Abstract
The product of the retinoblastoma tumor susceptibility gene (RB1) is a key regulator of cell proliferation and this function is thought to be central to its tumor suppressive activity. Several studies have demonstrated that inactivation of pRB not only allows inappropriate proliferation, but also undermines mitotic fidelity, leading to genome instability and ploidy changes. Such properties promote tumor evolution and correlate with increased resistance to therapeutics and tumor relapse. These observations suggest that inactivation of pRB may contribute to both tumor initiation and progression. Further characterization of pRB’s role in chromosome segregation will provide insight into processes that are misregulated in human tumors and may reveal new therapeutic targets to kill or stall these chromosomally unstable lesions. Here, we review the evidence that pRB promotes genome stability and discuss the mechanisms that likely contribute to this effect.
Aneuploidy and CIN drive tumorigenesis
Genetic instability and the development of aneuploidy have long been linked to the acquisition of invasive and metastatic characteristics [1, 2], with the percent of aneuploid tumors increasing with both histological differentiation and tumor size to near 100% in advanced tumors [3–6]. Such genomic changes may be beneficial to cancer development. For example, genome instability, which includes chromosomal and subchromosomal alterations such as inversions, deletions, mutations, duplications and translocations of large chromosomal segments, has the potential to drive mutations in oncogenes and tumor suppressor genes. Aneuploidy, a state in which a cell exhibits an abnormal number of chromosomes, is a hallmark of many cancer cells. Several different defects can cause aneuploidy, including defects in mitosis that promote chromosome segregation errors. Underlying defects in mitotic chromosome segregation can result in a consistent elevated rate of gains and losses of whole chromosomes, a process that is known as chromosomal instability (CIN) [7]. Although cells can become aneuploid without displaying CIN, CIN necessarily results in aneuploidy. While aneuploidy has been shown to be detrimental to growth in otherwise normal cells and organisms [8–10], CIN can promote loss of heterozygosity (LOH), uncovering heterozygous mutations, thereby providing selective growth advantage in tumor cells [11].
The majority of solid tumors exhibit structural and numerical chromosome aberrations, with evidence of aneuploidy in even very early, benign lesions [12, 13]. This is illustrated by high-throughput genomic profiling studies showing that most human tumors display abnormalities in the number of whole chromosomes or chromosome arms [14–16]. Additionally, many tumors have been shown to be chromosomally unstable [7]. CIN and aneuploidy have been proposed to promote the evolution of tumor cells such that these genomic changes appear to promote metastasis and chemotherapeutic resistance, and correlate with poor patient prognosis [17–24]. Importantly, recent studies show that aneuploidy and CIN can have a causal role in tumorigenesis and relapse [10, 11, 25].
CIN is the result of an underlying defect in mitotic fidelity and several mechanisms that result in whole chromosome missegregation have been described. These include defects in bipolar spindle formation, errors in chromosome–spindle association, failed chromosome cohesion, and defects in the spindle assembly checkpoint [9, 11, 25–32]. In addition, studies of CIN cell lines suggest that merotely, an erroneous kinetochore attachment where a single kinetochore associates with microtubules from both spindle poles (box 1), is a dominant mechanism for chromosome missegregation in tumor cells [9, 28, 30, 33]. Consistent with this link between merotely, CIN and tumorigenesis, tetraploidy has been shown to be a sufficient precursor to chromosomal instability and tumorigenesis and this is thought to be due in large part to the presence of extra centrosomes that promote merotelic attachments [28, 30, 34, 35]. However, the majority of solid tumors exhibit a near diploid karyotype, making it unlikely that tetraploidy is an initiating factor in CIN or tumorigenesis in these cells [36].
Box 1. Merotelic kinetochore attachments.
Form when microtubules from two spindle poles interact with a single kinetochore
Formation is promoted by aberrant spindle geometry (multipolar spindle, supernumerary centrosomes) and/or centromere deformation (decreased cohesion or condensation, mis-localization/reduction of kinetochore proteins, etc), and/or hyper-stable kinetochore-microtubules attachments
Do not signal the spindle assembly checkpoint and therefore allow anaphase progression prior to correction of erroneous attachments
Often result in lagging chromosomes which may missegregate and/or be resolved into micronuclei during the subsequent G1
Correction is influenced by a number of factors that regulate kinetochore-microtubule stability and mitotic timing
Proposed to be a major mechanism of chromosome segregation errors in CIN cells
For cells to maintain CIN, they must additionally acquire tolerance of the resulting aneuploidy. In response to ploidy changes, non-transformed cells exhibit a strong proliferative defect [8, 9, 37], and in order to propagate their aneuploid genotype tumor cells must overcome or bypass such an effect. Although numerous pathways have been described that can contribute to chromosome segregation errors and tolerance of subsequent ploidy changes, the underlying molecular mechanism(s) behind such defects remains unclear. Given the broad array of factors that apparently contribute to mitotic fidelity and proliferation of aneuploid cells, it is possible that different subsets of defects exist in different tumors. Alternatively, a common defect may contribute to the susceptibility of tumor cells to both mis-segregate chromosomes and become tolerant of the subsequent aberrant ploidy. Indeed recent data has implicated the pRB protein pathway in both the generation of CIN and the tolerance of aneuploidy, raising the possibility that pRB inactivation may contribute to many of the changes seen in tumor cells. Here, we discuss some of the evidence linking pRB to the maintenance of genome stability.
Loss of pRB promotes aneuploidy and chromosome instability
The retinoblastoma tumor susceptibility gene (RB1) was one of the first tumor suppressor genes to be discovered and most, if not all, tumors acquire lesions in the pRB pathway. While the loss of pRB function has been implicated in the development of numerous cancers [38], the significance of RB1 inactivation is perhaps best illustrated by the pediatric cancer, retinoblastoma, for which the gene was named. In these tumors, the inactivation of both copies of RB1 is rate-limiting for tumorigenesis and is thought to be an initiating event [39–41].
