Abstract
The main pathogenic process underlying dialysis-related amyloidosis (DRA) is the accumulation of β-2-microglobulin (β2m) as amyloid fibrils in the musculoskeletal system, and some evidence suggests that Cu(II) may play a role in β2m amyloid formation. Cu(II)-induced β2m fibril formation is preceded by the formation of discrete, oligomeric intermediates, including dimers, tetramers, and hexamers. In this work, we use selective covalent labeling reactions combined with mass spectrometry to investigate the amino acids responsible for mediating tetramer formation in wild-type β2m. By comparing the labeling patterns of the monomer, dimer, and tetramer, we find evidence that the tetramer interface is formed by the interaction of D strands from one dimer unit and G strands from another dimer unit. This covalent labeling data along with molecular dynamics calculations enable the construction of a tetramer model that indicates how the protein might proceed to form even higher order oligomers.
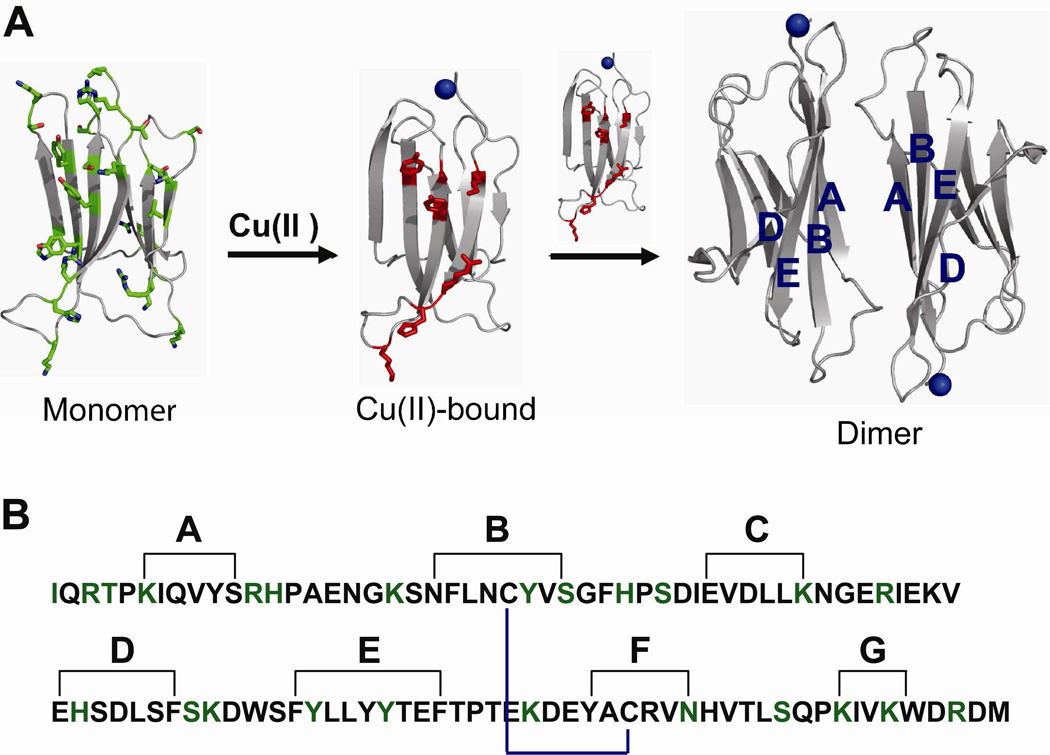
β-2-microglobulin (β2m) is the non-covalently bound light chain of the class I major histocompatibility complex (MHC-I) (1) and can accumulate as amyloid fibrils in the musculoskeletal system as a complication of long-term hemodialysis, leading to a condition known as dialysis-related amyloidosis (DRA). β2m has 99 residues (~12 kDa) and adopts an immunoglobulin fold with seven β strands (2), forming a β-sandwich in its native state (Figure 1). One β sheet is formed by strands A, B, D and E, and the other consists of strands C, F and G. A disulfide bond between Cys25 and Cys80 connects strands B and F in the folded state of the protein.
Figure 1.
(A) Ribbon representation of monomeric β2m (PDB ID: 2D4F), Cu(II)-bound β2m, and the formation of the dimer by stacking of two antiparallel ABED sheets. Amino acids modified by the covalent labels are shown as green sticks. The amino acids found in the dimer interface (ABED β-sheet) are shown as red sticks. (B) Amino acid sequence of β2m showing strand nomenclature (2). Black lines show amino acids on each β strand. The internal disulfide bond is shown in blue. The amino acids probed by the covalent labels are shown in green.
