Summary
A limited set of cell-cell signaling pathways presides over the vast majority of animal developmental events. The typical raison d'etre for signal transduction is to control the transcription of protein-coding genes. However, with the recent appreciation of microRNAs, growing attention has been paid towards understanding how signaling pathways intertwine with microRNA-mediated regulation. This review highlights recent studies that uncover unexpected modes of microRNA regulation by cell signaling pathways. Not only can miRNA transcription be positively or negatively regulated by cell signaling, the TGF-β/BMP pathways and Ras/MAPK pathways have now been shown to directly influence microRNA biogenesis to mediate substantial cellular phenotypes.
Introduction
Fundamental to the organized development of all multicellular organisms is the ability of cells to communicate with each other. In animals, a handful of fundamental cell signaling systems are used reiteratively to determine cell fates and pattern tissues, including the Notch, Hedgehog, Wnt, TGF-β/BMP, receptor tyrosine kinase (RTK), Jak/STAT, nuclear receptor and Hippo pathways [1,2]. The typical view of these cell signaling pathways is to transduce inputs from the cell surface to the nucleus, to alter the transcriptional status of protein-coding target genes. However, other outputs of the fundamental cell signaling systems have been catalogued, including direct regulation of cytoskeletal dynamics, cell adhesion, cell polarity, and/or cell death. Since many diseases and cancers involve deregulation of these core developmental signaling pathways, a comprehensive view of their action is necessary.
The recognition of an extensive class of ~22 nucleotide (nt) RNAs generated from endogenous hairpin transcripts, collectively known as microRNAs (miRNAs), changed the playing field for understanding gene regulatory mechanisms [3]. The founding miRNAs were recognized in the 1990s upon cloning of the C. elegans temporal identity mutants lin-4 and let-7 [4,5], but the generality and the regulatory reach of the miRNA network was not appreciated until this past decade. Many animal genomes encode hundreds of miRNAs, and evidence suggests that they regulate a majority of protein-coding transcripts [6]. It may come as no surprise then, that there are many compelling links between developmental cell signaling pathways and miRNAs. We and others have reviewed how components of many signal transduction pathways are regulated by miRNAs [7,8]. Here, we highlight recent advances on how cell signaling regulates miRNAs, with an emphasis on unexpected intersections of cell signaling with miRNA biogenesis.
Basics of miRNA biogenesis and function
While a diversity of miRNA biogenesis schemes have been reported, a canonical mechanism governs the production of the majority of animal miRNAs [9]. Most miRNAs are transcribed by RNA polymerase II, as part of non-coding genes or from introns of protein-coding genes. Primary miRNA (pri-miRNA) transcripts contain one or more local hairpins that are cleaved by the nuclear RNase III enyzme Drosha and its dsRBD partner DGCR8. Drosha exists in multiple complexes, with larger complexes containing additional factors such as the RNA helicases p68 and p72 [10]; the latter are involved in maturation of a subset of miRNAs [11]. Drosha cleavage releases ~55-70 nt hairpins known as pre-miRNAs, which are exported to the cytoplasm and cleaved again by a Dicer RNase III enzyme to yield ~22 nt miRNA duplexes. One strand is preferentially incorporated into an Argonaute (AGO) protein, which serves as the core of an effector complex that is guided by the small RNA to targets [12]. Animal miRNAs can repress targets via surprisingly short complements of ~7 nt to the 5' ends of miRNAs (preferentially nucleotides 2-8), inducing mRNA destabilization and/or inhibiting productive translation [13]. Comparative genomics provides compelling evidence for purifying selection operating on tens of thousands target sites distributed amongst a majority of protein-coding genes [6], and both transcriptome [14,15] and proteome studies [16,17] provide experimental evidence that individual miRNAs can directly repress hundreds of targets.
