Abstract
The persistence of Shiga toxin-producing E. coli O157:H7 in the environment poses a serious threat to public health. However, the role of Shiga toxins and other virulence factors in the survival of E. coli O157:H7 is poorly defined. The aim of this study was to determine if the virulence factors, stx 1, stx 2, stx 1–2, and eae in E. coli O157:H7 EDL933 play any significant role in the growth of this pathogen in rich media and in soils. Isogenic deletion mutants that were missing one of four virulence factors, stx 1, stx 2, stx 1–2, and eae in E. coli O157:H7 EDL933 were constructed, and their growth in rich media and survival in soils with distinct texture and chemistry were characterized. The survival data were successfully analyzed using Double Weibull model, and the modeling parameters of the mutant strains were not significantly different from those of the wild type. The calculated Td (time needed to reach the detection limit, 100 CFU/g soil) for loamy sand, sandy loam, and silty clay was 32, 80, and 110 days, respectively. It was also found that Td was positively correlated with soil structure (e.g. clay content), and soil chemistry (e.g. total nitrogen, total carbon, and water extractable organic carbon). The results of this study showed that the possession of Shiga toxins and intimin in E. coli O157:H7 might not play any important role in its survival in soils. The double deletion mutant of E. coli O157:H7 (stx 1 − stx 2 −) may be a good substitute to use for the investigation of transport, fate, and survival of E. coli O157:H7 in the environment where the use of pathogenic strains are prohibited by law since the mutants showed the same characteristics in both culture media and environmental samples.
Introduction
Escherichia coli O157: H7 was initially identified as an important human pathogen in 1982 during an investigation into a food-borne disease outbreak in the United States [1]. Since then, an increasing number of E. coli O157:H7 outbreaks have been reported in the United States. It is estimated that in the United States E. coli O157: H7 alone is responsible for a total of 73,480 cases of disease per year, among which, there are more than 1,800 cases of hospitalizations and 52 deaths. Evidence has shown that E. coli O157:H7 is one of the most commonly isolated bacterial pathogens from meat and fresh produce after Campylobacter, Salmonella, and Shigella spp [2]. In addition to the USA, many large outbreaks of E. coli O157:H7 infections have also been reported in many countries making E. coli O157:H7 an increasing public health concern worldwide. The infectious threshold of E. coli O157:H7 is very low, and ingestion of 10 cells may be enough to cause severe gastrointestinal illness [3]. The typical clinical symptoms of E. coli O157:H7 infections are watery diarrhea and hemorrhagic colitis [1], which can progressively develop into life-threatening hemolytic uremic syndrome (HUS) [4], [5].
Outbreaks of E. coli O157: H7 infections are always traced back to consumption of food that has been directly or indirectly contaminated by manure/water containing E. coli O157:H7. Animals including deer, horses, dogs, and birds [6], [7], [8], [9] are known to be E. coli O157:H7 carriers. However, cattle are thought to be the main carrier of E. coli O157:H7 [10], [11]. E. coli O157:H7 in the environment may originate from farms where manure amendments are used as fertilizer. The pathogen could be mobilized through irrigation water, providing an opportunity for the pathogen to spread out into its secondary reservoir, typically water and soil. The persistence and regrowth in these habitats may increase the potential for the pathogen to enter into the food chain and thereby constitute a public health risk. There have been some cases of infection from direct contact with E. coli O157:H7 contaminated soil, and more cases of food poisoning caused by or consumption of vegetables grown in soils contaminated by E. coli O157:H7 [12], [13].
The survival of E. coli O157:H7 in water [14], [15], [16], [17], [18], [19], manure and manure slurry [20], [21], [22], manure-amended soil [23], [24], [25], and sediment [26], [27], is well documented with sporadic reports in natural soils [28], [29]. More direct results could be obtained by applying pathogenic strain in the survival experiments [30], however, most of the studies used nonpathogenic E. coli O157:H7 strains [22], [24], [31], [32] due to environmental safety and regulations. This raises the question on how well those indirect results can represent results using pathogenic strains for comparison. Therefore, additional evidence is needed to clearly understand the role of stx genes and other virulence factors in the survival of pathogenic E. coli O157:H7 in the environment. Previous work [20] showed that there was a similar survival pattern between a Shiga toxin negative E. coli O157:H7 strain and a Shiga toxin positive E. coli O157:H7 strain. However, these strains were not isogenic, and the minor differences in survival might be attributed to other factors, such as the differences in their genomic DNA. Indeed, the variability in growth and survival of E. coli in soils has been shown to be strain-dependent [28].
In the current study, we chose E. coli O157:H7 EDL933 as the model pathogenic E. coli since its genome has been fully sequenced and annotated [42]. E. coli O157:H7 EDL933 and its isogenic mutant derivatives that are missing one of the following virulence factors, stx 1, stx 2, stx 1–2, and eae, were constructed, their growth in rich medium and survival in soils compared to that of the wild type parental strain (Fig. 1). We hypothesized that since all of the strains are isogenic the results will provide insights into the role of stx and eae genes in the survival of E. coli O157:H7 in soils. Additionally, the survival of the E. coli O157:H7 EDL933 in soils will correlate with the survival of pathogenic E. coli O157:H7 strains in the environment.
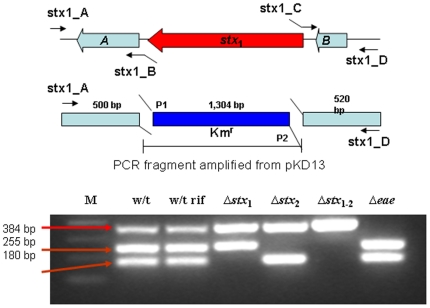
Figure 1. Construction of stx and eae mutants (top) and multiplex PCR confirmation of the mutant constructed (bottom).
M represents 100 bp λDNA ladder.
