128, 137, 140, 165, 187 |
| THR | 148, 158, 203 | 31 | |
| H1oo | SER | 8, 13 | - |
| THR | - | - | |
| H1.X | SER | - | |
| THR | - | - | |
Figure 3.
Graphical representation of potential for O-β-GlcNAc modification in Ser and Thr residues in the different subtypes of linker histone H1 in Homo sapiens. Green vertical lines show the potential of Ser/Thr residues for O-β-GlcNAc modification and light blue horizontal wavy lines show threshold for modification potential.
Identification of False-Negative Sites
The Ser and Thr residues which were not predicted to be O-β-GlcNAc modified but have very high potential for phosphorylation and very close to threshold value are known as false-negative sites (FN-sites). All the Ser and Thr residues which were predicted false-negatively with high conservation status and phosphorylation potential among different subtypes of linker histone H1 are given in Table 3.
Possible proposed YinYang sites within different subtypes of linker histone H1
The possible proposed Yin Yang sites for the interplay of phosphorylation and O-β-GlcNAc modification are given in Table 3. These Yin Yang sites are proposed on the basis of conservation status of Ser/Thr residues in each subtype of linker histone H1. The Ser/Thr residues are also proposed for the possible interplay of phosphorylation and O-β-GlcNAc modification on the basis of their similarity with other species. These Ser/Thr residues which are predicted "by similarity" are not yet experimentally known in Homo sapiens but these are known in other species of vertebrates.
Discussion
Human linker histones have more than eight sub-types, all consisting of a highly conserved globular domain and less conserved N- and C-terminal tails. The sequence of terminal tails of different subtypes of linker histone H1 within a species is much less conserved but the sequence of terminal tails of a specific subtype is well conserved among different species [58]. In addition to heterogeneity of their primary structures, the histone tails are also post-translationally modified under various biological conditions [59]. The proportion of linker histone H1 subtypes varies in a tissue- and species-species manner [60], and the expression of each subtype varies throughout development and differentiation [61]. Studies of the structure of different subtypes of linker histone H1 and their interaction with the nucleosome and their roles in controlling gene activity indicate that linker histones have both an essential architectural function and an important task in regulating transcription [2]. The precise functions and modifications of linker histones are not yet fully understood, but it is known that different linker histone variants are preferentially localized to particular chromosomal domains. The sequences within the globular domain of linker histone H1 are thought to be responsible for the differential effect of overproduction of different linker histone variants on gene expression [62], while the N- and C-terminal domains of linker histone H1 are responsible for the condensation of chromatin [63]. The N-terminal of linker histone H1 binds with linker DNA [64] and C-terminal of linker histone H1 has binding affinity with core histones [58]. Different linker histone H1 subtypes have different chromatin condensing abilities [65]. All linker histone H1 subtypes differ not only in primary sequence but also in turnover rate, timing of synthesis during development and extent of phosphorylation and they also have the potential to add a great deal of flexibility to chromatin structure and transcriptional activation [66]. Linker histone H1 is required for longitudinal compaction of replicated chromosome. Enrichment of linker histone H1 onto chromatin required passage through interphase, when DNA replication takes place. Thus, linker histone H1 contributes to chromosome condensation in vertebrates [67]. In mouse depletion of linker histone H1 caused chromatin structure changes which include decreased global nucleosome spacing, reduced chromatin compaction and decreased in certain histone modifications like methylation [68]. In vitro experiments showed that linker histone H1 represses transcriptional promoters and factors by condensing the chromatin material [69] but in vivo studies showed that linker histone H1 does not function as a global transcriptional repressor, but instead participates in complexes that either activate or repress specific genes [70]. Differences between linker histone H1 subtypes for both binding and the capacity to aggregate polynucleosome into condensed structure implies functional differences between the different linker histone H1 subtypes during cell cycle and development of organism [71]. Sub-fractions of H1 histones differ in their effectiveness in condensing DNA fibers into ordered aggregates. Furthermore, each of linker histone H1 variant has differences in their binding capacity with DNA [72].
