Abstract
Oscillating gene expression is a mechanism of patterning during development in both plants and animals. In vertebrates, oscillating gene expression establishes the musculoskeletal precursors (somites), while in plant roots it establishes the position of future organs (lateral roots). Both mechanisms constitute a specialized type of biological clock that converts temporal information into precise spatial patterns. Similarities, differences, and their functionality in organisms that evolved independently are discussed.
Key words: development, patterning, gene expression, oscillation, time, biological clock
Introduction
Most living organisms have developed the ability to measure time by using biological clocks. All biological clocks are time-keeping mechanisms that can inform organisms about when a biological process is required. However, they differ in the nature of the information they provide. Based on this information or their activity, biological clocks can be classified into two types. As precise chronometers, one type allows species from different kingdoms to deal with periodic changes in the environment. A second type is able to mark the pace of certain developmental events in a highly reproducible fashion.
Well-known cases of the first type of clocks are circadian rhythms, which are present in almost all eukaryotes and in some bacteria.1,2 These circadian oscillators allow organisms to anticipate the coming day or night, changing of seasons as well as periodic changes in light and temperature. This anticipation ensures that peaks of particular physiological responses occur at appropriate times, thus providing a competitive advantage in fitness and survival. Regulation of these physiological processes is mainly achieved through regulation of expression patterns of genes involved in growth, metabolism and the cell cycle. It has also been proposed that such regulation might involve gating of physiological and environmental responses to certain times of the day, likely by enhancing sensitivity to different hormones or signaling pathways.3–6
Development is dynamic and occurs progressively in expanding space and over time. A second type of biological clocks regulates the spatiotemporal formation of developmental patterns. Examples are the segmentation clock of vertebrates, which is involved in positioning the precursors of the musculoskeletal structures of the trunk (somites) along the dorsal spine,7–10 and the recently discovered Arabidopsis thaliana root clock, which is involved in the positioning of lateral roots (LR) along the primary root axis.11 A major question in developmental biology is what controls the timing of patterning events during development.12 Both the root and the segmentation clock, which involve oscillating gene expression, offer an evolutionarily convergent solution to the problem of how to position repeating units along an elongating axis.11,13 These mechanisms therefore integrate temporal and spatial information for proper patterning and development.
Oscillating Gene Expression in the Root and the Segmentation Clocks
In the vertebrate segmentation clock, oscillating gene expression is required to establish the periodic spatial pattern of somites along the tail.14 The first evidence of oscillating gene expression in positioning LRs along the plant primary root came from the observation of the dynamic expression of the DR5 marker gene.11,15 Fusion of DR to the Luciferase reporter allowed examination of its expression in real time.11 Expression of DR5 shows cyclic pulses of expression over a region of the Arabidopsis root tip termed the oscillation zone (OZ). Remarkably, the DR5 oscillation resembles that of some of the oscillating genes described in the chicken and mouse somitogenesis clock, e.g., c-Hairy1 or Lunatic fringe (Lfng).13 In a similar fashion to c-Hairy 1 or Lfng (Fig. 1), at the beginning of the oscillation cycle, DR5 expression is observed at the part of the OZ closer to the root tip and, over time, DR5 expression increases and moves further from the root tip. At the end of the cycle, DR5 expression is located at end of the OZ and shuts down. A new cycle then begins. Following each DR5 oscillation, the expression of DR5 is observed again, but this time outside of the OZ as a static point of expression termed a prebranch site. Lineage analyses of prebranch sites revealed that LRs were exclusively generated at these sites. The DR5 oscillation therefore reports the selection of subsets of cells that become competent to generate new organs (LRs).
Figure 1.
Comparison of the expression patterns of the oscillating genes in the vertebrate segmentation clock (A) (adapted from Pourquie14 and Dequeant13) and in the root clock (B). Both the presomitic mesoderm and the root elongate from top to bottom in this schematic, as indicated by the arrow, while gene expression propagates in the opposite direction over time (as depicted from left to right). Gene expression oscillations in two opposite phases occur at the peak of the respective oscillations in the oscillation zones (green frames) as represented by Lunatic fringe and Axin2 in the segmentation clock (A) and by the marker gene DR5 and Auxin Response Factor 7 (ARF 7) in the root clock (B).