Materials and Methods
Bacterial strains, construction and growth of mutants
The bacteria and plasmids used in this study are listed in Table 1. In order to facilitate the enumeration of E. coli O157:H7 EDL933 on selective media, the E. coli O157:H7 wild type was tagged with nalidixic acid in addition to rifampicin resistance, and its growth curve in LB (Luria-Bertani) broth was found to be identical to that of the non-tagged wild-type strain.
Table 1. Bacterial strains and plasmids.
| Strain or plasmid | Relevant characteristics | Source or reference |
| strains | ||
| E. coli DH5α | General laboratory strain | Gibco-BR |
| E. coli S17-1 | General laboratory strain | Simon et al. 1983 |
| E. coli EDL933 | wild type | ATCC 43895 |
| E. coli EDL933 | rifampicin tagged, Rifr | This study |
| E. coli EDL933 | stx1(del), , Km, Kmr | This study |
| E. coli EDL933 | stx2(del), , Cm, Cmr | This study |
| E. coli EDL933 | stx1–2(del), , KmCm, KmrCmr | This study |
| E. coli EDL933 | eae(del), , Km, Kmr | This study |
| plasmids | ||
| pWM91 | Suicide vector, Apr | Metcalf et al., 1996 |
| pKD3 | plasmid carrying Cm resistance cassette, Cmr | Datsenko and Wanner, 2000 |
| pKD4 | plasmid carrying Km resistance cassette, Kmr | Datsenko and Wanner, 2000 |
Rifr, rifampicin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol; Apr, ampicillin resistance.
Mutants lacking Stx1, Stx2, and Eae were generated by allelic exchange protocol [33]. The flanking regions were amplified by PCR with specific primers (Table 1), among which primers B and C (e.g. stx1_B, stx1_C) have the linkers at the 5′ end that are complimentary to primers P1 and P2 [14], respectively, for crossover PCR. The kanamycin (Km) cassette was amplified from pKD4 (GenBank accession #, AY048743.1) and the chloramphenicol (Cm) cassette was amplified from pKD3 (GenBank accession #, AY048742.1) using the universal primer set consisting of forward primer P1 and reverse primer P2. Three-way crossover PCR was performed using the flanking regions and Km or Cm cassette as templates, and primers A and D (e.g. stx1_A, stx1_D) were used in this process. The PCR product was then cloned into pWM91 digested with XcmI (T-vector). The resulting plasmid was transformed into E. coli S17-1 λ pir, and then introduced into EDL933 by transconjugation. Recombinants resulting from double crossover events were obtained by sacB and sucrose positive selection. All the mutant strains and the wild type strain were separately stored under −80°C on cryoprotective beads in MicroBank microbial storage tubes (Pro-Lab Diagnostics, Ontario, Canada).
The stx and eae mutants, together with the wild type strain were inoculated into 100 ml of LB broth, and grew under 37°C with a rotation rate of 250 rpm. The optical density at 610 nm (OD610 nm) was monitored using a VIS-UV spectrophotometer (Pharmacia Biotech Inc. NJ). The OD610 nm was plotted against incubation time, and the apparent growth rate (k, h−1) was calculated using the following equation,
where OD 1, OD 2 are the optical density measured at time t1 and time t2, respectively, k is the apparent growth rate (h−1).
Multiplex PCR confirmation of mutants
Multiplex PCR was performed on the mutants and the wild type to confirm the deletions of stx 1, stx 2, stx 1–2, and eae genes in their genomes. PCR was performed using Ready-to-Go PCR beads with the three primer pairs (Table 2) targeting stx 1, stx 2, and eae gene [34]. Thermocycler protocol included an initial denaturation at 95°C for 10 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 40 s, and a final extension at 72°C for 5 min. The PCR product was resolved by electrophoresis on a 1.0% agarose gel. The gel was then stained with ethidium bromide, visualized and photographed using a gel imaging system (Bio-Rad Lab., Irvine, CA). The PCR products with the correct sizes were cloned into TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to manufacture's protocol, and the resulting plasmids were sequenced. DNA sequence analysis was performed using DNAStar software (Lasergene, Madison, WI). Database searches were conducted with identified open reading frames (ORFs) by using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov) to confirm the deletion of the corresponding gene(s).
Table 2. Primers for mutants' construction and multiplex PCR.
| Primers ID | Nucleotide sequence (5′ end to 3′ end) | Predicted product size (bp) | Source or reference |
| stx1_A | GGGTCCGGACGGTCATATGT | 827 | This study |
| stx1_B | gaagcagctccagcctacacTCAGTGAAAATAGCAGGCGC | ||
| stx1_C | ctaaggaggatattcatatGACCCCCTGAAGGACGGCGTTTT | 814 | This study |
| stx1_D | CACCCATTGCCGCCGGATTT | ||
| stx2_A | CATGCTGATGATGCTGGGAGTG | 781 | This study |
| stx2_B | gaagcagctccagcctacacGCGCGTTGTACTGGATTCGA | ||
| stx2_C | ctaaggaggatattcatatGAACCTGATTCGTGGTATGTGGG | 801 | This study |
| stx2_D | TGGATCAGGGCTGTCGAATG | ||
| eae_A | GCAATAACCAAATCATATCCGC | 852 | This study |
| eae_B | gaagcagctccagcctacacAACCACCCCGGCTAAAATATGT | ||
| eae_C | ctaaggaggatattcatatGCTCGAGTTTTTCAGGGGTAGCA | 799 | This study |
| eae_D | TCCAGCATAGGGACCGTGCA | ||
| P1 | GTGTAGGCTGGAGCTGCTTC | 1463 or 1014 | Datsenko and Wanner, 2000 |
| P2 | CATATGAATATCCTCCTTAGTTCC | ||
| stx1_F | ATAAATCGCCATTCGTTGACTAC | 180 | Paton and Paton, 1998 |
| stx1_R | AGAACGCCCACTGAGATCATC | ||
| stx2_F | GGCACTGTACTGAAACTGCTCC | 255 | Paton and Paton, 1998 |
| stx2_R | TCGCCAGTTATCTGACATTCTG | ||
| eae_F | GACCCGGCACAAGCATAAGC | 384 | Paton and Paton, 1998 |
| eae_R | CCACCTGCAGCAACAAGAGG |
Collection, characterization, and inoculation of soils samples
Dello loamy sand, Arlington sandy loam, and Willow silty clay were collected from Santa Ana River bed, fallow field at the University of California-Riverside, and Mystic Lake dry bed, California, respectively (Table 3). Arlington sandy loam is a typical agricultural soil found in Riverside, CA, while the other two soils are typical soil types used for cattle production in eastern and western Riverside County, USA. Permit was obtained from the University of California Riverside to collect the Arlington sandy loam. The soil from the Mystic Lake dry bed has high clay content (71%), and the soil from Santa Ana River bed has high sand content (99%). The texture and chemistry of the three soils are listed in Table 3. Soil samples were collected, sieved (2 mm), put into plastic bags, and stored at 4°C in dark. Soil properties characterized included, clay, silt, and sand content, water content, water holding capacity (WHC), soil organic carbon (OC), and total nitrogen (T-N) [35]. Soil microbial biomass carbon (MBC) was extracted by the chloroform-fumigation-extraction method [36], and water extractable organic carbon (WEOC) was measured by a total organic carbon analyzer (TOC-500, Shimadzu Corp., Kyoto, Japan) according to the method by Liang et al. [37]. The assimilable organic carbon (AOC) fraction in WEOC was determined using a luminous bacterium strain, Vibrio harveyi (Ma et al., unpublished).