Hale et al. showed that phosphorylation of linker histone H1 provides a signal for the disassembly of higher order chromatin structure during cell cycle [73]. Linker histone H1 phosphorylated in a cell-cycle dependent manner, in G1 phase levels of H1 phosphorylation are usually lowest and then rise continuously during S and G2 phase. The M-phase where chromatin is highly condensed shows the maximum no. of phosphorylated sites [74]. The phosphorylation of linker histone H1 subtypes occurs on specific Ser and Thr residues during cell cycle in the presence of different protein kinases [75]. Interphase phosphorylation occurs mainly on Ser residues while during mitosis, Thr phosphorylation takes place [76]. The C-terminal domain of linker histone H1 not only makes up half of the linker histone molecule, but also has the abundant lysine/arginine residues and (S/T)PXK consensus sequences (phosphorylation motifs) [77]. The relative contributions of linker histone H1 binding amino acids and the (S/T)PXZ or (S/T)PXK motifs are examined. The presence of (S/T)PXK phosphorylation sites in histone H1.4 and H1.5 suggest that these DNA-binding motifs have greater influence on the binding affinities. The short C- terminal domain of linker histone H1.5 to the length of histone H1.2 results in a significant reduction in the binding of the H1.5 protein which demonstrates that the (S/T)PXK motifs are not the sole determinants of the affinity of histone H1 binding [78,79]. It is also very interesting to know that phosphorylation of linker histone also found in N-terminal regions where no (S/T)PXK consensus sequence found and so there is no absolute cell cycle specific site for phosphorylation [80]. Linker histone phosphorylation mainly depends upon their specific subtypes which occur during cell cycle at different residues. Linker histone H1.5 phosphorylated in both the C- and N-terminal regions while linker histone H1.2, H1.3 and H1.4 exclusively phosphorylated in the C-terminal regions [81].
Linker histones not only regulate gene expression and transcription but also have roles in ageing, DNA repair and apoptosis which suggest their importance in maintaining chromatin and genomic integrity [82]. These regulations are in response to changes in the ionic environment by electrostatic interactions between DNA, histone proteins, and free ions [6]. Decondensation of chromatin mediated through phosphorylation of linker histone that weakens the electrostatic interactions between the negatively charged DNA and positively charged C-terminal tails of linker histone subtypes and vice versa [83]. During mitosis linker histone H1.1 phosphorylated on two residues Thr-152 and Ser-182 [79], histone H1.2 phosphorylate on Ser-172, histone H1.3 phosphorylate on Ser-188, histone H1.4 phosphorylate on three residues including two Ser residues 171 and 186, and one Thr residue 145 while linker histone H1.5 phosphorylate on four residues, two Ser 17 and 172, and two Thr 137 and 154 [73]. Linker histone H1.T phosphorylates on three residues Ser-177, Thr-158 and 159 while H1.X also phosphorylates three residues Ser-2, 31 and 33 [83]. There is no experimental data available about the phosphorylated sites of other two remaining linker histone subtypes H1.F and H1.0 in mammals. It is found that during interphase, phosphorylation of Ser residues occurs while during mitosis Thr residues are phosphorylated. This shows the dual effect of linker histones phosphorylation during cell cycle; firstly during interphase the phosphorylation of Ser residues of all subtypes of linker histone H1 promotes DNA replication, transcription and gene regulation and then during mitosis phosphorylation of Thr residues of linker histone H1.4, H1.5 and H1.T may be required for recruiting proteins that are involved in condensation mechanism by unknown mechanism [84].
Our results of NetPhos K 1.0 for the prediction of phosphorylation potential of all Ser and Thr residues (which are experimentally known and described above and also involved in phosphorylation in different subtypes of linker histone H1) showed that these residues are phosphorylated by different kinases during cell cycle as shown in Table 2. These experimentally verified residues are conserved in all subtypes of linker histones in mammals and we can conclude that these phosphorylated sites can be present on linker histones of other mammals "by similarity" where these phosphorylation sites are not yet experimentally known. O-β-GlcNAc modification can occur on these Ser and Thr residues where kinases are involved in phosphorylation as it is well known that kinases and OGT can compete for same site modification [85]. This shows a possibility for interplay between phosphorylation and OGT on these residues. YinOYang 1.2 prediction results had shown that all subtypes of linker histone H1 of mouse have high potential for O-linked glycosylation (Figure 3). The proteins modified by O-β-GlcNAc are more concentrated on condensed chromatin as compared with transcriptionally active regions [86] thus the O-β-GlcNAc modification acts in a reciprocal manner to phosphorylation. Chromatin and several transcription factors are also found to be modified by OGT [87].