DR5 is also a marker for the transcriptional readout of auxin response or signaling, although it can also respond to other plant hormones.16 One hypothesis was that periodic local accumulation of auxin was responsible for the DR5 oscillation. This would represent, then, a patterning mechanism more analogous to patterning leaves and/or flowers along the plant stem.17 To test this hypothesis, additional auxin responsive promoters were examined but these failed to oscillate. Furthermore, exogenous auxin treatments localized to the OZ failed to initiate a new DR5 oscillation.11 This indicated that DR5 oscillations were not generated by fluctuations in auxin content. In contrast, auxin has been described to form a gradient at the root tip,18 although this observation cannot predict changes over time as it is the result of average measurements for many different roots at single time points. Stronger evidence supporting a static auxin gradient at the root tip would require development of a specific non-destructive sensor for this hormone that allowed more precise measurement of local changes in auxin over time.
The DR5 oscillation represents a broad oscillation in gene expression throughout the developing root. Microarray analyses of the OZ revealed approximately 2,000 genes with a similar oscillatory pattern as DR5, and about 1,400 oscillating in an opposite phase or anti-phase oscillatory pattern. No genes were found to oscillate in other phases.11 Remarkably, microarray analyses of the presomitic mesoderm in mouse also revealed two sets of genes oscillating in opposite phases.13,19 Real time imaging of oscillating transcription factor genes fused to the Luciferase reporter in roots showed that their expression propagated across the OZ, and confirmed oscillations in two opposite phases, analogous to gene expression through the presomitic mesoderm during somitogenesis.11,13 In the somitogenesis clock, both oscillatory phases of gene expression are required for somite patterning.7 Remarkably, in roots, mutations in genes coding for transcription factors oscillating both in phase and anti-phase showed irregular positioning and reduced numbers of prebranch sites, indicating that both phases of oscillations are also required.11 The oscillating genes in Arabidopsis and mice are clearly different, but both species use two sets of genes propagating along an axis and oscillating in opposite phases to convert time into a spatial developmental pattern. Because the last common ancestor between plants and animals was a unicellular organism, both time-keeping systems had to have evolved independently, thus their similarity suggests an evolutionary convergence on a similar mechanism.
Oscillating genes in the segmentation clock mainly fall into three different signaling pathways: Notch, β-catenin/Wnt and Fibroblast Growth Factor (FGF). Genes in the Notch and FGF pathway oscillate in the same phase, while genes in the β-catenin/Wnt pathway oscillate in the opposite phase.13 In contrast, only a few genes oscillating in the root were found to belong to a known signaling pathway, the auxin signaling pathway. It is possible that other genes, not yet assigned to any specific signaling pathway or belonging to unknown signaling pathways, also oscillate in the same or in opposite phases to the genes in the auxin signaling pathway.
In Arabidopsis, formation of prebranch sites by periodic oscillations in gene expression appears to act as a biological clock. One characteristic normally associated with biological clocks is that the period is buffered against variation in temperature.2 In the Arabidopsis root, changes in temperature do not have significant effects on the rate of prebranch site production; a new prebranch site is formed every 6 h over a range of temperatures (18–24°C).11 This indicates that there is temperature compensation for oscillating gene expression and subsequent pre-branch site formation. By contrast, there is not temperature compensation in the period of the somitogenesis clock. In zebra fish, the period and amplitude of the gene oscillations becomes shorter and slower, respectively, which results in fewer somites formed per hour, although of the same size.20,21 As vertebrate embryogenesis is also decelerated by low temperatures, maintenance of somite size is likely more critical for animals given the fact that they have a fixed body plan. In contrast, a fixed production rate of prebranch sites might optimize exploration of the soil environment by the root. Ultimately, both clocks use temporal information to establish robust spatial patterns.
Establishing Patterns in Development
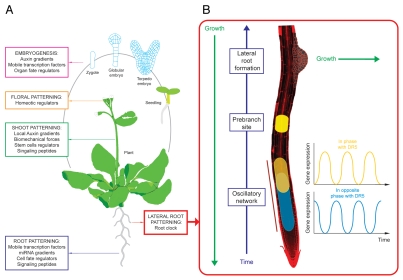
The development of multicellular organisms relies on proper organ patterning and growth. However, coordination of these developmental processes requires that certain sets of cells adopt a specific character or fate while other cells need to adopt different fates. As cells within organs or embryos need to be organized in space, this constitutes a three dimensional problem in which cells must both generate a functional spatial pattern and also simultaneously divide for proper organ growth. In animals, where cells move during embryogenesis, patterning is normally resolved by extracellular morphogen gradients. A classical example is the dorsoventral patterning of the amphibian embryo that is orchestrated by gradients originated from two groups of cells located at opposite ends: the dorsal or Spemann's organizer and the ventral center.12 In plants, cells are constrained in files and cannot move. Patterning under these constraints may also be established by gradients22,23 but alternatively may be established by exchanging positional information among cells, which determines cell fate (Fig. 2A). Recent research in Arabidopsis during root postembryonic development has shown that the transcription factor SHORT-ROOT (SHR) moves from the vasculature, where it is made, to the adjacent layer to activate specific cell cycle genes involved in the formative divisions that generate the endodermis.24,25 Conversely, the lack of SHR or its movement results in a default program by which cells are able to proliferate but are not specified as endodermis.25,26 SHR is also involved in crosstalk between the endodermis and the central vascular cylinder that results in patterning of the water conducting cell types centripetally. This patterning requires SHR movement to the endodermis, where SHR activates the expression of the microRNA MIR165, then movement of MIR165 back to the vasculature specifies the water conducting cells in a dose-dependent manner.27
Figure 2.
