Abstract
A multifaceted strategy using a composite of anti-cancer nanotherapeutic and natural biomaterials silk fibroin (SF) and chitosan (CS) blend scaffolds was investigated for the treatment of a tissue defect post-tumor resection by providing local release of the therapeutic and filling of the defect site with the regenerative bioscaffolds. The scaffold-emodin nanoparticle composites were fabricated and characterized for drug entrapment and release, mechanical strength, and efficacy against GILM2 breast cancer cells in vitro and in vivo in a rat tumor model. Emodin nanoparticles were embedded in SF and SFCS scaffolds and the amount of emodin entrapment was a function of the scaffold composition and emodin loading concentration. In vitro, there was a burst release of emodin from all scaffolds during the first 2 days though it was detected even after 24 days. Increase in emodin concentration in the scaffolds decreased the overall elastic modulus and ultimate tensile strength of the scaffolds. After 6 weeks of in vivo implantation, the cell density (p < 0.05) and percent degradation (p < 0.01) within the remodeled no emodin SFCS scaffold was significantly higher than the emodin loaded SFCS scaffolds, although there was no significant difference in the amount of collagen deposition in the regenerated SFCS scaffold. The presence and release of emodin from the SFCS scaffolds inhibited the integration of SFCS into the adjacent tumor due to the formation of an interfacial barrier of connective tissue that was lacking in emodin-free SFCS scaffolds. While no significant difference in tumor size was observed between the in vivo tested groups, tumors treated with emodin loaded SFCS scaffolds had decreased presence and size and similar regeneration of new tissue as compared to no emodin SFCS scaffolds.
Keywords: Silk fibroin, Emodin, Scaffold, Breast cancer cells, Tissue regeneration
Introduction
Local recurrence in 2–7.5% of breast cancers post-mastectomy defects is attributed to inadequate resection of tumor, undetected multifocal tumor, and lack of post-surgical radiation therapy.14 In addition, local recurrence is shown to be present exactly at the initial location of the primary tumor and found to be the first site of therapeutic failure.19,28 To overcome these issues, we developed the dual therapy approach by embedding an anti-cancer drug into a biodegradable tissue-regenerative scaffold in order to treat the mastectomy defect site (Fig. 1a). While the drug inhibits the proliferation of tumor cells, the scaffold fills the defect after tumor removal and supports the regeneration of local tissue within its controlled microenvironment.
Figure 1.
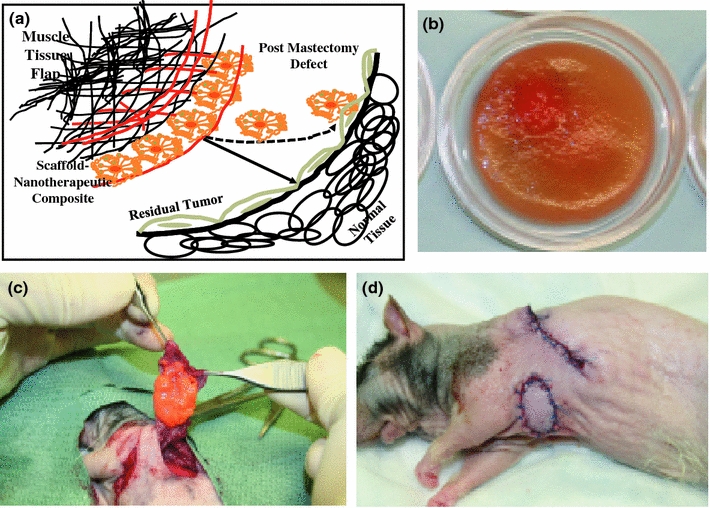
(a) A schematic of the cross-section of silk fibroin and chitosan blend (SFCS)–emodin nanoparticle–latissimus dorsi muscle (LDM) flap composite applied at the tumor resected breast tissue site for the delivery of a nanotherapeutic to treat residual tumor overlaying the normal breast tissue and reconstruct the defect site with the SFCS scaffold via the process of tissue regeneration. (b) SFCS scaffold (25:75 blend) loaded with emodin in a circular petri dish mold (average diameter: 2.6 ± 0.1 cm and thickness: 1.3 ± 0.04 mm). (c) At the time of treatment of the tumor resected breast tissue site, emodin-loaded SFCS scaffold was sutured on the underside of the LDM flap. (d) Wound closures after scaffold–nanoparticle–flap composite implantation at the tumor bearing breast tissue site
Natural biodegradable polymers, silk fibroin (SF) and chitosan (CS), were chosen to mimic the extracellular matrix (ECM) and to regenerate the native tissue similar to regeneration of other tissue types.3,8,27 SF protein polymer is derived from the silkworm Bombyx mori and is fibrous, highly permeable to oxygen and water, exhibits high strength with flexibility, has relatively low thrombogenicity, low inflammatory response, and supports cell adhesion and growth.2,13 CS is a deacetylated product of chitin and provides good wound healing properties, compressibility, and water storage capacity.22,30 Three-dimensional (3D) scaffolds comprising of various fractions of SF and CS were prepared and their structural and mechanical properties were examined previously.13 The SFCS scaffold was shown to support the regeneration of abdominal wall musculofascial defect,8 formation of critical-sized bone in an in vivo sheep model,27 and the wound healing of a dermal wound using stem cells.3
Scaffolds composed of either SF33 or CS17 and embedded with microparticles have been reported to be used for the delivery of biological agents. SF spheres31,34 and CS microcapsules1 have also been used previously as drug carriers. Collagen-CS scaffolds loaded with angiogenin were developed in order to promote angiogenesis in artificial dermis.29 A liposomal CS scaffold/human fibrin gel composite system was studied for the delivery of low-molecular weight hydrophilic drugs such as Tirofiban.32 Similar studies involving SF-derived curcumin nanoparticles have been conducted in order to provide long-term therapy against cancerous cells.12
In this study, anti-cancer drug ‘emodin’ was chosen as a therapeutic since it blocks phosphorylation of Her2/neu, which is over-expressed in many breast cancers.5,36 Emodin has shown impressive activity with low toxicity in vivo. 36,37 Recent study has also shown emodin-induced apoptosis in BCap-37 breast cancer cells through mitochondrial signaling pathway.15
We have previously shown the fabrication and characterization of SF-derived liposomal emodin.5,10 These particles were shown to exhibit high efficacy against Her2/neu over-expressing breast cancer cells due to targeting of multiple pathways.5,10 Since liposomes promote site-specific delivery with controlled and sustained release,16,25 emodin-loaded liposomes were entrapped within SFCS scaffold for this study. It was hypothesized that the liposomal emodin-loaded SFCS scaffolds when combined with a tissue flap as a composite would deliver anti-tumor therapy and reconstructs the mastectomy defect site simultaneously.
In this study, emodin-loaded liposomal nanoparticles were embedded in various blends of SF and CS scaffolds and characterized. Drug entrapment, release, and efficacy against breast cancer cells were analyzed in vitro. Efficacy of the emodin-loaded scaffolds was evaluated in an in vivo rat breast cancer model, where emodin-loaded SFCS scaffolds and a tissue flap were used to reconstruct the resected defect. Scaffold degradation, regeneration of new tissue, and tumor size were evaluated as end points at 6 weeks. Overall, the efficacy of locally delivered emodin and the regeneration/reconstruction with the SFCS scaffold of the resected tumor defect site was shown to be an effective treatment modality.
