Abstract
P120 catenin (p120ctn) is an adherens junction protein recognized to regulate barrier function, but emerging evidence indicates that p120ctn may also exert control on other cellular functions such as transcriptional suppression of genes. We investigated the hypothesis that loss of p120ctn in human endothelial cells activates transcription of pro-inflammatory adhesion molecules. For study, siRNA targeted to p120ctn was transfected into brain microvascular (HBMECs) or pulmonary artery endothelial cells (HPAECs) for 24–120 hr, which depleted 50–80% of endogenous p120ctn. This loss of p120ctn resulted in increased promoter reporter activity of transcription factors, NFκB, AP-1, and Kaiso, as well as of target genes, MMP-1 and ICAM-1. Real-time RT-PCR analysis indicated that the mRNA for ICAM-1, VCAM-1, and E- and P-selectins were all upregulated during the period of 24–120 hr of p120ctn depletion, although the time-course and extent of the expression profiles differed. The upregulated mRNA of adhesion molecules corresponded with increased PMN adhesion to the EC surface and elevated ICAM-1 protein expression. We further explored the role of ERK1/2 as a potential signaling mechanism responsible for regulation of transcriptional activities by p120ctn. Results indicated that loss of p120ctn increased phosphorylated ERK1/2, and a MEK1 inhibitor (PD98059) prevented NFκB nuclear translocation. This implicates ERK1/2 in signaling the NFκB activation induced by p120ctn loss. The findings provide strong evidence that deficiency in p120ctn expression in endothelial cells is a potent stimulus for transcriptional upregulation of multiple adhesion molecules. We conclude that p120ctn functions to suppress transcription, which is an important and novel regulation in vascular endothelium.
Keywords: ICAM-1, adhesion molecules, neutrophil adhesion, NFκB, inflammation, ERK1/2
INTRODUCTION
Emerging evidence indicates that p120 catenin (p120ctn), a critical protein of the adherens junction (AJ), has the ability to regulate endothelial function beyond its role in vascular endothelial permeability. Not only does p120ctn directly bind cadherins of the AJ to control cell-cell adhesion (Davis et al., 2003; Iyer et al., 2004; Xiao et al., 2003), it can also bind transcription factors [i.e., Kaiso and Glis2 (Daniel and Reynolds, 1999; Hosking et al., 2007)], signaling molecules such as RhoA (Anastasiadis et al., 2000; Castano et al., 2007), protein kinases (Piedra et al., 2003) and protein phosphatases (Holsinger et al., 2002; Keilhack et al., 2000; Zondag et al., 2000), indicating a formidable array of binding partners and underscoring the potential multiple cellular activities regulated by p120ctn. Yet, to date, the significance of these emerging roles of p120ctn in endothelial physiology and pathophysiology remains unknown. Recently, we investigated p120ctn and one of its binding partners, the transcription factor Kaiso, in regulation of transcription in endothelial cells (ECs). The study found that p120ctn suppresses Kaiso-dependent transcription activity in human vascular ECs (Zhang et al., 2010), providing the first evidence of a transcriptional role of p120ctn in endothelium.
Recent reports link inappropriate expression of p120ctn with enhanced pro-inflammatory activities of several tissues. The experimentally-induced loss of p120ctn expression (i.e., using RNAi or knock-out mice) in turn increased cytokine levels (Perez-Moreno et al., 2006; Wang et al., 2010), increased leukocyte extravasation (Perez-Moreno et al., 2006; Smalley-Freed et al., 2010), and induced activation of transcription factor NFκB (Perez-Moreno et al., 2006; Wang et al., 2010). Various pathological conditions such as lipopolysaccharide-induced bronchial epithelial injury (Wang et al., 2010), ulcerative colitis and Crohn’s disease (Karayiannakis et al., 1998), and cancers (van Hengel and van Roy, 2007) are associated with reduced levels of p120ctn. These observations suggest that deficiencies of p120ctn could critically contribute to the inflammatory injury of tissues under pathological conditions. The mechanistic link between p120ctn loss and the inflammatory response is unknown, but the mitogen-activated protein kinase (MAPK) family members are possible intermediaries in signaling as several reports have found that ERK1/2 positively regulates NFkB activity (Jiang et al., 2001; Kurland et al., 2003).
In vascular endothelium, less is known about the involvement of p120ctn in regulation of inflammatory responses. The induction of an endothelial inflammatory phenotype by p120ctn deficiency is critically significant since there is abundant evidence that junctions of ECs become disrupted and disorganized in response to a host of pathological agents. A recent report indicates that p120ctn depletion can exacerbate endothelial inflammatory responses in a lipopolysaccharide (LPS) murine model of lung injury (Wang et al., 2011). However, it remains unclear whether p120ctn deficiency alone is sufficient to trigger pro-inflammatory activities in ECs. Our recent evidence clearly suggests that p120ctn is functioning as a cofactor in transcriptional suppression in ECs (at least with Kaiso) (Zhang et al., 2010), and therefore its loss could in turn release the suppression. In this study, we investigated the hypothesis that decreased levels of p120ctn in ECs activate transcription of pro-inflammatory adhesion molecules. The study was made using an experimental down-regulation of endogenous p120ctn expression in human vascular ECs.
MATERIALS and METHODS
Materials
General reagents were obtained as follows: EGM-2 Bulletkit (Lonza; Walkersville, MD); Trizol, RPMI 1640, DMEM, L-glutamine 100× solution, penicillin-streptomycin 100× solution, sodium pyruvate, phosphate-buffered saline (PBS), Hank’s Balanced Salt Solution (HBSS), MEM nonessential amino acids, MEM Vitamins, ProLong Gold Anti-Fade reagent with DAPI (4',6-diamidino-2-phenylindole); and Lipofectamine 2000 (Invitrogen Corp.; Carlsbad, CA); fetal bovine serum (FBS) (Hyclone; Logan, UT); human epidermal growth factor (EGF), hydrocortisone, endothelial cell growth supplement (ECGS), heparin, Protease inhibitor cocktail, phenylmethylsulfonyl fluoride (PMSF), leupeptin, pepstatin A and aprotinin (Sigma Chemical Co.; St. Louis, MO); ECL Kit (Amersham Pharmacia Biotech; Piscataway, NJ); Luciferase Activity kit, RQ1 RNase-free DNase (Promega Corporation; Madison, WI); Coomassie Plus Protein Assay (Pierce; Rockford, IL); Immu-Mount mounting gel (Shandon; Pittsburgh, PA); High Capacity cDNA Reverse Transcription Kit, SYBR Green PCR Master Mix (Applied Biosystems; Foster City, CA); MAPK/ERK kinase 1 (MEK1) inhibitor PD98059 (Calbiochem, Gibbstown, NJ); and Target-Plus SMARTpool siRNA, siCONTROL Non-Targeting siRNA, Dharmafect (Dharmacon, Lafayette, CO).
The following were antibodies used: Monoclonal anti-p120ctn antibody (Ab) (BD Transduction Laboratories; San Jose, CA); monoclonal anti-ICAM-1 Ab (Santa Cruz Biotechnology; Santa Cruz, CA); anti-β-actin Ab (Sigma Chemical Co.; St. Louis, MO); rabbit polyclonal anti-NFκB p65 CT Ab (Upstate, Lake placid, NY); monoclonal anti-phospho-ERK1/2 (Thr202/Tyr204) and polyclonal anti-ERK1/2 Ab (Cell Signaling Technology; Beverly, MA); anti-mouse peroxidase-conjugated (HRP) Ab (Amersham Pharmacia Biotech; Piscataway, NJ); donkey F(ab')2 rhodamine red-X-conjugated anti-IgG antibody (Jackson Immunoresearch Laboratories Inc.; West Grove, PA); and anti-mouse and anti-rabbit IRDye Ab (LI-COR Biosciences; Lincoln, NE).
Methods
Cell culture
Human pulmonary artery endothelial cells (HPAECs) were maintained in culture and used experimentally at up to population doubling 13 (Lonza). HPAECs were grown in EGM-2 base growth medium with the following supplements: FBS, hydrocortisone, hFGF-B, VEGF, R3-IGF-1, ascorbic acid, hEGF, GA-1000, heparin. Human brain microvascular ECs (HBMECs) were used in population doublings 17–30 and cultured as previously described (Qiao et al., 2006).
Targeted silencing of endogenous p120ctn with siRNA
The knockdown of p120ctn expression in ECs was made using a mixture of four siRNA oligonucleotide duplexes (siRNA-p120ctn) made commercially based on the human p120ctn gene sequence, accession number NM_001331: i) GAAUGUGAUGGUUUAGUUUU, ii) UAGCUGACCUCCUGACUAAUU, iii) GGACCUUACUGAAGUUAUUUU, and iv) GAGGUAAGCUCGCCGGAAAUU). For transfection, ECs were grown in appropriate culture dishes and treated with siRNA-p120ctn duplexes pre-mixed in Dharmafect1 siRNA transfection reagent at a ratio of 1 µl of Dharmafect1 to 0.2 pmol siRNA for up to 120 hr as indicated. During incubation with siRNA, overt cell toxicity was monitored by phase microscope observations. Following incubation, the cells were used for different assays as described. The siCONTROL Non-Targeting siRNA (NT), which has no known mRNA targets served as control for specificity of knockdown.
Promoter reporter activity
Reporter constructs
The luciferase reporter plasmids containing full-length (−1393) ICAM-1 (intercellular adhesion molecule-1) promoter [p(−1393)ICAM1-Luc], three copies of AP-1/Ets elements (pAP1-Luc) or five copies of the consensus NFκB binding elements (pNFκB-Luc) were generously provided by Dr. Kenneth A. Roebuck (Roebuck et al., 1995). The wild-type (wt) kaiso binding site (KBS) luciferase reporter plasmid, pwtKBS-Luc, was a kind gift from Dr. Juliet M. Daniel (McMaster University, Canada) (Spring et al., 2005). The MMP-1 (matrix metalloproteinase-1) promoter luciferase reporter plasmid [p(-562)MMP-1-Luc] was provided by Dr. Hee-Jeong Im, Department of Biochemistry, Rush University Medical Center, Chicago, IL.