Table 3. Soil texture and chemistry.
| Soil type | Sand (%) | Silt (%) | Clay (%) | Bulk density (g/cm) | WHC (%) | pH | T-N (g/kg) | OC (g/kg) | WEOC (mg/kg) | MBC ((mg/kg) | AOC ((mg/kg) |
| Dello loamy sand | 99.1 | 0.2 | 0.7 | 1.67 | 17 | 7.1 | 0.07 | 0.58 | 10 | 11 | 0.20 |
| Arlington sandy loam | 70.9 | 20.8 | 8.3 | 1.54 | 21 | 7.2 | 0.61 | 5.40 | 44 | 56 | 0.90 |
| Willow silty clay | 3.7 | 49.1 | 47.2 | 1.51 | 63 | 7.2 | 1.61 | 20.4 | 242 | 278 | 4.94 |
WHC, water holding capacity; T-N, total nitrogen; OC, organic carbon; MBC, microbial biomass carbon; AOC, assimilable organic carbon.
One cryoprotective bead from MicroBank microbial storage tube containing E. coli O157:H7 was aseptically transferred to a 15 ml tube containing 5.0 ml LB broth and incubated at 37°C for 18 h. From the overnight culture, a 1.0 ml aliquot was transferred into a 250 ml flask containing 100 ml LB broth, and incubated at 37°C for 18 h to achieve early stationary phase. Stationary phase cells were used because in the natural environment, the majority of bacteria exist in this condition [38]. The cells were harvested by centrifugation at 3500 g (Beckman, Brea, CA), washed three time using phosphate buffer (10 mM, pH 7.2), and finally resuspended in sterile deionized water. The wash step was essential to remove the nutrient, typically organic carbon from the LB broth, since E. coli O157 is able to grow at low carbon concentrations in freshwater [39].
Cell from stock cultures were streaked on LB agar (without antibiotics), and incubated 37°C overnight. Single colonies were picked and restreaked onto LB agar with appropriate antibiotics. Single colonies were streaked onto SMAC (sorbitol MacConkey) agar supplemented with BCIG (5-bromo-4-chloro-3-indoxyl-ß-D-glucuronide) (Lab M, Lancashire, UK). The isolated colonies were inoculated into 100 ml LB broth with appropriate antibiotics (Table 1), and incubated at 37°C for about 16 h. The overnight culture were harvested by centrifugation at 4°C, washed three times with phosphate buffer (10 mM, pH 7.2), resuspended in sterile deionized water, and inoculated into soil samples. Cell concentrations in soils were about 0.5×107 CFU per gram soil (g/dw) according to Franz et al. [22]. Briefly, the cell suspension was thoroughly mixed with soil in a plastic bag and 500 gram of the inoculated soil was transferred to a top perforated plastic bag for air exchange. The same amount of non-inoculated soil was put into another plastic bag, which was used as uninoculated control, with deionized water added instead of cell suspension. The experiment use triplicate bags of soils. The plastic bags were weighed and incubated at 10°C in darkness. Moisture content of the soil sample were adjusted to 60% water holding capacity (WHC), and water concentration was maintained during the course of experiment by adding additional deionized water weekly to obtain the original weight. Antibiotics were added into the agar media at the following concentrations, kanamycin (Km), 50 µg/ml; chloramphenicol (Cm), 25 µg/ml; rifampicin (Rif), 100 µg/ml; and nalidixic acid (Nal), 25 µg/ml.
Sampling and enumeration
The inoculated soils were sampled periodically to determine the survival of the wild-type and mutant strains over time. At each point, two samples (1.0 g) of each triplicate bag was removed from the middle of the soil sample and put into pre-weighed dilution tubes. The tubes containing soil samples were weighted to calculate the exact size of soil sample. A 5.0 ml of 0.1% peptone buffer (Lab M, Lancashire, UK) was added to the test tube containing the soil sample, and the soil was thoroughly mixed with the buffer by inverting the tube several times and then vortexed for 2×20 s. The resulting soil paste (cell suspension) was then subjected to 10-fold serial dilutions. Fifty µl of the two highest dilutions were plated in duplicate on SMAC/BCIG agar with appropriate antibiotics for enumeration. The inoculated SMAC agar plates were incubated at 37°C for 16 h, and the results expressed as log colony forming units per gram dry weight (CFU g/dw). The detection limit of the plating method was approximately 100 CFU g/dw. Our preliminary experiments showed that the average cell recovery rate of the method was from 90 to 110% of the theoretical value.