Materials and Methods
Preparation of Emodin-Loaded Liposomal Nanoparticles
Emodin (Sigma-Aldrich, St. Louis, MO) was weighed and dissolved in t-butanol at 1 mg/mL as described before.10 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC, Avanti Polar Lipids Inc., Alabaster, AL) was separately dissolved in t-butanol at 5 mg/mL and 5% of Tween 20 solution (10% v/v prepared in t-butanol) was added to DMPC solution. Emodin and DMPC solutions were then mixed together, frozen at −80 °C overnight and lyophilized. Dried DMPC and emodin mixture was suspended in phosphate buffer saline (PBS) at 2.86 mg/mL emodin concentration and mixed gently. Then, the emodin-loaded liposomes were sonicated using water sonicator (70 W, 42 kHz, Branson Ultrasonic Co., Danbury, CT) for various times (5, 30, and 60 min). Sonication was used to reduce the liposome size24 and to enhance the emodin entrapment.
Entrapment efficiency of emodin within liposomes was calculated by centrifuging liposomal emodin solution for 5 min at 1000 rpm.18 The pellet containing the free emodin was collected and the absorbance at 442 nm was measured using UV Spectrophotometer (ThermoSpectronics, Rochester, NY).
Dynamic Light Scattering (DLS)
The liposome size was analyzed using DLS. The measurements were performed on a Brookhaven Instruments Inc. 90 plus Particle Size Analyzer (Brookhaven Instruments Inc., Holtsville, NY). The emodin-loaded liposome particles were dispersed in filtered PBS buffer and volume based analysis was performed in duplicates for 2 min each to measure the mean diameter.
Preparation of Scaffolds with Entrapped Liposomal Emodin
The preparation of SF solution and its blend with CS has been described previously.9 Briefly, raw silk (Sao Paulo, Brazil) was degummed in 0.25% (w/v) sodium carbonate and 0.25% (w/v) sodium dodecylsulfate (Sigma-Aldrich) for 1 h at 100 °C to remove the sericin. The dried silk 10% (w/v) was dissolved in calcium nitrate tetrahydrate and methanol solution (molar ratio of 1:4:2 Ca:H2O:MeOH) at 65 °C. In order to make SFCS blends, CS was dissolved in 2% acetic acid and added to the SF in different proportions of SF and CS at 75:25, 50:50, and 25:75% (v/v). Pure SF solution and SFCS solution were dialyzed against deionized water.
Emodin-loaded liposomes were prepared as above after sonication for 5 min. Three different amounts of emodin-loaded liposomal nanoparticles (1.7, 2.8, and 4.0 mg emodin content) were mixed with 5 mL of SFCS blend solution or pure SF solution. The mixture was then poured into 35 × 10 mm petri dishes, frozen overnight at −80 °C, and lyophilized for 3 days. The dry scaffolds were then crystallized with 4 mL of 50:50 methanol:1 N sodium hydroxide solution to crystallize SF and to neutralize the pH of the acidic CS. The samples with pure SF were crystallized with 50:50 methanol:water. Before characterization, the scaffolds (Fig. 1b) were washed three times with PBS to remove excess methanol and sodium hydroxide, then sterilized in ethanol overnight, and again washed three times with PBS to remove the ethanol.
Emodin Entrapment and Release from Scaffolds
The entrapment of emodin in the final scaffold was calculated by subtracting the amount of released emodin during the crystallization, sterilization, and PBS washing steps. The released emodin contents were analyzed by measuring absorbance at 442 nm.
To measure the emodin release, small punches (6 mm diameter) of the emodin-loaded scaffold were placed in 1 mL of PBS in a 1.5 mL centrifuge tube at 37 °C incubator shaker. The absorbance was measured at different time points up to 24 days to analyze the released emodin.
Mechanical Testing
Uniaxial tensile test was performed using EnduraTEC mechanical tester (Bose, Eden Prairie, MN). The circular scaffold samples were first trimmed to a more rectangular shape and cut into three strips before being tested with a 225 N load cell at a strain rate of 0.1 mm per second. The stress–strain data were collected to calculate the elastic modulus (EM) and ultimate tensile strength (UTS).
Scanning Electron Microscopy
The scaffolds were fixed in 2% paraformaldehyde and 3% glutaraldehyde solution and imaged using JSM-590 scanning electron microscope (JEOL, USA, Inc., Peabody, MA) as described previously.30 Briefly, the samples were treated with a series of buffers, coated with aluminum alloy and imaged at an accelerating voltage of 5 kV. The SEM images were later analyzed using ImageJ software.
Cell Culture
Breast cancer cell line GILM2 was obtained from lung metastasis and developed in the laboratory of Dr. Janet Price at The University of Texas M. D. Anderson Cancer Center.21 The cells were cultured in Dulbecco Modified Eagles Medium with F-12 (Invitrogen, Grand Island, NY) and supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% antibiotics (Invitrogen).
MTT Assay for Cell Viability
Breast cancer cells were cultured in 96-well plates at 5000 cells per well overnight. Emodin at various concentrations (20–100 μM) was suspended in the culture medium and added to the cell-seeded wells. Also, the emodin-loaded scaffolds (4.0 mg emodin) and SFCS control scaffolds 6 mm (diameter) punches and placed in the wells. Large amounts of culture medium (300 μL) were used in the wells so that the scaffold would float and minimize cell suffocation at the bottom of the well. After 4 days of incubation, the Cell Quanti-MTT cell viability assay kit (BioAssay Systems, Hayward, CA) was used to determine the viability of cells remaining in each well. Briefly, 15 μL of MTT reagent was added to 80 μL of culture medium in each well. After 4 h of incubation, MTT solubilization solution (100 μL) was added to each well and plates were placed on a shaker for 1 h. The absorbance was then measured at 570 nm using MRX Microplate Reader (Dynex Technologies, Guernsey, Channel Islands, Great Britain).
In Vivo Implantation of Scaffold Composite
Nude rats underwent surgical manipulations with the approval of the Institutional Animal Care and Use Committee (IACUC) at The University of Texas M. D. Anderson Cancer Center. Human breast cancer cell lines GILM2 were injected into the L4 mammary fat pad of nude rats (200–300 g) to develop tumors. The tumor from a single rat was transferred to more rats when tumor mass grew over 50 mm3 based on the ellipsoid volume calculation7 in order to expand the number of tumor bearing rats. At the time of tissue flap composite reconstruction surgery, most of the tumor volume of 50 mm3 was resected and a volume of 10 mm3 remained for treatment with emodin eluting tissue flap composite. Animals were divided in three groups as reconstruction involved flap tissue only (7 rats), SFCS scaffold (8 rats), and emodin-loaded SFCS scaffold (8 rats). Nude rats were anesthetized with isoflurane (0.5%) and oxygen (2 L/min) by mask. The animal was placed on a rubber heating pad and normal body temperature maintained at 37 °C. The latissimus dorsi muscle flap (LDMF) that is commonly used to cover the wound in breast tumor reconstruction was raised from the back of the rats and placed over the resected breast tumor site with the SFCS scaffolds (with or without emodin) sutured and sandwiched between the LDMF and the tumor site (Fig. 1c). The surgical site was closed and animals were secured in their cages under normal conditions until harvest (Fig. 1d).