Inactivation of pRB impacts many cellular processes. For example, pRB-deficient cells have altered regulation of G1 checkpoints, changes in the control of cell cycle exit (differentiation, senescence, quiescence), and altered levels of autophagy, apoptosis, angiogenesis, and metastatic potential (reviewed in [38]). Many of these effects are thought to be a consequence of altered gene expression. The most intensively studied function of pRB is its ability to repress transcription of E2F-regulated genes, a role that enables it to regulate the expression of many genes that are needed in cell cycle progression and cell proliferation.
More recently, a series of studies have highlighted an additional consequence of RB inactivation that seems likely to impact tumorigenesis. In numerous in vivo and in vitro model systems, loss of pRB activity enhances genomic instability. These studies have linked the functional inactivation of pRB to various types of genomic change, including endoreduplication, increases in ploidy on both the chromosomal and subchromosomal (local amplifications, chromosome arm gains and losses) levels, and consistently high rates of chromosome segregation errors resulting in whole chromosome missegregation (CIN), as well as tolerance of such genomic variations [42–49] (Table 1). One potential reason for these changes is that the inactivation of pRB leads to defects during mitosis. Indeed, the mitotic defects of pRB-deficient cells have been characterized in detail and although less dramatic than those in G1 regulation that are evident earlier in the cell cycle, these subtle changes undermine the fidelity of chromosome segregation. The loss of pRB results in supernumerary centrosomes, centromeric defects, and formation of micronuclei. Remarkably, many of these changes are consistent with the formation of merotelic kinetochore attachments during mitosis (Box 1). It is well established that merotelic attachments promote whole chromosome missegregation and that frequent occurrence of such erroneous attachments, as is found in chromosomally unstable tumor cell lines, can result in aneuploidy (reviewed in [31]). Together this suggests that pRB loss of function leads to CIN in tumors by promoting merotelic kinetochore attachment (Table 2). Currently, there is scant evidence that pRB acts directly in mitosis. Instead, it seems probable that the loss of pRB function causes changes during earlier stages of the cell cycle that subsequently influence chromosome segregation. As described below, there is not one connection between pRB and mitosis; instead the mitotic defects seem likely to be the cumulative effect of several types of change resulting from the inactivation of pRB.
Table 1.
pRB Loss Undermines Genome Stability: an Overview
| Described effect on Aneuploidy, CIN and/or DNA Damage response | Type of Lesion/model | Date | Ref(s) |
|---|---|---|---|
| Cell cycle analysis of Rb deficient fibroblasts shows they have an increased incidence of aneuploidy | Primary fibroblasts isolated from Rb mutant mouse embryos | 1996 | [91] |
| Rb loss or inactivation allows for re-replication following drug induced mitotic arrest | Rb−/− mouse embryonic fibroblasts (MEFs); E7 expressing fibroblasts, p16−/− and p21−/− MEFs | 1997/1998 | [92] [93] |
| Loss of Rb, but not p107 or p130 impairs G1/S checkpoint response to damaging inducing agents | Rb−/− MEFs | 1998 | [94] |
| Loss of the Rb family prevents appropriate G1 arrest in response to DNA damage | Rb−/−; p107−/−; p130−/− Triple Knockout (TKO) MEFs | 2000 | [95, 96] |
| pRB interacts with Hec1 and hsRB expression in a yeast temperature sensitive hec1 allele suppresses segregation errors | Heterologous yeast system expressing human pRB and human Hec1 | 2000 | [55] |
| Deficiency of Rb causes increased loss of a marker gene | Rb−/− mouse embryonic stem (ES) cells with an inserted chromosomal marker | 2002 | [97] |
| Rb null cells exhibit increased levels of aneuploidy | Rb−/− MEFs | 2002 | [98] |
| pRB depletion leads to upreguation of the mitotic spindle assembly checkpoint protein Mad2 and near diploid aneuploidy | Rb−/− MEFs, mouse and human cell lines expressing pRB-targeting short hairpins or E1A expression constructs | 2004 | [44] |
| Loss of Rb leads to increased ploidy and failure of the DNA damage checkpoint response | Conditional Rb knockout in Mouse Adult Fibroblasts (MAFs) | 2004 | [99] |
| Cells lacking Rb function exhibit increased double strand breaks and compromised cell cycle arrest following genotoxic stress. | Conditional Rb knockout in (MAFs) | 2004/2005 | [76, 80] |
| Rb loss leads to increased ploidy following serum starvation | Rb−/− and Rb−/−p107−/− MEFs | 2005 | [100] |
| Rb loss leads to increased levels of aneuploidy and tetraploidy, independent of p53 status | Rb1−/−, p107−/−; p130−/−, and TKO MEFs | 2005 | [62] |
| Loss of pRB leads to E2F1-mediated accumulation of DNA double strand breaks | Acute depletion of pRB in human fibroblast and osteosarcoma cell lines | 2005/2006 | [101, 102] |
| Acute loss of Rb induces centrosome amplification and aneuploidy | Conditional Rb deficient MEFs | 2006 | [45] |
| Disruption of Rb LXCXE pocket reduces pericentromeric H4K20me3 and induces ploidy changes | Rb1ΔL/ΔL, Rb1−/− mouse ES cells | 2006 | [46] |
| Acute and sustained liver specific Rb loss promotes ploidy changes | Liver-specific conditional Rb knockout mice | 2005/2007 | [48, 103] |