Highly dose-sensitive nature of cell signaling pathways
The bulk of miRNA targets are rather subtly repressed at the transcript and protein level, leading to the notion that miRNAs serve on the whole to finely tune target levels [6]. However, the quantitative strength of miRNA-mediated repression does not necessarily predict phenotypically relevant gene regulation, since this must also take into account the biological function of the target genes [18]. While the level of many genes can be manipulated over a wide range without apparent effect to the organism, slight changes in the activity of some genes suffice to cause substantial phenotypes. In particular, members of cell signaling pathways are frequently dose-sensitive. This feature has been exploited in genetic screens for new signaling components. Specifically, in a background that is sensitized for pathway activity, one can easily identify loci for which loss of one gene copy enhances or suppresses the starting phenotype.
This strategy was first used to uncover components of RTK signaling during photoreceptor specification, including the small GTPase Ras1 [19]. Notably, activating mutations of Ras are amongst the most common features of human tumors. Later, an activated Drosophila Ras1 modeled on the common human V12 mutation was used for saturation-level screening of ~850,000 mutants [20]. This yielded an impressive collection of ~300 dominant enhancer and ~600 dominant suppressor mutations, indicating that heterozygosity can frequently alter the consequences of Ras1 overactivity. The Ras1 modifiers proved to identify most components of this signaling pathway, including an extensive kinase cascade that is engaged by active Ras, culminating in the mitogen activated protein kinase (MAPK, also known as ERK).
Another conserved cell signaling pathway is mediated by ligands of the TGF-β/BMP family. The most broadly used homolog in Drosophila is Decapentaplegic (Dpp), which directs tissue patterning and growth throughout many developmental settings. Screening for dominant modifiers of dpp mutants revealed many such loci, including the Mothers against dpp (Mad) and Medea loci [21]. Mad and Medea proved to encode related transcription factors, of which Mad is more broadly required for Dpp signaling; similarly, its orthologs Smad1/2/3/5/8 (the regulatory Smads, or R-Smads), are the major nuclear effectors of mammalian TGF-β/BMP signaling; Medea is classified with mammalian Smad4 as "co-Smads" [22]. A third example of a highly dose-sensitive cell signaling cascade is the Notch pathway [23]. Not only are phenotypic outputs of Notch signaling highly amenable to genetic modification [24], three core pathway components in Drosophila (the receptor Notch, the ligand Delta, and the nuclear corepressor Hairless) are haploinsufficient; that is loss of one allele confers fully penetrant morphological defects. This is not peculiar to insects, since haploinsufficient phenotypes have also been observed for mammalian Notch pathway components [25,26].
The dose-sensitive nature of the fundamental cell signaling pathways suggests that they may be enriched for compelling instances of miRNA targeting. In fact, studies conducted prior to the formal recognition of miRNAs revealed that multiple target genes of the Drosophila Notch pathway were critical targets of miRNA-mediated repression. In particular, two large families of Notch targets encoding bHLH repressor genes and Bearded genes bear conserved ~7 nt motifs termed Brd boxes (AGCUUUA), GY boxes (GUCUUCC) and K boxes (cUGUGAUa) in their 3' UTRs [27,28]. Their significance was hinted at by gain-of-function alleles in Bearded family and bHLH-R genes associated with loss of 3’ UTRs [29,30]. Indeed, these motifs mediated negative post-transcriptional regulation, including reduction of steady state transcript levels concomitant with loss of poly-A tails. More strikingly, mutation of these motifs within genomic transgenes recapitulated aspects of the original gain-of-function phenotypes. Years later with the first cloning of Drosophila miRNAs [31], it was shortly noticed that many bore perfect Watson-Crick complementarity to Brd, GY, or K boxes, specifically at the 5’ ends of the miRNAs [32]. Thus, the study of post-transcriptional regulation of Notch signaling laid a groundwork for understanding key features of miRNA:target interactions. More recent genetic studies now reveal an impact of miR-8/200 family members on Notch-mediated tumorigenesis [33,34]. Altogether, these observations provide strong rationale to study functional connections between cell signaling pathways and miRNAs in detail.