Transfection protocol
Promoter reporter plasmid DNA was transfected into ECs based on a modified protocol as previously described (Lum et al., 1999). ECs were grown in 12-well tissue culture dishes for transfection at ~70% confluence under antibiotic-free conditions. The cells were incubated for 3–4 hr with plasmid DNA pre-mixed with Lipofectamine 2000 reagent at a 1:3 ratio (µg DNA: µl Lipofectamine). Empty vector (pGL3) transfection control was included using the same volume of Lipofectamine 2000 without DNA. Following incubation, the cells were washed and replaced with complete medium containing 2× FBS and incubated overnight. After washing twice with Ca2+-free and Mg2+-free PBS, the cells were collected in reporter lysis reagent (passive lysis buffer) for luciferase activity assay.
Luciferase reporter assay
After the transfected ECs were collected in passive lysis buffer, the ECs were clarified by centrifugation and the supernatant was collected for analysis. Sample aliquots in duplicates were assayed for reporter activity as relative light units in a luminometer (Zylux FB12, Maryville, TN) according to manufacturer’s protocol. Remaining aliquots were analyzed for protein concentration, which was used for normalization of luciferase activity. Protein concentration was determined using a BCA kit.
RNA Collection and RT-PCR
Total RNA from cell lysate was isolated using Trizol according to manufacturer’s instructions and RNA concentration determined by absorbance at 260 nm. Two µg RNA was treated with RQ1 RNase-free DNase prior to reverse transcription reaction. Synthesis of cDNA was performed using the high capacity cDNA Archive Kit. Primer pairs were designed using Integrated DNA Technologies (Table 1). Real-time RT-PCR was performed using SYBR green PCR master mix kit according to manufacturer’s specifications. The amplification conditions were: Step 1: 50 °C 2 min, 95 °C 10 min; step 2: 40 cycles of the following: 95 °C 15s, 60 °C 1 min; step 3: 95 °C 15 s, 60 °C, 1 min, then 60 °C (+0.5, 10 s), 80 cycles to final 95°C.
Table 1.
Primer pairs for adhesion molecules and GAPDH
| Accession | forward primer | reverse primer | |
|---|---|---|---|
| ICAM-1 | NM_000201 | 5’ AAGGTGACCGTGAATGTGCTCT 3’ | 3’ ATTGGCGGTCGCCTTCTAGTT 5’ |
| E-selectin | NM_000450 | 5’ CCTTCCTGCCAAGTGGTAAA 3' | 3’ CAAACCGTGACACACGTTCA 5' |
| P-selectin | NM_003005 | 5’ GTCAACTACCGTGCCAACCT 3' | 3' CAGACTATTACCCACCCTGC 5' |
| VCAM-1 | NM_001078 | 5’ AGTTGAAGGATGCGGGAGTA 3' | 3’ AGGACTCGAAGAGCACGAGA 5' |
| GAPDH | NM_002046 | 5’ ATGGCAAATTCCATGGCACCG 3’ | 3’ TAGTGGTAGAAGGTCCTCGCT 5’ |
Indirect immunofluorescence
ECs were grown to confluence on glass cover slips precoated with fibronectin (5 µg/mL). After appropriate experimental treatment, ECs were fixed and permeabilized using fresh fixative buffer [90% PEM buffer (0.1 M Pipes, pH 6.6, 1 mM EGTA and 1 mM MgSO4), 10% paraformaldehyde, and 0.5% Triton-X 100] for 1 hr at room temperature. For detection of cell surface proteins, ECs were fixed in fixative buffer in the absence of Triton-X 100. ECs were then washed in HBSS with Ca2+ and Mg2+ (HBSS+) and incubated in primary Ab (1 hr, 37°C) in HBSS+. After washing with HBSS+, the ECs were incubated with fluorescently labeled secondary Ab (1 hr, 37°C) diluted in HBSS for the appropriate period of time in the dark. The ECs were washed with HBSS+, rinsed in double-distilled H2O (ddH2O), and mounted onto slides with Prolong Anti-Fade Mounting Gel with DAPI. Images were recorded using a Zeiss Axiovert 100 confocal microscope and image processing performed using Axiovision software. In order to compare intensity of ICAM-1 fluorescence between experimental groups, the same exposure time was used to capture each image in a given experiment. In general, ten randomly chosen fields, each with an area of 0.035 mm2, were photographed for each slide. Six or seven slides were prepared for each experimental group. A negative control without the addition of primary antibody was included in order to assess the contribution of non-specific fluorescence.
Western blot
Following the experimental protocol, ECs were collected and prepared for routine Western blot analysis. After determining protein concentration by BCA, an equal amount of total protein of each sample was loaded on an 8% sodium dodecyl sulfate (SDS) polyacrylamide gel, electrophoresed (100 V, 1 hr), and transferred (100 V, 1 hr) onto nitrocellulose membrane. The resultant membrane was blocked with 5% nonfat dry milk in Tris buffered saline (TBS) for 1 hr at room temperature. The membrane was then incubated in primary Ab diluted in 1% nonfat dry milk in TBS with 0.2% Tween-20 (Ab diluent) for 1 hr at room temperature. After washing in TBS with 0.05% Tween-20 (TBS-T), the membrane was incubated with a secondary Ab conjugated with infrared fluorescent dye diluted in Ab diluent in the dark for 1 hr at room temperature. Protein was detected and quantified with an Odyssey Imaging System (Licor). For study of ERK1/2 phosphorylation, an appropriate anti-IgG secondary antibody conjugated with horseradish peroxidase was used, and the bands were detected using the ECL kit.
PMN adhesion assay
Isolation and labeling of polymorphonuclear leukocytes (PMNs)
Blood from healthy human volunteers was collected according to the approved IRB protocol (#08080201-IRB01-CR02) and layered onto lymphocyte separation medium (LSM) (Biowhitaker). After centrifugation at 300 g for 30 min, the PMN-enriched pellet was re-suspended in a hypotonic buffer (1:1 sterile water:HBSS+) to lyse red blood cells. This cell suspension was centrifuged at 250 g for 10 min, and the lysing procedure was repeated. The final cell pellet was re-suspended in HBSS+ and the PMNs were counted. The PMNs were labeled with 5 µM calcein/acetomethoxy (AM) (Invitrogen) as previously described by us (Huang et al., 2007; Lum et al., 1994) and re-suspended to a final concentration of 107 PMN/mL in HBSS+ for adhesion assay described below.
HPAECs were plated in 96-well dishes and treated according to the experimental groups. The HPAECs were washed with HBSS+, and calcein-labeled PMNs were added at a ratio of 10:1 (PMN:EC) and incubated for 3 hr at 37 °C. Fluorescence was measured with a TRIAD multimode reader (485 nm excitation, 535 nm emission) (Dynex; Chantilly, VA) before and after removing non-adherent PMNs by thorough washing. Percent PMN adhesion was calculated as fluorescence of PMNs (post-wash/pre-wash × 100).
Statistical analysis
Single sample data were analyzed by the two-tailed, independent Student’s t test (SPSS 15.0, Excel).