Histological Examination
After 6 weeks the animals were euthanized by an overdose of isoflurane. The reconstructed site containing the tissue flap–SFCS therapeutic composite and the underlying breast tissue were excised and harvested for histological examination. The excised tissue was cut in 2–6 parallel sections and fixed in 10% formalin, embedded in paraffin and sectioned (4–6 μm thickness). The sections were stained with hematoxylin–eosin (H&E) to assess scaffold degradation, tumor presence, and cellular analysis. The samples were also stained with Movat pentachrome, which stained cell nuclei and elastic fibers as black, collagen as yellow, muscle as red and glycosaminoglycans as blue color. All the stained sections were examined under a light microscope (Olympus 1X70, Olympus, Center Valley, PA) and imaged using a colored CCD camera (Retiga 4000R, QImaging, Surrey, Canada). Images were later analyzed in the ImageJ software to calculate the cell number and the area of tumor, scaffold, and collagen in a tissue section.
Statistical Analysis
The data were compared statistically using GraphPad Instat 3 program and significance level was chosen as p < 0.05. Also, one-way ANOVA with post hoc Tukey test was performed for pair-wise comparisons. All data was represented as mean ± standard error of mean.
Results
Effect of Sonication on Liposome Size and Emodin Entrapment
The size of the liposomes with no sonication showed a biomodal distribution, where 11% of the total fraction was between 131 and 150 nm (134 ± 5 nm) size and 89% between 361 and 435 nm (397 ± 13 nm) size as shown by the size distribution (Fig. 2a). After sonication of 5 min, the biomodal distribution remained although the size was significantly (p < 0.001) reduced to 79% of the total liposomal fraction between 31 and 44 nm (37 ± 3 nm) and the remaining at 302 ± 24 nm (Fig. 2b). Increasing the sonication time beyond 5 min did not decrease the liposomal size any further (Fig. 2c).
Figure 2.
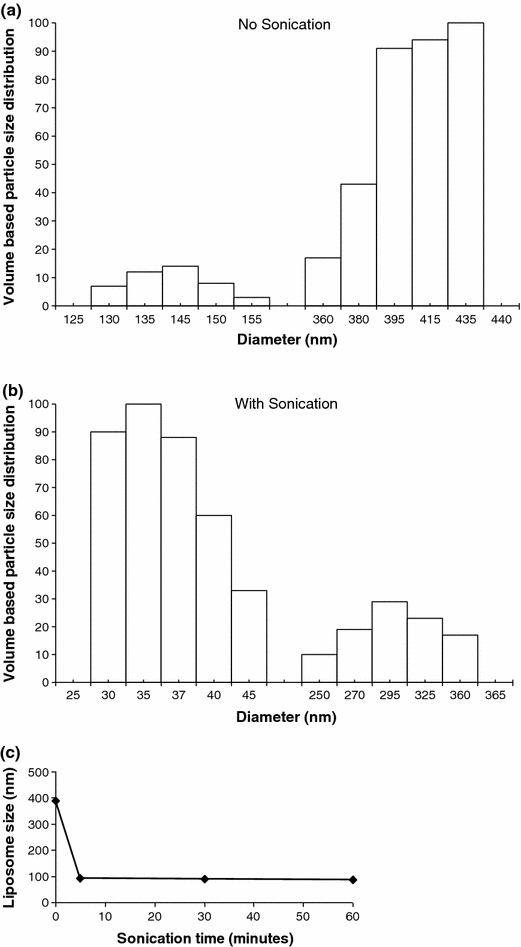
The size of the emodin-loaded liposomes was measured using dynamic light scattering. (a) The volume based size distribution of the emodin-loaded liposomes without sonication results in higher frequency of particles in >360 nm range. (b) Sonication of liposomal emodin after 5 min of sonication results in higher frequency of nanoparticles ranging in size from 30 to 45 nm. (c) Mean diameter of liposomal emodin after sonication of 5 min decreased and the size was not affected by further exposure to sonication up to 60 min
Characteristics of Liposomal Emodin-Loaded Scaffolds
Three different concentrations of emodin (1.7, 2.8, and 4.0 mg emodin/scaffold) were used to prepare the emodin-loaded SFCS and SF scaffolds. While the medium emodin loading concentration of 2.8 mg was used based on previously developed formulation of liposomal emodin that showed high efficacy against breast cancer cells,16 the concentration of 1.7 and 4.0 mg were studied as lower and upper limits for drug loading in order to minimize fragility of the SF based scaffolds and maximize emodin entrapment. The diameter and the thickness of all the scaffolds were fairly consistent among the different blends. The average diameter of emodin-loaded scaffold was 2.6 ± 0.1 cm and thickness was 1.3 ± 0.04 mm (Fig. 1b).
Emodin Entrapment and Release from Scaffolds
The emodin entrapment within liposomes was significantly higher (p < 0.01) after sonication (>99%) compared to no sonication (83.2 ± 4.9%) as shown in Fig. 3a. The entrapment efficiency of 100% suggests that there was no free emodin in the suspension. There was no difference in entrapment efficiencies with various sonication times. Emodin entrapment efficiencies were found to be the highest for SF (80–87%) as compared to the three SFCS blends (42–55%) as shown in Fig. 3b. Absolute emodin entrapment in SF scaffolds significantly increased with initial loading concentration (4.0 mg > 2.8 mg > 1.7 mg), although percent (%) entrapment efficiency (measured with respect to the initial emodin loading concentration) decreased with increased initial loading concentration (Fig. 3c). Percent emodin entrapment efficiency was significantly less (p < 0.05) for 4.0 mg initial loading concentration vs. the 1.7 mg concentration. For all three emodin loading concentrations, addition of CS (25–75%) to the SF based scaffolds significantly reduced the % entrapment efficiency and the amount of absolute emodin loading in the SFCS scaffolds as compared to the same emodin concentration in pure SF scaffolds. For all three SFCS blends, increase in absolute emodin loading concentration from 1.7 to 4.0 mg increased the amount of loaded drug significantly as evidenced by statistically significant differences between 1.7 mg concentration and 4.0 mg concentration, though there was no difference in % emodin entrapment efficiency due to initial emodin loading concentrations within the SFCS blends.
Figure 3.
