Abstract
Both anabolism and catabolism of the amino acids released by starvation-induced autophagy are essential for cell survival, but their actual metabolic contributions in adult animals are poorly understood. Herein, we report that, in mice, liver autophagy makes a significant contribution to the maintenance of blood glucose by converting amino acids to glucose via gluconeogenesis. Under a synchronous fasting-initiation regimen, autophagy was induced concomitantly with a fall in plasma insulin in the presence of stable glucagon levels, resulting in a robust amino acid release. In liver-specific autophagy (Atg7)-deficient mice, no amino acid release occurred and blood glucose levels continued to decrease in contrast to those of wild-type mice. Administration of serine (30 mg/animal) exerted a comparable effect, raising the blood glucose levels in both control wild-type and mutant mice under starvation. Thus, the absence of the amino acids that were released by autophagic proteolysis is a major reason for a decrease in blood glucose. Autophagic amino acid release in control wild-type livers was significantly suppressed by the prior administration of glucose, which elicited a prompt increase in plasma insulin levels. This indicates that insulin plays a dominant role over glucagon in controlling liver autophagy. These results are the first to show that liver-specific autophagy plays a role in blood glucose regulation.
Key words: amino acid, autophagy, liver, gluconeogenesis, insulin, phosphoenolpyruvate carboxykinase
Introduction
Macroautophagy (hereafter referred to as autophagy) is a major catabolic process that is responsible for the turnover of cell constituents, including cell organelles and cytosolic proteins.1–3 Autophagy is a membrane-dynamic phenomenon, in which a unique double membrane collectively and nonselectively surrounds cytoplasmic components to form an autophagosome. The outer membrane of the autophagosome then fuses with the lysosome membrane, and the sequestered substrates in the lumen and inner autophagosomal membranes are degraded by lysosomal hydrolases to release sugars, fatty acids, glycerol, amino acids, etc.1,2 More than 20 autophagy-related (ATG) genes participate in the molecular process of autophagy. During the last decade, several molecular and cellular biological approaches have revealed versatile roles for autophagy in cytoplasmic quality control, innate immunity, antigen presentation, carcinogenesis, etc.3–7
One unequivocal characteristic of autophagy is that it proceeds at low rates under nutrient-rich conditions (constitutive or basal autophagy), but is strongly enhanced under nutrient-deprivation conditions (starvation-induced autophagy). Recent investigations using liver- and brain-specific autophagy-deficient mice have demonstrated that the cumulative effects of impaired basal autophagy are diverse pathological symptoms, such as ubiquitin/p62-positive inclusion formation and the stacking of abnormal membrane whirls and layers.8–11 Thus, basal autophagy makes a substantial contribution to cellular quality control.
On the other hand, starvation-induced autophagy has been implicated in metabolic compensation, which is also referred to as the recycling of the sugars, fatty acids and amino acids that are released as a result of autophagic degradation. These compounds can be catabolized via the citric acid cycle to generate adenosine triphosphate (ATP). Mice deficient in Atg5 died within 24 h after birth because they could not overcome nutrient- and energy-insufficiency. Adenosine monophosphate (AMP-)-activated protein kinase, which is activated by an elevated AMP/ATP ratio through the phosphorylation of an α-subunit, was phosphorylated in Atg5-deficient, but not in wild-type, neonatal mice,12 indicating severe impairment of ATP production as a result of insufficient autophagy. In another case, specific inhibition of autophagy, which was induced by IL-3 withdrawal in Bax−/−/Bak−/− bone marrow cells by 3-methyladenine, caused a remarkable decline in cellular ATP levels, which was partially restored by supplying methylpyruvate, which is a membrane-permeable form of pyruvate.13 Meanwhile, amino acids can also be used anabolically for de novo protein synthesis. In the yeast Saccharomyces cerevisiae, amino acids generated via starvation-induced autophagy could be reused for de novo synthesis of starvation-adaptive proteins such as HSP26 and argininosuccinate synthetase.14 Autophagic protein degradation triggered by fertilization provides abundant amino acids, which are necessary for protein synthesis during early oocyte development.15
In adult animals, starvation-induced autophagy takes place predominantly in the liver. Earlier studies using perfused rat livers and in vivo experiments using mice have revealed that autophagic protein degradation, which proceeds at a rate of ∼1.5% of total liver protein/hour under nutrient-rich conditions, is enhanced approximately two- to three-fold during starvation,16 resulting in the loss of nearly 40% of total liver protein during a 48 h starvation period.17 The conversion of glucogenic amino acids generated during liver autophagy to glucose via gluconeogenesis has long been proposed as a potential metabolic contribution. Using perfused rat liver, glucagon, which significantly stimulates autophagic protein degradation,18 has enhanced intracellular utilization of glucogenic amino acids through its effect on gluconeogenesis.19 Because the liver is the major organ that produces and supplies blood glucose, the utilization of glucogenic amino acids for glucose production may be an important contribution of liver autophagy. However, little is known about the mechanism of the autophagy-dependent release of glucogenic amino acids for glucose production.
To clarify this issue, we systematically investigated the fate of amino acids produced by liver autophagic protein degradation by comparing normal wild-type and liver-specific conditional autophagy (Atg7)-deficient mice.20 We found that a significant portion of the released amino acids was converted into glucose via hepatic gluconeogenesis to maintain blood glucose levels, while the rest of the amino acids were released into circulation to maintain plasma amino acids in wild-type mice. By contrast, in liver-specific Atg7-deficient mice, the inability to release amino acids via autophagic proteolysis resulted in a significant reduction in blood glucose levels during starvation.
Results
Autophagy is induced after 24 h of starvation under synchronous fasting-initiation conditions.