R-Smads: Transcription factors in the TGF-β pathway moonlight as miRNA biogenesis factors
Many studies have documented the regulation of TGF-β/BMP signaling by miRNAs [35-38], as well as the transcriptional control of miRNAs by TGF-β/BMP signaling [39,40]. However, recent work illuminates an unexpected influence of TGF-β/BMP signaling on miRNA biogenesis. Studies of the ability of TGF-β and BMPs to induce a contractile phenotype in vascular smooth muscle cells, revealed that both ligands caused rapid, post-transcriptional, upregulation of mature miR-21 [41]. This miRNA subsequently represses programmed cell death protein 4 (PDCD4), an inhibitor of smooth muscle contractile genes. Functional knockdown of miR-21 activity prevented the ability of these ligands to induce the contractile phenotype, indicating that miR-21 is an important effector of TGF-β/BMP signaling.
Surprisingly, the mechanism of miR-21 induction involved the formation of a direct protein-protein interaction between different R-Smads (Smad 1/3/5) and the RNA helicase p68, which associates with Drosha/DGCR8 to promote cleavage of specific pre-miRNA transcripts, including pri-mir-21 and pri-mir-199a (Figure 1). Interestingly, they observed some ligand specificity to the response, in that Smad1 was recruited to pri-mir-21 upon BMP4 stimulation, whereas Smad2 and Smad3 were recruited upon TGF-β stimulation; Smad4 (the co-Smad) was not recruited to pri-miRNAs by either stimulus [41].
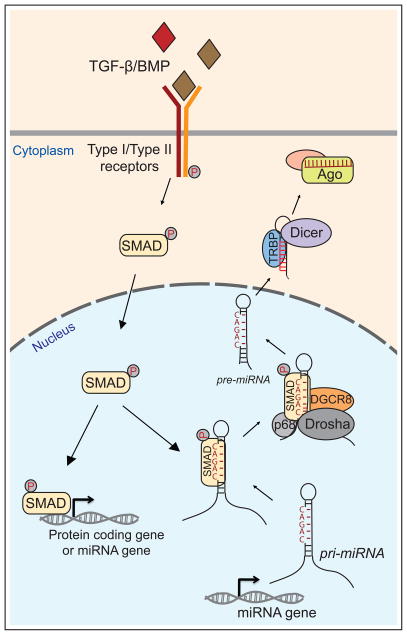
Figure 1.
Control of transcription and miRNA biogenesis by TGF-β/BMP signaling. At the core of this pathway, ligand binding to Type I/II heterodimeric receptors induces phosphorylation and nuclear translocation of SMAD transcription factors, which directly regulate the expression of protein-coding and miRNA genes. Mammalian SMAD proteins can also bind to the double-stranded stems of certain pri-miRNAs that bear CAGAC motifs, and promote their cleavage by the RNase III enzyme Drosha and its dsRBD partner DGCR8. This enhances the production of CAGAC-bearing pre-miRNAs, which are exported to the cytoplasm, cleaved by Dicer, and loaded into Argonaute (Ago) complexes to repress target genes.
Recent follow-up work expanded the generality of this response, and illuminated the underlying mechanism [42]. miRNA profiling following BMP4 and TGF-β treatment revealed elevation of the levels of 20 mature miRNAs, including the originally studied miR-21 and miR-199a. Analysis of their hairpin sequences revealed that 17 contained a CAGAC motif located ~10 bp from the terminal loop. Curiously, this is very similar to the known DNA binding site of Smads, and the corresponding miRNA hairpin motif was termed the RNA Smad binding element (R-SBE). Mutational analysis showed that CAGAC motifs were necessary for Smad-mediated enhancement of Drosha processing (Figure 1). More significantly, such a motif was also sufficient to bring a non-targeted pri-miRNA under Smad control. Finally, they demonstrated that the known DNA binding MH1 domain of Smad was sufficient to interact with double stranded R-SBE, while the MH2 domain of Smad interacted with p68. In summary, TGF-β/BMP stimulation induces recruitment of R-Smads into R-SBE containing pri-miRNAs to enhance Drosha processing of a broad set of pri-miRNAs (Figure 1).