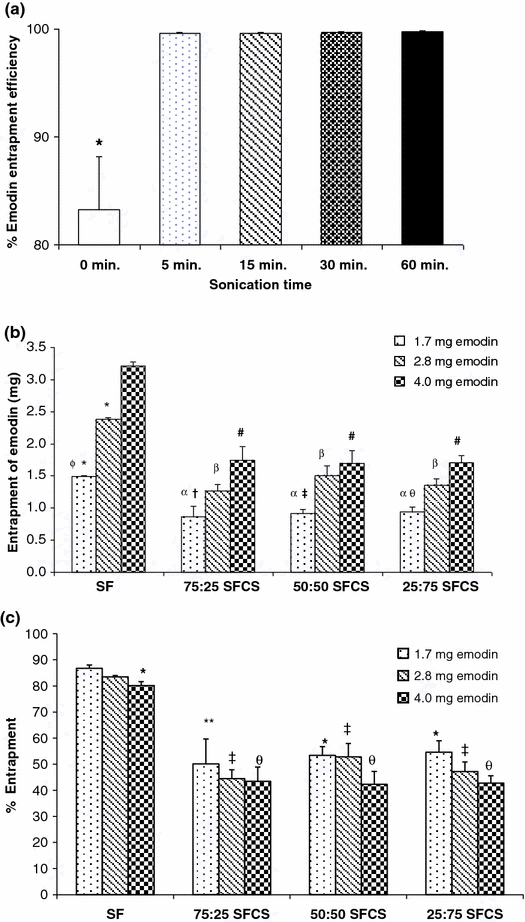
Emodin loading efficiency within the liposomes and embedding within the SF and SFCS scaffold blend formulations. (a) Entrapment efficiency of emodin within the DMPC liposomes increased after sonication of 5 min and remained at 100% efficiency with increased sonication time n = 3, *p < 0.01 vs. all other sonication times. (b) Absolute emodin entrapment within the liposomal nanoparticles embedded in SF and SFCS scaffold blends (n = 3). *p < 0.001 vs. SF, 4.0 mg emodin, φ p < 0.001 vs. SF, 2.8 mg emodin, α p < 0.05 vs. SF, 1.7 mg emodin, β p < 0.001 vs. SF, 2.8 mg emodin, # p < 0.001 vs. SF, 4.0 mg, † p < 0.05 vs. 75:25 SFCS, 4.0 mg emodin, ‡ p < 0.05 vs. 50:50 SFCS, 4.0 mg emodin, θ p < 0.01 vs. 25:75 SFCS, 4.0 mg emodin, emodin. (c) Percent entrapment of emodin within the liposomal nanoparticles embedded in SF and SFCS scaffold blends n = 3, *p < 0.05, **p < 0.01 vs. SF, 1.7 mg emodin, ‡ p < 0.001 vs. SF, 2.8 mg emodin, θ p < 0.001 vs. SF, 4.0 mg emodin. All data reported as mean ± SEM
While there was a burst release of emodin from scaffolds during the first 2 days (Fig. 4), released emodin amounts were detected (0.1–1.0 μg/mL) even after 24 days. SF scaffolds showed highest emodin release as compared to all SFCS blends (only 25:75 SFCS is shown in Figs. 4a, 4b). However, there was no difference in release between various blends for the same starting emodin concentration (data not shown). As shown in Fig. 4b for 25:75 SFCS, % emodin release was slightly higher for lower starting emodin concentration (1.7 mg) than higher concentrations (2.8 and 4.0 mg). However, the absolute amount of emodin release increased with an increase in starting emodin concentration (lowest for 1.7 mg and highest for 4.0 mg emodin). Similar trends were found for other SFCS blends and pure SF.
Figure 4.
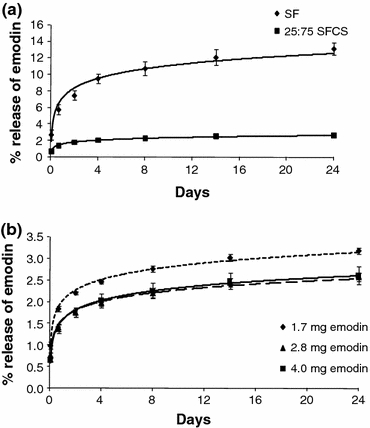
Representative curves for the emodin release from liposomal nanoparticle and SF based scaffolds. (a) Percent emodin released from SF and 25:75 SFCS scaffolds for emodin loading concentration of 4.0 mg shows higher amounts of emodin released from SF scaffolds as compared to the SFCS scaffolds (n = 3). (b) Comparison of percent emodin released from 25:75 SFCS with three different emodin loading concentrations (n = 3). All data reported as mean ± SEM
Mechanical Strength of Liposomal Emodin-Loaded Scaffolds
Representative stress vs. strain curves are shown for no emodin SF, no emodin 25:75 SFCS, and 1.7 mg emodin 25:75 SFCS (Fig. 5a). The EM and the UTS for all scaffolds with and without emodin were calculated from stress–strain curves. As shown in Fig. 5b, the elastic moduli of the SFCS controls (no emodin) were significantly lower than the SF control (no emodin) (p < 0.001). Addition of emodin to SF scaffolds for all concentrations reduced the EM significantly (p < 0.01). SFCS scaffolds for all three blends with 4.0 mg of emodin had significantly lower EM (p < 0.05 for 75:25 SFCS, p < 0.01 for 50:50 SFCS, and 25:75 SFCS) as compared to no emodin SFCS controls for their respective blends. While 75:25 SFCS scaffolds showed significantly lower EM vs. no emodin controls only at 2.8 and 4.0 mg (p < 0.05 for both) concentrations, 25:75 SFCS showed significantly lower EM for all three emodin loading concentrations.
Figure 5.
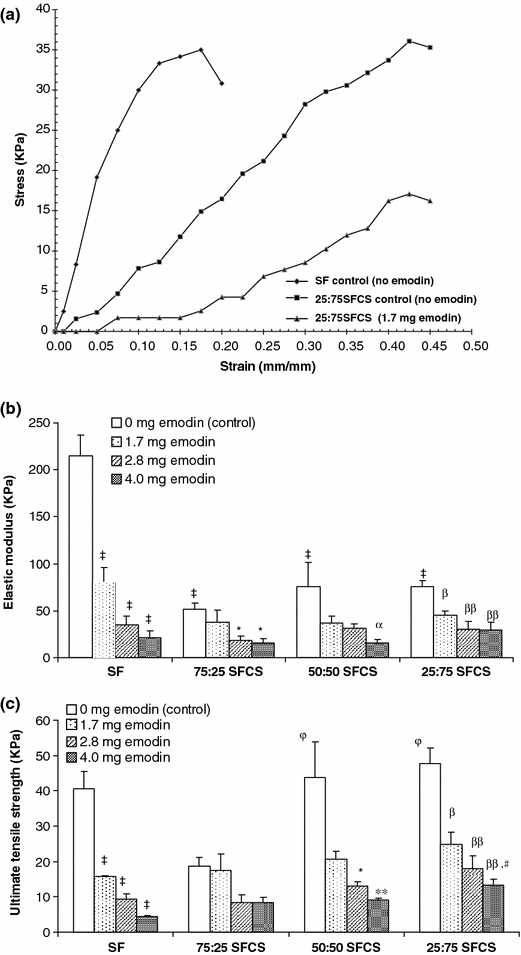
Mechanical properties of SF and SFCS scaffolds with and without emodin loading for the three concentrations were determined using uniaxial tensile testing. (a) Representative stress vs. strain curves for SF scaffolds (no emodin), 25:75 SFCS scaffolds (no emodin), and 25:75 SFCS scaffolds, 1.7 mg emodin are shown. (b) Comparison of the elastic modulus of SF and SFCS scaffolds with and without emodin nanoparticle loading (n = 6). ‡ p < 0.001 vs. SF no emodin control, *p < 0.05 vs. 75:25 SFCS no emodin control, α p < 0.01 vs. 50:50 SFCS no emodin control, β p < 0.05 and ββ p < 0.001 vs. 25:75 SFCS no emodin control. (c) Comparison of ultimate tensile strength (UTS) of SF and SFCS scaffolds with and without emodin nanoparticle loading (n = 6). ‡ p < 0.001 vs. SF no emodin control, *p < 0.05 and **p < 0.01 vs. 50:50 SFCS no emodin control, φ p < 0.05 vs. 75:25 SFCS no emodin control, β p < 0.01 and ββ p < 0.001 vs. 25:75 SFCS no emodin control, # p < 0.01 vs. SF (4.0 mg emodin). All data reported as mean ± SEM
Increase in emodin loading concentration resulted in significant decrease in UTS for SF scaffolds (p < 0.001) (Fig. 5c). The addition of 25% CS to pure SF control scaffolds appeared to decrease the UTS, although not significantly. On the other hand, increasing the amount of CS from 25 to 75% increased the UTS (p < 0.05), while showing no significant difference from the pure SF no emodin control scaffolds. Increase in emodin loading concentration had no effect on the UTS of 75:25 SFCS, significantly decreased the UTS for 50:50 SFCS only at 2.8 and 4.0 mg emodin concentrations, and decreased UTS for 25:75 SFCS at all emodin loading concentrations as compared to the respective no emodin blend controls (Fig. 5c). In addition, the UTS of SF scaffolds was lower than 25:75 SFCS scaffolds at 4.0 mg emodin loading concentration (p < 0.01).