RESULTS
A.) P120ctn depletion by siRNA-p120ctn
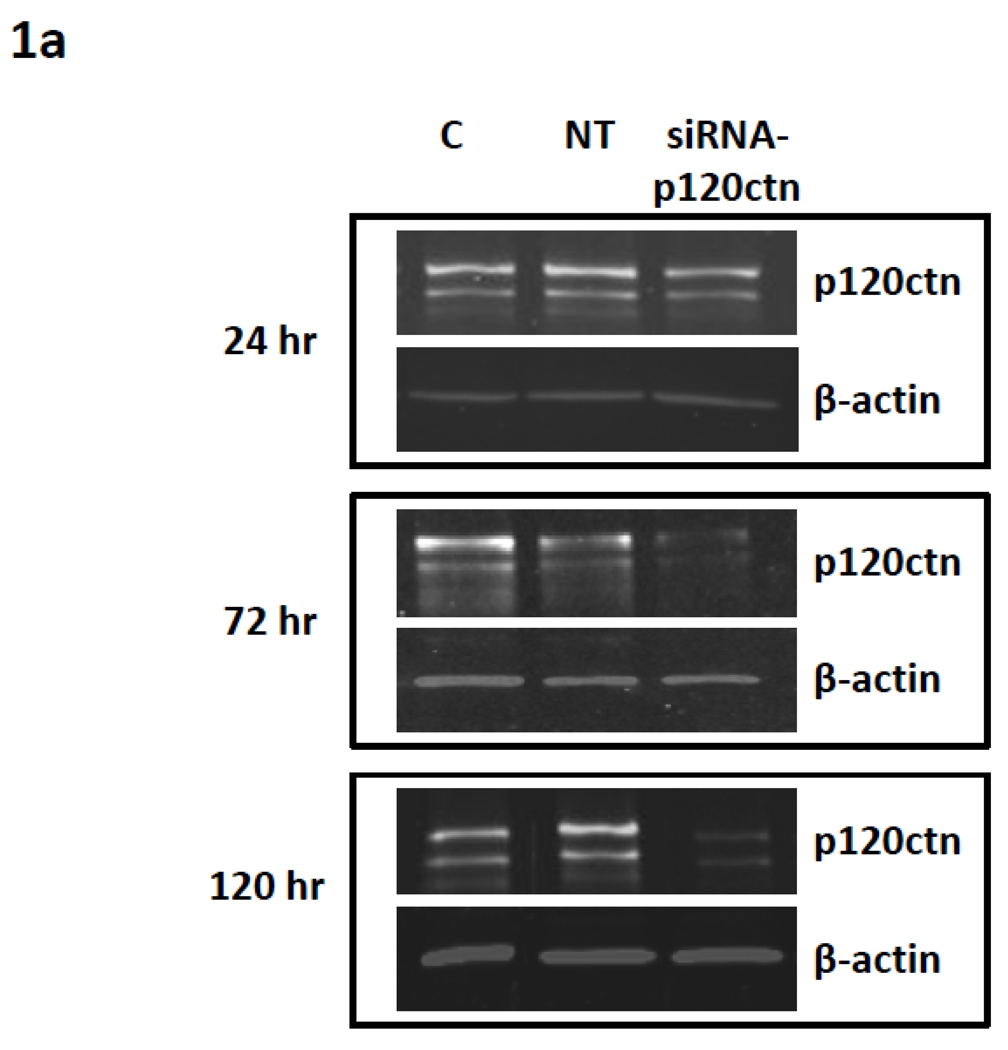
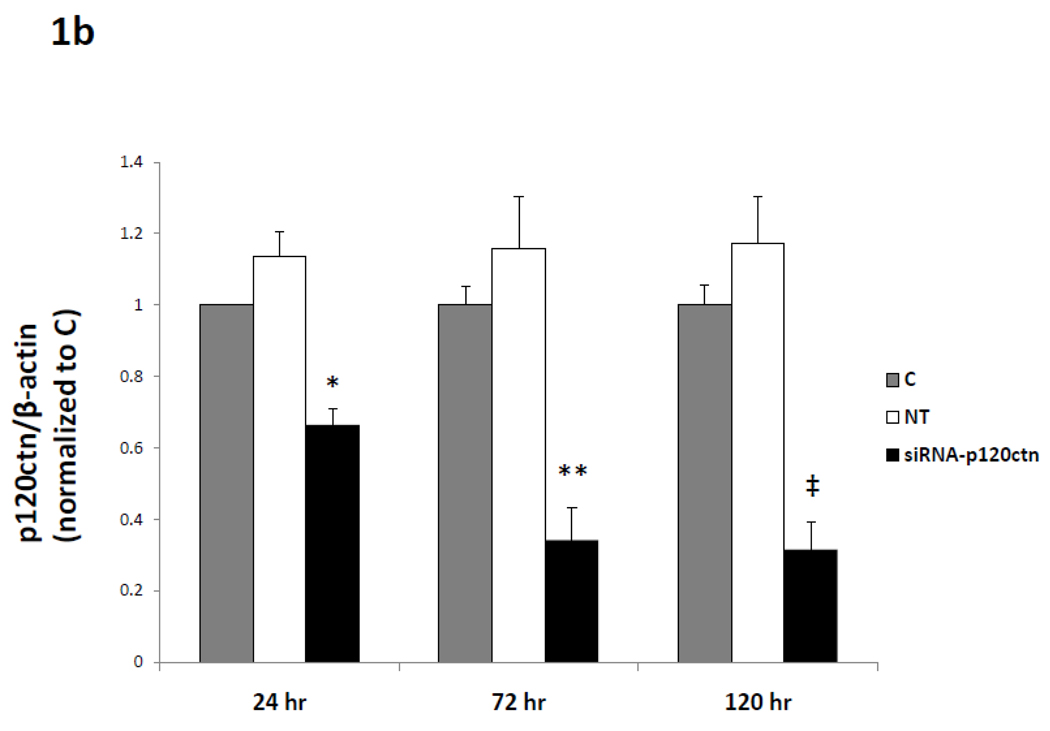
To investigate the hypothesis that decreased levels of p120ctn in ECs activate pro-inflammatory gene expression, we used commercially made siRNA duplexes targeted to p120ctn to induce endogenous p120ctn depletion in HPAECs and HBMECs. The ECs were incubated with siRNA-p120ctn duplexes for 24, 72, or 120 hr and extent of p120ctn knockdown was assessed by Western Blot analysis (Materials and Methods). Control groups included NT and untreated control. Within 24 hr of siRNA treatment, the level of p120ctn was significantly reduced 30–40% (Fig. 1a,b). Continuous siRNA treatment to 72 or 120 hr further depleted p120ctn to 60–70% relative to the control groups (Fig. 1a,b). Both HPAECs (Fig. 1a,b) and HBMECs (not shown) presented similar degrees of p120ctn knockdown after 48–72 hr treatment with siRNA- p120ctn, a finding we have previously reported (Zhang et al., 2010).
Fig. 1. Effects of targeted siRNA on p120ctn depletion.
ECs were treated with either siRNA-p120ctn, non-targeting siRNA (NT), or left untreated (control) for 24 hr, 72 hr, or 120 hr, and cells were collected for analysis by a) Western blot and b) quantification of band fluorescence intensity. P120ctn protein was normalized to β-actin and reported relative to untreated control (C). *p < 0.01, **p<0.0015, and ‡p<0.005 compared to NT. For 24 hr, N = 3 for all groups. For 72 hr, N = 5 for C, N = 7 for NT, and N = 3 for siRNA-p120ctn. For 120 hr, N = 10 for C and NT, N = 7 for siRNA-p120ctn.
B.) Loss of p120ctn increased promoter reporter activity
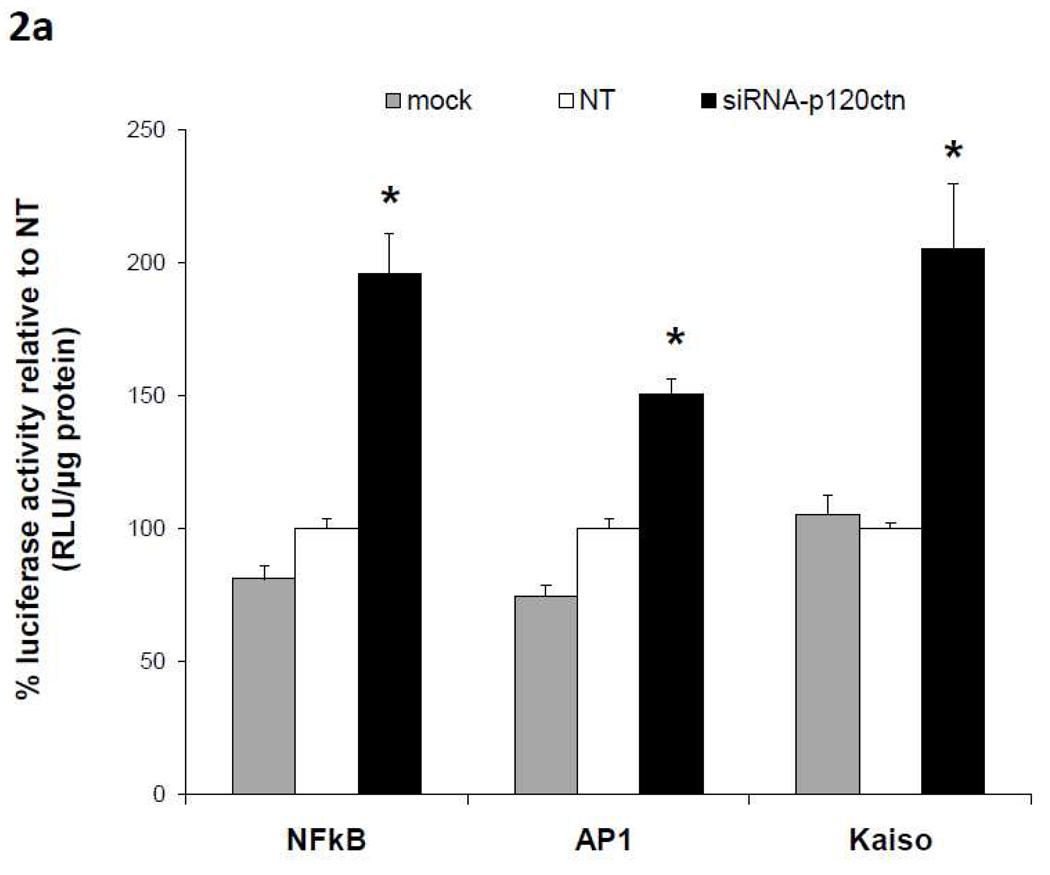
We next determined whether the decreased p120ctn expression activates transcriptional activity by measuring the effects of p120 depletion on activity of transcription factors NFκB, AP-1, and Kaiso. In the following studies, HBMECs were treated with siRNA targeted to p120ctn, or NT or mock for 24 hr knockdown of endogenous p120ctn. Following siRNA treatment, the ECs were then transfected with promoter reporter luciferase plasmids containing binding elements for NFκB, AP-1, or Kaiso for an additional 24 hr, and collected for assay of luciferase activity (Materials and Methods). Reporter activity was normalized to protein concentration and reported relative to NT control. The results show that the loss of p120ctn significantly induced reporter luciferase activities driven by NFκB, AP-1, or Kaiso (Fig. 2a).
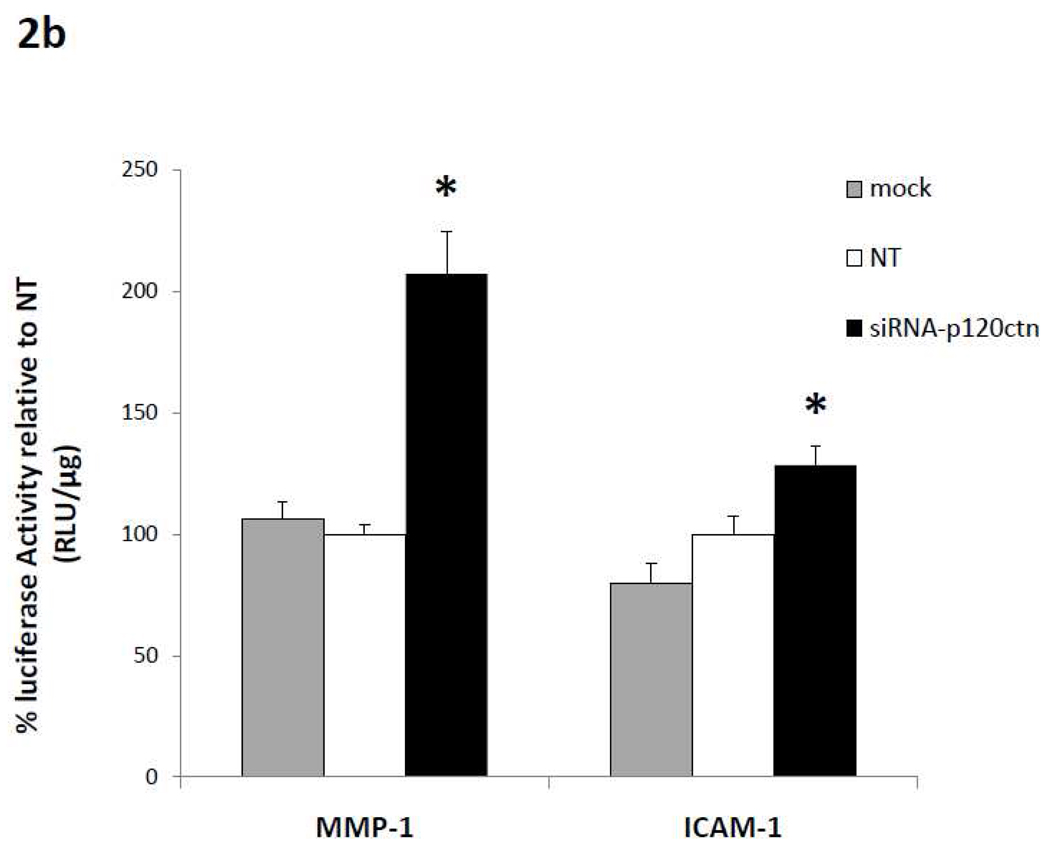
Fig. 2. Loss of p120ctn activated promoter reporter luciferase activity.