The first step was to induce autophagy in mice by withdrawing their diet according to the conventional procedure. However, various tissue parameters, including the autophagosomal membrane marker microtubule-associated protein 1A/1B light chain 3 (LC3), fluctuated excessively among individual mice, implying that autophagic induction was not synchronized in the mice examined. To synchronize autophagic induction in all mice of an experimental group, a starvation/feeding/re-starvation regimen was conducted, as described previously in reference 21. First, wild-type mice were fasted for 24 h and then fed a pelleted laboratory diet for 2 h to suppress autophagy to a minimal level. Subsequently, the mice were re-fasted to induce autophagy. As depicted below, this experimental manipulation was used to synchronize autophagic induction in all mice, enabling the statistical analysis of the experimental data.
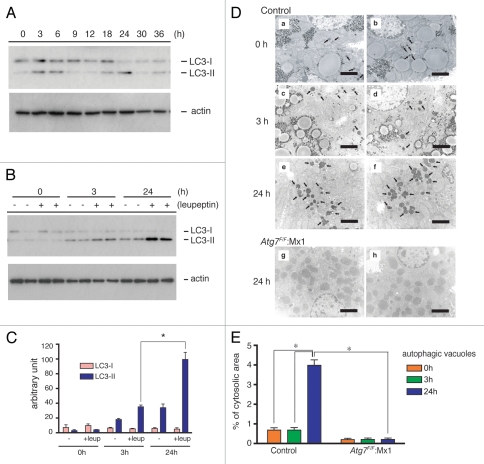
Initially, endogenous LC3 levels in liver samples collected at various times after re-starvation were examined by immunoblotting. LC3 exists in two different forms: LC3-I, a soluble cytosolic form; and, LC3-II, a phosphatidylethanolamine-conjugated form, which is specifically recruited onto autophagosomal membranes.22 The LC3-II level increased transiently after both 3–6 h and 18–24 h of starvation. With the exception of these two periods, LC3-II was barely detectable (Fig. 1A), which suggests transient autophagic induction during these periods. Next, to investigate whether the increase in LC3-II truly meant induction or enhancement of autophagic flux (i.e., autophagosome formation, autophagosome maturation into autolysosomes and lysosomal degradation of the sequestrated materials), leupeptin, a lysosomal cysteine proteinase inhibitor, was injected into mice intraperitoneally. Intraperitoneal (i.p.) injection of leupeptin suppresses autophagic flux at the lysosomal degradation step in the liver, leading to the accumulation of autophagosomes/autolysosomes,21,23 which increases the level of LC3-II.24 Therefore, the i.p. injection of leupeptin allowed us to easily monitor autophagy flux in the liver by estimating the level of autophagosomes/autolysosomes and/or LC3-II. As shown in Figure 1B, the leupeptin injection led to a marked increase in LC3-II levels in the livers of mice that had been fasted for 24 h, while there was only a slight increase in the livers of mice fasted for 3 h. Quantitative densitometry revealed that the LC3-II level after 24 h of starvation was about three times higher than it was after 3 h of starvation when mice were treated with leupeptin (Fig. 1B and C). To confirm autophagic induction after a 24 h starvation period, autophagic vacuoles (autophagosomes plus autolysosomes) were observed and counted by electron microscopy. Liver specimens were isolated from leupeptin-treated mice that had been starved for either 3 h or 24 h (for details, see Materials and Methods). As shown in Figure 1D and E, the number of vacuoles in the liver after 24 h of starvation was increased approximately seven-fold compared with 3 h of starvation, occupying 4% of the total cytoplasmic area of hepatocytes. Such vacuoles were not observed in the liver-specific Atg7-deficient mice, Atg7F/F: Mx1 (Fig. 1D and E).20 These results indicate that induction of autophagy in the livers occurred in response to 24 h of starvation, but not 3 h.
Figure 1.
Starvation-dependent changes in LC3-II levels and autophagic vacuoles in wild-type livers. (A and B) Postnuclear supernatants of the livers (10 µg protein) isolated from mice starved for the indicated periods were analyzed by immunoblotting analysis. (A) LC3-1 and LC3-II in the livers of starved mice without leupeptin administration. LC3-I, soluble form; LC3-II, PE -conjugated form. (B) LC3-1 and LC3-II in the livers of leupeptin-treated, starved mice. Leupeptin was injected as described in Materials and Methods. The data are representative of three separate experiments. (C) Densitometric analyses of LC3-I and LC3-II levels between leupeptin-administered (+leup) and nonadministered (−) livers isolated from nonstarved (0 h), 3 h and 24 h starved mice. Each value is the mean ± SEM of data from at least three mice. *p < 0.005. (D) Electron micrographs of liver samples isolated from wild-type mice starved for 0 h (a and b), 3 h (c and d) and 24 h (e and f), respectively and liver-specific Atg7-deficient mice starved for 24 h (e and h). All mice were administered leupeptin 1 h before dissection. Note that the autophagic vacuoles (arrows in a–f) markedly increased in 24-h starved wild-type mice (e and f) compared with 0 h and 3 h starved wild-type mice (a–d). Autophagic vacuoles were not detected in Atg7-deficient mouse livers starved for 24 h (g and h). Bars, 2 µm. (E) The volume density (%) of autophagic vacuoles in the cytoplasmic area was calculated as described in the Methods. Each value is the mean ± SEM of 40 parts. *p < 0.001.
Liver autophagy is induced in response to a reduction in plasma insulin levels.