| Rb loss leads to deregulation of DNA synthesis and elevated ploidy | Conditional Rb knockout in MAFs | 2007 | [49] |
| pRB inactivation compromises the G2/M DNA damage checkpoint | Stable pRB depletion in U2OS cells | 2007 | [104] |
| Whole chromosome gains and losses are coincident with progression from retinoma to retinoblastoma | Genetic RB1 mutation/human tumor samples | 2008 | [40] |
| Rbf1 loss leads to defects in condensin II chromatin association and chromatin condensation, and abnormal chromosome segregation | Drosophila Rbf1 mutants | 2008 | [60] |
| Acute depletion of pRB leads to supernumerary centrosomes, formation of micronuclei and aneuploidy. Depletion of CENPA partially suppresses the number of aneuploid cells. | Acute siRNA pRB depletion in human fibroblasts and HCT116 tumor cells | 2009 | [42, 105] |
| Suggests a functional link between pRB and the kinetochore protein Hec1 influences chromosome segregation | Transient and stable depletion of pRB in HCT116 tumor cells | 2010 | [53] |
| pRB loss causes defects in chromosome cohesion and condensation, centromere dysfunction, and promotes high rates of chromosome missegregation | Acute and stable depletion of pRB in human epithelial cells; Drosophila Rbf1 mutants | 2010 | [106] |
| An Rb1 mutant that retains it’s ability to regulate E2F exhibits defects in chromatin condensation, promotes more aggressive tumors, and accelerates loss of heterozygosity in a tumor model | Rb1ΔL/ΔL; TRP53+/− and Rb1ΔL/ΔL; TRP53−/− knock in mouse tumor models and MEFs and mouse ES cells | 2010 | [107] |
| Loss of the Rb family of proteins compromises the G2 DNA damage checkpoint, allowing cells to enter mitosis with unrepaired damage, thereby promoting genome instability and aneuploidy | TKO MEFs | 2010 | [78] |
| Replication induced DNA damage due to pRB pathway defects is caused by nucleotide deficiency and leads to genome instability | Primary keratinocytes infected with E6/E7 expression vector; Cyclin E overexpression in BJ fibroblasts | 2011 | [79] |
Table 2.
Mitotic defects identified following pRB pathway lesions are consistent with the presence of merotelic kinetochore attachments
| Mitotic Delay and Increased Mitotic Index | [42, 44, 47] |
| Supernumerary Centrosomes | [45, 57, 58] |
| Micronuclei Formation | [42, 43] |
| Changes in Heterochromatin Formation (i.e. Centromeric Regions) | [43, 46, 47, 60–62, 108, 109] |
| Changes in expression, recruitment and/or localization of kinetochore or centromere proteins | [44, 53–55, 105, 110] |
| Changes in Mitotic Spindle Assembly Checkpoint Control | [44, 53] |
| Lagging chromosomes during anaphase | [47, 105] |
E2F-dependent mechanisms promoting CIN
The loss of pRB deregulates E2F. Comparison of gene expression data shows a significant overlap between the changes associated with CIN and the changes that occur in pRB-deficient cells, raising the possibility that the CIN signature may be, at least in part, a consequence of pRB misregulation [50–52]. Well-characterized targets of E2F include multiple genes whose products are required for accurate chromosome segregation during mitosis, supporting the idea that one of the ways that pRB contributes to the maintenance of genome stability is through its regulation of E2F. Consistent with this idea, recent work has shown that upregulation of Mad2, one such E2F target that is deregulated by the inactivation of pRB, is sufficient to induce chromosome missegregation [25, 44]. In addition, the expression level and/or localization of several structural components of the kinetochore are also misregulated following pRB depletion [53–55]. Importantly, upregulation of at least one of these proteins, Hec1, has been linked to chromosome segregation errors [56].
A majority of solid tumors possess extra centrosomes, the presence of which can induce chromosome missegregation [28]. Centrosome amplification has been shown to result from E2F-dependent misregulation of several genes following RB loss [45, 57] and this may contribute to the chromosomal instability seen in pRB-depleted cells. However, it is not clear that all cells lacking pRB generate extra centrosomes, and in at least some cells that do, extra centrosomes are soon lost while chromosome missegregation continues [28, 45, 58]. While CIN is typically believed to be a cause of aneuploidy, recent data has raised the possibility that aneuploidy itself may be a cause of CIN, such that CIN becomes a self-propagating phenomenon [59]. It is possible, therefore, that transient defects, such as the presence of extra centrosomes, which can occur following pRB-depletion may be sufficient to initiate perpetual chromosomal instability.
E2F-independent functions of pRB that suppress CIN
Our recent work shows that pRB loss leads to structural defects at the centromeric region of chromosomes, changes that appear to result from a failure to properly recruit cohesin and condensin II components to chromatin [47]. The resulting centromeric defects, which manifest during mitosis as hyperstretched kinetochores, promote erroneous merotelic kinetochore attachments that lead to chromosome missegregation. In a related study, it was found that an pRB mutant (RbΔL) that retains its ability to interact with E2F likewise has defects in chromosomal stability, and reported that regulation of chromatin compaction via an interaction between pRB and the condensin II complex has a role in tumor suppression [43].