The Ser and Thr residues of linker histone H1 which are know to be experimentally phosphorylated and also showed positive potential for O-β-GlcNAc modification are Ser-188 of H1.3, Ser-186 and Thr-145 of H1.4, Ser-17 of H1.5 and Ser-177 of linker histone H1.T. NetPhos 2.0 prediction results showed that there are many Ser and Thr residues which are not yet experimentally verified but have high potential for phosphorylation, same as; YinOYang 1.2 also predicted such type of residues to have high potential for O-β-GlcNAc modification (Table 1). These predicted sites can also be phosphorylated by different kinases (Table 2) and act as possible Yin Yang sites for O-β-GlcNAc modification (Table 3). The remaining Ser and Thr residues of linker histone subtypes which are conserved in different species and either known or predicted to be phosphorylated, showed negative potential for O-β-GlcNAc modification but are very close to threshold value are known as false-negative Yin Yang (FN-Yin Yang) sites (Table 3). These conserved sites can be accessed by different kinases so that these sites have also strong possibility for OGT access and thus can also act as source of interplay for phosphorylation and O-β-GlcNAc [54-56]. The binding of DNA with nucleosome can be increased with the mutation of Ser and Thr phosphorylation sites to alanine residues at different subtypes of linker histone H1 [22]. This phenomenon has showed that these Ser and Thr residues are involved in transcription and gene regulation during cell cycle through interplay of phosphorylation and O-β-GlcNAc modification.
The above discussion reveals that all the conserved phosphorylated residues which show positive potential for O-β-GlcNAc modification or predicted as FN-Yin Yang sites as shown in Table3 may involved in modulating the functions through interplay between phosphorylation and O-β-GlcNAc modification among different subtypes of linker histone H1. These linker histone H1 subtypes phosphorylated on specific Ser residues at N-terminal region; enhance the process of DNA replication, transcription and gene regulation by decondensation of chromatin material during interphase. We propose that this decondensation process can be blocked by O-β-GlcNAc modification on these specific Ser residues which may result in chromatin condensation and repress transcription of DNA. Secondly the interplay between phosphorylation and O-β-GlcNAc modification on Thr residues during mitosis may activate proteins which are involved in condensation mechanism. Thus we can conclude that phosphorylation in different subtypes of linker histone H1 on proposed Ser/Thr residues is involved in decondensation of chromatin structure which leads to transcription regulation and gene expression, whereas the O-β-GlcNAc modification occurring on the same Ser/Thr residues may involved in condensation of chromatin. As histone O-GlcNAcylation is concomitant with DNA condensation, hyperthermia has been shown to sensitize tumor cells to radiotherapy. Although the mechanism for this sensitization has not been elucidated, it has been suggested that prior treatment with heat affects the cellular response to DNA damage induced by ionizing radiation and changes in histone O-GlcNAcylation might be another potential mechanism for radio-sensitization [47].
Abbreviations
PTMs: post-translational modifications; Ser: Serine; Thr: Threonine; O-β-GlcNAc: N-acetylglucosamine; OGT: O-GlcNAc transferases; PUGNAc: O-2-acetamide-2-deoxy-D-glucopyranosylideneamino-N-phenylcarbamate.
Competing interests
All authors have no any kind of institutional or financial competing interests.
Authors' contributions
NN, SN, SQ and MASM collected and analyzed data. WA and KS design the study and wrote the manuscript. All authors read and confirmed the final manuscript.