HBMECs were treated with siRNA-p120ctn for 48 hr, and were transfected with a luciferase reporter plasmid (Material and Methods) containing response elements for a) transcription factors NFκB, AP-1, or kaiso, or b) gene targets p(−1393)ICAM1-Luc or p(−562)MMP-1-Luc. Mock = transfection agent, NT = non-targeting siRNA. Luciferase activity was measured as relative light units (RLU) and normalized to protein, and reported as % relative to NT. * p < 0.005 compared to either control groups NT or Mock. N = 6 for NFκB; N = 4 for AP-1; for kaiso, N = 11 for NT and siRNA, N = 3 for mock; N = 13 for p(−562)MMP-1-Luc and N = 8 for p(−1393)ICAM1-Luc.
Since NFκB and AP-1 are recognized transcription factors of a host of pro-inflammatory genes, the study was extended to include promoter activity of two potential target genes, MMP-1 and ICAM-1. Similarly as described above, following treatment with siRNA-p120ctn, NT or mock for 24 hr, ECs were transfected with p(-562)MMP-1 or p(−1393)ICAM-1 reporter plasmids for 24 hr, and luciferase activity was determined. We found that the decreased p120ctn expression significantly increased both (−562)MMP-1 and (−1393) ICAM-1 promoter luciferase activities relative to control groups (Fig. 2b).
C.) Loss of p120ctn increased expression of pro-inflammatory adhesion molecules
Both NFκB and AP-1 are critically important in the transcription of adhesion molecules such as ICAM-1 (Collins et al., 1995; Hubbard and Rothlein, 2000; Roebuck et al., 1995), E-selectin (Collins et al., 1995; Montgomery et al., 1991), P-selectin (regulation by NFκB only) (Pan and McEver, 1995), and VCAM-1 (Ahmad et al., 1998; Collins et al., 1995). Additionally, a preliminary analysis of ICAM-1 sequences identified a possible KBS site ~2 Kb upstream of the transcriptional start site. Therefore, we next determined whether loss of p120ctn in ECs upregulated expression of these adhesion molecules as detected by real-time RT-PCR. Since our results indicated that treatment with siRNA significantly reduced p120ctn protein as early as 24 hr, we determined the effects of p120ctn loss from 24 hr up to 120 hr. The results are as follows:
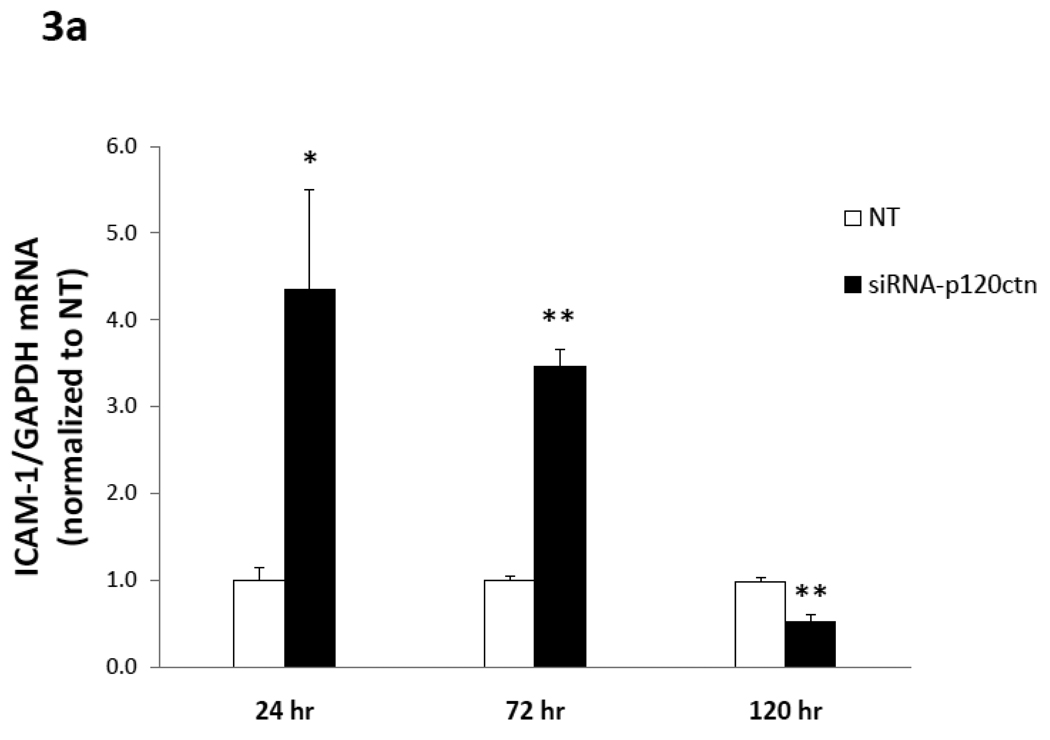
a) ICAM-1 mRNA
The results indicated that ICAM-1 mRNA was increased 4-fold with respect to NT after 24 hr of p120ctn knockdown in HPAECs, and this increase abated slightly to 3.5-fold at 72 hr (Fig. 3a). However, after 120 hr knockdown, ICAM-1 mRNA was decreased below the NT control (~50%) (Fig. 3a). We also determined whether the upregulated ICAM-1 mRNA in response to loss of p120ctn was EC type-specific. For study, HBMECs were transfected with siRNA-p120ctn or NT control for 72 hr, and ICAM-1 mRNA was measured. Results indicated that loss of p120ctn in HBMECs increased ICAM-1 two-fold in HBMECs with respect to NT (p < 0.01; N = 3). The findings indicated that the increased ICAM-1 mRNA induced by loss of p120ctn was not limited to one EC type, albeit the degree of upregulation in HBMECs was not as great as in HPAECs. For this reason, HPAECs were used for most of the studies of adhesion molecule expression.