Interestingly, there are several lines of evidence indicating that these mitotic defects and chromosome segregation errors are separable from the regulation of E2F-dependent transcription. Overexpression of E2F1 is insufficient to produce centromeric defects seen following pRB depletion [47]. In addition, because the pRbΔL protein retains its ability to interact with E2F, it is competent to repress E2F targets like Mad2 [43, 46]. Moreover the mitotic defects of RbΔL/ΔL mice were evident in ES cells that lack normal programs of E2F regulation [43]. Studies in Drosophila have revealed an extensive co-localization between RBF1 and the Condensin II protein dCAP-D3 on polytene chromosomes that is independent of E2F/DP regulation [60]. This, together with evidence of physical interactions between pRB/RBF and Condensin II proteins, suggests that pRB has an E2F–independent function that promotes the normal recruitment of Condensin II components to chromatin. Together these studies suggest that changes in the recruitment of condensin II and cohesin complexes, resulting from either pRB loss or mutation, has functional consequences on both chromosome structure and genome stability.
pRB regulates programs of gene expression and many of the proteins that have been discovered to associate with pRB are transcription factors or chromatin associated proteins that impact transcription [38, 61–63]. Fitting with this trend, although best known for their roles in regulation of mitotic chromosome structure, recent studies have suggested that both the cohesin and condensin complexes have additional roles in regulation of gene expression. Effects of cohesin on gene expression have been observed in numerous organisms (reviewed in [64, 65]) and, unlike the role of cohesin in mitosis, these effects are independent of cell cycle progression [64, 66, 67]. In addition, changes in gene expression have been noted in both mouse models and in human diseases associated with cohesin mutations, where surprisingly, effects on mitotic chromosome cohesion are minimal [68, 69]. Recent studies have found that cohesin physically and functionally interacts with the mediator complex, an important transcriptional regulator, at active genes [70]. Cohesin has also been shown to be recruited by, and to influence the activity of, the transcriptional regulator CTCF [71–73]. Likewise, Condensin II complex’s effects on chromatin compaction have been suggested to alter the accessibility of chromatin to the transcriptional machinery, and potentially to regulate recruitment of various transcription regulators [74]. Indeed, both cohesin and condensin have been shown to influence enhancers, silencers and insulators of gene expression [75]. These observations raise the intriguing possibility that the connections between pRB proteins and cohesin and condensin II complexes may reflect a shared role in transcriptional control. Currently however, the loci that are targets of this co-regulation, or the context in which this occurs, are not known.
Elevated levels of DNA damage in pRB mutant cells
In addition to structural changes to chromatin, several studies have reported that loss of pRB renders cells more prone to DNA damage, suggesting that pRB family members may be generally required to maintain genome integrity (Table 1). Just as there are several ways in which the loss of pRB can influence progression through mitosis, there are several ways in which the inactivation of pRB may lead to the accumulation of DNA damage. For example, pRB is important for the maintenance of DNA damage-induced cell cycle arrest during G1 [76, 77]. In addition, although cells lacking pRB family members are able to initiate G2 arrest in response to DNA damage, they are not always competent to maintain the arrest and instead enter mitosis with broken chromosomes [78]. Other links stems from the importance of pRB in regulation of replication and in controlling gene expression. Recent work shows that misregulation of the pRB pathway leads to nucleotide deficiency, and consequent replication-induced DNA damage [79]. Also, since pRB and pRB-related proteins regulate the expression of DNA damage repair factors [77, 80] they may also indirectly affect the efficiency of repair processes. An additional connection is suggested by the recent evidence that pRB affects the distribution of cohesin complexes. Cohesin complexes have been shown to play a role in regulating efficient repair of double strand breaks (DSBs) [81], so this may represent an additional way in which pRB may influence the DNA damage response.
Finally, we note that pRB interacts with many chromatin regulators and, in addition to their roles in transcriptional control (reviewed in [63]), several of these regulators are important for the formation of heterochromatin and, like cohesin and condensin II complexes, may have effects on both centromeric and telomeric structure [61, 62]. The loss or mutation of pRB leads to changes in chromatin structure, both in the local vicinity of promoter regions and in the organization of chromosomal structures. It is possible that the compound effect of all of these changes may make chromatin more vulnerable to damage. For example, general changes in chromosome architecture may predispose the cell to chromosome fragility and/or segregation errors. Such global effects would be difficult to attribute to any specific gene or binding partner.
pRB’s regulation of genome stability: an additional tumor suppressive role?
pRB is a multifunctional protein. In addition to its role as a negative regulator of the G1 to S transition, pRB has also been shown to promote cellular differentiation, modulate cell fate decisions, be important for oncogene-induced senescence, and affect cellular sensitivity to apoptosis (reviewed in [38]). Currently it is unclear which of these activities are most important for tumor suppression and, depending on the context, their relative importance is likely to vary. There is increasing evidence that CIN and aneuploidy have causative roles in tumorigenesis and in the evolution of cancer cells. Given the extensive changes seen in pRB-deficient cells it seems likely that pRB’s role in maintaining genome stability also contributes to its tumor suppressive activity [43].
Mutation of RB1 is a rate-limiting event in the development of most retinoblastomas. Recent studies suggest that homozygous mutation of RB1 leads to the appearance of benign retinomas that subsequently progress to retinoblastoma [40]. The role of pRB in E2F-regulated promotion of cell cycle progression, as well as promotion of differentiation and senescence explain why loss of pRB activity would be beneficial at the initial stages of tumor development. It is generally thought that there is a temporal aspect of tumor evolution in which cells gradually acquire numerous mutations [82]. The presence of chromosomal instability would promote such evolution and, indeed, the malignant progression from retinoma to retinoblastoma has been correlated with greatly increased levels of aneuploidy and genomic instability [40].