Survival data
Survival of E. coli O157:H7 was modeled by fitting the experimental data to the double Weibull survival model proposed by Coroller et al. [40] using GInaFiT version 1.5 developed by Dr. Annemie Geeraerd at Katholieke Universiteit, Leuven, Belgium [41]. The double Weibull survival model was constructed based on the hypothesis that the population is composed of two subpopulations differing in their capability on resistance to stress, and deactivation kinetics of both subpopulations follows a Weibull distribution. The size of the surviving population can be calculated using equation 1,
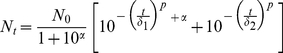 |
(1) |
| (2) |
Where N is the number of survivors, N 0 is the inoculum size; t is the time; p is the shape parameter, when p>1 a convex curve is observed; when p<1 a concave curve is observed, when p = 1 a linear curve is observed. The scale parameter, δ, represents the time needed for first decimal reduction; f, varying from 0 to 1, is the fraction of subpopulation 1 in the population. Another parameter, α, varying from negative infinity to positive infinity, is obtained by logit transformation of f as shown in equation 2. The strong correlation between the scale (δ) and the shape (p) parameters makes the double Weibull model to fit most of the shapes of deactivation curves. Previous study proved that the double Weibull model can successfully describe a biphasic shape with nonlinear decrease, which can not be described by other survival models [40]. Additionally, when δ 1 = δ 2, the double Weibull model can be simplified into a single Weibull model, and the survival curve can be described by only three parameters. A very important and useful parameter, Td (time needed to reach detection limit, 100 CFU g/dw) can also be calculated when using GInaFiT to fit the experimental survival data.
Statistical analysis
Analysis of variance (ANOVA) was performed to investigate the differences in growth in rich medium, and the survival in soils using SPSS 16.0 software package (Chicago, IL).
Results
Mutant construction and confirmation
The genome of E. coli O157:H7 EDL933 has been fully sequenced and annotated [42], which makes it possible to knock out the genes of interest. The multiplex PCR assay (Fig. 1) clearly showed that the wild type strains displayed three bands representing the amplicons from eae, stx 2, and stx 1 genes, from top to bottom, with predicted sizes of 384, 255, and 180 bp, respectively. For the mutant derivatives, there was one band missing for Δstx 1, Δstx2, and Δeae, and two bands missing for the double mutant construct Δstx 1–2, compare to the wild type strain. Figure 1 clearly showed that the virulence factors were successfully deleted as evidenced by the missing of the corresponding bands on the agarose gel.
Growth in rich medium and survival in soils
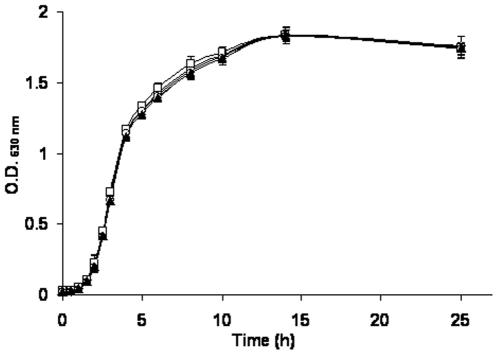
The growths of the mutant strains in LB broth were compared with that of the wild type (Fig. 2). The results showed that the E. coli O157:H7 EDL933 mutant derivatives growth was not significantly different from that of the wild type. Overall, there was about 1.5 h lag time followed by an exponential phase (5 h from incubation), then stationary phase (8 h post inoculation), and no decay phase was observed until 25 h of growth in LB broth under 37°C. The calculated apparent growth rates (r) of wild type, Δstx 1, Δeae, Δstx 2, and Δstx 1–2 were as following, 0.46±0.02, 0.46±0.03, 0.46±0.03, 0.47±0.04, and 0.46±0.03.
Figure 2. Growth curves of wild type strain (•) and its derivative mutants strains, Δstx 1 (◊), Δeae (▴), Δstx 2 (□), and Δstx 1–2 (○).
The data represent the average of triplicate measurements.
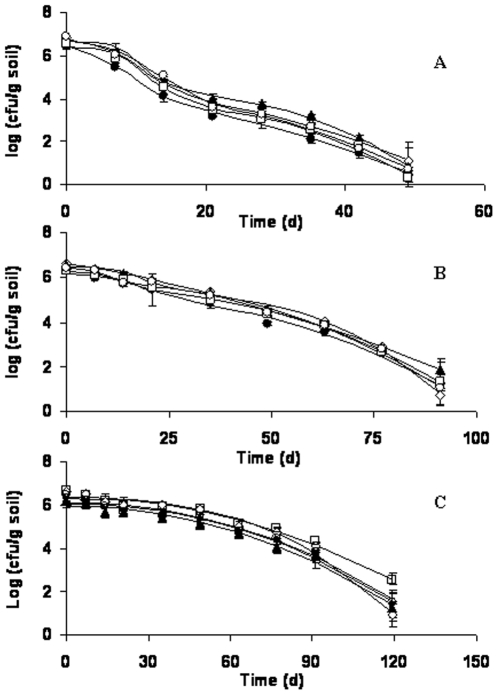
The wild type strain and the mutant strains were inoculated in soils to test their survival at 10°C. The results (Fig. 3) showed that within the same soil, there were no significant differences in deactivation profiles between the mutant and the wild type strains. It was also observed that the survival varies greatly in different soils. The cells survived shortest (32 day) in loamy sand with less nutrients (Fig. 3A), longest survival (113 day) was found in silty clay soil where there are more finer particles and more nutrient (e.g., organic carbon, nitrogen) (Fig. 3C), while the survival length was intermediate (82 day) in sandy loam soil. In loamy sand (Fig. 3A), there was a sharp decline of cell population within the first two weeks post inoculation, followed by a steady decrease until cell concentration dropped below detection limit. In sandy loam (Fig. 3B), a similar trend was also observed, a quick drop during first two weeks followed by a progressive decline. While in the silty clay (Fig. 3C), cells survived longer, because the cell concentrations did not decline significantly until four weeks post inoculation. Here after, cells started a very slow decline and dropped below detection limit (100 CFU g dw−1) after 113 days.