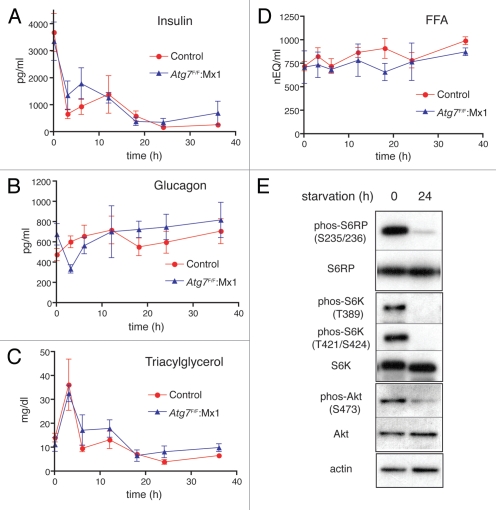
What kinds of hormones and/or metabolites induce liver autophagy after 24 h of starvation? Liver autophagy is suppressed by amino acids and insulin, while it is enhanced by glucagon.16,18,25,26 Hence, the concentrations of insulin, glucagon, triacylglycerol and free fatty acids were measured in plasma collected after various periods of starvation. Insulin, though exhibiting striking fluctuations during 0–18 h, finally decreased to its lowest level after approximately 24 h of starvation (Fig. 2A). Meanwhile, plasma glucagon remained at a high and stable level throughout the starvation periods (Fig. 2B). Neither triacylglycerol nor free fatty acids in the plasma exhibited fluctuations corresponding to the induction of liver autophagy during a 24 h starvation period (Fig. 2C and D). These results suggest that the reduction in insulin plays a principal role in the induction of liver autophagy.
Figure 2.
Changes in plasma insulin, glucagon, triacylglycerol and free fatty acids in wild-type and Atg7-deficient mice during starvation. At the times indicated after initiation of starvation, plasma samples were taken from wild-type (red circle) and Atg7-deficient (blue triangle) mice and the concentrations of insulin (A), glucagon (B), triacylglycerol (C) and free fatty acids (D, FFA) were determined. Note that insulin fell to its lowest level after a 24 h starvation period. Each value is the mean ± SEM of data from at least three mice. (E) Phosphorylated states of S6 ribosomal protein, p70 S6 kinase and Akt in nonstarved and 24 h starved wild-type livers. Postnuclear supernatants isolated from non-starved and 24 h starved mice were subjected to immunoblotting using antibodies specific for phosphorylated S6 ribosomal protein [phos-S6RP (S235/236)], S6 ribosomal protein (S6RP), phopsphorylated P70 S6 kinase [phos-S6K (T389) and phos-S6K (T421/S424)], P70 S6 kinase (S6K), phosphorylated Akt [phos-Akt (S473)], Akt and actin, respectively. The same pattern was also confirmed with Atg7-deficient livers before and after starvation.
Insulin binding to insulin receptors causes consecutive activation of class I-PI3-kinase and Akt, which eventually activates mTor.27,28 Activated mTor suppresses autophagy, while inactivation of mTor induces autophagy.29–33 To examine whether inactivation of mTor occurred in the livers of mice starved for 24 h, at which time insulin fell to its lowest level, the phosphorylation of signal molecules, including Akt, S6K and ribosomal S6, which are phosphorylated in an insulin-dependent manner, was investigated. As shown in Figure 2E, dephosphorylation of these molecules was observed at 24 h, implying that liver autophagy was induced through the inactivation of the Akt/mTor pathway in response to a reduction in plasma insulin.
Transient and robust amino acid release (amino acid surge) in liver, blood and skeletal muscles through liver autophagy.
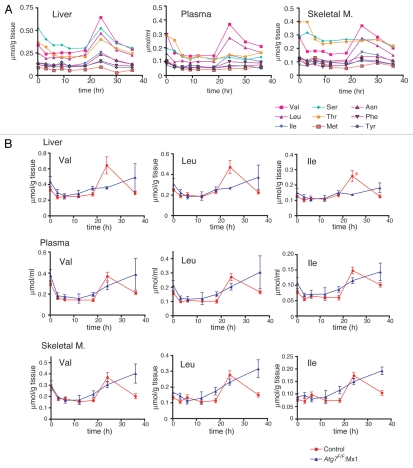
Because autophagy was induced after 24 h of starvation (Fig. 1), the levels of free amino acids in the liver should increase as a result of autophagic protein degradation. Thus, the concentration of each of the 20 amino acids (except for two acid-labile amino acids, cysteine and tryptophan) was examined in liver, plasma and skeletal muscle isolated from control wild-type mice at various times after the induction of starvation. During the first 18 h of starvation, the levels of some amino acids in the liver decreased gradually, while the levels of other amino acids remained constant (Fig. 3A and liver and Sup. Fig. 1). It was interesting that a drastic and transient increase in amino acids in the liver occurred after 24 h of starvation, followed by a gradual decrease until 36 h (Fig. 3A and liver). Among the 18 amino acids measured, the increase was most evident for nine, including the branched-chain amino acids (BCAAs). The clear BCAA response most likely occurred because BCAAs are primarily catabolized extrahepatically in muscle.34 This transient release of amino acids, which has been designated an “amino acid surge,” coincided with a starvation-dependent increase in LC3-II and autophagic vacuoles (Fig. 1). This result suggests that the amino acid surge was truly implicated in liver autophagy following 24 h of starvation. Intriguingly, a similar transient increase in amino acid levels was observed concomitantly in plasma and skeletal muscle (Fig. 3A and plasma, skeletal muscle). This observation raises the possibility that the increase in amino acids in plasma and skeletal muscle resulted from liver autophagic protein degradation. Specifically, amino acids produced via liver autophagic degradation might be excreted into circulation and transported to skeletal muscles. An alternative possibility is that plasma amino acids might be supplied by muscle autophagy that occurred in coordination with liver autophagy.
Figure 3.