Patterning throughout the life cycle of the Arabidopsis thaliana plant includes hormone gradients, fate regulators, signaling molecules and the root clock (A). Positioning of new lateral roots is mediated by an oscillatory network at the root tip (the root clock) that establishes prebranch sites (B). Prebranch sites subsequently develop into new lateral roots.
When development calls for repeating structures to be positioned along an axis, for instance LRs along the primary root or vertebrae along the spine, patterning appears to be reduced from a three-dimensional spatial problem to a one-dimensional problem. In these cases organisms are organized in space but positioning of the repeating structures mainly occurs along one dimension (i.e., roots or spines). However, in order to solve this patterning problem, organisms must first resolve the problem of measuring time. Remarkably, it appears that two types of organisms (plants and vertebrates) have independently evolved similar mechanisms to measure time. In vertebrates, and also probably in Arabidopsis, this is achieved through self-sustained oscillations in gene expression or autonomous oscillators driving periodic pulses of gene expression (Fig. 2B).11,13 Once a time-keeping mechanism has evolved, it can be used to position repeating units (LR) along a growing axis. Spatial elongation of the axis separates the repeating structures, which are positioned at periodic intervals by the time-keeping mechanism. However, positioning of LR (or other developmental units) at regular spatial intervals only occurs if the growth rate of the axis is constant. Thus, changes in primary root growth rate can result in LRs that are closer to one another if the growth rate is reduced or vice versa.11
Alternatively, the position of repeating structures can be achieved by intracellular gradients that establish differential gene expression. A classic example of this is the Bicoid gradient in Drosophila that patterns the different body segments before cellularization.12 A potential problem is that the number of segments cannot be easily changed without major structural or genetic changes. In contrast, a time-keeping oscillator in an elongating axis represents a less evolutionary constrained solution, where the number of units can vary by changing the ratio between the pace of the oscillation and the growth of the axis. An excellent example of the flexibility of this developmental mechanism can be seen in vertebrates with different numbers of somites. In corn snakes (Pantherophis guttatus), axis growth as measured by the number of cell divisions in the growth zone is similar to that of vertebrates with fewer somites (i.e., ∼21 cell generations and 315 somites in corn snakes versus 17 cell generations and 65 somites in mice). These differences result from the fact that gene oscillations occur almost four times faster relative to the growth of the tail in snakes, which results in a much greater number of somites, although of smaller size.28 A similar mechanism might be responsible for the variable number of LRs observed among different plant species.
An oscillatory-based mechanism may be limited to patterning only strictly repeating or very similar developmental units (i.e., somites or LRs). All LRs are identical; however, there are slight differences in size and shape between somites. A different case can be observed during development of most arthropods. The three anterior or naupliar segments are patterned simultaneously at the blastoderm stage, and then the posterior segments are successively patterned from a localized growth zone. Segmentation of the spider Cupiennius salei and the centipede Strigamia maritime requires the orthologs of some of the central players in the vertebrate segmentation clock. These genes appear to show dynamic expression in these arthropods, which suggests the existence of underlying oscillatory mechanisms.12 Given the fact that the posterior segments are reasonably similar, especially in species such as centipedes, it is possible that a clock and/or oscillating gene expression represents a less evolutionarily constrained mechanism that would allow rapid changes in segment number between species. However, patterning of the relatively different anterior segments (the antennal, intercalary and mandibular segments) might require a mechanism based on gradients. Notably, plants use gradients of the plant hormone auxin to pattern the main body axis and organs during embryogenesis and pattern the shoot postembryonically,17,22,23,29 while an oscillatory mechanism operates to position identical LRs along the primary root during postembryonic development.11
Keeping Track of Time: Oscillators and Oscillating Gene Expression