Figure 3. Survival of the wild type (•) and its mutant derivatives, Δstx 1 (◊), Δeae (▴), Δstx 2 (□), and Δstx 1–2 (○), in loamy sand (3A), sandy loam (3B), and silty clay (3C).
The data represent the average of triplicate experiments.
Modeling of survival data
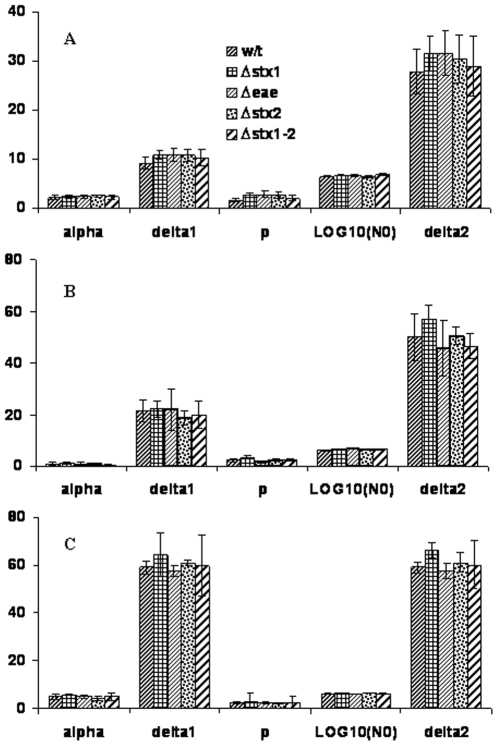
To accurately compare the survival kinetics between the wild type and mutant strains, survival data were modeled using a double Weibull equation as shown in Fig. 4. Similar modeling parameters (α, δ, and p) from mutant strains and the wild type strain were calculated when they were inoculated into the same soil. However, more variations in these parameters were observed from different soils, especially the δ values. When these strains were characterized in loamy sand and sandy loam soils, distinct δ 1 and δ 2 were observed indicating that the two subpopulations behave differently in both soils. The subpopulation with greater δ value declines slower than the one with smaller δ value. In contrast, almost identical δ 1 and δ 2 values were calculated from the survival data in silty clay soil indicating that the two subpopulations of cells in this soil likely behave similarly, thus the survival data in silty clay might be simplified into one Weibull model that can be described by only three parameters, α, δ and p. The initial sharp decrease in cell numbers in loamy sand soil might largely be attributed to the faster decline of subpopulation with smaller δ. However, with the time, the subpopulation with greater δ dominated the cell population, leading to a slower and steadier decline of the cell concentrations. A similar trend was also observed in sandy loam soil. However, in silty clay soil, the cell concentrations did not change until 3 weeks post inoculation. This was followed by a steady decrease in cell concentrations until the population dropped below the detection limit (57–66 days).
Figure 4. Double Weibull Model parameters of wild type strain and its mutant derivatives in loamy sand (4A), sandy loam (4B), and silty clay (4C).
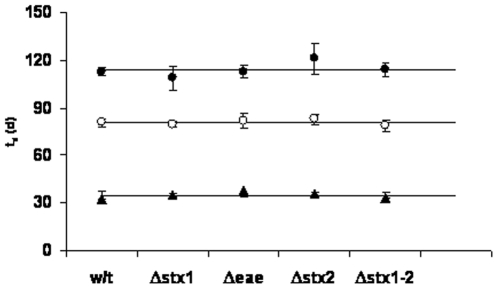
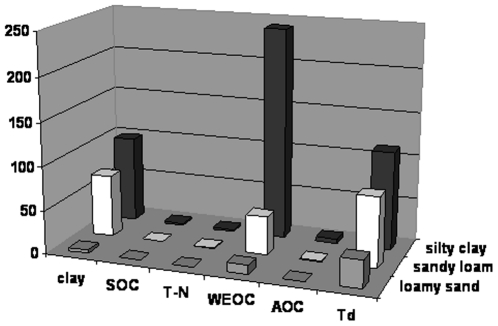
The time to reach detection limit (Td) between the wild type and mutant strains in the same soil was not significantly different (P = 0.05) (Fig. 5). Td values in soils follow the order of, silty clay>sandy loam>loamy sand, which is consistent with the order of fine particle and nutrient levels in the soils. The effect of soil properties on the survival of E. coli O157:H7, and the time that it takes for the pathogen to reach detection limit was determined (Fig. 6). The results showed that with the increase in clay content, total organic carbon, total nitrogen, and water extractable organic carbon, there was a corresponding increase in Td values.
Figure 5. Td values calculated from the double Weibull model for wild type and its mutants derivatives in loamy sand (▴), sandy loam (○), and silty clay (•).
The data represent the average of triplicate modeling of the raw survival data.
Figure 6. Effects of clay content (%), soil organic carbon (OC, %), total nitrogen (T-N, %), water extractable organic carbon (WEOC, mg/kg) Td.
Gray, white, and black columns represent loamy sand, sandy loam, and silty clay, respectively.
Discussion
The most significant finding of this work is Shiga toxins and intimin have no influence on the survival of pathogenic E. coli O157:H7 EDL933 in the three soils. The Shiga toxins stx 1, stx 2 genes, and eae gene in E. coli O157:H7 have been intensively investigated [43], [44]. Shiga toxins might induce an advantage in E. coli O157:H7 survival in the environment [44], [45], [46]. However, the role of these genes in survival of the pathogenic E. coli O157:H7 is still not completely understood [20]. Most of the previous survival studies used nonpathogenic E. coli O157:H7 strains [30], [24], [31], [32], and the survival data based on pathogenic strains in the environment are not available due to regulations and safety concerns [27], [28]. The typical nonpathogenic E. coli O157:H7 strain widely used in the literature include a green fluorescence protein labeled strain (E. coli O157:H7 B6-914 GFP-91) constructed by Fratamico et al. [47] and a bioluminescent construct (E. coli O157:H7 Tn5, luxCDABE) by Ritchie et al. [31], both of which have been shown to survive in soil for months. Researchers used these nonpathogenic strains in the survivals studies on the assumption that they behave the same with the pathogenic strains. Therefore, comparative studies relating the survival of pathogenic E. coli O157:H7 strains to nonvirulent strains are needed to make a firm conclusion. The study by Kudva and colleagues [7] revealed that identical or very similar survival patterns were observed within a Shiga toxin positive E. coli O157:H7 strain (ATCC 43894) and Shiga toxin negative E. coli O157:H7 strain (ATCC 43888), indicating that Shiga toxins might have little or no influence on E. coli O157:H7 survival in manure and manure slurry. However, the strains used in their analysis were not isogenic, and factors other than Shiga toxins may have contributed to the minor survival differences observed in that study. In the present study, we have constructed a cluster of mutant derivatives from E. coli O157:H7 EDL933, with one of the following virulence factors deleted, stx 1, stx 2, stx 1–2, and eae. The indistinguishable growth curves between the mutants and the wild type strains in rich medium, in combination with their similar survival profiles in three different soils, offer strong evidence that the Shiga toxin genes and eae gene do not likely play important role in the survival of E. coli O157:H7 in soils.