The concept that cell signaling pathways can influence miRNA biogenesis is broadened by studies of the nuclear receptor, estrogen receptor alpha (ERalpha). Upon activation by the steroid hormone estrogen, ERalpha can also associate with the Drosha complex and inhibit the processing of certain miRNAs [43]; the molecular mechanism remains to be elucidated. More generally, the notion that transcription factors might have dual DNA- and RNA-binding capacities is food for thought [44]. For example, the eminent p53 tumor suppressor exerts its transcriptional role not only by the coordinate regulation of many protein-coding genes, but also by direct activation of tumor suppressor locus mir-34 [45,46]. On the other hand, p53 can also promote the cleavage of certain pri-miRNAs by a mechanism similar to the Smads, i.e. via recruitment to the Drosha microprocessor complex via direct interaction with p68 [47]. Notably, p53 has been documented to have RNA-binding activity [48,49], although it is not yet known whether this is involved in its ability to modulate miRNA processing. Such studies open a window onto the expanding possibilities for post-transcriptional regulation of miRNA processing [50].
Ras signaling: multiple mechanisms to affect miRNA transcription and biogenesis
Studies of Ras/MAPK signaling reveal many functional connections to miRNAs. One of the most compelling early observations was that the let-7 miRNA directly targets Ras in both C. elegans and mammals [51], suggesting that this miRNA could be a tumor suppressor. This has since been shown to be the case [52,53], in no small part due to the fact that let-7 can target many genes, including additional oncogenes such as HMGA2 [54,55]. Other miRNAs have also been shown to repress Ras members, suggestive of other tumor suppressor activities [56–59] (Figure 2).
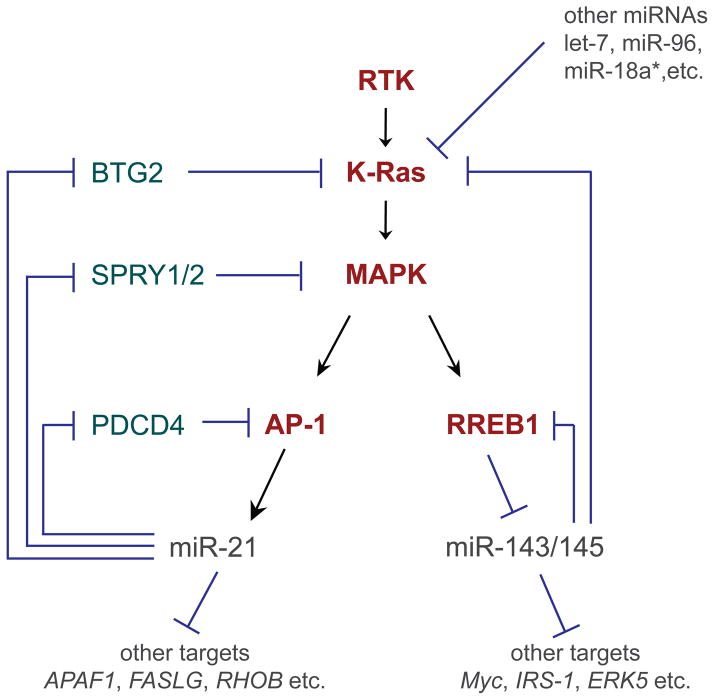
Figure 2.
Extensive feedback and feedforward loops between Ras signaling and miRNAs. In this simplified view of receptor tyrosine kinase (RTK) signaling, extracellular stimulation activates a Ras small GTPase (e.g. K-ras) and a kinase cascade including MAP kinase (MAPK), which regulates downstream transcriptional activators such as AP-1 and transcriptional repressors such as RREB1. In addition to regulating the expression of protein-coding genes, these transcription factors also regulate miRNA genes. AP-1 directly activates mir-21, an oncogenic miRNA that blocks multiple inhibitors of Ras signaling, including BTG2, SPRY1/2, and PDCD4, thereby establishing a potent feed-forward loop. RREB-1 directly represses the mir-143/145 cluster, which in turn have tumor suppressor activity by feedback repression of Kras and RREB1. These miRNAs also have other targets, many of which contribute to their oncogenic or tumor suppressive activities; as well, other miRNAs regulate Ras pathway components.