As part of normal cell turnover, β2m is released from MHC-I and carried to the kidney where it is usually degraded. Upon renal failure, serum levels of β2m increase up to ~60 times above their normal levels of about 0.1 µM, and the protein aggregates into insoluble amyloid deposits (3,4). An elevated level of β2m, however, is not unique to renal failure patients and is not sufficient to trigger fibrillogenesis (5,6). β2m amyloid formation must therefore result from factors particular to hemodialysis. These causative factors are not definitively known, but several approaches to generate β2m amyloid fibrils in vitro have been established. These include incubation under acidic conditions (pH < 3.6) (7), removal of the first six N-terminal amino acids (8), mixing with collagen at pH = 6.4 (9), sonication with sodium dodecyl sulfate at pH = 7.0 (10), and incubation with stoichiometric amounts of Cu(II) under physiological conditions (11,12).
We have become interested in Cu(II) as a causative factor for several reasons. It has been argued that Cu(II) might initiate β2m fibril formation in vivo because of the elevated Cu(II) concentrations in dialysate (11). The in vitro conditions necessary to stimulate β2m fibril formation in the presence of Cu(II) are also more similar to physiological conditions than other methods used to stimulate β2m fibril formation. Moreover, a recent study indicates that Cu(II) plays a catalytic role in causing β2m fibril formation (13). This latter observation is important because large systemic increases in Cu(II) concentrations are therefore not necessary. While these observations do not confirm a role for Cu(II) in vivo, β2m does represent yet another protein system for which Cu(II) can stimulate the formation of amyloid fibrils (14–18). Finally, from a biochemical perspective, adding Cu(II) is a discrete way to trigger amyloid formation so that the intermediates that precede the fibrils can be more easily studied.
Previous work has shown that Cu(II)-induced β2m amyloid formation is preceded by the formation of discrete, oligomeric intermediates, including dimers, tetramers, and hexamers (13,19). Several studies have attempted to characterize these pre-amyloid oligomers, but this task is very challenging because the oligomers are present as a mixture of species and, as intermediates, are only transiently populated. One successful approach to obtain structural information about the oligomers has been to create β2m mutants that are stable enough to crystallize as oligomers (20,21). In this way, it has been found that a P32A mutant of β2m forms a dimer in the absence of Cu(II) and an H13F mutant forms a hexamer in the presence of Cu(II). These crystal structures provide high resolution atomic-level information about possible β2m oligomer structures, but they do not provide a complete picture about oligomer structure because these mutants do not ultimately form amyloid fibrils.
As a complement to these crystallographic studies, we have used a protein surface mapping approach in conjunction with mass spectrometry (MS) to study the oligomeric intermediates of β2m. The method involves covalent modification of amino acid side chains that are exposed to solvent and the identification of the modified residues by MS. Because amino acids are usually buried upon formation of new protein-protein interfaces, residues involved in mediating these interactions are much less reactive and can be identified from differential modification patterns. This approach is directly applicable to wild-type β2m under amyloid-forming conditions, and the specificity of MS allows us to obtain structural information even when a mixture of β2m forms is present. We recently used this method to gain insight into the structure of the dimer formed by wild-type β2m and found that the dimer interface is formed by the anti-parallel stacking of ABED β-sheets from two β2m monomers (22).
In the current study, we have applied this covalent labeling/MS approach to characterize the tetrameric form of β2m that follows formation of the dimer. Through these measurements, we find evidence that the tetramer is formed by the interaction of D strands of one dimer unit and G strands of another dimer unit. Using the covalent labeling data along with molecular dynamics calculations, we are then able to build a model of the tetramer that suggests how the protein can continue to form higher order oligomers. Our results demonstrate the potential power of covalent labeling and MS for studying oligomer-forming proteins, and they help develop a model for the pre-amyloid tetramer of β2m that could be used to design inhibitors of β2m oligomerization and fibril formation.