In the current study, the survival data were successfully modeled by the double Weibull model. Different models were fitted into the survival data, but the best fit was obtained by applying double Weibull model. Since double Weibull model was based on the assumption that there are two subpopulations, and they differs in level of resistance to stress, and the survival of both subpopulations follow a Weibull distribution. Subpopulation with smaller δ die off faster compared to the other subpopulation with greater δ. In loamy sand, and sandy loam soils, distinct δ values, i.e. δ1≠δ2, were obtained for the two subpopulations, indicating that the two subpopulation exhibit different resistant capability in both soils. On the other hand, almost identical δ values, i.e. δ1≈δ2, were observed for the two subpopulations in silty clay soil, implying that the two subpopulations show a similar survival behavior in silty clay soil.
The persistence of E. coli O157:H7 is highly dependent on soil types, since distinct persistence time (Td) of this pathogen varies significantly in different soils in terms of soil chemistry and texture. The longest survival was observed in silty clay soil, while the shortest survival was found in loamy sand soil. The results of soil characterization revealed that the silty clay soil is most abundant in clay, organic carbon, total nitrogen, and water extractable organic carbon, while the least abundant of those fractions is found in loamy sand soil. The variation in Td was best explained by the clay content in soils, since Td was closely correlated with the clay content. This agrees with the fact that the pathogens survived longer in finer-textured (clayey) than in courser (sandy) soils under similar environmental conditions [48]. Colonization of soil particles and aggregates is thought to be critical for the inoculated bacteria to survive in soil [49]. Finer textured soils (clayey) compared to coarser textured soils (sandy) may provide protective pore spaces to improve the survival of soil bacteria [50]. Indeed, the survival of a bacterial pathogen in 23 soils types was found to be positively correlated with soil clay content, in addition to other factors [51]. Indeed, greater survival of E. coli in sediment rich in clay (>25%) has been observed [25].Similarly, survival of E. coli O157:H7 was primarily determined by the soil texture, with prolonged survival associated with more clay particles compared with sand particles [22], [52], [53]. In addition to soil texture, soil chemistry characteristics, such as organic carbon, total nitrogen, and water extractable organic carbon, were also found to be positively related to survival of E. coli O157:H7. In our study, the availability of nutrient, such as nitrogen and organic carbon in soil were found to correlate with the pathogen survival in soils. Recently, Franz et al. [22] showed that the survival of E. coli O157 in 36 soils can best be explained by dissolved organic carbon and the ratio of dissolved organic carbon to microbial biomass carbon. In addition to soil texture and soil chemistry, biological factors cannot be neglected when interpreting the survival data of E. coli O157:H7 in soils. Overall, soils that are rich in clay or organic carbon might be a good secondary medium for extended persistence of E. coli O157:H7. Special attention should be paid to such soils when evaluating the environmental risk associated with E. coli O157:H7. The studies by the above authors and a recent review [54], to the best of our knowledge, have produced the most up to date data on survival of E. coli O157:H7 in soil. The review showed that temperature, soil structure, and microbial communities are the most important factors affecting survival. These authors showed from their previous studies [32] that the survival of E. coli O157:H7 was inversely proportional to the diversity of the microbial community established through differential fumigation and regrowth activities. Niche dependency strategy has also been suggested as a mechanism for E. coli O157:H7 survival in the open environment [55] rather than the biphasic growth model tested in this study. This argument is based on nutrient availability as the most important physiological factor for survival of E. coli O157 in nutrient-limited environment. However, we did not test this phenomenon in this study, but further studies in our laboratory will be looking at this in the nearest future.
In summary, stx 1, stx 2, and eae genes conferred in E. coli O157:H7 EDL933 did not play any direct role in survival of this pathogen in soil because the isogenic mutant strains showed indistinguishable survival profiles in three soils with distinct soil chemistry. The survival results obtained based on the non-pathogenic isogenic E. coli O157 strains from this study might be safely extrapolated to be equivalent to data obtained from pathogenic strains since the survival data from pathogenic strains in the environment are not available due to regulations and safety concerns. However, other conditions should be considered, e.g., genes other than stx and eae that might be important in E. coli O157 survival in the environment. Best management practices (BMPs) and good agricultural practices (GAPs) must be followed when leafy greens are grown in soils with high clay and organic carbon contents to reduce the risk of such soils being contaminated with E. coli O157:H7.