In an alternative view, recent work by several groups highlights the idea that not all tumor progression is gradual and that occasional isolated events can occur that greatly advance tumor evolution in a single step (punctuated equilibrium). One example of this is cytokinesis failure and the generation of a tetraploid cell. In the context of p53 mutations, tetraploidy has been shown to be initiating for tumor formation, and the subsequent presence of extra centrosomes promotes CIN through the formation of merotelic attachments [28, 34]. A second example of such a disastrous event is the recently described chromothripsis, in which a single chromosome is shattered and then haphazardly pieced back together, resulting in massive rearrangements, deletions and amplifications along a single chromosome [83], potentially leading to oncogene amplifications or tumor suppressor deletions. That usually only one chromosome is involved suggests that the affected chromosome is spatially separated. This may occur by resolution of chromosome bridges following cytokinesis, as proposed by the authors, or alternatively by the formation of micronuclei, which occasionally result following merotelic attachment. Interestingly, work by David Pellman and colleagues shows that chromatin located in micronuclei accumulate damage (personal communication). Merotely can also give rise to lagging chromosomes during anaphase that are positioned under the cytokinetic furrow. Subsequent furrow ingression can cause chromosome breakage [84, 85]. In support of this idea, recent work shows that DNA double strand breaks are apparent following merotelic attachments [86] and exciting new work by Medema and colleagues show that following cytokinesis, cells predisposed to forming merotelic attachments exhibit increased levels of DNA damage and subsequent chromosomal abnormalities (personal communication). This suggests that the increase in merotelic attachments that occur when pRB is lost may lead not only to the missegregation of whole chromosomes, but may also predispose afflicted chromosomes to catastrophic damage, increasing the chance of tumorigenic mutations.
Although increased severity of chromosomal changes and aneuploidy correlate with tumor progression, the generation of aneuploidy by increasing chromosome missegregation rates alone is growth inhibitory in culture [8, 9, 37], and results in few tumors in only a subset of tissues, and late in life, when examined in mouse models [10, 87, 88]. The fact that many other tumors are able to tolerate such ploidy changes and continue to propagate with an ever-changing genome indicates that they have acquired specific adaptive mechanisms and these can perhaps be targeted to halt tumor growth [89]. Recent work has linked growth arrest in newly aneuploid cells to metabolic abnormalities [90] and activation of the p53 pathway[59]. A series of studies have shown that pRB loss is sufficient for cells to acquire ploidy changes and to remain competent to proliferate (see Table 1). This suggests that corruption of pRB pathway activity is likely to be a significant factor contributing to tolerance of aneuploidy. Understanding how pRB activity contributes to genome stability, ensuring accurate chromosome segregation and the intolerance of ploidy changes, could prove useful in devising therapeutic approaches that target aneuploid cancers.
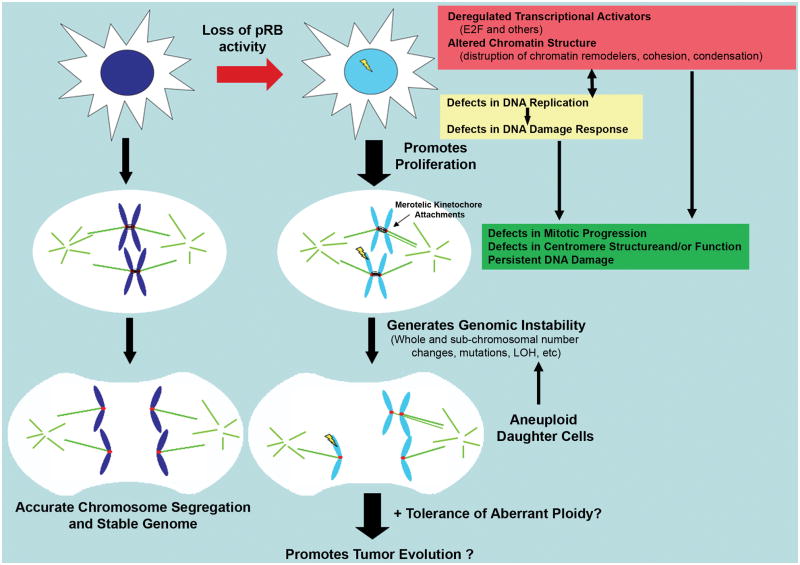
These observations illustrate the point that the inactivation of pRB has the potential to cause multiple types of changes (Figure 1). For example, the links to condensin and cohesin connect pRB to the organization of chromosome structure, gene regulation, and DNA damage responses and repair. Potentially, each of these roles may have a variety of consequences for tumor cells. The centromeric dysfunction and merotelic attachment seen when pRB is lost can result in aneuploidy, chromosome instability, and genomic instability, and each of these phenomena presents a different set of risks for additional copy gains or losses and/or mutation of oncogenes and tumor suppressor genes.
Figure 1. Complex role of pRB in maintenance of genomic stability.
Functional inactivation of pRB has been shown to result in numerous changes, including transcriptional changes (local effects: regulation of specific transcriptional activators: E2F, etc; broad effects: regulation of heterochromatin formation, including telomeric and centromeric regions; and global effects: chromosome condensation and cohesion levels [purple box, top right]). Each of these defects in turn influence DNA replication, mitotic fidelity and the DNA damage response machinery through the transcriptional regulation of important players (yellow and green Boxes; DNA damage represented by yellow lightning bolt). In addition, transcription-independent interactions of pRB with components of many of these pathways have been identified, raising the possibility that pRB’s influence on these downstream processes may be independent of its role in transcription (yellow and green Boxes). Importantly, defects in each of these cellular processes have been shown to result in genomic instability. In addition, pRB loss has been implicated in tolerance of aneuploidy, thereby allowing the propagation of these newly aneuploid cells. The molecular mechanisms behind many of these effects remain unclear and the complex interplay between different effectors of pRB loss makes it difficult to tease apart functional relevance. The importance of individual effects of pRB inactivation under different cellular contexts may in part explain the varying degree of genomic instability seen in tumors possessing pRB pathway lesions.