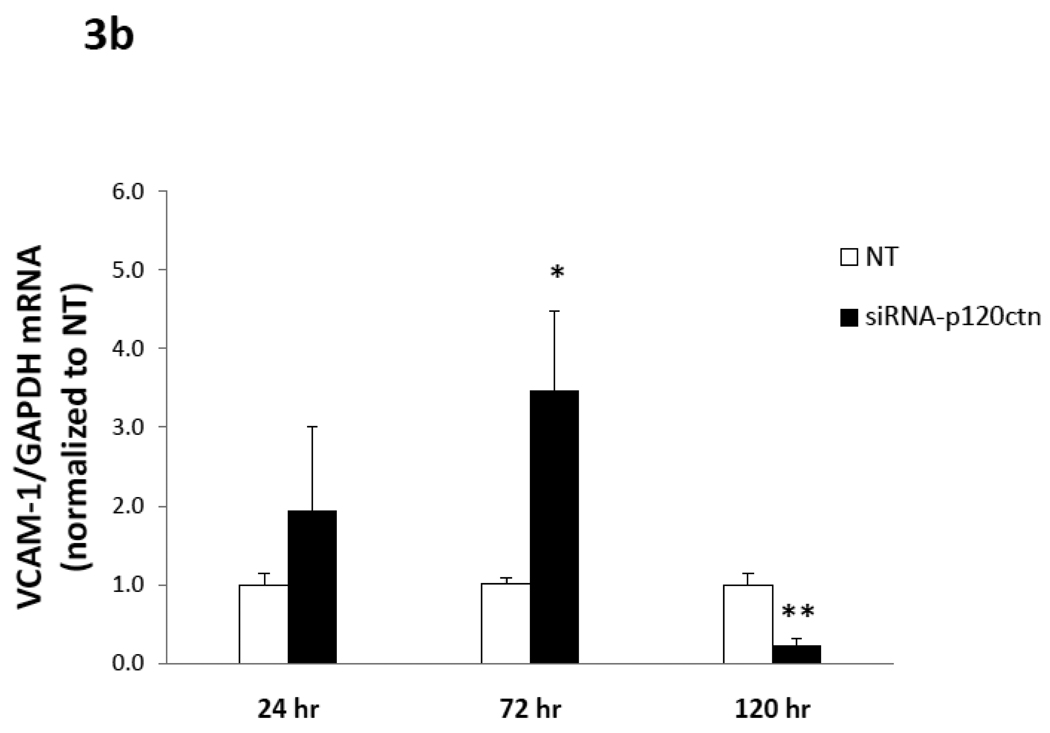
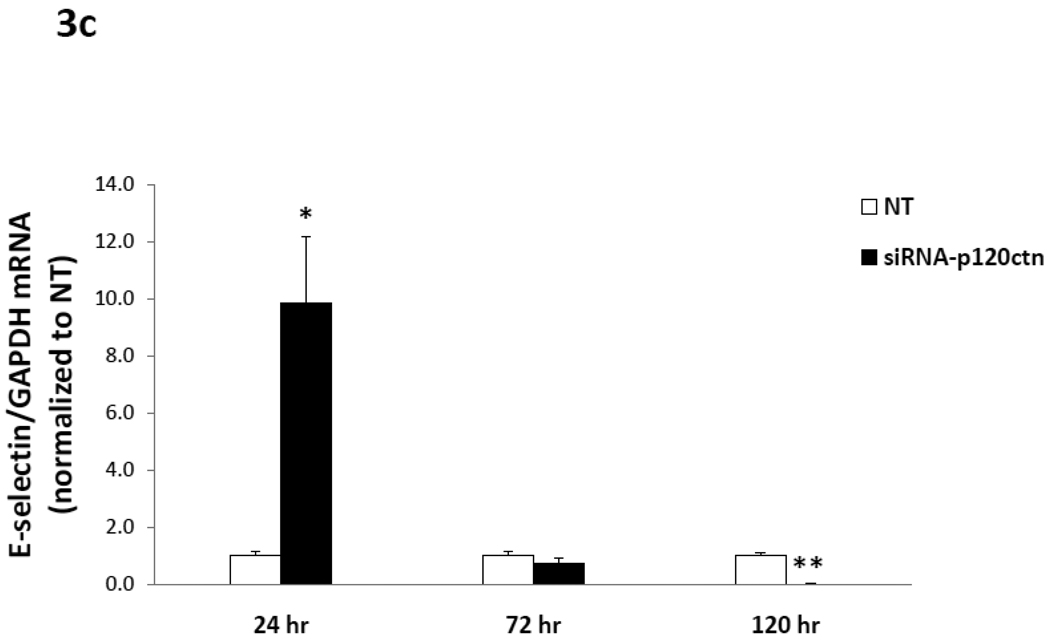
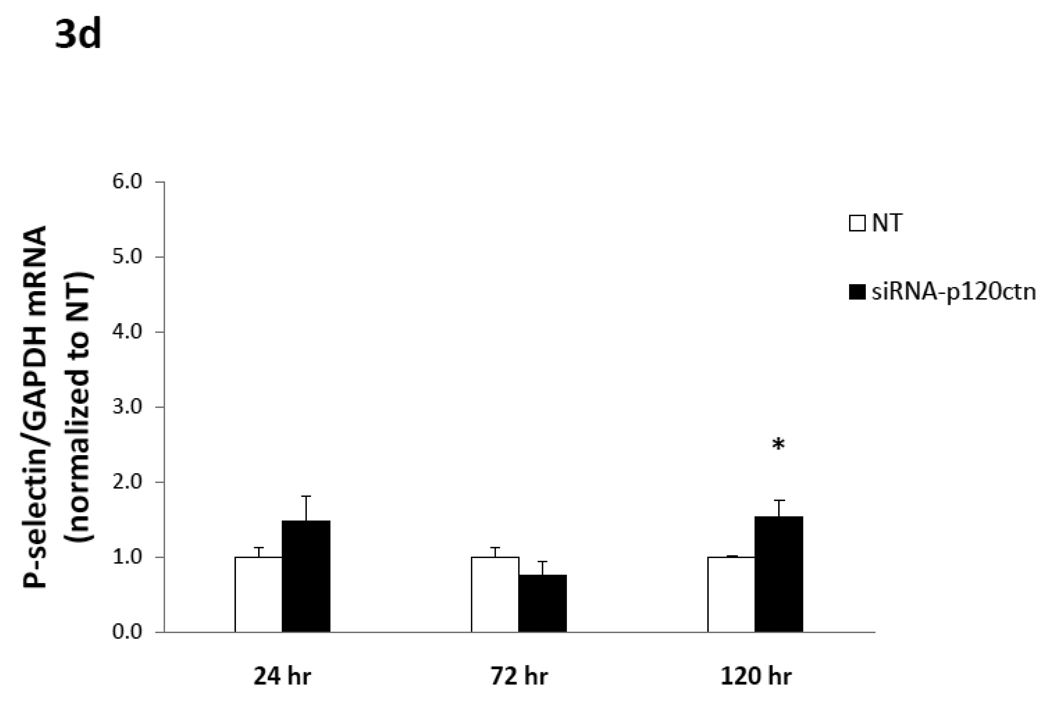
Fig. 3. Time-course of adhesion molecule mRNA expression induced by p120ctn loss.
The mRNA of ICAM-1, VCAM-1, E- and P-selections were determined by real-time RT-PCR from HPAECs treated with siRNA-p120ctn or non-targeting control (NT) for 24, 72, or 120 hr. The mRNA of each adhesion molecule was normalized to GAPDH housekeeping mRNA and reported relative to NT as follows: a) ICAM-1, b) VCAM-1, c) E-selectin, and d) P-selectin. Note that the bar is not visible for the siRNA-p120ctn group for E-selectin at 120 hr knockdown because the standard error was near zero. N for knockdown at 24, 72, and 120 hr was respectively: ICAM-1 (N = 6, 10, and 7); VCAM-1 (N = 6, 13, and 7); P-selectin (N = 6, 10, and 10); *p < 0.05 and **p < 0.005 compared to NT control.
b) VCAM-1 mRNA
VCAM-1 mRNA was significantly upregulated at 72 hr of p120ctn knockdown. This peak increase was ~4-fold over the NT control, and VCAM-1 mRNA levels then subsided to below baseline levels at 120 hr (Fig. 3b).
c) mRNA for E- and P-selectins
The results indicated that by 24 hr of p120ctn knockdown, E-selectin was upregulated 10-fold over NT (Fig. 3c). E-selectin mRNA levels then returned towards baseline levels at 72 hr, and finally decreased below the NT control by 120 hr knockdown (Fig. 3c). In contrast, P-selectin mRNA was not changed following 24 or 72 hr of p120ctn knockdown, and was upregulated 80% only after 120 hr of p120ctn knockdown (Fig. 3d).
In summary, the downregulation of endogenous p120ctn expression in ECs was a sufficient signal to upregulate multiple pro-inflammatory adhesion molecules, which were differentially regulated with respect to duration of p120ctn knockdown.
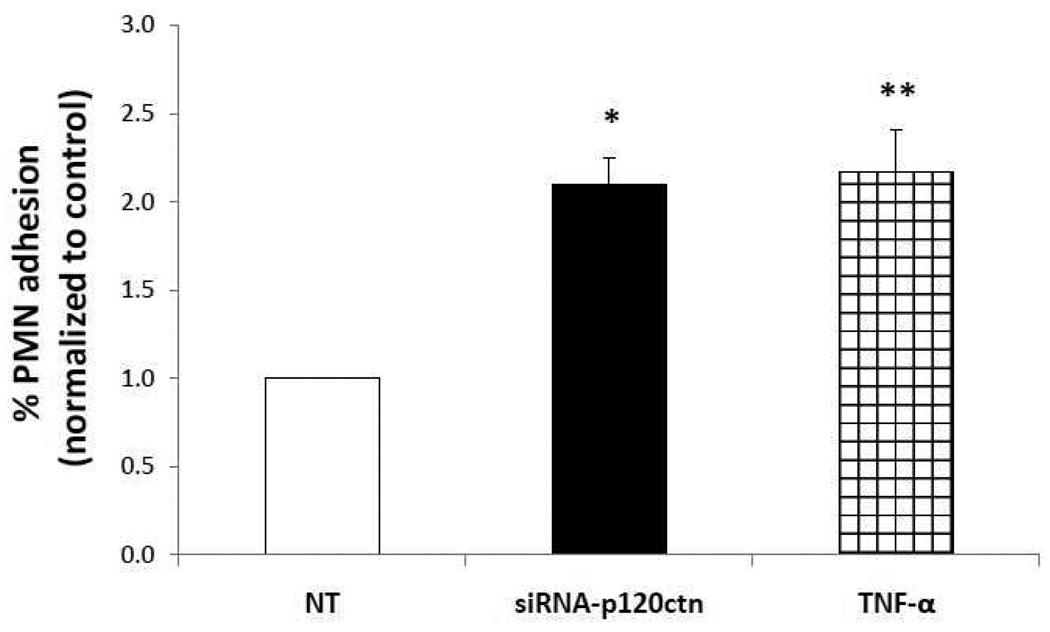
D.) Loss of p120ctn increased PMN adhesion to ECs
The upregulated adhesion molecules strongly suggest that loss of p120ctn expression would in turn regulate leukocyte adhesion to the ECs. For study, we evaluated the effects of p120ctn knockdown at 72 hr since our data indicated that mRNA (at least for ICAM-1 and E-selectin) was already increased by 24 hr, and thus their protein levels ought to be increased at 72 hr of knockdown. Following p120ctn knockdown in HPAECs, calcein-labeled human PMNs were added to determine percent PMN adhesion (Materials and Methods). Tumor necrosis factor-α (TNF-α)-activated HPAECs served as a positive control (12 hr treatment; 25 µM). Results indicated that PMN adhesion more than doubled in siRNA-p120ctn-treated HPAECs with respect to NT, with the extent of PMN adhesion comparable to the TNF-activated HPAECs (Fig. 4). These findings provide functional corroboration that p120ctn loss promoted pro-inflammatory activity in ECs.
Fig. 4. Loss of p120ctn promoted PMN adhesion to EC surface.
Following treatment with siRNA-p120ctn for 72 hr, HPAECs were incubated with calcein-labeled PMNs for 3 hr (Materials and Methods). PMN adhesion was assessed by measuring fluorescence of adherent PMNs after thorough washing. % PMN adhesion was calculated as: fluorescence after washing/fluorescence before washing × 100, and is reported relative to NT. N = 4; *p < 0.001 and **p < 0.025 compared to NT. TNF-α (25 µg/mL; 12 hr) was used as a positive control.