We first consider new advances in the regulation of miRNA transcription by Ras/MAPK signaling. Investigation of miRNA responses to oncogenic K-ras showed that the mir-143/145 cluster was consistently downregulated in mammalian and fish model systems, suggesting a highly conserved mechanism [60]. This was not just a correlation, since re-expression of mir-143/145 at physiological levels suppressed K-ras-mediated transformation. The mechanism by which K-ras signaling represses mir-143/145 transcription involves RREB1, a transcription factor downstream of Ras, directly via RREB1 binding sites in the mir-143/145 promoter (Figure 2). In a further twist, miR-143/145 themselves directly repress K-ras and RREB1 via their 3' UTRs. This establishes a mutually exclusive, feed-forward paradigm by which K-ras promotes an oncogenic state [60]. Reciprocally, miR-143/145 target other pro-growth factors such as Myc, Insulin Receptor Substrate 1, and ERK5, and to enforce an anti-proliferative state. The fact that loss of mir-143/145 is crucial for K-ras-mediated transformation suggests its potential as an anti-cancer therapy.
Going in the other direction, Ras signaling induces the transcription of certain miRNAs. Amongst its targets is mir-21, which is directly activated by the downstream transcription factor AP-1 [61,62]. The expression of miR-21 was earlier shown to be universally elevated in hundreds of human solid tumors across a panel of tissue origins [63], and a variety of tests in cancer cell lines suggested it to have oncogenic activity, since it represses a number of tumor suppressor genes [62,64], inhibits apoptosis, and can compromise the DNA damage-induced cell cycle checkpoint [65]. Recently, the cancer relevance of miR-21 has been addressed in animal models. In vivo overexpression of mir-21 enhanced lung tumorigenesis in concert with activated K-ras, but perhaps more significantly, the genetic deletion of mir-21 suppressed K-ras-driven tumors [62]. As with miR-143/145, oncogenic miR-21 may involve a feed-forward loop, since amongst its direct targets are multiple repressors of Ras signaling including Btg2 (which reduces the active GTP-bound Ras state), Sprouty genes (which are MAPK inhibitors) and PDCD4 (an inhibitor of AP-1) [62] (Figure 2).
Beyond these typical (although certainly complex and intertwined) mechanisms of miRNA-regulated cell signaling and signaling-regulated miRNA transcription, a biochemical approach recently revealed an unexpected influence of MAPK signaling on miRNA biogenesis. As with many other proteins, many components of the miRNA processing pathway are subject to post-translational modification. For example, there exist hyperphosphorylated forms of TRBP, a dsRBD cofactor for Dicer [66]. Identification of 4 serine phosphorylation sites provided the opportunity to investigate functional alterations exhibited by phospho-mutant and phospho-mimetic variants. These studies showed that phosphorylation increases the level of stable TRBP protein, which in turn enhances both general miRNA processing and miRNA-mediated silencing (Figure 3).