Authors' information
Ahmad W (M Phil Chemistry) is Research Officer at CEMB, University of the Punjab, Shabbiri K (M Phil Chemistry) and Qaiser S are Lecturers at GC University Lahore. Mughal MAS (MPhil Chemistry) is teaching assistant at GC University Lahore while Nazar N and Nazar S are BSc (Hons) students at GC University, Lahore.
Contributor Information
Waqar Ahmad, Email: waqarchemist@hotmail.com.
Khadija Shabbiri, Email: khadija_shabbiri@hotmail.com.
Noreen Nazar, Email: noreen869@yahoo.com.
Shazia Nazar, Email: shazianazar857@yahoo.com.
Saba Qaiser, Email: saba_qaiser@hotmail.com.
Mirza Abid Shabbir Mughal, Email: me_aabi@yahoo.com.
Acknowledgements
The authors are very grateful to Dr. Nasir-Ud-Din and Ishtiaq Ahmad from Institute of Molecular Sciences and Bioinformatics for their cooperation.
References
- Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding-wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin O, Greally JM, Sokoultchi AI, Bouhassira EE. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci USA. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- Ausio J. Histone H1 and evolution of sperm nuclear basic proteins. J Biol Chem. 1999;274:31115–31118. doi: 10.1074/jbc.274.44.31115. [DOI] [PubMed] [Google Scholar]
- Bednar J, Horowitz AR, Grigoryev AS, Carruthers ML, Hansen CJ, Koster JA, Woodcock LC. Nucleosomes, linker DNA, and Linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Cell Biol. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Dillon N. Heterochromatin structure and function. Biol Cell. 2004;96:631–637. doi: 10.1016/j.biolcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW. ISWI Regulates Higher-Order Chromatin Structure and Histone H1 Assembly In Vivo. PLoS Biol. 2007;5:e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, Kaneda Y. Linker histone variants control chromatin dynamics during early embrogenesis. PNAS. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarg B, Green A, Soderkvist P, Helliger W, Rundquist L, Lindner HH. Characterization of sequence variations in human histone H1.2 and H1.4 subtypes. FEBS J. 2005;272:3673–3683. doi: 10.1111/j.1742-4658.2005.04793.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Horikoshi M. Cloning of the cDNA encoding a novel subtype of histone H1. Gene. 1996;173:281–285. doi: 10.1016/0378-1119(96)00020-0. [DOI] [PubMed] [Google Scholar]
- Khochbin S. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene. 2001;271:1–12. doi: 10.1016/S0378-1119(01)00495-4. [DOI] [PubMed] [Google Scholar]
- Kasinsky HE, Lewis JD, Dacks JB, Ausio J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- O'Neill LP, Turner BM. Histone H1.4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. pp. 3946–57. 195. [DOI] [PMC free article] [PubMed]
- Santisteban MS, Arents G, Moudrianakis EN, Smith MM. Histone octamer function in vivo: mutations in the dimmer-tetramer interfaces disrupt bith gene activation and repression. EMBO J. 1997;16:2493–2506. doi: 10.1093/emboj/16.9.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Kruger S, Mann M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics. 2007;6:72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi IA, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Talasz H, Helliger W, Puschendorf B, Lindener H. In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry. 1996;35:1761–1767. doi: 10.1021/bi951914e. [DOI] [PubMed] [Google Scholar]
- Kaludov NK, Pabon-Pena L, Seavy M, Robinson G, Hurt MM. A mouse histone H1 variant, H1b, binds preferetionally to a regulatory sequence within a mouse H3.2 replication-dependent histon gene. J Biol Chem. 1997;272:15120–15127. doi: 10.1074/jbc.272.24.15120. [DOI] [PubMed] [Google Scholar]
- Parseghian MH, Luhrs KA. Beyond the walls of the nucleus: the role of histones in cellular signaling and innate immunity. Biochem Cell Biol. 2006;84:589–604. doi: 10.1139/o06-082. [DOI] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Murray AM. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/S0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- Geng Y, Whoriskey W, Park MY, Bronson RT, Medema RH, Li T, Weinberg RA, Sicinski P. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell. 1999;97:767–777. doi: 10.1016/S0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