Taken together, the current information shows that pRB loss promotes defects in mechanisms of chromosome segregation, instigating changes in whole chromosome copy number as well as more complex subchromosomal changes. The loss of pRB causes a consistent, low level of chromosome missegregation (CIN) and this effect is likely to involve both E2F dependent and E2F-independent pathways. pRB pathway lesions also impair the DNA damage response pathway. Through these changes, disregulation of the pRB pathway promotes cell cycle progression and mitotic failure, resulting in genomic instability and aneuploidy (Figure 1). Data showing the prevalence of merotelic attachments in CIN tumor cells, together with new findings suggesting that such erroneous attachments can lead to both whole chromosome segregation defects and the accumulation of DNA damage, highlight the potential importance of pRB’s influence on mitotic fidelity.
The details are important
A recurring theme in the RB literature is the observation that pRB’s role is highly context dependent. pRB can interact with many different proteins, and both its biological function and binding partners vary greatly. Among the challenges for future studies of the link between pRB and genomic instability is the need to determine which type(s) of tumor cells are most vulnerable to genetic change following the inactivation of pRB, and to discover which of the molecular consequences of pRB inactivation drive genomic instability in these cells.
While the loss of pRB function promotes genomic changes through misregulation of chromosome structure, the tolerance and effect of such structural changes may manifest differently in tumors of different origin. The effects of pRB loss are very likely to be strongly influenced by mutations in other tumor suppressors or oncogenes. For example, loss of pRB can result in p53-dependent cell death. In addition, p53 has roles in the DNA damage checkpoint and regulation of response to aneuploidy. Therefore, a tumor is more likely to benefit from genomic instability and chromosome mis-segregation imparted by loss of pRB when the p53-pathway has also been functionally inactivated.
A second complication is that pRB can be functionally compromised in tumor cells by several different types of changes. In addition to the loss or mutation of RB1, the mutation or lowered expression of p16INK4A or the overexpression of Cyclin/Cdks alter the phosphorylation state of pRB. Currently it is unclear how the changes in genome stability resulting from RB1 mutation compare with those that occur in cells where pRB is deregulated by cdk-phosphorylation. In addition to targeting pRB, G1 cdks phosphorylate two pRB related proteins, p107 and p130, that, in certain cell types, can compensate for some effects resulting from the loss of pRB. Differences in the type of lesion in the pRB pathway, as well as cell type-dependent differences in the degree of functional overlap/redundancy between pocket protein family members (p107 and p130) and potential mechanisms of suppression/compensation may contribute to the variation in type and degree of genomic instability seen in the vast array of tumors that have lost pRB function.
CIN and aneuploidy are thought to have causative roles in tumorigenesis and tumor evolution, and the gene expression signature following pRB loss is able to predict poor survival in some human cancer [48], suggesting that pRB’s role in maintaining genome stability may be an additional important tumor suppressive function of this remarkable protein. Many questions remain to be answered (Box 2). It will be important to determine the impact of pRB’s role in genome stability and how the genomic changes resulting from pRB inactivation of contribute to tumor growth, evolution, and response to treatment in various cancer models. Recent work has shown that enhancement of microtubule dynamics is one strategy to suppress CIN in tumor cells [26] and a better understanding of the mechanism(s) by which the loss of RB leads to chromosomal instability may uncover additional novel therapeutic targets. On the one hand these defects may represent an “Achilles heel” that can be specifically enhanced in tumor cells. Alternatively, it may be possible to suppress these changes, helping to stabilize the cancer genome and impairing tumor evolution, effects that that may enhance the potency of traditional therapeutics. For these ideas to become a reality, it will likely be important to better define the context in which pRB inactivation leads to genomic instability, and the situations in which this instability changes the biology of the tumor.
Box 2. Outstanding Questions.
Is genomic instability resulting from mitotic defects a consequence of pRB loss of function in transcription, replication, DNA damage repair, chromatin structural changes at the telomere and centromere, mitotic progression, or a combination of these changes?
What aspect(s) of genomic instability is influenced by pRB’s role in transcription (E2F-dependent and otherwise)?
Does pRB affect mitotic fidelity directly at the time of cell division, or are pRB defects generated earlier in the cell cycle and then manifest during mitosis?
Is pRB regulation of cohesin and condensin II the result of a direct physical interaction, or mediated through pRB’s regulation of histone modification?
Under what context does pRB loss of function promote genome instability? (i.e. cooperation with other mutations or LOH, type of pRB pathway lesion, etc)
To what extent does tumor type/tissue of origin influence pRB’s role in genome stability?
How does pRB’s role in maintenance of genome stability influence tumor growth and evolution?
Acknowledgments
We thank Drs Pellman and Medema for sharing their unpublished work, Neil Ganem for critical reading of the manuscript, and apologize to our colleagues whose work was not cited due to space constraints. A.L.M is supported by a post-doctoral fellowship from the American Cancer Society. N.J.D is the MGH Cancer Center Saltonstall Foundation Scholar and is supported by funding from NIH (R01GM81607 and R01CA64402) and AstraZenica.