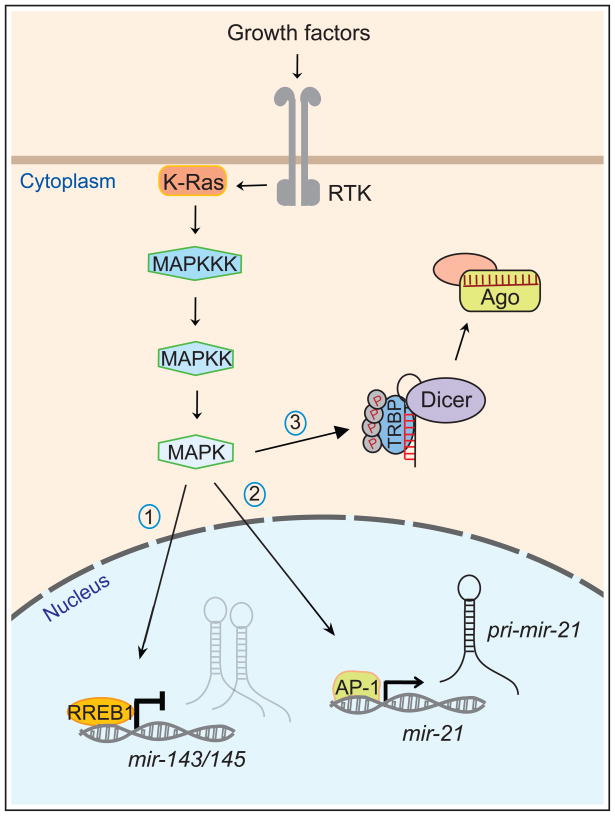
Figure 3.
Control of transcription and miRNA biogenesis by RTK/Ras signaling. Stimulation of receptor tyrosine kinases (RTK) by extracellular growth factors results in activation of the small GTPase Ras and a downstream kinase cascade including MAP kinase kinase kinase (MAPKKK), MAPKK and MAPK. Translocation of MAPK to the nucleus mediates the activity of downstream transcription factors, including (1) RREB1, a repressor of mir-143/145 and (2) AP-1, an activator of mir-21. Naturally, there are also many protein-coding genes whose transcription is regulated by RTK/Ras signaling. In addition, activation of MAPK results in phosphorylation of TRBP, a dsRBD cofactor of the Dicer RNase III enzyme. Phospho-TRBP mediates enhanced biogenesis of most miRNAs, including many growth-promoting miRNAs, although its activation is also associated with lower levels of the tumor suppressive miRNA let-7. For simplicity, Drosha cleavage of pri-mir-21 is not shown. Saj and Lai Highlights A set of fundamental cell signaling pathways controls most aspects of animal development. These pathways regulate expression of protein-coding genes and non-coding genes, including miRNAs. Several cell signaling pathways also directly regulate miRNA biogenesis at a post-transcriptional level.
The relevant kinase was found to be MAPK/ERK, which associates with TRBP, and whose chemical inhibition blocked phosphorylation of TRBP [66]. As well, this reaction could be reconstituted in vitro using recombinant TRBP, Erk2, and an activated form of the kinase upstream of Erk activation, MAP kinase kinase (MAPKK). Perhaps more importantly, this work assigned the miRNA generating machinery as a functional effector of MAPK signaling in promoting cell proliferation and survival. Phosphomimetic TRBP could promote both of these properties, while the presence of phospho-mutant TRBP could partially block the mitogenic effects of activating MAPK. Curiously, global profiling of miRNA expression changes induced by phosphomimetic TRBP showed a general increase in miRNA levels, including many pro-growth miRNAs, but repression of the tumor-suppressive miRNA let-7. Therefore, the differential coordination of miRNA biogenesis caused by MAPK-mediated phosphorylation of the Dicer cofactor TRBP represents a phenotypically substantial aspect of MAPK signaling.
These selected examples illustrate the varied ways in which EGFR/Ras/MAPK signaling can affect miRNA expression: they can be modulated by transcriptional activation or repression, and the activity of a core miRNA biogenesis component (TRBP) can be enhanced by signaling, with complex effects on miRNA biogenesis (Figure 3). These findings highlight the intricacy of gene regulatory programs unleashed by Ras signaling.
Concluding remarks
It is clear that beyond the association of protein-coding components of cell signaling pathways as miRNA targets, the control of miRNA transcription by signaling pathways is also of great importance. More surprisingly, we now appreciate that several key cell signaling systems can directly regulate miRNA biogenesis at post-transcriptional levels. In general, one can imagine diverse possibilities for the regulation of miRNA biogenesis, including at the level of Drosha cleavage, at nuclear export of pre-miRNAs, Dicer cleavage, Argonaute loading, removal from Argonaute, and probably other steps not currently appreciated. The study of post-transcriptional control of miRNA biogenesis has proven to be a rich field of study the past few years [50], and will undoubtedly continue to grow in the future.