Efficacy of Emodin Against GILM2 Breast Cancer Cells
Emodin dose dependant effects were tested in cell culture and the cell numbers were calculated from a standard curve between cell concentration and fluorescence readings using MTT assay. With no exposure to emodin, the number of GILM2 cells increased from 5000 to 12,000–18,000 per well by 4 days and were significantly (p < 0.001) higher than emodin exposed cells (Fig. 6a). As the emodin exposure concentration was increased from 20 to 100 μM, the cancer cell viability decreased significantly (p < 0.05). There was a dose dependant effect of emodin on GILM2 breast cancer cell efficacy.
Figure 6.
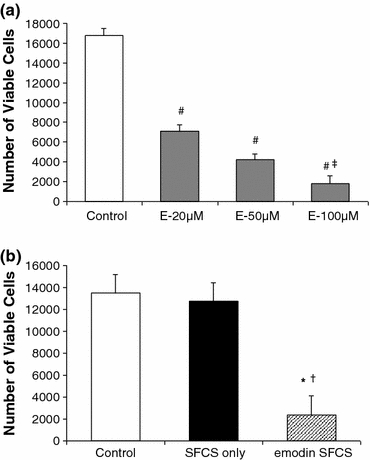
(a) Cell viability of GILM2 breast cancer cells measured using the MTT assay after 4 days of exposure to liposomal emodin nanoparticles. The increase in the emodin concentration decreased the number of viable cells (n = 3), # p < 0.001 vs. control, ‡ p < 0.05 vs. 20 μM emodin concentration. (b) Comparison of GILM2 breast cancer cell viability after exposure to liposomal emodin 25:75 SFCS, 4.0 mg emodin scaffolds with no emodin controls with and without the 25:75 SFCS scaffold (n = 3), *p < 0.01 vs. control, † p < 0.05 vs. SFCS only. All data reported as mean ± SEM
GILM2 breast cancer cell efficacy was also evaluated using 4.0 mg emodin 25:75 SFCS scaffolds (Fig. 6b). Exposure of GILM2 cells to emodin-loaded SFCS scaffolds significantly decreased the number of viable cells as compared to cells in culture dishes (no SFCS, no emodin, p < 0.01) and cells exposed to SFCS scaffolds only (no emodin p < 0.05).
In Vivo Efficacy of LDMF–Emodin SFCS Composite
The LDM flap tissue used for reconstruction in the in vivo model was divided into three layers: skin, connective tissue, and muscle (Figs. 7a, 7b). The tumor showed distinguished dense tissue with a very high concentration of cells, which interfaced with the muscle tissue flap, in the case of flap reconstruction only. At 6 weeks, the defect reconstruction with SFCS scaffold only (no emodin) resulted in the residual SFCS scaffold seamlessly integrating adjacent to the muscle site (Fig. 7b) and the tumor site (Fig. 7c), whereas the emodin-loaded SFCS scaffold appeared to integrate with a distinct interface between the tumor and the regenerated scaffold tissue (Fig. 7d). The tumor appeared to be isolated yet connected with the emodin-loaded SFCS scaffold via a new tissue interface. The release of emodin from the SFCS appears to block the tumor invasion into the neighboring regenerated SFCS scaffold. Both scaffolds integrated well at the muscle tissue flap interface evidenced by invasion of cells from the muscle and formation of new blood vessels (see arrows at the interface) at the interface (Fig. 7e). By 6 weeks, remodeling and regeneration of the SFCS scaffold was evidenced by cellular infiltration and new matrix deposition in both scaffold groups (Fig. 7f). Deposition of new matrix was evidenced by areas of deep yellow positive staining for collagen and light blue staining for glycosaminoglycans in regions of remodeling scaffold (Figs. 7e, 7f). The degraded scaffold fibrils were dispersed throughout the remodeled/regenerated tissue and stained in shades of red/magenta with H&E/Movat staining, respectively.
Figure 7.
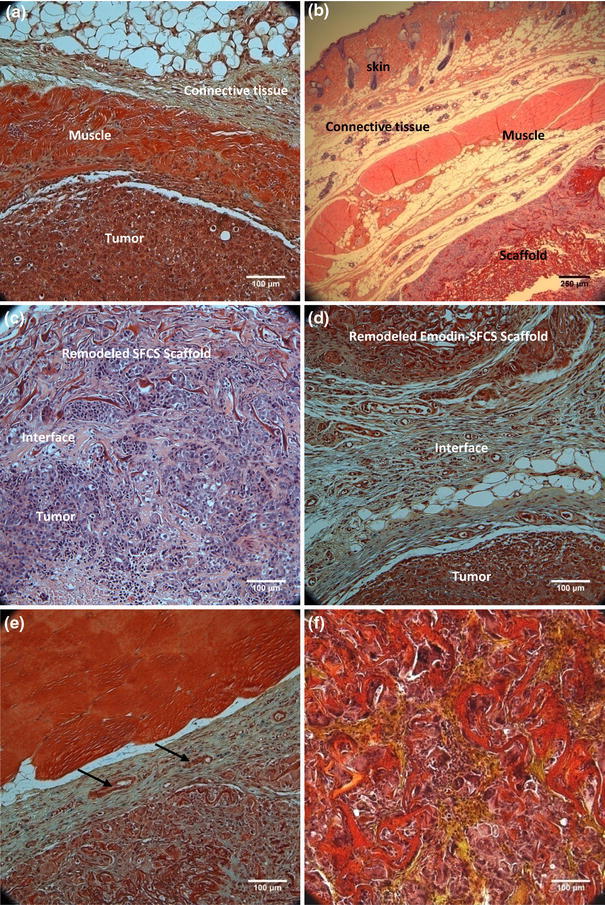
Histological microsections of the reconstructed GILM2 breast tumor site after 6 weeks of implantation of 25:75 SFCS-emodin nanoparticle-muscle flap composite stained with either H&E or Movat stains. (a) A Movat stained cross-section of the breast tumor site reconstructed with the LD muscle tissue flap only. (b) A H&E stained cross-section of the tumor site reconstructed with the SFCS scaffold shows the muscle flap components (skin, connective tissue, muscle) interfaced and well-integrated with the partially degraded emodin-loaded SFCS scaffold that contains the interspersed fibrils embedded in the regenerated tissue. (c) A H&E stained cross-section of tumor scaffold interface that was seamlessly integrated when reconstructed with SFCS scaffold-LDM flap composite with no emodin loading. (d) A Movat stained cross-section of remodeled emodin-loaded SFCS scaffold interfaced with a tumor via layers of connective tissue. (e) A Movat stained cross-section showing the blood vessels (see arrows) formed at the interface of the muscle and the emodin loaded SFCS scaffold. (f) A Movat stained cross-section of the SFCS scaffold showing partial degradation and remodeling of the scaffold. The remainder scaffold fibrils (red) are surrounded by collagen (yellow) deposited in the regenerated tissue
Quantification of In Vivo Therapeutic and Remodeling Response
Histological sections with H&E staining were evaluated for quantification of tumor presence and relative tumor size. Fewer slides (25%) showed tumor in the emodin-loaded SFCS scaffold group compared to flap only (40%) and SFCS scaffold only (33%) group. Although, no significant difference was observed in tumor size between groups, the emodin-loaded SFCS scaffold group showed decreased tumor presence and size (Table 1). Tumor was evident in 25% of the emodin-loaded SFCS scaffolds and the size of these tumors was slightly smaller than the SFCS scaffold only group.