Glossary
- Cohesin
A protein complex that associates with chromatin and is best known for its role in holding sister chromatids together. Cohesin is loaded on chromatin concurrent with replication and is maintained there until mitosis when it is removed sequentially from chromosome arms in response to phosporylation, and subsequently from the centromeric region by cleavage upon anaphase onset
- Condensin
A protein complex structurally similar to cohesin whose chromatin association is cell cycle regulated. Mammalian cells have 2 described condensin complexes Condensin I and II, which differ in a subset of their components. Chromatin association of condensin complexes drives coiling of interphase chromatin and compaction prior to mitosis known as condensation
- Centromere/Kinetochore
The primary constriction during mitosis where sister chromatids are held together. The centromere is a specialized condensed region of each chromosome upon which the proteinaceous kinetochore structure is built. The kinetochore mediates association between the chromosome and microtubule fibers of the mitotic spindle to allow for chromosome movement and ensure accurate segregation during cell division
- Centrosome
The microtubule organizing center of the cell. The centrosome is duplicated in preparation for cell division, with each centrosome forming one of two mitotic spindle poles. Presence of extra centrosomes is implicated in multipolar spindle formation, merotelic attachment and chromosome segregation errors
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 3.Hemmer J, Kreidler J. Flow cytometric DNA ploidy analysis of squamous cell carcinoma of the oral cavity. Comparison with clinical staging and histologic grading. Cancer. 1990;66:317–320. doi: 10.1002/1097-0142(19900715)66:2<317::aid-cncr2820660220>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 4.Hemmer J, Nagel E, Kraft K. DNA aneuploidy by flow cytometry is an independent prognostic factor in squamous cell carcinoma of the oral cavity. Anticancer Res. 1999;19:1419–1422. [PubMed] [Google Scholar]
- 5.Auer GU, Heselmeyer KM, Steinbeck RG, Munck-Wikland E, Zetterberg AD. The relationship between aneuploidy and p53 overexpression during genesis of colorectal adenocarcinoma. Virchows Arch. 1994;424:343–347. doi: 10.1007/BF00190554. [DOI] [PubMed] [Google Scholar]
- 6.Bomme L, Bardi G, Pandis N, Fenger C, Kronborg O, Heim S. Cytogenetic analysis of colorectal adenomas: karyotypic comparisons of synchronous tumors. Cancer Genet Cytogenet. 1998;106:66–71. doi: 10.1016/s0165-4608(98)00047-8. [DOI] [PubMed] [Google Scholar]
- 7.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 8.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bardi G, Parada LA, Bomme L, Pandis N, Willen R, Johansson B, Jeppsson B, Beroukas K, Heim S, Mitelman F. Cytogenetic comparisons of synchronous carcinomas and polyps in patients with colorectal cancer. Br J Cancer. 1997;76:765–769. doi: 10.1038/bjc.1997.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverstein MJ. Ductal carcinoma in situ of the breast. Br J Surg. 1997;84:145–146. [PubMed] [Google Scholar]
- 14.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 15.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2000;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 17.Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML, Werts A, Haak P, Vande Woude GF. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci U S A. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi OP, Isola J. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–1604. [PubMed] [Google Scholar]
- 19.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 20.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 21.Choi CM, Seo KW, Jang SJ, Oh YM, Shim TS, Kim WS, Lee DS, Lee SD. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer. 2009;64:66–70. doi: 10.1016/j.lungcan.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Heilig CE, Loffler H, Mahlknecht U, Janssen JW, Ho AD, Jauch A, Kramer A. Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med. 2010;14:895–902. doi: 10.1111/j.1582-4934.2009.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–3266. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- 24.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, Hardcastle T, Lee A, Roy R, East P, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 28.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 30.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome Res. 2010 doi: 10.1007/s10577-010-9179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 35.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 38.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corson TW, Gallie BL. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer. 2007;46:617–634. doi: 10.1002/gcc.20457. [DOI] [PubMed] [Google Scholar]
- 40.Dimaras H, Khetan V, Halliday W, Orlic M, Prigoda NL, Piovesan B, Marrano P, Corson TW, Eagle RC, Jr, Squire JA, et al. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet. 2008;17:1363–1372. doi: 10.1093/hmg/ddn024. [DOI] [PubMed] [Google Scholar]
- 41.Knudson AG., Jr Genetic predisposition to cancer. Cancer Detect Prev. 1984;7:1–8. [PubMed] [Google Scholar]
- 42.Amato A, Lentini L, Schillaci T, Iovino F, Di Leonardo A. RNAi mediated acute depletion of retinoblastoma protein (pRb) promotes aneuploidy in human primary cells via micronuclei formation. BMC Cell Biol. 2009;10:79. doi: 10.1186/1471-2121-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2009;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 45.Iovino F, Lentini L, Amato A, Di Leonardo A. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer. 2006;5:38. doi: 10.1186/1476-4598-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isaac CE, Francis SM, Martens AL, Julian LM, Seifried LA, Erdmann N, Binne UK, Harrington L, Sicinski P, Berube NG, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26:3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2009;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, Wikenheiser-Brokamp K, Boivin GP, Lee JS, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 49.Srinivasan SV, Mayhew CN, Schwemberger S, Zagorski W, Knudsen ES. RB loss promotes aberrant ploidy by deregulating levels and activity of DNA replication factors. J Biol Chem. 2007;282:23867–23877. doi: 10.1074/jbc.M700542200. [DOI] [PubMed] [Google Scholar]
- 50.Black EP, Hallstrom T, Dressman HK, West M, Nevins JR. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc Natl Acad Sci U S A. 2005;102:15948–15953. doi: 10.1073/pnas.0504300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 52.Markey MP, Angus SP, Strobeck MW, Williams SL, Gunawardena RW, Aronow BJ, Knudsen ES. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–6597. [PubMed] [Google Scholar]