Transient increase in free amino acids in the liver, plasma and skeletal muscle during starvation. (A) Plasma and tissue samples were collected from control wild-type mice starved for the indicated periods and were processed for amino acid analyses as described in Materials and Methods. The concentrations of nine amino acids, including the BCAAs, are expressed as µmol/g wet tissue (liver and skeletal muscles) or µmol/ml of plasma. (B) Comparison of time-dependent changes in BCAA concentrations between wild-type liver (red circle) and Atg7-deficient liver (blue triangle) during starvation. BCAA of liver, plasma and skeletal muscle taken from control and liver-specific Atg7-deficient mice at the indicated times during starvation were determined. Each value is the mean ± SEM of data from at least three mice.
To test these possibilities, starvation-dependent changes in amino acid levels in the liver, plasma and skeletal muscle were compared between control wild-type mice and liver-specific autophagy (Atg7)-deficient mice. As the BCAA data clearly show, the amino acid surge, not only in the liver but also in plasma and skeletal muscles, was inhibited by liver autophagy-deficiency (Fig. 3B). A similar profile was obtained for the other amino acids (Sup. Figs. 2 and 3). These results indicate that the transient increase in free amino acids in plasma and skeletal muscle of wild-type mice starved for 24 h was due to liver autophagic proteolysis.
Amino acid production via liver autophagy is essential for the maintenance of blood glucose levels.
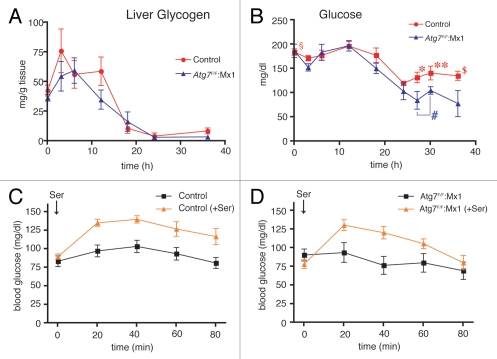
We next focused on the metabolic fate of free amino acids produced by liver autophagy (amino acid surge). Based on the aforementioned data (Figs. 1–3), it was postulated that the released amino acids should have been metabolized to compensate for a shortage of some key metabolite(s) during 24 h of starvation. Since the liver is known to supply blood glucose and hepatic glycogen is a source of glucose, it would be reasonable to assume that the glucogenic amino acids generated in the liver through autophagy were converted into glucose and that the resulting glucose was released into the blood to maintain blood glucose levels. To examine this hypothesis, changes in liver glycogen content were examined during starvation. As shown in Figure 4A, the liver glycogen content fluctuated to some extent during 3–12 h of starvation, but then it steadily decreased to its lowest level in both the wild-type and Atg7-deficient mice. After 24 h of starvation, liver glycogen was almost completely depleted (Fig. 4A), indicating that the source of blood glucose in hepatocytes was completely consumed.
Figure 4.
Activation of liver gluconeogenesis in association with starvation-induced autophagic protein degradation. Changes in liver glycogen levels (A) and plasma glucose concentration (B) were determined. At various times after starvation, liver and blood samples were taken from wild-type (red circle) and liver-specific conditional Atg7-deficient mice (blue triangle). Each value is the mean ± SEM of data from at least three mice. Numbers of mice used were: wild-type mice starved for 0 (n = 10), 3 (n = 11), 6 (n = 7), 12 (n = 7), 18 (n = 7), 24 (n = 5), 27 (n = 10), 30 (n = 6) and 36 h (n = 10), respectively and liver-specific Atg7-deficient mice starved for 0 (n = 7), 3 (n = 5), 6 (n = 3), 12 (n = 3), 18 (n = 3), 24 (n = 4), 27 (n = 5), 30 (n = 9) and 36 h (n = 3), respectively. The statistical significance and insignificance in the differences between wild-type and mutant mice at 0 (§p = 0.854), 27 (*p = 0.0340), 30 (**p = 0.0311) and 36 ($p = 0.0238) h of starvation and the statistical insignificance (#p = 0.482) in the plasma glucose levels of liver-specific Atg7-deficient mice between two periods of starvation (27 and 30 h) are shown. (C and D) Recovery of blood glucose concentration by oral administration of a glucogenic amino acid, serine. Wild-type (C) and liver-specific Atg7-deficient mice (D) starved for 24 h were orally administered serine (30 mg) at 24 h of starvation and blood glucose concentration was determined with a glucometer for the subsequent 80 min as described in Materials and Methods. Each value is the mean ± SEM of data. Wild-type (no treatment; n = 12, serine administered; n = 14) (C) and liver-specific Atg7-deficient mice (no treatment; n = 6, serine administered; n = 8) were examined. As total blood was used for the determination, the glucose level was significantly lower than the plasma glucose level determined in (B).
Changes in blood glucose levels were examined next. Blood glucose concentrations remained nearly unchanged from the initial values (∼180 mg/dl) in both wild-type and liver-specific Atg7-deficient mice until 12 h of starvation, followed by a gradual decrease between 12–24 h in both wild-type and liver-specific Atg7-deficient mice (Fig. 4B). Strikingly, whereas the blood glucose concentration in wild-type mice increased again—to two-thirds of the initial concentration ∼120 mg/dl—in accordance with the period of amino acid surge in wild-type mice, the concentration in mutant mice continued to decrease to ∼70 mg/dl (Fig. 4B). These data suggest that the amino acids released in the wild-type livers were converted into glucose to maintain the blood glucose concentration.
To determine whether oral administration of amino acids to starved mice could restore blood glucose concentrations, both control wild-type mice and liver-specific Atg7-deficient mice starved for 24 h were orally administered one regimen of representative glucogenic amino acid serine (30 mg per animal), after which blood glucose concentrations were monitored for 80 min. Figure 4C and D show that the blood glucose concentration in both wild-type and mutant mice was transiently increased to greater than 120 mg/dl between 20 and 40 min after serine administration, followed by a gradual decrease to initial concentrations. These data indicate that both wild-type and Atg7-deficient livers starved for 24 h possess comparable activities of converting glucogenic amino acids to glucose. The data also support the hypothesis that the amino acids generated by liver autophagy were converted into glucose in the livers of starved wild-type mice and that this glucose was released into the blood.