Generation of oscillatory rhythms has been shown to minimally require one negative feedback loop able to carry the oscillating element back to the starting point with a delay in time.30 This delay can be the time required for the element (protein) to be produced or the time necessary for the mRNA to travel to the cytoplasm, produce the protein, and for the protein to come back to the nucleus and repress its own mRNA expression. In this case, both the mRNA and protein levels show periodic oscillations. Addition of a positive feedback loop prevents the system from reaching a homeostatic steady state and, therefore, performs sustained oscillations. Oscillating expression of genes that regulate a specific biological process can then evolve the appropriate periodicity in order to integrate temporal information into such regulation. In terms of gene regulatory networks this can be formulated using three elements organized as a negative feedback loop, or as a combination of short positive- and negative-feedback loops.30 Remarkably, the oscillator of the circadian clock of the cyanobacterium Synechococcus elongates forms by positive and negative phosphorylation feedback loops of just three proteins Kai A, Kai B and Kai C.1 In multicellular organisms, circadian oscillators contain a greater number of elements that form more intricate negative and positive feedback loops and might be regulated at different molecular levels.3 An example of this complexity is captured by recent mathematical models of the Arabidopsis circadian oscillator that include up to four loops and rates of mRNA and protein degradation.31
In the mouse segmentation clock, all the oscillating pathways (Notch, β-catenin/Wnt and FGF) form negative feedback loops that can be potentially used to generate oscillations.7 For example, the oscillating transcription factor Hes7, in the Notch pathway, represses its own transcription and is necessary for the oscillation of other genes, such as the glycosyl-transferase Lfng. Lfng, in turn, represses the activity of the Notch1 intracellular domain (NCID). Activation of Notch signaling releases NCID from the membrane and NCID then recruits various transcriptional regulators and activates the expression of Hes7.7,9 Modeling of the Hes7 negative feedback loop predicts sustained oscillations for a short-lived Hes7 protein, which has been validated in vivo.32 Mouse mutants with a longer half-life of Hes7 showed disorganized gene oscillations of Lfng and severe somite fusion. However, oscillations of genes in the β-catenin/Wnt pathway still occurred, indicating that this Notch oscillator is not the pace-maker of the somitogenesis clock. Similarly, negative feedback loops in β-catenin/Wnt and FGF have been proposed to originate oscillations in the somitogenesis clock, but mutants in the FGF pathway or constitutive activation of the β-catenin/Wnt pathway also failed to prevent oscillations of genes in the other signaling pathways.7 Thus, it appears that oscillations in the somitogenesis clock are driven by a complex oscillator involving genes in the Notch, β-catenin/Wnt and FGF pathways, or alternatively by an independent pacemaker. Modeling has shown that negative feedback loops can produce oscillations in each one of the three signaling pathways separately, but that common intermediates can couple the Notch, Wnt and FGF oscillators and lead to synchronized oscillations.33
In the novel root clock, future work might also show a complex oscillator and/or oscillations also involving genes in different signaling pathways. Interestingly, signaling in some of the pathways involved in the somitogenesis clock does not appear to regulate oscillating gene expression in all cases. For instance, Hes7 oscillations are still observed after inactivation of Notch signaling, but constitutive Notch activation results in steady Hes7 expression.7 Likewise in the root clock, oscillations of Auxin-Response Factor 7 (ARF7), a protein in the auxin signaling pathway, occur independently of auxin activation after supplementation of this hormone.11 However, it is formally possible that ARF7 oscillations might be affected by inactivation of the auxin signaling pathway. Future work will address the nature of the oscillations in roots.
Concluding Remarks
Patterning requires the selection of subsets of cells to adopt distinct fates. Over the last few years, many studies in plants and animals have shown diverse developmental mechanisms patterning the main body parts and organs during embryonic and postembryonic development. Some of these mechanisms include gradients, while others involve specialized biological clocks that convert temporal information into precise spatial patterns. The root clock and the somitogenesis or segmentation clock fall into this category. Some advantages of using time-keeping mechanisms to generate patterns are robustness, reproducibility, and the ability to compensate for environmental conditions and thus maintain the functionality of the patterns (i.e., prebranch production rate or somite size). These features likely provide a competitive advantage not only in fitness and survival, but also in the evolutionary flexibility to adopt new patterns without major structural changes (i.e., greater number of LRs and a more compact root system or vice versa). Interestingly, oscillating gene expression underlies both the root and somitogenesis clocks. Future work might reveal the existence of common or disparate principles underlying the generation of periodic rhythms of gene expression in roots and tails, as well as the properties of the genetic programs that select cells to become a new LR or somite.