We also touched upon the principle that miRNA genes are often involved in feed-forward or feed-back loops within a given signaling system, and cross-regulatory loops between different signaling systems undoubtedly occurs. For example, the transcription and biogenesis of miR-21 is under complex control by Ras/MAPK and TGF-β/BMP signaling. Since the studies to date were in different cell systems, it might be that these are separate regulatory events. On the other hand, it seems eminently possible that these signaling pathways may cooperate in some settings. Going beyond the study of individual regulatory interactions, systems approaches will be needed in the future to fully understand the complexity of gene regulatory networks initiated by signaling pathways, at both transcriptional and post-transcriptional levels [67].
A final key point to the studies discussed regards the phenotypic contribution of miRNAs and miRNA pathway components to signaling-mediated phenotypes, especially in light of the fact that miRNAs are often perceived to have subtle or fine-tuning effects. During induction of the contractile phenotype in vascular smooth muscle cells by BMP and TGF-β signaling, mammalian miR-21 is a critical and non-redundant effector gene. During oncogenic Ras/MAPK signaling, the post-translational activation of the miRNA biogenesis factor TRBP by phosphorylation, the transcriptional repression of mir-143/145, and the transcriptional activation of mir-21, all make substantial phenotypic contributions. In fact, recent studies of an inducible mir-21 transgene showed that continuous miR-21 activity is needed for maintenance of an oncogenic state, providing first in vivo evidence of tumor addiction to a miRNA [68]. These findings provide promise that the knowledge of miRNA modulation under conditions of dysfunctional cell signaling, so frequently known to be causal for disease and cancer, may eventually lead to new therapeutic strategies.
Saj and Lai Highlights.
A set of fundamental cell signaling pathways controls most aspects of animal development.
These pathways regulate expression of protein-coding genes and non-coding genes, including miRNAs.
Several cell signaling pathways also directly regulate miRNA biogenesis at a posttranscriptional level.
Acknowledgments
We apologize to the authors of many relevant works that could not be accommodated in this short review. We thank Peter Smibert, Alex Flynt, Qi Dai and Katsutomo Okamura for helpful discussions. Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Starr Cancer Consortium (I3-A139) and the NIH (R01-GM083300).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19 :491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002:16. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 3.Lai EC. microRNAs: runts of the genome assert themselves. Curr Biol. 2003;13 :R925–936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- •4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- •5.Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. Refs 4 and 5 describe the first miRNAs, lin-4 and let-7, which were cloned from C. elegans developmental mutants. These landmark studies provided prescient insights into miRNA biogenesis and target recognition. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 8.Hagen JW, Lai EC. microRNA control of cell-cell signaling during development and disease. Cell Cycle. 2008;7:2327–2332. doi: 10.4161/cc.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 12.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2010;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 15.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 16.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 18.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 20.Karim FD, Chang HC, Therrien M, Wassarman DA, Laverty T, Rubin GM. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics. 1996;143:315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raftery LA, Twombly V, Wharton K, Gelbart WM. Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 24.Kankel MW, Hurlbut GD, Upadhyay G, Yajnik V, Yedvobnick B, Artavanis-Tsakonas S. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics. 2007;177:2493–2505. doi: 10.1534/genetics.107.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Turkoz A, Jackson EN, Corbo JC, Engelbach JA, Garbow JR, Piwnica-Worms DR, Kopan R. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest. 2011;121:800–808. doi: 10.1172/JCI43114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krebs LT, Shutter JR, Tanigaki K, Honjo T, Stark KL, Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •27.Lai EC, Burks C, Posakony JW. The K box, a conserved 3' UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- •28.Lai EC, Posakony JW. The Bearded box, a novel 3' UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. Refs 27 and 28 describe some of the first miRNA target genes, from studies of post-transcriptional repression of Notch target genes by 7 nt 3' UTR motifs. These studies provided early insight into the seed-complementarity rule, as well as reduction of target transcript levels by miRNAs. [DOI] [PubMed] [Google Scholar]