Pre-administration of glucose to starved mice significantly suppresses the amino acid surge through inhibition of autophagy.
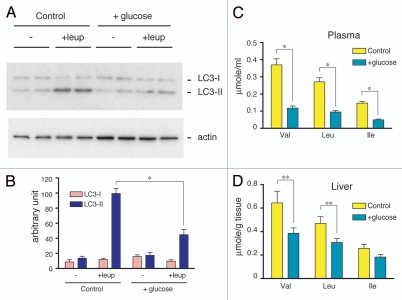
Because the amino acid surge had metabolic effects on blood glucose via gluconeogenesis in the livers of animals subjected to 24 h of starvation, it was interesting to examine whether or not prior administration of glucose to starved mice affects the amino acid surge or autophagic protein degradation. We orally administered glucose to wild-type mice after both 18 h and 21 h of starvation. Blood glucose reached a maximal concentration (∼400 mg/dl) at 21.5 h, followed by a gradual decrease to a stable concentration at 24 h (Sup. Fig. 4A). As shown in Figure 5A and B, glucose administration to control mice significantly inhibited accumulation of LC3-II in the liver in the presence of leupeptin. This result indicates that autophagy was significantly suppressed by glucose administration. Moreover, the amino acid surge that is typically detected by the release of BCAA was also markedly suppressed in the liver and plasma (Fig. 5C and D). Under these conditions, plasma insulin increased transiently 30 min after the second glucose administration, while glucagon levels remained constant (Sup. Fig. 4B and C). These results clearly indicate that the amino acid surge (i.e., liver autophagy) was reversibly controlled by blood glucose levels.
Figure 5.
Suppression of the amino acid surge by glucose administration. (A) Suppression of leupeptin-induced accumulation of LC3-II by glucose administration. Wild-type mice were orally administered 0.2 g of an aqueous glucose solution or tap water as a control after 18 and 21 h of starvation. Then, leupeptin was injected intraperitoneally into mice after 23 h of starvation. Plasma and livers were collected from 24 h starved mice. Postnuclear supernatants (10 µg protein) of the livers were subjected to immunoblotting analysis. The data shown are representative of three separate preparations. (B) Quantitative densitometry of endogenous LC3-I and LC3-II levels in postnuclear supernatants of livers from control or glucose-administered mice with or without leupeptin treatment. The data are means ± SEM of at least three different preparations. (C and D) Effect of glucose administration on the BCAA concentration in the plasma (C) and liver (D) were analyzed using an amino acid analyzer, as described in Materials and Methods. Each value is the mean ± SEM of data from four mice. *p < 0.001, **p < 0.05.
Discussion
Starvation-induced liver autophagy is essential for the maintenance of blood glucose levels.
Recent studies that showed activation of AMP-activated protein kinase in Atg5-null mice neonates and reduced cellular ATP levels under autophagy inhibition clearly demonstrated that an important contribution of amino acids, which are the major products released by starvation-induced autophagy, is catabolism for ATP production.12,13 Another important contribution of the amino acids released by autophagy is de novo protein synthesis, as demonstrated for yeast autophagy and oocyte-specific autophagy.14,15
The data presented in the present study highlight a third potential metabolic use of the amino acids produced by liver autophagic protein degradation, i.e., the conversion of amino acids to glucose via hepatic gluconeogenesis. The rationale for this conclusion is based on the following observations: (1) whereas blood glucose levels were maintained within the normal range after the amino acid surge (24–36 h) provided by liver autophagic protein degradation in wild-type mice (Fig. 4B), blood glucose levels continued to decrease and leveled off at one-half the normal concentration in liver-specific autophagy-deficient mice (Fig. 4B); and (2) when glucogenic amino acid, serine, was administered to wild-type and liver-specific Atg7-deficient mice starved for 24 h, blood glucose concentrations increased within 20 min (Fig. 4F), showing that both wild-type and autophagy-deficient livers possess an equal ability to convert glucogenic amino acids to glucose via gluconeogenesis.
In wild-type livers, abundant amino acids were released promptly and transiently at 24 h of starvation. Out of 18 amino acids determined, 11 exhibited clear surge responses: valine, leucine, isoleucine, serine, threonine, methionine, asparagine, phenylalanine, tyrosine, lysine and arginine (Sup. Fig. 1). With the exceptions of leucine and lysine, the remaining nine have the potentiality to be metabolized in order to produce glucose via gluconeogenesis. Importantly, serine, a potential glucogenic amino acid, which can be converted to pyruvic acid via a one-step serine dehydratase reaction, is a major amino acid, which shows a surge response (Fig. 3A). In contrast, alanine, another important glucogenic amino acid, shows a subtle increase (Sup. Fig. 1). Nevertheless, alanine levels in wild-type livers were significantly higher than those of Atg7-deficient livers at 24 and 36 h of starvation. The lack of a surge response in alanine may indicate a faster conversion of alanine to pyruvic acid due to much higher alanine transferase activity compared with serine dehydratase activity.35 Taken together, these results indicate that the amino acids produced through liver autophagy were efficiently converted into glucose under starvation conditions.
Liver autophagy is induced concomitantly with a reduction in plasma insulin.