- Ashrafi M, Bathaie SZ, Taqhikhani M, Moosavi-Movahedi AA. The effect of carotenoids obtained from saffron on histone H1 structure and H1-DNA interaction. Int J Biol Macromol. 2005;36:246–252. doi: 10.1016/j.ijbiomac.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JR, Patzlaff JS, Vallis AJ. Evidence that the endogenous histone H1 phosphatase in HeLa mitotic chromosomes is protein phosphatase 1, not protein phosphatase 2A. J Cell Sci. 1996;109:1437–1447. doi: 10.1242/jcs.109.6.1437. [DOI] [PubMed] [Google Scholar]
- Zhao M, Sutherland C, Wilson DP, Deng J, Macdonald JA, Walsh MP. Identification of the linker histone H1 as a protein kinase Cepsilon-binding protein in vascular smooth muscle. Biochem Cell Biol. 2004;82:538–546. doi: 10.1139/o04-053. [DOI] [PubMed] [Google Scholar]
- Dou Y, Gorovsly MA. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol Cell. 2000;6:225–231. doi: 10.1016/S1097-2765(00)00024-1. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Almouzni G, Ura K, Pruss D, Hayes JJ. Transcription factor access to DNA in the nucleosome. Cold Spring Harb Symp Quant Biol. 1993;58:225–235. doi: 10.1101/sqb.1993.058.01.027. [DOI] [PubMed] [Google Scholar]
- Chadee DN, Taylor WR, Hurta RA, Allis CD, Wright JA, Davie JR. Increased phosphorylation of histone H1 in mouse fibroblasts transformed with oncogenes or constitutively active mitogen-activated protein kinase kinase. J Biol Chem. 1995;270:20098–20105. doi: 10.1074/jbc.270.34.20098. [DOI] [PubMed] [Google Scholar]
- Comtesse N, Maldner E, Meese E. Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase. Biochem Biophysc Res Commun. 2001;283:634–640. doi: 10.1006/bbrc.2001.4815. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry. 2000;39:11609–11620. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- O'Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi R, Iyer SPN, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmelt G, Wakeham A, Elia A, Sasaki T, Plyte S, Potter J, Yang Y, Tsang E, Ruland J, Iscove NI, Dennis JD, Mak TW. Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J. 2000;19:5092–5104. doi: 10.1093/emboj/19.19.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Miller MW. Use of galactosyltansferase to assess the biological function of O-linked N-acetyl-d-glucosamine: a potential role for O-GlcNAc during cell division. Exp Cell Res. 2000;263:243–253. doi: 10.1006/excr.2000.5110. [DOI] [PubMed] [Google Scholar]
- Dong DLY, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta_D-glucosaminidase from rat spleen cytosol. J Biol Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- Haltiwanger RS, Grove K, Philipsberg GA. Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O = GlcNAc-beta-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-D-glucopyranosylidene) amino-N-phenycarbamate. J Biol Chem. 1998;273:3611–3617. doi: 10.1074/jbc.273.6.3611. [DOI] [PubMed] [Google Scholar]
- Kim TH, Park JY, Lee SH, Chang HI. Possible glycosylation of H1 histone in Murine liver nucleus. Korean Biochem J. 1994;27:240–244. [Google Scholar]
- Salwson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GE. Perturbations in O-linked β-N-Acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- Kaleem A, Hoessli DC, Ahmad I, Walker-Nasir E, Nasim A, Shakoori AR, Nasir-ud-Din. Immediate-early gene regulation by interplay between different post-translational modifications on human histone H3. J Cell Biochem. 2008;103:835–851. doi: 10.1002/jcb.21454. [DOI] [PubMed] [Google Scholar]