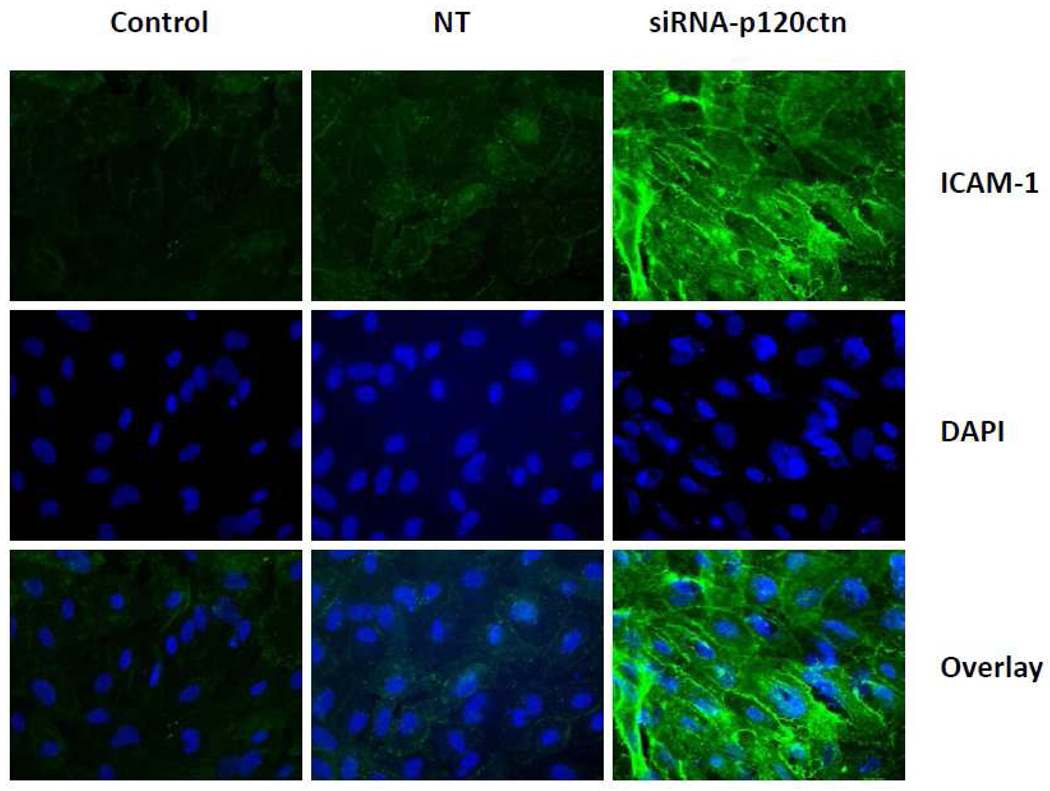
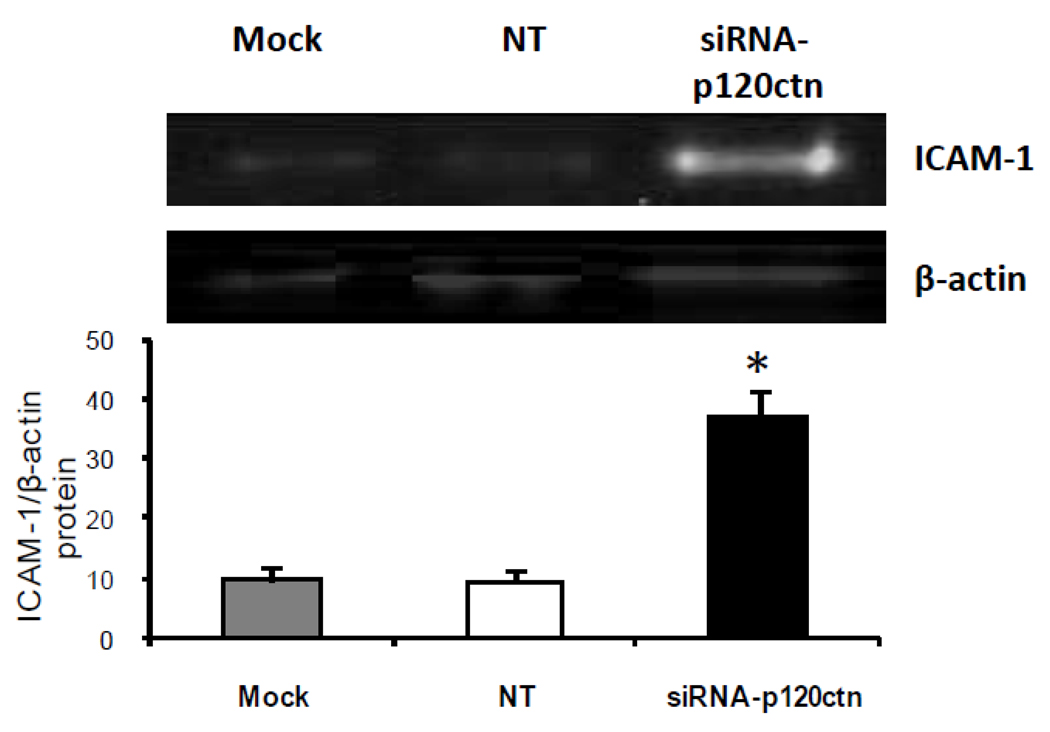
We further determined whether this increased PMN adhesion was related to increased ICAM-1 protein levels in ECs since tight PMN adhesion to the cell surface is dependent predominantly on ICAM-1 rather than either VCAM-1 or the selectins. For study, we determined whether the surface pool of ICAM-1 was increased in response to the 72 hr knockdown of p120ctn in HPAECs. Immunofluorescent analysis results indicated that loss of p120ctn increased surface ICAM-1 expression, which was predominantly localized at the cell periphery (Fig. 5). In general, the EC morphology was similar among the experimental groups, suggesting that p120ctn knockdown or siRNA per se did not impair cellular integrity. We also investigated whether the total ICAM-1 protein was elevated after 72 hr siRNA treatment. Western blot analysis indicated that ICAM-1 protein increased 3-fold over control (Fig. 6).
Fig. 5. Loss of p120ctn increased ICAM-1 expression on EC surface.
HPAECs were treated with siRNA-p120ctn for 72 hr and surface ICAM-1 was visualized by immunofluorescence (Materials and Methods); nuclei were visualized by DAPI. Representative images show the following experimental groups: control (no treatment), non-targeting siRNA (NT), or siRNA-p120ctn; N = 3.
Fig. 6. ICAM-1 protein expression is upregulated after p120ctn knockdown.
HPAECs were treated with siRNA-p120ctn for 72 hr and collected for Western blot analysis for ICAM-1. Fluorescence intensity of proteins bands was quantified using Odyssey software (Licor) and ICAM-1normalized to β-actin. Mock = Dharmafect1 transfection agent alone, NT = non-targeting siRNA. *p < 0.025 compared to mock and NT; N = 3.
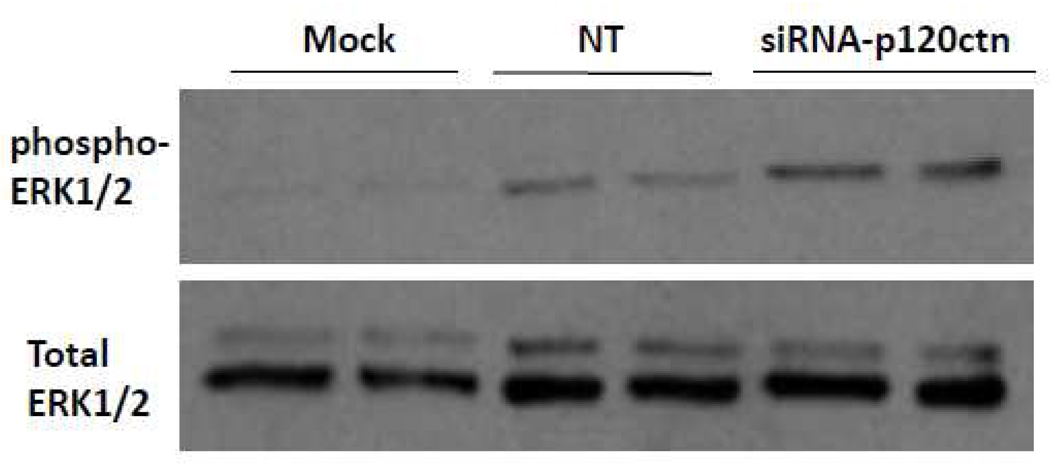
E.) P120ctn knockdown activated NFκB in an ERK1/2-dependent manner
ERK1/2 are MAPKs recognized as important signal transduction pathways in response to hormones, cytokines and other extracellular cues which relay to the nucleus, ultimately activating immediate early genes (Murphy and Blenis, 2006). We postulate that the loss of p120ctn may activate ERK1/2 signaling to regulate gene transcription events in ECs. For study, the effects of 48 hr of siRNA-p120ctn treatment in HBMECs on activation of ERK1/2 was assessed by Western blot detection of ERK1/2 phosphorylation (Materials and Methods.) The phospho-ERK1/2 Ab used binds to phosphorylated activation loop residues Thr202/Tyr204 and Thr185/Tyr187 of ERK1 and ERK2, respectively (Domina et al., 2000; Hayne et al., 2000). The results indicated that p120ctn knockdown increased the amount of phosphorylated ERK1/2 relative to control groups Mock and NT (Fig. 7).
Fig. 7. ERK1/2 is activated after p120ctn knockdown.
HBMECs were treated with siRNA-p120ctn for 48 hr and collected for determination of ERK1/2 activation by Western blot detection of phosphorylated ERK1/2. The phospho-ERK1/2 Ab used is specific for phosphorylated tyrosine activation loop residues of ERK1/2. Mock = Dharmafect1 transfection agent, NT = non-targeting siRNA. N = 3.