Liver autophagy is controlled differently by plasma amino acids and hormones, such as insulin and glucagon.16,18,25,26 The suppressive effects of amino acids on autophagy have been investigated mostly in perfusion experiments using isolated rat livers or cultured hepatocytes.16,25,36,37 However, either loss of insulin action or stimulation by glucagon appears to play a more important role in autophagy induction in vivo, because plasma amino acid concentrations do not fluctuate enough in vivo to induce or suppress autophagy, in contrast with ex vivo experiments.18,25
The data from the present study show that autophagy was induced concomitantly with a fall in plasma insulin levels, as well as with depletion of liver glycogen, after 24 h of starvation. As plasma glucagon was maintained at a high and stable level, the drop in insulin appears to be a key event in the induction of autophagy. These data are consistent with the previously reported result that administration of streptozotocin to rats, which depressed plasma insulin, induced significant liver autophagic degradation.38 This result indicates that loss of insulin action is a sufficient signal for the induction of liver autophagy. This mechanism is further supported by the observation that Akt, P70k-S6kinase and the S6 ribosomal subunits were dephosphorylated, showing that the mTor/S6-kinase pathway was inactivated in the livers of mice subjected to 24 h of starvation, whereas they were phosphorylated or activated in the livers of fed mice (Fig. 2E).
It is particularly interesting that pre-administration of glucose to mice after 18 and 21 h of starvation inhibited autophagy, resulting in a significant reduction in the amino acid surge (Fig. 5). During the course of this experiment, glucagon levels remained constant (Sup. Fig. 4C), whereas insulin increased promptly, but transiently, to high levels, which surpassed the normal level observed in fed mice (Fig. 2). These results indicate that the glucose-induced increase in plasma insulin exerted a strong suppressive effect on autophagy in mice. Thus, insulin plays a principal role in controlling liver autophagic protein degradation, which is closely connected to the maintenance of blood glucose levels.
A portion of the amino acids released in the liver is excreted into the blood.
In parallel with the amino acid surge in the liver, free amino acids in plasma and skeletal muscles increased transiently in wild-type mice. As this increase was absent in liver-specific Atg7-deficient mice, the data indicate that in addition to use by the liver, a portion of the released amino acids was excreted from the liver into circulation for use in peripheral tissues, such as skeletal muscle (Fig. 3A and B). During 24 h of starvation, liver weight was reduced by ∼25% of its initial weight to ∼1 g. At this time point, the total blood volume was about 2 ml, which means that the plasma volume was about 1.5 ml. Therefore, we speculate that in the midst of the amino acid surge, the net release of amino acids in the liver was actually much greater than that depicted in Figure 3A. It is interesting that, in autophagy-deficient liver, a delayed slow increase in amino acids occurred between 24 and 36 h of starvation (Fig. 3B and Sup. Fig. 1). A similar increase was also observed in both plasma and skeletal muscles (Fig. 3B and Sup. Figs. 2 and 3). Interestingly, the increase began at earlier periods of starvation (∼12 h) in skeletal muscles than in liver. Immunoblotting data show that LC3-II in skeletal muscles of liver-specific autophagy-deficient mice increased transiently between 3 and 12 h of starvation, whereas a similar but subtle increase in LC3-II was found in control wild-type muscles between 12 and 24 h (Sup. Fig. 5). In contrast, MuRF1, a representative ubiquitin E3 ligase in skeletal muscle, is more abundantly present in wild-type muscles than in the muscles of liver-specific Atg7-deficient mice (Sup. Fig. 5). These results may indicate that amino acids released by muscle autophagy are secreted to the circulation to be transported to the liver in liver-specific Atg7-deficient mice.
In summary, the present study is the first to show that the amino acids released as a result of starvation-induced autophagic proteolysis in the liver of mice are metabolized, in part, via hepatic gluconeogenesis to glucose, which is excreted into the circulation to maintain blood glucose concentrations. The amino acids were also excreted from the liver into the blood and transported to skeletal muscle. In view of the fact that the liver is virtually the only organ that supplies blood glucose, the use of amino acids to produce blood glucose can be considered a liver-specific metabolic contribution of starvation-induced autophagy.
Materials and Methods
Animals.
C57BL/6J mice were used as the wild-type mice. Liver-specific conditional Atg7 knockout mice (Atg7F/F :Mx1 mice) were generated as described previously in reference 20. To delete Atg7 from the liver, Cre expression in the liver was induced by i.p. injection of polyinosinic acid-polycytidilic acid (pIpC, Sigma-Aldrich, P1530). Specifically, 0.3 ml of a pIpC solution (1 mg/ml in water) was injected intraperitoneally three times at 48-h intervals. Atg7 is a long-lived protein with a half-life of about one week.20 Complete deletion of Atg7 protein in the liver was verified using immunoblotting analyses. In each experiment described herein, the conditional Atg7-knockout mice were used at 10 days after the last pIpC injection. All mice were maintained in an environmentally controlled room (lights on 8:00 to 20:00) and were fed a pelleted laboratory diet and tap water ad libitum, unless otherwise stated.
For synchronous induction of autophagy in the liver, mice previously fasted for 24 h were fed a pelleted laboratory diet for 2 h (20:00–22:00) in the dark to suppress autophagic activity to a minimum. The diet was then withdrawn and the mice were again starved. In separate experiments, it was confirmed that the stomach and intestine were filled with digested diet at the end of the 2 h feeding period. For leupeptin administration, a leupeptin solution (1.25 mg/ml in 0.15 M NaCl) was injected intraperitoneally at a dose of 15 mg/kg body weight 1 h prior to euthanasia. In the experiments shown in Figure 4C and D, 0.3 ml of serine (100 mg/ml) was orally administered to wild-type and liver-specific Atg7-deficient mice starved for 24 h. The experimental protocol was approved by the Ethics Review Committee for Animal Experimentation of Juntendo University.
Analysis of free amino acids of plasma and tissues.