Table 1.
Quantitative analysis of tumor size, scaffold degradation and cell density in the tissues after 6 weeks of in vivo implantation
| Groups | Tumor presence based on slide | Tumor size per microsection (mm2) | Cell density in scaffolds | Collagen in remodeling scaffold (%) | Scaffold degradation (%) |
|---|---|---|---|---|---|
| Flap only | 40% (6/15) | 1.02 ± 0.43 | na | na | na |
| SFCS scaffold only | 33% (7/21) | 1.47 ± 0.46 | 3238 ± 152 (n = 23) | 9.0 ± 1.8 (n = 6) | 94.3 ± 0.8 (n = 30) |
| Emodin-loaded SFCS scaffold | 25% (5/20) | 0.97 ± 0.57 | 2733 ± 118† (n = 28) | 7.8 ± 1.5 (n = 10) | 87.6 ± 2.1* (n = 25) |
*p < 0.01 and † p < 0.05 vs. SFCS scaffold only
Differences in cell density were evaluated and compared within the groups (Table 1). The cell density in the remodeled SFCS scaffold (3238 ± 152 cells/mm2) was significantly higher (p < 0.05) than the emodin-loaded SFCS scaffold (2733 ± 118 cells/mm2). There was no significant difference between the two SFCS scaffold groups in the amount of area occupied by newly deposited collagen. Scaffold degradation was quantified by processing original images as shown in Fig. 8. By 6 weeks, scaffold degradation in the emodin-loaded SFCS scaffold (87.6 ± 2.1%) was significantly (p < 0.01) less than no emodin SFCS scaffold (94.3 ± 0.8%).
Figure 8.
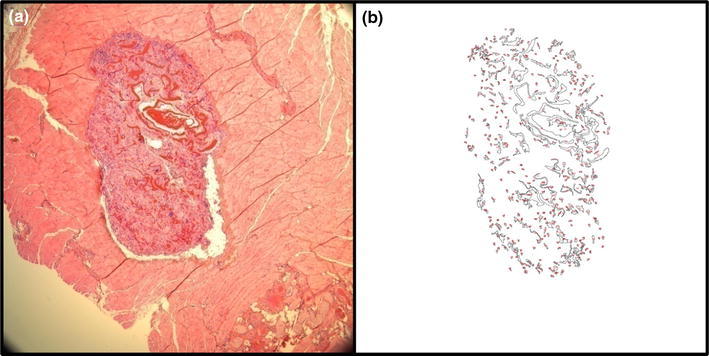
The degradation of SFCS scaffold was quantified using image processing in Image J software. An example of the (a) original H&E image and (b) the image after processing shows the area covered by non-degraded scaffold
Discussion
The emodin-loaded SFCS scaffold composite was designed for the treatment of breast tissue defects that occur due to tumor resection. In the in vitro part of this study, the composite was successfully prepared by embedding emodin nanoparticles within various blends of SF and CS scaffolds. Emodin entrapment was a function of initial emodin loading concentration and the SF to CS blend ratios which also affected the overall mechanical properties of the resultant scaffolds. The efficacy of emodin-loaded SFCS scaffold composites was tested in vitro and in an in vivo rat GILM2 breast tumor model, which showed an effect on the wound healing response of the tumor with and without emodin-loaded SFCS scaffolds and regeneration of the defect site with the SFCS scaffold.
The liposomal encapsulation of emodin has been previously developed by our group and thus served as a starting point for embedding emodin in the SFCS scaffolds.5,10 Initially, the entrapment of emodin within the SFCS scaffolds was tested without liposomal encapsulation, although the entire drug washed out in the crystallization and sterilization steps. The hydrophobic nature of emodin and its affinity towards organic solvents results in the drug leaching out during the crystallization and sterilization steps. Therefore, the emodin-loaded SFCS scaffolds consisted of liposomal emodin rather than free emodin embedded in the SFCS matrix.
The process of sonication was performed to reduce the liposomal emodin size to less than 100 nm24 and to enhance the entrapment of emodin within the liposomes. The sonication of only 5 min was sufficient to decrease the average liposome size (<100 nm) and to enhance the emodin entrapment within liposomes. Entrapment significantly improved by sonication that generated smaller liposomes with higher surface area. Although, sonication significantly reduced the average size of the liposomal emodin, a small amount of particles in the 200–300 nm range could have affected the reduction in tensile strength observed for SF and SFCS blends with increasing emodin loading concentration. Once encapsulated within the SF or SFCS scaffolds, the nanoparticles were visible on the surface of the scaffold. Similar observation of thin SF layer on PLGA microparticles embedded in SF scaffolds has been observed previously.33
Final emodin entrapment within the scaffolds was significantly higher for pure SF compared to all SFCS blends although there was no difference between SFCS blends, which indicates that CS does not favor the entrapment of liposomal emodin. CS at biologically relevant pHs holds an inherently negative charge due to the lone pair of electrons on the free amines11 and this may result in lower entrapment of hydrophobic emodin as compared to SF and its blends, where the hydrophobic peptides such as alanine and glycine that form the crystalline core of SF may have facilitated the higher entrapment of hydrophobic emodin, especially in pure SF scaffolds. Regardless, the SFCS scaffolds were able to retain 42–55% of initial emodin loading concentration. The higher starting emodin concentrations resulted in higher entrapped emodin for all SFCS blends.
Percent emodin release was higher for pure SF than SFCS blends, which may be a function of higher initial entrapment of emodin in SF vs. SFCS. Under in vitro conditions, the emodin release was slow from the scaffolds because of the barriers of the liposomal wall and the scaffold that emodin would have to extravasate during release. The release process would be facilitated in vivo by the wound healing microenvironment that results in the degradation of the scaffold with time. The release mechanism of emodin from SF-coated liposomes has been described previously as diffusion limited.10 It may be speculated that the emodin release from SFCS scaffolds is primarily diffusion controlled similar to previously reported curcumin release from SFCS particles.12 Nevertheless, under in vivo conditions, the actual release rates are also a function of the amount of degradation of the scaffolds with time. The in vivo release of emodin from the liposomes in SFCS scaffolds is synergistically facilitated by diffusion and degradation of SFCS with time.