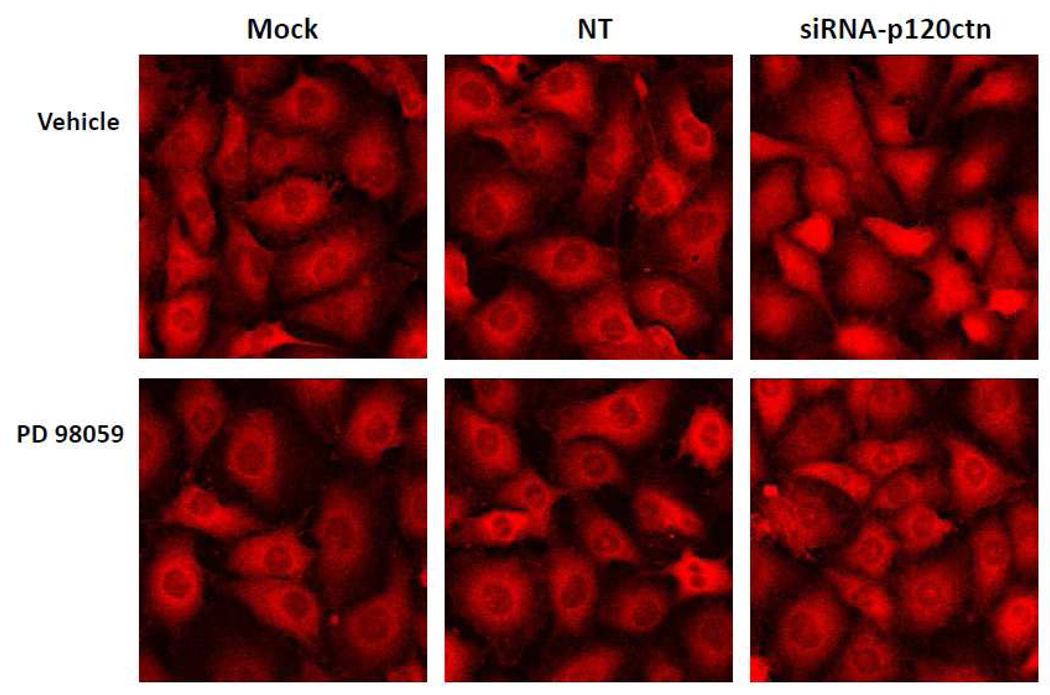
Since ERK1/2 can positively regulate NFkB activity (Jiang et al., 2001; Kurland et al., 2003), we investigated whether the increase in NFκB activity in response to p120ctn loss could be regulated through ERK1/2. Using immunofluorescent localization of the human p65 subunit of NFκB, we found that 48 hr knockdown of p120ctn resulted in increased nuclear localization of the p65 subunit in HBMECs (Fig. 8, top right panel); whereas in NT and Mock groups, the p65 subunit remained predominantly in the perinuclear region. This finding further confirmed the activated NFκB reporter activity (Fig. 2a). Treatment with a MEK1 inhibitor (PD98059; 50 µM) abrogated the NFκB translocation induced by p120ctn knockdown (Fig. 8, bottom panels), whereas the vehicle control showed no inhibition (Fig. 8, top panels). These findings suggest that ERK1/2 is a downstream signaling pathway activated by the loss of p120ctn, providing a potential mechanistic link between p120ctn loss and NFκB activation.
Fig. 8. NFκB nuclear translocation in response to p120ctn knockdown is ERK1/2-dependent.
HBMECs were treated with siRNA-p120ctn for 48 hr and NFκB was visualized by immunofluorescence (Materials and Methods). The primary Ab was directed against the p65 subunit of NFκB. MEK1 inhibitor PD98059 (bottom panels; 50 µM) or DMSO vehicle control (top panels) was added to cells 6 hr after initiation of siRNA treatment. Representative images are shown. Mock = Dharmafect1 transfection agent, NT = non-targeting siRNA. N = 2.
DISCUSSION
The key finding from this study is that decreased p120ctn expression in human ECs was a potent signal that activated transcription factor promoter reporter activities, upregulated adhesion molecule expression, increased adhesion for PMNs, and activated ERK1/2. This strongly suggests that appropriate p120ctn expression operates to suppress endothelial pro-inflammatory potential.
The finding that loss of p120ctn activated promoter reporter activities of Kaiso, NFκB and AP-1 suggests that p120ctn negatively regulated these transcription factors. NFκB is known for its regulation of pro-inflammatory genes, including a host of adhesion molecules and cytokines such as TNF-α and interleukin-8 (Barnes and Karin, 1997), whereas AP-1 regulates a host of genes controlling the cell cycle, apoptosis, as well as inflammation (Hess et al., 2004). However, little is known regarding target genes regulated by Kaiso in tissues. A handful of Kaiso-regulated genes have been reported, including those that regulate inflammation (matrilysin (Spring et al., 2005)) and proliferation (Fos and Myc (van Roy and McCrea, 2005)), but whether Kaiso also regulates these genes in vascular endothelium is unknown.
The specific mechanisms by which p120ctn suppresses transcription factors remain to be fully elucidated. We previously found that p120ctn binds directly with the transcription factor Kaiso in ECs, and appears to function as a cofactor in transcription repression (Zhang et al., 2010). In the current study, the loss of p120ctn activated Kaiso, NFκB and AP-1 activities, clearly indicating that regulation of transcriptional events was not limited to Kaiso. While the binding of Kaiso to p120ctn has been mapped (Daniel et al., 2002), there is no evidence to-date that p120ctn interacts directly with NFκB or AP-1. However, our data indicates that p120ctn suppresses NFκB activation through ERK1/2, implicating this signaling pathway by which p120ctn suppresses transcription factor activity and thereby, the adhesion molecule expression. This suggests that ERK1/2 and possibly other MAPK family members are important candidates in ECs for future investigation as molecular targets of p120ctn-regulated transcription.
Based on the finding of increased transcription factor activity, it was not surprising that we also observed upregulated mRNA for ICAM-1, VCAM-1 and the selectins, although the temporal profiles of expression were different among the adhesion molecules during the time periods of p120ctn knockdown. While both ICAM-1 and E-selectin were significantly upregulated following 24 hr of p120ctn loss, only ICAM-1 (but not E-selectin) remained elevated through 72 hr. On the other hand, both VCAM-1 and P-selectin showed a delayed increase at 72 hr and 120 hr of p120ctn knockdown, respectively. Further, the mRNA levels (for ICAM-1, VCAM-1, and E-selectin) all returned to below control levels by 120 hr, with the exception of P-selectin, which was increased at this time point. These results indicate that loss of p120ctn in ECs induced the upregulation of multiple adhesion molecules.
The temporal profiles of expression mirror the pattern of expression induced by inflammatory agents, implicating at least in part possible similar transcriptional regulation. For example, ICAM-1 and VCAM-1 are upregulated in both early and late phase response to TNF-α, IL-1, and LPS, whereas E-selectin is upregulated only in the early phase response (Lorenzon et al., 1998; Sluiter et al., 1993). In the current study, ICAM-1 and E-selectin were also observed to increase early (by 24 hr of p120ctn loss), and at the later time period of 72 hr of p120ctn loss, both ICAM-1 and VCAM-1 were increased. Less is known of transcriptional regulation of P-selectin, but its EC surface expression is mostly due to translocation of pre-existing P-selectin rather than transcription and synthesis (Sluiter et al., 1993).
P120ctn was found to regulate transcriptional activity in ECs from two diverse vascular tissue types, human brain and human pulmonary artery. Both HPAECs and HBMECs responded to loss of p120ctn with increased ICAM-1 mRNA, although the extent of the increase varied. It is widely recognized that different EC types are different functionally, biochemically, and morphologically (Stevens et al., 2001), and therefore it is not unexpected that the ICAM-1 response differed between the two cell types. Our observations, however, suggest that pathological p120ctn deficiencies could impact globally on vascular endothelium of diverse tissues in the context of inflammatory responses.
The loss of p120ctn by 72 hr significantly increased PMN adhesion and ICAM-1 protein expression to the EC surface, further corroborating the pro-inflammatory activity indicated by the upregulated mRNA of the adhesion molecules. Interestingly, the extent of the increased PMN adhesion was comparable to that elicited by the positive control, the potent multifunctional mediator TNF-α. Immunofluorescent analysis indicated that p120ctn knockdown increased surface expression of ICAM-1 predominantly at the peripheral cell margins, suggesting that the localized increases of ICAM-1 likely function to coordinate regulation of PMNs near EC junctions. Our findings in ECs were consistent with observations reported in other tissues. For example, p120ctn depletion alone promoted PMN adhesion in the mouse small intestinal and colonic epithelial cells (Smalley-Freed et al., 2010) as well as in mouse skin keratinocytes (Perez-Moreno et al., 2008). In contrast, a recent report showed that p120ctn depletion alone in rat lung microvascular ECs did not increase either PMN adhesion nor ICAM-1 expression, but rather only enhances the LPS-mediated responses (Wang et al., 2011). It is unclear as to reasons behind these different findings in ECs, but it may be attributed to differences in the experimental design (i.e., our study used human ECs from pulmonary artery and brain, whereas in the recent publication, rat lung microvascular ECs were used (Wang et al., 2011). Nonetheless, the overall results obtained from other tissues as well as ECs provide strong evidence that p120ctn is a critical suppressor of inflammatory activities in vascular endothelium.
In conclusion, results from this study provide strong evidence that deficiency in p120ctn expression in ECs activated a robust transcriptional response that increased expression of multiple adhesion molecules, and point to a novel role of p120ctn functioning to suppress transcription in vascular endothelium. The well recognized effects of pathological agents in disruption of endothelial AJs underscores an important potential pathogenic role of p120ctn in vascular-based diseases. As such, important future studies are needed to i) define the upstream control of p120ctn expression, particularly by pathological mediators, and ii) delineate p120ctn-mediated downstream signaling mechanisms (i.e., MAPK and RhoA) which regulate transcription factors, particularly AP-1, NFkB, and kaiso.