Plasma samples were mixed with 50% TCA to give a final concentration of 3.3%. After centrifugation at 10,000x g and 4°C for 20 min, the free amino acids in the supernatants were analyzed using a L8500 amino acid analyzer (Hitachi, Ltd.). For determination of free amino acid concentrations in tissue, liver and muscle samples were homogenized with five volumes of 10% TCA and centrifuged at 10,000x g and 4°C for 20 min. The supernatants (0.5 ml) were mixed with an equal volume of 0.02 N HCl and analyzed using a L8500 amino acid analyzer.
Analytical methods.
Mouse livers were homogenized with five volumes of 10 mM Hepes-NaOH (pH 7.4) containing 0.25 M sucrose and a protease inhibitor cocktail (Roche Diagnostics, 1836170), using a motor-driven glass/Teflon homogenizer (5 up-down strokes at 800 rpm). The liver homogenates were centrifuged at 500x g and 4°C for 5 min to obtain postnuclear supernatants. The postnuclear supernatants were separated by SDS-PAGE according to the published method in reference 39. Separated polypeptides were analyzed by immunoblot analyses according to the published procedure,40 with the exception that Super Signal West Pico Chemiluminescent Substrate (34080) or Supersignal West Dura Extended Duration Substrate (34075) (Thermo Fisher Scientific, Inc.,) was used as the substrate for the horseradish peroxidase-conjugated secondary antibodies. The antibodies against Atg7 and LC3 were prepared as described previously in reference 20. The antibodies against S6 ribosomal protein (2317), phospho-S6 ribosomal protein (Ser235/236, 2211), p70 S6 kinase (2708), phospho-p70 S6 kinase (Thr 389, 9205), phospho-p70 S6 kinase (Thr421/Ser424, 9204), Akt (9272), phospho-Akt (Ser473, 9271) and MuRF1 (4305) were purchased from Cell Signaling Technology. The antibody against actin (clone C4, MAB1501) was obtained from Millipore Corporation. For quantitative densitometry, immunoblotting data were scanned and analyzed using a calibrated densitometer (BioRad Laboratories, GS-800) with a 200 mm resolution at the highest sensitivity setting. The scanned raw data were processed for quantification using the gel analysis software Quantity One (BioRad Laboratories).
The glycogen levels of liver and muscle were measured using the Lo method.41 The concentrations of insulin and glucagon in plasma were determined using an ELISA kit (Shibayagi Co., Ltd., AKRIN-011) and a glucagon enzyme-immunoassay kit (Yanaihara Institute Inc., YK090), respectively. Plasma glucagon levels determined by an ELISA kit are reportedly one order of magnitude larger than that determined by radioimmuno assay.42,43 The concentrations of glucose, non-esterified fatty acids and triacylglycerol in plasma were determined using an automatic biochemical analyzer JCA-BM8000 (JEOL Ltd.,) with a hexokinase-coupled spectrophotometric assay kit (Sino-Test Corporation, 326023042 and 326023059), an acyl-CoA synthetase/acyl-CoA oxidase-coupled colorimetric assay kit (Eiken Chemical Co., Ltd., G-HE99) and a lipoprotein lipase-coupled colorimetric assay kit (Wako Pure Chemical Industries, 412-37494 and 418-37594), respectively. In the experiments shown in Figure 4C and D, total blood samples were collected via tail tip bleeds and examined for glucose concentration using a glucometer (Glutest Pro; Sanwa Kagaku Kenkyusho). The protein concentration was determined using a BCA protein assay, according to the manufacturer's protocol (Thermo Fisher Scientific Inc., 23228).
Electron microscopy.
Autophagic vacuoles (autophagosomes plus autolysosomes) exist in a rapid equilibrium between autophagosome formation and the subsequent maturation/degradation of autophagosomes; only a small number of autophagic vacuoles can be detected, unless leupeptin is administered to the animals.16,44,45 In preliminary experiments, it was confirmed that in the absence of leupeptin administration, the number of vacuoles was too low to assess statistical significance in the quantification of autophagic vacuoles. In the livers of mice that had been administered leupeptin, many more autophagic vacuoles were counted, so that quantification of the vacuoles was feasible. Liver-specific conditional Atg7 knockout mice and control littermates were administered leupeptin (15 mg/kg) 1 h before use.
The livers were fixed by cardiac perfusion using 0.1 M phosphate buffer (pH 7.4) containing 2.5% glutaraldehyde. They were post-fixed with 1% OsO4, embedded in Epon812 and sectioned. Morphometric analyses were performed according to the published procedure in reference 46. Specifically, for each liver slice, 20 digital electron micrographs were taken with systematic random sampling at a primary magnification of 3,000×. These original electron micrographs were enlarged 2.6-fold and printed on projection paper. The cytoplasmic (perikaryal) volume fractions of electron-dense organelles that corresponded to autophagic vacuoles, mitochondria and peroxisomes were calculated by point counting, using a double-lattice test system with 1.5 cm spacing.
Statistical analysis.
All data are means ± SEM. Statistical differences between the two groups were evaluated using a two-tailed Student's t test. A p value of less than 0.05 was considered statistically significant.
Acknowledgements
This work was supported in part by a Grant-in-aid for Scientific Research (C) (16590253 to J.E.), Grants-in-aid for Scientific Research (B) (19390079 to E.K. and 14380308 to T.U.), a Grant-in-aid for Scientific Research on Priority Areas (18076005 to I.T., M.K., T.U.), a Grant-in-aid for “High-Tech Research Center” Project for Private Universities: matching fund subsidy (E.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and a Research Grant from the Takeda Science Foundation (T.U.). The authors thank Yoshinori Hosokawa and Toshio Kumasaka, Department of Human Pathology, Juntendo University School of Medicine for their technical assistance with the electron microscopy and James L. McDonald, Scientific Editorial Services, for language editing of the text.