Acknowledgments
Mention of trademark or propriety products in this manuscript does not constitute a guarantee or warranty of the property by the United States Department of Agriculture and does not imply its approval to the exclusion of other products that may also be suitable.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National Research Initiative/National Institute of Food and Agriculture Agreement No. 2008-35201-18709 and the 206 Manure and Byproduct Utilization Project of the United States Department of Agriculture-Agriculture Research Service. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Riley L, Remis RS, Helgerson SD, McGee HB, Wells JG, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:981–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 2.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffin PM, Tauxe RV. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 4.Carter AO, Borczyk AA, Carlson JA, Harvey B, Hockin JC, et al. A severe outbreak of Escherichiacoli O157:H7.associated hemorrhagic colitis in a nursing home. N Engl J Med. 1987;317:1496–500. doi: 10.1056/NEJM198712103172403. [DOI] [PubMed] [Google Scholar]
- 5.Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. Escherichia coli cytotoxin, haemolytic–uraemic syndrome, and haemorrhagic colitis. Lancet. 1983;2:1299–1300. doi: 10.1016/s0140-6736(83)91167-4. [DOI] [PubMed] [Google Scholar]
- 6.Chapman PA, Siddons CA, Cerdan-Malo AT, Harkin MA. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol Infect. 1997;119:245–250. doi: 10.1017/s0950268897007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudva IT, Hatfield PG, Hovde CJ. Characterisation of Escherichia coli O157:H7 and other shiga toxin-producing E. coli serotypes isolated from sheep. J Clin Microbiol. 1997;35:892–899. doi: 10.1128/jcm.35.4.892-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice DH, Hancock DD, Besser TE. Verotoxigenic E. coli colonisation of wild deer and range cattle. Vet Rec. 1995;137:524. doi: 10.1136/vr.137.20.524. [DOI] [PubMed] [Google Scholar]
- 9.Wallace JS, Cheasty T, Jones K. Isolation of verocytotoxin producing Escherichia coli O157 from wild birds. J Appl Microbiol. 1997;82:399–404. doi: 10.1046/j.1365-2672.1997.00378.x. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong GL, Hollingsworth J, Morris JG., Jr Emerging foodborne pathogens, Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. 1996;18:29–51. doi: 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- 11.Chase-Topping M, Gally D, Low C, Mathews L, Woolhouse M. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nature Rev Microbiol. 2008;6:904–912. doi: 10.1038/nrmicro2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cieslak PR, Barrett TJ, Griffin PM, Gensheimer KF, Beckett G, et al. Escherichia coli O157:H7 infection from a manured garden. Lancet. 1993;342:367. doi: 10.1016/0140-6736(93)91509-k. [DOI] [PubMed] [Google Scholar]
- 13.Morgan GM, Newman C, Palmer SR, Allen JB, Shepherd W, et al. First recognized community outbreak of haemorrhagic colitis due to verotoxin-producing Escherichia coli O157 in the UK. Epidemiol Infect. 1988;101:83–91. doi: 10.1017/s0950268800029241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artz RRE, Killham K. Surivival of Escherichia coli O157:H7 in private drinking water wells influences of protozoan grazing and elevated copper concentration. FEMS Microbiol Lett. 2002;216:117–122. doi: 10.1111/j.1574-6968.2002.tb11424.x. [DOI] [PubMed] [Google Scholar]
- 16.Avery LM, Williams AP, Killham K, Jones DL. Survival of Escherichia coli O157:H7 in waters from lakes, rivers, puddles and animal-drinking troughs. Sci Total Environ. 2007;389:378–385. doi: 10.1016/j.scitotenv.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 17.McGee P, Bolton DJ, Sheridan JJ, Earley B, Kelly G, et al. Survival of Escherichia coli O157:H7 in farm water, its role as a vector in the transmission of the organism within herds. J Appl Microbiol. 2002;93:706–13. doi: 10.1046/j.1365-2672.2002.01752.x. [DOI] [PubMed] [Google Scholar]
- 18.Ravva SV, Sarreal CZ, Duffy B, Stanker LH. Survival of Escherichia coli O157:H7 in wastewater from dairy lagoons. J Appl Microbiol. 2006;101:891–902. doi: 10.1111/j.1365-2672.2006.02956.x. [DOI] [PubMed] [Google Scholar]
- 19.Watterworth L, Rosa B, Schraft H, Topp E, Leung KT. Survival of various ERIC-genotypes of Shiga toxin-producing Escherichia coli in well water. Water, Air, Soil Pollut. 2006;177:367–382. [Google Scholar]
- 20.Bolton DJ, Byrne CM, Sheridan JJ, McDowell DA, Blair IS. The survival characteristics of a non-pathogenic strain of Escherichia coli O157:H7. J Appl Microbiol. 1999;86:407–411. doi: 10.1046/j.1365-2672.1999.00677.x. [DOI] [PubMed] [Google Scholar]
- 21.Kudva IT, Blanch K, Hovde CJ. Analysis of Escherichia coli O157:H7 survival in ovine or bovine manure and manure slurry. Appl Environ Microbiol. 1998;64:3166–3174. doi: 10.1128/aem.64.9.3166-3174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams AP, McGregor KA, Killham K, Jones DL. Persistence and metabolic activity of Escherichia coli O157:H7 in farm animal faces. FEMS Microbiol Lett. 2008;287:168–173. doi: 10.1111/j.1574-6968.2008.01310.x. [DOI] [PubMed] [Google Scholar]
- 23.Franz E, Semenov AV, Termorshuizen AJ, de Vos OJ, Bokhorst JG, et al. Manure-amended soil characteristics affecting the survival of E. coli O157:H7 in 36 Dutch soils. Environ Microbiol. 2008;10:313–327. doi: 10.1111/j.1462-2920.2007.01453.x. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Morgan J, Doyle MP. Fate of Escherichia coli O157:H7 in Manure-Amended Soil. Appl Environ Microbiol. 2002;68:2605–2609. doi: 10.1128/AEM.68.5.2605-2609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenov A, Franz E, van Overbeek L, Termorshulizen AJ, et al. Estimating the stability of Escherichia coli O157:H7 survival in manure-amended with different management histories. Environ Microbiol. 2008;10:1450–1459. doi: 10.1111/j.1462-2920.2007.01558.x. [DOI] [PubMed] [Google Scholar]