- Kamemura K, Hayes BK, Comer FI, Hart GW. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: alternative glycosylation/phosphorylation of THR-58, a known mutational hot spot of c-Myc in lymphomas, is regulated by mitogens. J Biol Chem. 2002;277:19229–19235. doi: 10.1074/jbc.M201729200. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. The emerging significance of O-GlcNAc in cellular regulation. Chem Rev. 2002;102:431–438. doi: 10.1021/cr000406u. [DOI] [PubMed] [Google Scholar]
- Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. J Biol Chem. 2010;285:34460–34468. doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AltschuL SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DJ, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–46780. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunk S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunk S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Diella F, Cameron S, Gemund C, Linding R, Via A, Kuster B, Sicheritz-Ponten T, Blom N, Gibson TJ. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;22:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Hoessli DC, Walker-Nasir E, Rafik SM, Shakoori AR, Nasir-ud-Din. Oct-2 DNA binding transcription factor: functional consequences of phosphorylation and glycosylation. Nucleic Acids Res. 2006;34:175–184. doi: 10.1093/nar/gkj401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleem A, Hoessli DC, Haq IU, Walker-Nasir E, Butt A, Iqbal Z, Zamani Z, Shakoori AR, Nasir-ud-Din. CREB in long-term potentiation in hippocampus: role of post-translational modifications-studies In silico. J Cell Biochem. 2011;112:138–146. doi: 10.1002/jcb.22909. [DOI] [PubMed] [Google Scholar]
- Ahmad I, Khan TS, Hoessli DC, Walker-Nasir E, Kaleem A, Shakoori AR, Nasir-ud-Din. In silico modulation of HMGN-1 binding to histones and gene expression by interplay of phosphorylation and O-GlcNAc modification. Protein Pept Lett. 2008;15:193–199. doi: 10.2174/092986608783489607. [DOI] [PubMed] [Google Scholar]
- Baldi P, Brunak S. MIT Press. 2. Cambridge, MA; 2002. Bioinformatics: The machine learning Approach. [Google Scholar]
- Goytisolo FA, Gerchman SE, Yu X, Rees C, Graziano V, Ramakrishan V, Thomas JO. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- Lennox RW. Differences in evolutionary stability among mammalian H1 subtypes. Implications for the roles of H1 subtypes in chromatin. J Biol Chem. 1984;259:669–672. [PubMed] [Google Scholar]
- Khochbin S, Wolffe AP. Developmentally regulated expression of linker-histone variants in vertebrates. Eur J Biochem. 1994;225:501–510. doi: 10.1111/j.1432-1033.1994.00501.x. [DOI] [PubMed] [Google Scholar]
- Helliger W, Lindner H, Grubl-Knosp O, Puschendorf B. Alteration in proportions of histone H1 variants during the differentiation of murine erythroleukaemic cells. Biochem J. 1992;288:747–51. doi: 10.1042/bj2880747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT, Gunjan A, Alexander BT, Sittman DB. Differential effect of H1 variant overproduction on gene expression is due to differences in the central globular domain. Nucleic Acids Res. 1997;25:5003–5009. doi: 10.1093/nar/25.24.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharath MM, Ramesh S, Chandra NR, Rao MR. Identification of a 34 amino acid stretch within the C-terminus of histone H1 as the DNA-condensing domain by site-directed mutagenesis. Biochemistry. 2002;41:7617–7627. doi: 10.1021/bi025773+. [DOI] [PubMed] [Google Scholar]
- Takeshima H, Suetake I, Tajima S. Mouse Dnmt3a Preferentially Methylates Linker DNA and Is Inhibited by Histone H1. J Mol Biol. 2008;383:810–821. doi: 10.1016/j.jmb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Lucia De, Faraone-Mennella MR, D'Erme M, Quessada P, Caiafa P, Farina B. Histone-induced condensation of rat testis chromatin: testis-specific H1 versus somatic H1 variants. Biochem Biophys Res Commun. 1994;198:9–32. doi: 10.1006/bbrc.1994.1005. [DOI] [PubMed] [Google Scholar]