ACKNOWLEDGMENTS
This work was supported by National Heart Lung and Blood Institute Grant HL093715 (HL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Ahmad M, Theofanidis P, Medford RM. Role of activating protein-1 in the regulation of the vascular cell adhesion molecule-1 gene expression by tumor necrosis factor-alpha. J. Biol. Chem. 1998;273:4616–4621. doi: 10.1074/jbc.273.8.4616. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- Castano J, Solanas G, Casagolda D, Raurell I, Villagrasa P, Bustelo XR, Garcia dH, Dunach M. Specific phosphorylation of p120-catenin regulatory domain differently modulates its binding to RhoA. Mol. Cell Biol. 2007;27:1745–1757. doi: 10.1128/MCB.01974-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T, Read MA, Neish AS, Whitley MZ, Thanos D, Maniatis T. Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokine-inducible enhancers. FASEB J. 1995;9:899–909. [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domina AM, Smith JH, Craig RW. Myeloid cell leukemia 1 is phosphorylated through two distinct pathways, one associated with extracellular signal-regulated kinase activation and the other with G2/M accumulation or protein phosphatase 1/2A inhibition. J. Biol. Chem. 2000;275:21688–21694. doi: 10.1074/jbc.M000915200. [DOI] [PubMed] [Google Scholar]
- Hayne C, Tzivion G, Luo Z. Raf-1/MEK/MAPK pathway is necessary for the G2/M transition induced by nocodazole. J. Biol. Chem. 2000;275:31876–31882. doi: 10.1074/jbc.M002766200. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Holsinger LJ, Ward K, Duffield B, Zachwieja J, Jallal B. The transmembrane receptor protein tyrosine phosphatase DEP1 interacts with p120(ctn) Oncogene. 2002;21:7067–7076. doi: 10.1038/sj.onc.1205858. [DOI] [PubMed] [Google Scholar]
- Hosking CR, Ulloa F, Hogan C, Ferber EC, Figueroa A, Gevaert K, Birchmeier W, Briscoe J, Fujita Y. The transcriptional repressor Glis2 is a novel binding partner for p120 catenin. Mol. Biol. Cell. 2007;18:1918–1927. doi: 10.1091/mbc.E06-10-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Mehta D, Predescu S, Kim KS, Lum H. A novel lysophospholipid- and pH-sensitive receptor, GPR4, in brain endothelial cells regulates monocyte transmigration. Endothelium. 2007;14:25–34. doi: 10.1080/10623320601177288. [DOI] [PubMed] [Google Scholar]
- Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic. Biol. Med. 2000;28:1379–1386. doi: 10.1016/s0891-5849(00)00223-9. [DOI] [PubMed] [Google Scholar]
- Iyer S, Ferreri DM, DeCocco NC, Minnear FL, Vincent PA. VE-cadherin-p120 interaction is required for maintenance of endothelial barrier function. Am. J. Physiol Lung Cell Mol. Physiol. 2004;286:L1143–L1153. doi: 10.1152/ajplung.00305.2003. [DOI] [PubMed] [Google Scholar]
- Jiang B, Brecher P, Cohen RA. Persistent activation of nuclear factor-kappaB by interleukin-1beta and subsequent inducible NO synthase expression requires extracellular signal-regulated kinase. Arterioscler. Thromb. Vasc. Biol. 2001;21:1915–1920. doi: 10.1161/hq1201.099424. [DOI] [PubMed] [Google Scholar]
- Karayiannakis AJ, Syrigos KN, Efstathiou J, Valizadeh A, Noda M, Playford RJ, Kmiot W, Pignatelli M. Expression of catenins and E-cadherin during epithelial restitution in inflammatory bowel disease. J. Pathol. 1998;185:413–418. doi: 10.1002/(SICI)1096-9896(199808)185:4<413::AID-PATH125>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Keilhack H, Hellman U, van Hengel J, van Roy F, Godovac-Zimmermann J, Bohmer FD. The protein-tyrosine phosphatase SHP-1 binds to and dephosphorylates p120 catenin. J. Biol. Chem. 2000;275:26376–26384. doi: 10.1074/jbc.M001315200. [DOI] [PubMed] [Google Scholar]
- Kurland JF, Voehringer DW, Meyn RE. The MEK/ERK pathway acts upstream of NF kappa B1 (p50) homodimer activity and Bcl-2 expression in a murine B-cell lymphoma cell line. MEK inhibition restores radiation-induced apoptosis. J. Biol. Chem. 2003;278:32465–32470. doi: 10.1074/jbc.M212919200. [DOI] [PubMed] [Google Scholar]
- Lorenzon P, Vecile E, Nardon E, Ferrero E, Harlan JM, Tedesco F, Dobrina A. Endothelial cell E- and P-selectin and vascular cell adhesion molecule-1 function as signaling receptors. J. Cell Biol. 1998;142:1381–1391. doi: 10.1083/jcb.142.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum H, Gibbs L, Lai L, Malik AB. CD18 integrin-dependent endothelial injury: effects of opsonized zymosan and phorbol ester activation. J. Leukoc. Biol. 1994;55:58–63. doi: 10.1002/jlb.55.1.58. [DOI] [PubMed] [Google Scholar]
- Lum H, Jaffe HA, Schulz IT, Masood A, RayChaudhury A, Green RD. Expression of PKA inhibitor (PKI) gene abolishes cAMP-mediated protection to endothelial barrier dysfunction. Am. J Physiol Cell Physiol. 1999;277:C580–C588. doi: 10.1152/ajpcell.1999.277.3.C580. [DOI] [PubMed] [Google Scholar]
- Montgomery KF, Osborn L, Hession C, Tizard R, Goff D, Vassallo C, Tarr PI, Bomsztyk K, Lobb R, Harlan JM. Activation of endothelial-leukocyte adhesion molecule 1 (ELAM-1) gene transcription. Proc. Natl. Acad. Sci. U. S. A. 1991;88:6523–6527. doi: 10.1073/pnas.88.15.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 2006;31:268–275. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Pan J, McEver RP. Regulation of the human P-selectin promoter by Bcl-3 and specific homodimeric members of the NF-kappa B/Rel family. J. Biol. Chem. 1995;270:23077–23083. doi: 10.1074/jbc.270.39.23077. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. p120-catenin mediates inflammatory responses in the skin. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno qM, Song W, Pasolli HA, Williams SE, Fuchs E. Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15399–15404. doi: 10.1073/pnas.0807301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia dH, Dunach M. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Mol. Cell Biol. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J, Huang F, Naikawadi RP, Kim KS, Said T, Lum H. Lysophosphatidylcholine Impairs Endothelial Barrier Function through the G Protein-Coupled Receptor, GPR4. Am. J Physiol Lung Cell Mol. Physiol. 2006;291:L91–L101. doi: 10.1152/ajplung.00508.2005. [DOI] [PubMed] [Google Scholar]
- Roebuck KA, Rahman A, Lakshminarayanan V, Janakidevi K, Malik AB. H2O2 and tumor necrosis factor-alpha activate intercellular adhesion molecule 1 (ICAM-1) gene transcription through distinct cis-regulatory elements within the ICAM-1 promoter. J. Biol. Chem. 1995;270:18966–18974. doi: 10.1074/jbc.270.32.18966. [DOI] [PubMed] [Google Scholar]
- Sluiter W, Pietersma A, Lamers JM, Koster JF. Leukocyte adhesion molecules on the vascular endothelium: their role in the pathogenesis of cardiovascular disease and the mechanisms underlying their expression. J. Cardiovasc. Pharmacol. 1993;22 Suppl 4:S37–S44. [PubMed] [Google Scholar]
- Smalley-Freed WG, Efimov A, Burnett PE, Short SP, Davis MA, Gumucio DL, Washington MK, Coffey RJ, Reynolds AB. p120-catenin is essential for maintenance of barrier function and intestinal homeostasis in mice. J. Clin. Invest. 2010;120:1824–1835. doi: 10.1172/JCI41414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring CM, Kelly KF, O'Kelly I, Graham M, Crawford HC, Daniel JM. The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp. Cell Res. 2005;305:253–265. doi: 10.1016/j.yexcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Stevens T, Rosenberg R, Aird W, Quertermous T, Johnson FL, Garcia JG, Hebbel RP, Tuder RM, Garfinkel S. NHLBI workshop report: endothelial cell phenotypes in heart, lung, and blood diseases. Am. J. Physiol Cell Physiol. 2001;281:C1422–C1433. doi: 10.1152/ajpcell.2001.281.5.C1422. [DOI] [PubMed] [Google Scholar]
- van Hengel J, van Roy F. Diverse functions of p120ctn in tumors. Biochim. Biophys. Acta. 2007;1773:78–88. doi: 10.1016/j.bbamcr.2006.08.033. [DOI] [PubMed] [Google Scholar]
- van Roy FM, McCrea PD. A role for Kaiso-p120ctn complexes in cancer? Nat. Rev. Cancer. 2005;5:956–964. doi: 10.1038/nrc1752. [DOI] [PubMed] [Google Scholar]
- Wang M, Li N, Li J, Ma Y, Li D, Qin L, Wang X, Wu R. Involvement of p120 in LPS-induced NF-kappaB activation and IL-8 production in human bronchial epithelial cells. Toxicol. Lett. 2010;195:75–81. doi: 10.1016/j.toxlet.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Wang YL, Malik AB, Sun Y, Hu S, Reynolds AB, Minshall RD, Hu G. Innate Immune Function of the Adherens Junction Protein p120-Catenin in Endothelial Response to Endotoxin. J. Immunol. 2011;186:3180–3187. doi: 10.4049/jimmunol.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J. Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, O'Donnell JJ, III, Holian O, Vincent PA, Kim KS, Daniel JJ, Lum H. P120 catenin represses transcriptional activity through Kaiso in endothelial cells. Microvasc. Res. 2010;80:233–239. doi: 10.1016/j.mvr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondag GC, Reynolds AB, Moolenaar WH. Receptor protein-tyrosine phosphatase RPTPmu binds to and dephosphorylates the catenin p120(ctn) J. Biol. Chem. 2000;275:11264–11269. doi: 10.1074/jbc.275.15.11264. [DOI] [PubMed] [Google Scholar]