- 53.Ferretti C, Totta P, Fiore M, Mattiuzzo M, Schillaci T, Ricordy R, Di Leonardo A, Degrassi F. Expression of the kinetochore protein Hec1 during the cell cycle in normal and cancer cells and its regulation by the pRb pathway. Cell Cycle. 2010;9:4174–4182. doi: 10.4161/cc.9.20.13457. [DOI] [PubMed] [Google Scholar]
- 54.Evans HJ, Edwards L, Goodwin RL. Conserved C-terminal domains of mCenp-F (LEK1) regulate subcellular localization and mitotic checkpoint delay. Exp Cell Res. 2007;313:2427–2437. doi: 10.1016/j.yexcr.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng L, Chen Y, Riley DJ, Chen PL, Lee WH. Retinoblastoma protein enhances the fidelity of chromosome segregation mediated by hsHec1p. Mol Cell Biol. 2000;20:3529–3537. doi: 10.1128/mcb.20.10.3529-3537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci U S A. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 58.Duensing S, Duensing A, Flores ER, Do A, Lambert PF, Munger K. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J Virol. 2001;75:7712–7716. doi: 10.1128/JVI.75.16.7712-7716.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22:1011–1024. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Cao M, Gonzalo S, Dean D, Blasco MA. A role for the Rb family of proteins in controlling telomere length. Nat Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 63.Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 64.Peric-Hupkes D, van Steensel B. Linking cohesin to gene regulation. Cell. 2008;132:925–928. doi: 10.1016/j.cell.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 65.Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 72.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 74.Legagneux V, Cubizolles F, Watrin E. Multiple roles of Condensins: a complex story. Biol Cell. 2004;96:201–213. doi: 10.1016/j.biolcel.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 75.Hagstrom KA, Meyer BJ. Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- 76.Bosco EE, Knudsen ES. Differential role of RB in response to UV and IR damage. Nucleic Acids Res. 2005;33:1581–1592. doi: 10.1093/nar/gki283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Genovese C, Trani D, Caputi M, Claudio PP. Cell cycle control and beyond: emerging roles for the retinoblastoma gene family. Oncogene. 2006;25:5201–5209. doi: 10.1038/sj.onc.1209652. [DOI] [PubMed] [Google Scholar]
- 78.van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24:1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide Deficiency Promotes Genomic Instability in Early Stages of Cancer Development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bosco EE, Mayhew CN, Hennigan RF, Sage J, Jacks T, Knudsen ES. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 2004;32:25–34. doi: 10.1093/nar/gkg919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mannini L, Menga S, Musio A. The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Hum Mutat. 2010;31:623–630. doi: 10.1002/humu.21252. [DOI] [PubMed] [Google Scholar]
- 82.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S, Barral Y. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 85.Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, Gerlich DW. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 86.Guerrero AA, Gamero MC, Trachana V, Futterer A, Pacios-Bras C, Diaz-Concha NP, Cigudosa JC, Martinez AC, van Wely KH. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc Natl Acad Sci U S A. 2010;107:4159–4164. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 88.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 89.Roschke AV, Kirsch IR. Targeting karyotypic complexity and chromosomal instability of cancer cells. Curr Drug Targets. 11:1341–1350. doi: 10.2174/1389450111007011341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Di Leonardo A, Khan SH, Linke SP, Greco V, Seidita G, Wahl GM. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 93.Khan SH, Wahl GM. p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res. 1998;58:396–401. [PubMed] [Google Scholar]
- 94.Harrington EA, Bruce JL, Harlow E, Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci U S A. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng L, Flesken-Nikitin A, Chen PL, Lee WH. Deficiency of Retinoblastoma gene in mouse embryonic stem cells leads to genetic instability. Cancer Res. 2002;62:2498–2502. [PubMed] [Google Scholar]
- 98.Lan Z, Sever-Chroneos Z, Strobeck MW, Park CH, Baskaran R, Edelmann W, Leone G, Knudsen ES. DNA damage invokes mismatch repair-dependent cyclin D1 attenuation and retinoblastoma signaling pathways to inhibit CDK2. J Biol Chem. 2002;277:8372–8381. doi: 10.1074/jbc.M108906200. [DOI] [PubMed] [Google Scholar]
- 99.Mayhew CN, Perkin LM, Zhang X, Sage J, Jacks T, Knudsen ES. Discrete signaling pathways participate in RB-dependent responses to chemotherapeutic agents. Oncogene. 2004;23:4107–4120. doi: 10.1038/sj.onc.1207503. [DOI] [PubMed] [Google Scholar]
- 100.Foijer F, Wolthuis RM, Doodeman V, Medema RH, te Riele H. Mitogen requirement for cell cycle progression in the absence of pocket protein activity. Cancer Cell. 2005;8:455–466. doi: 10.1016/j.ccr.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 101.Frame FM, Rogoff HA, Pickering MT, Cress WD, Kowalik TF. E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene. 2006;25:3258–3266. doi: 10.1038/sj.onc.1209352. [DOI] [PubMed] [Google Scholar]
- 102.Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–755. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- 103.Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 104.Eguchi T, Takaki T, Itadani H, Kotani H. RB silencing compromises the DNA damage-induced G2/M checkpoint and causes deregulated expression of the ECT2 oncogene. Oncogene. 2007;26:509–520. doi: 10.1038/sj.onc.1209810. [DOI] [PubMed] [Google Scholar]
- 105.Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McCabe MT, Davis JN, Day ML. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005;65:3624–3632. doi: 10.1158/0008-5472.CAN-04-2158. [DOI] [PubMed] [Google Scholar]
- 109.Knudsen ES, Sexton CR, Mayhew CN. Role of the retinoblastoma tumor suppressor in the maintenance of genome integrity. Curr Mol Med. 2006;6:749–757. doi: 10.2174/1566524010606070749. [DOI] [PubMed] [Google Scholar]
- 110.Chakraborty S, Khare S, Dorairaj SK, Prabhakaran VC, Prakash DR, Kumar A. Identification of genes associated with tumorigenesis of retinoblastoma by microarray analysis. Genomics. 2007;90:344–353. doi: 10.1016/j.ygeno.2007.05.002. [DOI] [PubMed] [Google Scholar]