Acknowledgments
We thank J. Van Norman, J. Petricka, C. Topp, R. Sozzani, M.P. Gonzalez-Garcia and H. Tsukagoshi for critical reading of the manuscript and helpful suggestions; and apologize to those, whose work we could not cover due to space limitations. Work in the Benfey lab on root morphogenesis and gene regulatory networks is funded by grants from the NIH, NSF and DARPA.
References
- 1.Dong G, Kim YI, Golden SS. Simplicity and complexity in the cyanobacterial circadian clock mechanism. Curr Opin Genet Dev. 2010;20:619–625. doi: 10.1016/j.gde.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jolma IW, Laerum OD, Lillo C, Ruoff P. Circadian oscillators in eukaryotes. Wiley Interdiscip Rev Syst Biol Med. 2010;2:533–549. doi: 10.1002/wsbm.81. [DOI] [PubMed] [Google Scholar]
- 3.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annu Rev Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 5.de Montaigu A, Toth R, Coupland G. Plant development goes like clockwork. Trends Genet. 2010;26:296–306. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Khapre RV, Samsa WE, Kondratov RV. Circadian regulation of cell cycle: Molecular connections between aging and the circadian clock. Ann Med. 2010;42:404–415. doi: 10.3109/07853890.2010.499134. [DOI] [PubMed] [Google Scholar]
- 7.Aulehla A, Pourquie O. Oscillating signaling pathways during embryonic development. Curr Opin Cell Biol. 2008;20:632–637. doi: 10.1016/j.ceb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Kageyama R, Niwa Y, Shimojo H. Rhythmic gene expression in somite formation and neural development. Mol Cells. 2009;27:497–502. doi: 10.1007/s10059-009-0068-1. [DOI] [PubMed] [Google Scholar]
- 9.Kageyama R, Niwa Y, Shimojo H, Kobayashi T, Ohtsuka T. Ultradian oscillations in Notch signaling regulate dynamic biological events. Curr Top Dev Biol. 2010;92:311–331. doi: 10.1016/S0070-2153(10)92010-3. [DOI] [PubMed] [Google Scholar]
- 10.Lewis J, Hanisch A, Holder M. Notch signaling, the segmentation clock and the patterning of vertebrate somites. J Biol. 2009;8:44. doi: 10.1186/jbiol145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329:1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis J. From signals to patterns: space, time and mathematics in developmental biology. Science. 2008;322:399–403. doi: 10.1126/science.1166154. [DOI] [PubMed] [Google Scholar]
- 13.Dequeant ML, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- 14.Pourquie O. The segmentation clock: converting embryonic time into spatial pattern. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- 15.De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, et al. Brassinolide induces IAA5, IAA19 and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 2003;133:1843–1853. doi: 10.1104/pp.103.030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 18.Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, et al. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell. 2009;21:1659–1668. doi: 10.1105/tpc.109.066480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dequeant ML, Ahnert S, Edelsbrunner H, Fink TM, Glynn EF, Hattem G, et al. Comparison of pattern detection methods in microarray time series of the segmentation clock. PLoS One. 2008;3:2856. doi: 10.1371/journal.pone.0002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang YJ, Aerne BL, Smithers L, Haddon C, Ish-Horowicz D, Lewis J. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- 21.Schroter C, Herrgen L, Cardona A, Brouhard GJ, Feldman B, Oates AC. Dynamics of zebrafish somitogenesis. Dev Dyn. 2008;237:545–553. doi: 10.1002/dvdy.21458. [DOI] [PubMed] [Google Scholar]
- 22.Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 23.Moller B, Weijers D. Auxin control of embryo patterning. Cold Spring Harb Perspect Biol. 2009;1:1545. doi: 10.1101/cshperspect.a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernou T, et al. Spatiotemporal regulation of cell cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 26.Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 27.Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez C, Ozbudak EM, Wunderlich J, Baumann D, Lewis J, Pourquie O. Control of segment number in vertebrate embryos. Nature. 2008;454:335–339. doi: 10.1038/nature07020. [DOI] [PubMed] [Google Scholar]
- 29.Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jonsson H, et al. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 2010;8:1000516. doi: 10.1371/journal.pbio.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard KE, Robertson FC, Dalchau N, Webb AA. Systems analyses of circadian networks. Mol Biosyst. 2009;5:1502–1511. doi: 10.1039/B907714f. [DOI] [PubMed] [Google Scholar]
- 32.Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, et al. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- 33.Goldbeter A, Pourquie O. Modeling the segmentation clock as a network of coupled oscillations in the Notch, Wnt and FGF signaling pathways. J Theor Biol. 2008;252:574–585. doi: 10.1016/j.jtbi.2008.01.006. [DOI] [PubMed] [Google Scholar]