- 26.Burton GA, Jr, Gunnison D, Lanza GR. Survival of pathogenic bacteria in various freshwater sediments. Appl Environ Microbiol. 1987;53:633–638. doi: 10.1128/aem.53.4.633-638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaLiberte P, Grimess DJ. Survival of Escherichia coli in lake bottom sediment. Appl Environ Microbiol. 1982;43:623–628. doi: 10.1128/aem.43.3.623-628.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell GR, Prosser J, Glover A, Killham K. Detection of Escherichia coli O157:H7 in soil and water using multiplex PCR. J Appl Microbiol. 2001;91:1004–1010. doi: 10.1046/j.1365-2672.2001.01465.x. [DOI] [PubMed] [Google Scholar]
- 29.Topp E, Welsh M, Tien Y-C, Dang A, Lazarovits G, et al. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol Ecol. 2003;44:303–308. doi: 10.1016/S0168-6496(03)00055-2. [DOI] [PubMed] [Google Scholar]
- 30.Vidovic S, Block HC, Korber DR. Effect of soil composition, temperature, indigenous microflora, and environmental conditions on the survival of Escherichia coli O157:H7. Can J Microbiol. 2007;53:822–829. doi: 10.1139/W07-041. [DOI] [PubMed] [Google Scholar]
- 31.Ritchie JM, Campbell GR, Shepherd J, Beaton Y, Jones D, et al. A Stable bioluminescent construct of Escherichia coli O157:H7 for hazard assessments of long-term survival in the environment. Appl Environ Microbiol. 2003;69:3359–3367. doi: 10.1128/AEM.69.6.3359-3367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Elsas JD, Hill P, Chroňáková A, Grekova M, Topalova Y, et al. Survival of genetically marked Escherichia coli O157:H7 in soil as affected by soil microbial community shifts. ISME J. 2007;1:204–214. doi: 10.1038/ismej.2007.21. [DOI] [PubMed] [Google Scholar]
- 33.Metcalf WW, Jiang WH, Daniels LL, Kim SK, Haldimann A, et al. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 34.Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx 1, stx 2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klute A. Methods of soil analysis, part 1, physical and mineralogical methods (2nd edition), American Society of Agronomy, Agronomy Monographs 9(1), Madison, Wisconsin. 1996 [Google Scholar]
- 36.Vance ED, Brookes PC, Jenkinson DS. An extraction method for measuring soil microbial biomass C. Soil Biol Biochem. 1987;19:703–707. [Google Scholar]
- 37.Liang BC, MacKenzie AF, Schnitzer M, Monreal CM, Voroney PR, et al. Management-induced change in labile soil organic matter under continuous corn in eastern Canadian soils. Biol Fertil Soils. 1998;26:88–94. [Google Scholar]
- 38.Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial cell. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 39.Vital M, Hammes F, Egli T. Escherichia coli O157 can grow in natural freshwater at low carbon concentrations. Environ Microbiol. 2008;10:2387–2396. doi: 10.1111/j.1462-2920.2008.01664.x. [DOI] [PubMed] [Google Scholar]
- 40.Coroller L, Leguerinel I, Mettler E, Savy N, Mafart P. General model, based on two mixed Weibull distributions of bacterial resistance, for describing various shapes of inactivation curves. Appl Environ Microbiol. 2006;72:6439–6502. doi: 10.1128/AEM.00876-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geeraerd AH, Valdramidis VP, van Impe JF. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Intl J Food Microbiol. 2005;102:95–105. doi: 10.1016/j.ijfoodmicro.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 42.Perna N, Plunkett G, III, Burland V, Mau B, Glasner JD, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 43.Besser RE, Griffin PM, Slustsker L. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infection disease. Annu Rev Med. 1999;50:355–367. doi: 10.1146/annurev.med.50.1.355. [DOI] [PubMed] [Google Scholar]
- 44.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 45.Lainhart W, Stolfa G, Koudelka GB. Shiga Toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J Bacteriol. 2009;191:5116–5122. doi: 10.1128/JB.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Loughlin E, Robins-Browne RM. Effect of Shiga toxins on eukaryotic cells. Microbes Infect. 2001;3:493–507. doi: 10.1016/s1286-4579(01)01405-8. [DOI] [PubMed] [Google Scholar]
- 47.Fratamico PM, Deng MY, Strobaugh TP, Palumbo SA. Construction and characterization of Escherichia coli O157:H7 strains expressing firefly luciferase and Green fluorescent protein and their use in survival Studies. J Food Prot. 1997;60:1167–1173. doi: 10.4315/0362-028X-60.10.1167. [DOI] [PubMed] [Google Scholar]
- 48.van Elsas JD, Dijkstra AF, Govaert JM, van Veen JA. Survival of Pseudomonas fluorescens and Bacillus subtilis introduced into two soils of different texture in field microplots. FEMS Microbiol Ecol. 1986;38:151–160. [Google Scholar]
- 49.Hattori T, Hattori R. The physical environment in soil microbiology, an attempt to extend principles of microbiology to soil microorganisms. CRC Crit Rev Microbiol. 1976;4:423–461. doi: 10.3109/10408417609102305. [DOI] [PubMed] [Google Scholar]
- 50.van Veen JA, van Overbeek LS, van Elsas JD. Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bashan Y. Enhancement of wheat root colonization and plant development by Azospirillum brasilense Cd following temporary depression of rhizosphere microflora. Appl Environ Microbiol. 1986;51:1067–1071. doi: 10.1128/aem.51.5.1067-1071.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mubiru DN, Coyne MS, Grove JH. Mortality of Escherichia coli O157:H7 in two soils with different physical and chemical properties. J Environ Qual. 2000;29:1821–1825. [Google Scholar]
- 53.Nicholson FA, Groves SJ, Chambers BJ. Pathogen survival during livestock manure storage and following land application. Bioresource Technol. 2005;96:135–143. doi: 10.1016/j.biortech.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 54.van Elsas JD, Semenov AV, Costa R, Trevors JT. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 2011;5:173–183. doi: 10.1038/ismej.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franz E, van Bruggen AHC. Ecology of E. coli O157:H7 and Salmonella enterica in the primary vegetable production chain. Crit Rev Microbiol. 2008;34:143–161. doi: 10.1080/10408410802357432. [DOI] [PubMed] [Google Scholar]