Emodin inhibited the growth of GILM2 cells in a dose-dependent manner as the highest dose (100 μM) caused minimum cell viability after 4 days of incubation. Previous studies have shown the emodin dose-dependent inhibition of BCap-37 breast cancer cells with 50% inhibition at 50 μM concentration by 48 h and 90% inhibition of MDA-MB 453 cells at 40 μM concentration with SF coated emodin-loaded liposomes.5,15 Emodin has been shown to cause breast cancer cell apoptosis through Her2/neu pathway5,36 or Bcl-2 mediated mitochondrial pathway.15 However, apoptosis of other cancer cells upon emodin exposure targeted other pathways such as caspase-3 cascade in promyeleukemic HL60 cells,6 JAK2/STAT3 pathway in myeloma cells,26 Bax and Fas pathway in lung carcinoma CH27 cells.20 Thus, there may be many possible mechanisms of GILM2 cell apoptosis upon exposure to emodin that were not investigated as part of this study.
In vivo implantation of emodin-loaded SFCS scaffolds resulted in integration at the tumor site via a connective tissue interface, reduced cell density, and slower degradation compared to SFCS scaffold only group. Lower cell density in the emodin-loaded scaffolds may have been due to the emodin release resulting in tumor cell apoptosis and inhibiting further tumor cell invasion of the remodeling SFCS scaffold. Overall reduction in cellular extravasation thus resulted in lower degradation of the scaffold and reduced integration with the surrounding tumor. Local delivery of a therapeutic via a poly(ester-carbonate)-collagen composite for the treatment of lung cancer has been reported to show higher efficacy in terms of lowering local recurrence as compared to intravenous administration of the therapeutic.35 In this study, the presence of tumor was low in the emodin-loaded SFCS group but there was no significant difference in the tumor size as compared to no emodin SFCS and flap only controls. While most rats had reduced tumor size, one of the rats in the emodin-loaded scaffold group had comparatively larger tumor residue that resulted in higher standard error and reduced statistical significance.
The presence of blood vessels and collagen within the SFCS scaffold after 6 weeks of implantation is evidence of the regenerative potential of the SFCS scaffolds regardless of the presence of the drug. Previous studies have shown tissue regeneration with SFCS scaffold in abdominal wall musculofascial repair model,8 bone regeneration model,27 and a wound healing model.3
In conclusion, liposomal emodin-loaded 25:75 SFCS scaffold composites were successfully prepared with good mechanical integrity, optimal drug entrapment and release over time, and reduced breast cancer cell viability in vitro. In the in vivo breast tumor model, the composite with entrapped drug resulted in reduced tumor presence, scaffold degradation, remodeling, and new tissue deposition. The drug loaded scaffold composite for cancer therapy and tissue regeneration has great promise for reconstruction of tumor defects in order to prevent recurrence and encourage long-term healing. SFCS composite has the flexibility for incorporating versatile therapeutics for the treatment of various tumors4,23 and guiding cellular response towards regeneration by incorporating small molecules/therapeutics, such as siRNA,4 for local delivery.
Acknowledgment
This project was funded by MD Anderson Cancer Center Development Office and the Department of Plastic Surgery Kyte Research Fund. We would like to thank Kenneth Dunner Jr. for processing the SEM samples at High Resolution Electron Microscopy Facility at MD Anderson Cancer Center (Institutional Core Grant #CA16672). We are also thankful to Dr. Rohan Bhavane for helping with DLS instrument and Arnie Jimenez for processing histology samples.
Conflicts of Interest
No potential conflicts of interest were disclosed.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Aiedeh K, Gianasi E, Orienti I, Zecchi V. Chitosan microcapsules as controlled release systems for insulin. J. Microencapsul. 1997;14:567. doi: 10.3109/02652049709006810. [DOI] [PubMed] [Google Scholar]
- 2.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Silk-based biomaterials. Biomaterials. 2003;24:401. doi: 10.1016/S0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 3.Altman AM, Yan Y, Matthias N, Bai X, Rios C, Mathur AB, Song YH, Alt EU. IFATS collection: human adipose-derived stem cells seeded on a silk fibroin–chitosan scaffold enhance wound repair in a murine soft tissue injury model. Stem Cells. 2009;27:250. doi: 10.1634/stemcells.2008-0178. [DOI] [PubMed] [Google Scholar]
- 4.Cheema SK, Chen E, Shea LD, Mathur AB. Regulation and guidance of cell behavior for tissue regeneration via the siRNA mechanism. Wound Repair Regen. 2007;15:286. doi: 10.1111/j.1524-475X.2007.00228.x. [DOI] [PubMed] [Google Scholar]
- 5.Cheema SK, Gobin AS, Rhea R, Lopez-Berestein G, Newman RA, Mathur AB. Silk fibroin mediated delivery of liposomal emodin to breast cancer cells. Int. J. Pharm. 2007;341:221. doi: 10.1016/j.ijpharm.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 6.Chen YC, Shen SC, Lee WR, Hsu FL, Lin HY, Ko CH, Tseng SW. Emodin induces apoptosis in human promyeloleukemic HL-60 cells accompanied by activation of caspase 3 cascade but independent of reactive oxygen species production. Biochem. Pharmacol. 2002;64:1713. doi: 10.1016/S0006-2952(02)01386-2. [DOI] [PubMed] [Google Scholar]
- 7.Feldman JP, Goldwasser R, Mark S, Schwartz J, Orion I. A mathematical model for tumor volume evaluation using two-dimensions. J. Appl. Quant. Methods. 2009;4:455. [Google Scholar]
- 8.Gobin AS, Butler CE, Mathur AB. Repair and regeneration of the abdominal wall musculofascial defect using silk fibroin–chitosan blend. Tissue Eng. 2006;12:3383. doi: 10.1089/ten.2006.12.3383. [DOI] [PubMed] [Google Scholar]
- 9.Gobin AS, Froude VE, Mathur AB. Structural and mechanical characteristics of silk fibroin and chitosan blend scaffolds for tissue regeneration. J. Biomed. Mater. Res. A. 2005;74:465. doi: 10.1002/jbm.a.30382. [DOI] [PubMed] [Google Scholar]
- 10.Gobin AS, Rhea R, Newman RA, Mathur AB. Silk-fibroin-coated liposomes for long-term and targeted drug delivery. Int. J. Nanomedicine. 2006;1:81. doi: 10.2147/nano.2006.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goy RC, de Britto D, Assis OBG. A review of the antimicrobial activity of chitosan. Polimeros: Ciencia e Tecnologia. 2009;19:241. [Google Scholar]