- 29.Leviten MW, Lai EC, Posakony JW. The Drosophila gene Bearded encodes a novel small protein and shares 3' UTR sequence motifs with multiple Enhancer of split Complex genes. Development. 1997;124:4039–4051. doi: 10.1242/dev.124.20.4039. [DOI] [PubMed] [Google Scholar]
- 30.Klämbt C, Knust E, Tietze K, Campos-Ortega J. Closely related transcripts encoded by the neurogenic gene complex Enhancer of split of Drosophila melanogaster. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 32.Lai EC. microRNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 33.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. Embo J. 2011;30:756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. Embo J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 37.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. Embo J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winbanks CE, Wang B, Beyer C, Koh P, White L, Kantharidis P, Gregorevic P. TGF-{beta} regulates miR-206 and miR-29 to control myogenic differentiation through regulation of histone deacetylase 4 (HDAC4) J Biol Chem. 2011 doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •42.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. Refs 41 and 42 describe post-transcriptional enhancement of Drosha-mediated pri-miRNA cleavage directed by SMAD proteins, which are mostly known as transcription factors. Along with refs 43 and 47, we now recognize growing possibilities for regulation of miRNA biogenesis by transcription factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O'Malley BW, Kato S. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •47.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 48.Miller SJ, Suthiphongchai T, Zambetti GP, Ewen ME. p53 binds selectively to the 5' untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor beta- and p53-mediated translational inhibition of cdk4. Mol Cell Biol. 2000;20:8420–8431. doi: 10.1128/mcb.20.22.8420-8431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riley KJ, Cassiday LA, Kumar A, Maher LJ., 3rd Recognition of RNA by the p53 tumor suppressor protein in the yeast three-hybrid system. Rna. 2006;12 :620–630. doi: 10.1261/rna.2286706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148:381–392. doi: 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, Liu J, Yu J, Chen J. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Luo XJ, Xiong AW, Zhang ZD, Yue S, Zhu MS, Cheng SY. MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via down-regulation of proto-oncogene N-ras. J Biol Chem. 2010;285:26599–26607. doi: 10.1074/jbc.M110.115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–959. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 59.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •60.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–2759. doi: 10.1101/gad.1950610. This study nicely illustrates the interplay of signaling pathways and miRNA expression in cancer states. Ras signaling represses expression of the tumor suppressor cluster mir-143/145, which in turn repress components of Ras signaling; re-expression of these miRNAs can suppress oncogenic Ras. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D'Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- •62.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. This study describes elegant genetic studies of a mouse mir-21 knockout, which demonstrate endogenous requirements for this miRNA for induction of tumorigenesis by K-Ras. This is accomplished by the repression of multiple negatively-acting components of Ras signaling by miR-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Connolly EC, Van Doorslaer K, Rogler LE, Rogler CE. Overexpression of miR-21 promotes an in vitro metastatic phenotype by targeting the tumor suppressor RHOB. Mol Cancer Res. 2010;8:691–700. doi: 10.1158/1541-7786.MCR-09-0465. [DOI] [PubMed] [Google Scholar]
- 65.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr, Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •66.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. This paper reveals that MAPK/Erk signaling directly modulates miRNA biogenesis by phosphorylation of TRBP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Avraham R, Sas-Chen A, Manor O, Steinfeld I, Shalgi R, Tarcic G, Bossel N, Zeisel A, Amit I, Zwang Y, et al. EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci Signal. 2010;3:ra43. doi: 10.1126/scisignal.2000876. [DOI] [PubMed] [Google Scholar]
- •68.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. This paper describes tumor addiction to miR-21, providing proof of principle that suppression of specific miRNAs may be a viable anti-cancer therapy. [DOI] [PubMed] [Google Scholar]