Abbreviations
- LC3
microtubule-associated protein 1A/1B light chain 3
- PE
phosphatidylethanolamine
- i.p.
intraperitoneal
- BCAA
branched-chain amino acid
- PEPCK
phosphoenolpyruvate carboxykinase
- pIpC
polyinosinic acid-polycytidilic acid
Supplementary Material
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano A, Nakagawa I, Yoshimori T. Autophagy in innate immunity against intracellular bacteria. J Biochem. 2006;140:161–166. doi: 10.1093/jb/mvj162. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu M, Ueno T, Waguri S, Uchiyama Y, Kominami E, Tanaka K. Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ. 2007;14:887–894. doi: 10.1038/sj.cdd.4402120. [DOI] [PubMed] [Google Scholar]
- 6.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 9.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 11.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 12.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 13.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 14.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 15.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 16.Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem. 1981;256:7652–7658. [PubMed] [Google Scholar]
- 17.Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci USA. 1983;80:2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schworer CM, Mortimore GE. Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc Natl Acad Sci USA. 1979;76:3169–3173. doi: 10.1073/pnas.76.7.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallette LE, Exton JH, Park Effects of glucagon on amino acid transport and utilization in the perfused rat ler. J Biol Chem. 1969;244:5724–5728. [PubMed] [Google Scholar]
- 20.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueno T, Muno D, Kominami E. Membrane markers of endoplasmic reticulum preserved in autophagic vacuolar membranes isolated from leupeptin-administered rat liver. J Biol Chem. 1991;266:18995–18999. [PubMed] [Google Scholar]
- 22.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kominami E, Hashida S, Khairallah EA, Katunuma N. Sequestration of cytoplasmic enzymes in an autophagic vacuole-lysosomal system induced by injection of leupeptin. J Biol Chem. 1983;258:6093–6100. [PubMed] [Google Scholar]
- 24.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 25.Mortimore GE, Poso AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- 26.Kadowaki M, Karim MR, Carpi A, Miotto G. Nutrient control of macroautophagy in mammalian cells. Mol Aspects Med. 2006;27:426–443. doi: 10.1016/j.mam.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 28.Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci USA. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 30.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 31.Mordier S, Deval C, Bechet D, Tassa A, Ferrara M. Leucine limitation induces autophagy and activation of lysosome-dependent proteolysis in C2C12 myotubes through a mammalian target of rapamycin-independent signaling pathway. J Biol Chem. 2000;275:29900–29906. doi: 10.1074/jbc.M003633200. [DOI] [PubMed] [Google Scholar]
- 32.Kanazawa T, Taneike I, Akaishi R, Yoshizawa F, Furuya N, Fujimura S, et al. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J Biol Chem. 2004;279:8452–8459. doi: 10.1074/jbc.M306337200. [DOI] [PubMed] [Google Scholar]
- 33.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 34.Shinnick FL, Harper AE. Branched-chain amino acid oxidation by isolated rat tissue preparations. Biochim Biophys Acta. 1976;437:477–486. doi: 10.1016/0304-4165(76)90016-7. [DOI] [PubMed] [Google Scholar]
- 35.Remesy C, Fafournoux P, Demigne C. Control of hepatic utilization of serine, glycine and threonine in fed and starved rats. J Nutr. 1983;113:28–39. doi: 10.1093/jn/113.1.28. [DOI] [PubMed] [Google Scholar]
- 36.Seglen PO, Gordon PB, Poli A. Amino acid inhibition of the autophagic/lysosomal pathway of protein degradation in isolated rat hepatocytes. Biochim Biophys Acta. 1980;630:103–118. doi: 10.1016/0304-4165(80)90141-5. [DOI] [PubMed] [Google Scholar]
- 37.Kovács AL, Grinde B, Seglen PO. Inhibition of autophagic vacuole formation and protein degradation by amino acids in isolated hepatocytes. Exp Cell Res. 1981;133:431–436. doi: 10.1016/0014-4827(81)90336-0. [DOI] [PubMed] [Google Scholar]
- 38.Lenk SE, Bhat D, Blakeney W, Dunn W., Jr Effects of streptozotocin-induced diabetes on rough endoplasmic reticulum and lysosomes of rat liver. Am J Physiol. 1992;263:856–862. doi: 10.1152/ajpendo.1992.263.5.E856. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo S, Russell JC, Taylor AW. Determination of glycogen in small tissue samples. J Appl Physiol. 1970;28:234–236. doi: 10.1152/jappl.1970.28.2.234. [DOI] [PubMed] [Google Scholar]
- 42.Maeda N, Funahashi T, Hibuse T, Nagasawa A, Kishida K, Kuriyama H, et al. Adaptation to fasting by glycerol transport through aquaporin 7 in adipose tissue. Proc Natl Acad Sci USA. 2004;101:17801–17806. doi: 10.1073/pnas.0406230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doi M, Yamaoka I, Nakayama M, Sugahara K, Yoshizawa F. Hypoglycemic effect of isoleucine involves increased muscle glucose uptake and whole body glucose oxidation and decreased hepatic gluconeogenesis. Am J Physiol Endocrinol Metab. 2007;292:1683–1693. doi: 10.1152/ajpendo.00609.2006. [DOI] [PubMed] [Google Scholar]
- 44.Furuno K, Ishikawa T, Kato K. Appearance of autolysosomes in rat liver after leupeptin treatment. J Biochem. 1982;91:1485–1494. doi: 10.1093/oxfordjournals.jbchem.a133840. [DOI] [PubMed] [Google Scholar]
- 45.Kovacs AL, Laszlo L, Kovacs J. Effect of amino acids and cycloheximide on changes caused by vinblastine, leupeptin and methylamine in the autophagic/lysosomal system of mouse hepatocytes in vivo. Exp Cell Res. 1985;157:83–94. doi: 10.1016/0014-4827(85)90154-5. [DOI] [PubMed] [Google Scholar]
- 46.Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–1728. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.