- Khadake JR, Rao MR. Condensation of DNA and chromatin by an SPKK-containing octapeptide repeat motif present in the C-terminus of histone H1. Biochemistry. 1997;36:1041–1051. doi: 10.1021/bi961617p. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Heald R. The long and the short of it: linker histone H1 is required for metaphase chromosome compaction. Cell Cycle. 2006;5:589–591. doi: 10.4161/cc.5.6.2581. [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, van Holde K. Histone H1 and transcription: still an enigma? J Cell Sci. 1992;103:889–895. doi: 10.1242/jcs.103.4.889. 1992. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Khochbin S, Dimitrov S. What do linker histone do in chromatin? Bioessays. 1997;19:249–255. doi: 10.1002/bies.950190311. [DOI] [PubMed] [Google Scholar]
- Talasz H, Sapojnikova N, Helliger W, Lindner H, Puschendorf B. In vitro binding of H1 histone subtypes to nucleosomal organized mouse mammary tumor virus long terminal repeat promotor. J Biol Chem. 1998;273:32236–32243. doi: 10.1074/jbc.273.48.32236. [DOI] [PubMed] [Google Scholar]
- Hill CS, Rimmer JM, Green BN, Finch JT, Thomas JO. Histone-DNA interactions and their modulation by phosphorylation of -Ser-Pro-X-Lys/Arg- motifs. EMBO J. 1991;10:1939–1948. doi: 10.1002/j.1460-2075.1991.tb07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale TK, Contreras A, Morrison AJ, Herrera RE. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha. Mol Cell. 2006;22:693–699. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Roth SY, Allis CD. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci. 1992;17:8–93. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- Ewen ME. Where the cell cycle and histone meet. Genes Dev. 2000;14:2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- Sarg B, Helliger W, Talasz H, Forg B, Lindner HH. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J Biol Chem. 2005;281:6573–6580. doi: 10.1074/jbc.M508957200. [DOI] [PubMed] [Google Scholar]
- Ponte I, Vidal-Taboada JM, Suau P. Evolution of the vertebrate H1 histone class: evidence for the functional differentiation of the subtypes. Mol Biol Evol. 1998;15:702–708. doi: 10.1093/oxfordjournals.molbev.a025973. [DOI] [PubMed] [Google Scholar]
- Th'ng JP, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J Biol Chem. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th'ng JP. The C-terminal domain of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- Gurley LR, Valdez JG, Buchanan JS. Characterization of the mitotic specific phosphorylation site of histone H1. Absence of a consensus sequence for the p34cdc2/cyclin B kinase. J Biol Chem. 1995;270:27653–27660. doi: 10.1074/jbc.270.46.27653. [DOI] [PubMed] [Google Scholar]
- Hadnagy A, Beaulieu R, Balicki D. Histone tail modifications and noncanonical functions of histones: perspectives in cancer epigenetics. Mol Cancer Ther. 2008;7:740–748. doi: 10.1158/1535-7163.MCT-07-2284. [DOI] [PubMed] [Google Scholar]
- Dou Y, Gorovsky MA. Regulation of transcription by H1 phosphorylation in Tetrahymena is position independent and requires clustered sites. Proc Natl Acad Sci USA. 2002;99:6142–6146. doi: 10.1073/pnas.092029599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Manm M. Global in vivo, and site specific phosphorylation dynamics in singling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Trollope AF, Sapojnikova N, Thorne AW, Crane-Robinson C, Myers FA. Linker histone subtypes are not generalized gene repressors. Biochim Biophys Acta. 2010;1799:642–652. doi: 10.1016/j.bbagrm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Haltiwanger RS, Busby S, Grove K, Li S, Mason D, Medina L, Moloney D, Philipsberg G, Scartozzi R. O-glycosylation of nuclear and cytoplasmic proteins: regulation analogous to phosphorylation. J Biochem Biophys Res Commun. 1997;231:237–242. doi: 10.1006/bbrc.1997.6110. [DOI] [PubMed] [Google Scholar]
- Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- Majumdar G, Harmon A, Candelaris R, Marteniz-Hernandez A, Raghow R, Solomon SS. O-glycosylation of Sp1 and transcriptional regulation of the calmodulin gene by insulin and glucagon. Am J Physiol Endocrinol Metab. 2003;285:584–591. doi: 10.1152/ajpendo.00140.2003. [DOI] [PubMed] [Google Scholar]