- 12.Gupta V, Aseh A, Rios CN, Aggarwal BB, Mathur AB. Fabrication and characterization of silk fibroin-derived curcumin nanoparticles for cancer therapy. Int. J. Nanomedicine. 2009;4:115. doi: 10.2147/IJN.S5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta V, Davis G, Gordon A, Altman AM, Reece GP, Gascoyne PR, Mathur AB. Endothelial and stem cell interactions on dielectrophoretically aligned fibrous silk fibroin–chitosan scaffolds. J. Biomed. Mater. Res. 2010;94:515. doi: 10.1002/jbm.a.32720. [DOI] [PubMed] [Google Scholar]
- 14.Howard MA, Polo K, Pusic AL, Cordeiro PG, Hidalgo DA, Mehrara B, Disa JJ. Breast cancer local recurrence after mastectomy and TRAM flap reconstruction: incidence and treatment options. Plast. Reconstr. Surg. 2006;117:1381. doi: 10.1097/01.prs.0000208116.86765.4a. [DOI] [PubMed] [Google Scholar]
- 15.Huang Z, Chen G, Shi P. Emodin-induced apoptosis in human breast cancer BCap-37 cells through the mitochondrial signaling pathway. Arch. Pharm. Res. 2008;31:742. doi: 10.1007/s12272-001-1221-6. [DOI] [PubMed] [Google Scholar]
- 16.Jain RK. Delivery of molecular medicine to solid tumors: lessons from in vivo imaging of gene expression and function. J. Control. Release. 2001;74:7. doi: 10.1016/S0168-3659(01)00306-6. [DOI] [PubMed] [Google Scholar]
- 17.Kim SE, Park JH, Cho YW, Chung H, Jeong SY, Lee EB, Kwon IC. Porous chitosan scaffold containing microspheres loaded with transforming growth factor-beta1: implications for cartilage tissue engineering. J. Control. Release. 2003;91:365. doi: 10.1016/S0168-3659(03)00274-8. [DOI] [PubMed] [Google Scholar]
- 18.Kunwar A, Barik A, Pandey R, Priyadarsini KI. Transport of liposomal and albumin loaded curcumin to living cells: an absorption and fluorescence spectroscopic study. Biochim. Biophys. Acta. 2006;1760:1513. doi: 10.1016/j.bbagen.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Langstein HN, Cheng MH, Singletary SE, Robb GL, Hoy E, Smith TL, Kroll SS. Breast cancer recurrence after immediate reconstruction: patterns and significance. Plast. Reconstr. Surg. 2003;111:712. doi: 10.1097/01.PRS.0000041441.42563.95. [DOI] [PubMed] [Google Scholar]
- 20.Lee HZ. Effects and mechanisms of emodin on cell death in human lung squamous cell carcinoma. Br. J. Pharmacol. 2001;134:11. doi: 10.1038/sj.bjp.0704205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lev DC, Kiriakova G, Price JE. Selection of more aggressive variants of the gI101A human breast cancer cell line: a model for analyzing the metastatic phenotype of breast cancer. Clin. Exp. Metastasis. 2003;20:515. doi: 10.1023/A:1025837631179. [DOI] [PubMed] [Google Scholar]
- 22.Madihally SV, Matthew HW. Porous chitosan scaffolds for tissue engineering. Biomaterials. 1999;20:1133. doi: 10.1016/S0142-9612(99)00011-3. [DOI] [PubMed] [Google Scholar]
- 23.Mathur AB, Gupta V. Silk fibroin-derived nanoparticles for biomedical applications. Nanomedicine. 2010;5:807. doi: 10.2217/nnm.10.51. [DOI] [PubMed] [Google Scholar]
- 24.Maulucci G, De Spirito M, Arcovito G, Boffi F, Castellano AC, Briganti G. Particle size distribution in DMPC vesicles solutions undergoing different sonication times. Biophys. J. 2005;88:3545. doi: 10.1529/biophysj.104.048876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metselaar JM, Mastrobattista E, Storm G. Liposomes for intravenous drug targeting: design and applications. Mini. Rev. Med. Chem. 2002;2:319. doi: 10.2174/1389557023405873. [DOI] [PubMed] [Google Scholar]
- 26.Muto A, Hori M, Sasaki Y, Saitoh A, Yasuda I, Maekawa T, Uchida T, Asakura K, Nakazato T, Kaneda T, Kizaki M, Ikeda Y, Yoshida T. Emodin has a cytotoxic activity against human multiple myeloma as a Janus-activated kinase 2 inhibitor. Mol. Cancer Ther. 2007;6:987. doi: 10.1158/1535-7163.MCT-06-0605. [DOI] [PubMed] [Google Scholar]
- 27.Rios CN, Skoracki RJ, Miller MJ, Satterfield WC, Mathur AB. In vivo bone formation in silk fibroin and chitosan blend scaffolds via ectopically grafted periosteum as a cell source: a pilot study. Tissue Eng. A. 2009;15:2717. doi: 10.1089/ten.tea.2008.0360. [DOI] [PubMed] [Google Scholar]
- 28.Schroer B, Linder R, Hoch J. Local recurrence of breast cancer. Isolated local recurrence located at the skin of the latissimus dorsi donor flap after breast reconstruction. Chirurg. 2003;74:145. doi: 10.1007/s00104-002-0569-3. [DOI] [PubMed] [Google Scholar]
- 29.Shi H, Han C, Mao Z, Ma L, Gao C. Enhanced angiogenesis in porous collagen-chitosan scaffolds loaded with angiogenin. Tissue Eng. A. 2008;14:1775. doi: 10.1089/ten.tea.2007.0007. [DOI] [PubMed] [Google Scholar]
- 30.VandeVord PJ, Matthew HW, DeSilva SP, Mayton L, Wu B, Wooley PH. Evaluation of the biocompatibility of a chitosan scaffold in mice. J. Biomed. Mater. Res. 2002;59:585. doi: 10.1002/jbm.1270. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Wenk E, Matsumoto A, Meinel L, Li C, Kaplan DL. Silk microspheres for encapsulation and controlled release. J. Control. Release. 2007;117:360. doi: 10.1016/j.jconrel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Wang SS, Yang MC, Chung TW. Liposomes/chitosan scaffold/human fibrin gel composite systems for delivering hydrophilic drugs—release behaviors of tirofiban in vitro. Drug Deliv. 2008;15:149. doi: 10.1080/10717540801952456. [DOI] [PubMed] [Google Scholar]
- 33.Wenk E, Meinel AJ, Wildy S, Merkle HP, Meinel L. Microporous silk fibroin scaffolds embedding PLGA microparticles for controlled growth factor delivery in tissue engineering. Biomaterials. 2009;30:2571. doi: 10.1016/j.biomaterials.2008.12.073. [DOI] [PubMed] [Google Scholar]
- 34.Wenk E, Wandrey AJ, Merkle HP, Meinel L. Silk fibroin spheres as a platform for controlled drug delivery. J. Control. Release. 2008;132:26. doi: 10.1016/j.jconrel.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Wolinsky JB, Liu R, Walpole J, Chirieac LR, Colson YL, Grinstaff MW. Prevention of in vivo lung tumor growth by prolonged local delivery of hydroxycamptothecin using poly(ester-carbonate)-collagen composites. J. Control. Release. 2010;144:280. doi: 10.1016/j.jconrel.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Chang CJ, Bacus SS, Hung MC. Suppressed transformation and induced differentiation of HER-2/neu-overexpressing breast cancer cells by emodin. Cancer Res. 1995;55:3890. [PubMed] [Google Scholar]
- 37.Zhang L, Lau YK, Xia W, Hortobagyi GN, Hung MC. Tyrosine kinase inhibitor emodin suppresses growth of HER-2/neu-overexpressing breast cancer cells in athymic mice and sensitizes these cells to the inhibitory effect of paclitaxel. Clin. Cancer Res. 1999;5:343. [PubMed] [Google Scholar]