MATERIALS AND METHODS
Materials
Human β-2-microglobulin (β2m) was obtained from Fitzgerald Industries International, Inc. (Concord, MA). Diethylpyrocarbonate (DEPC), 2,3-butandeione (BD), sulfo-N-hydroxysuccinimide acetate (NHSA), imidazole, dithiothreitol (DTT), copper-(II) sulfate (CuSO4), 3-morpholinopropanesulfonic acid (MOPS), potassium acetate, arginine, ubiquitin from bovine erythrocytes, equine heart cytochrome c, equine skeletal muscle myoglobin, chicken egg white ovalbumin, human hemoglobin, and bovine transferrin were purchased from Sigma-Aldrich (St. Louis, MO). Tris(hydroxymethyl)-aminomethane (Tris) was purchased from EM Science (Gladstone, NJ). Urea was purchased from Mallinckrodt Chemicals (Phillipsburg, NJ). Trypsin was from Promega (Madison, WI), and chymotrypsin was purchased from Roche Diagnostics (Indianapolis, IN). Centricon molecular weight cutoff (MWCO) filters were obtained from Millipore (Burlington, MA). Deionized water was prepared from a Millipore (Burlington, MA) Simplicity 185 water purification system.
Formation of β2m Oligomers and Fibrils
Reports have shown that discrete oligomers precede β2m amyloid fibrils when monomeric β2m is incubated under near-physiological conditions in the presence of Cu(II) (13,19). Amyloid fibrils were formed by incubation of 100 µM β2m in 200 mM potassium acetate, 500 mM urea, and 25 mM MOPS (pH 7.4) with 200 µM CuSO4 at 37 °C. This concentration of protein is higher than found in dialysis patients but was necessary to ensure the formation of oligomers in an experimentally reasonable time period. All components were equilibrated at 37 °C prior to Cu(II) addition, and immediately returned to 37 °C after mixing. The covalent labels were added to aliquots of incubated β2m taken at several time points after initiating the amyloid fibril formation reaction.
Carbethoxylation with DEPC
Stock solutions of DEPC were prepared in acetonitrile. The DEPC reactions of β2m were performed for 1 min at 37 °C and were initiated by adding 0.25 mM DEPC. The total reaction volume for the experiments was 30 µL, and the total amount of acetonitrile added was ~ 1.5 %. The reactions were quenched after 1 min by adding 10 mM imidazole.
Acetylation with NHSA
Stock solutions of NHSA were prepared in water. The labeling of β2m with 0.30 mM NHSA was carried out for 1 min at 37 °C. The total reaction volume for the experiments was 30 µL. The reactions were quenched by adding 10 mM TRIS.
BD Modification
Stock solutions of BD were prepared in water. The reactions of β2m with 35 mM BD were performed in the dark for 1 min at 37 °C. Reactions were carried out in the dark to avoid possible photoactivation of the label, which could enhance nonspecific reactions with residues other than arginine (23,24). The total reaction volume for the experiments was 30 µL. The reactions were quenched by adding 100 mM arginine.
Proteolytic Digestion
Before proteolytic digestion, all modified samples were purified using a 10,000 MWCO filter and reconstituted with deionized water to a final concentration of 300 µM. Purified β2m samples in 25 mM Tris-HCl (pH 7) and 1 mM CaCl2 were first reacted with 10 mM DTT at 37 °C for 45 min to reduce the disulfide bonds. This was followed by addition of acetonitrile and incubation at 37 °C for 45 min. Trypsin and chymotrypsin (1 µg/µL) were then added to DEPC-modified samples to yield a final enzyme/substrate ratio of 1:20. For the NHSA- and BD-modified β2m samples, only chymotrypsin (1 µg/µL) was added. Although trypsin is a reliable and robust protease, chymotrypsin was used instead because trypsin can no longer cleave proteins after acetylated lysine and modified arginine residues. All samples were digested at 37 °C for 16 h. The enzymes were inactivated by adding 2 µL of acetic acid, and the samples were immediately analyzed.
Instrumentation
The amount of modification was determined by removing an aliquot of the purified β2m and analyzing the samples using a Bruker amaZon quadrupole ion trap mass spectrometer (Billerica, MA) equipped with an electrospray ionization (ESI) source. The ESI source was operated at a spray voltage of 4.5 kV, and the capillary temperature was set at 220°C. The voltages for the transfer optics between the ESI source and the ion trap were optimized for maximum signal.
The proteolytic fragments were separated by an Agilent HP1100 (Wilmington, DE) HPLC system with a C18 column (15 cm × 2.1 mm, 5 µm particle size, Supelco, St. Louis, MO) for on-line analysis by MS and MS/MS. The fragments of NHSA- and BD-modified β2m were eluted using a linear gradient of methanol that increased from 5 to 70% over 20 min and 70 to 100% over the final 2 min at a flow rate of 0.25 mL/min. For the fragments of DEPC-modified β2m, a linear gradient of methanol that increased from 5 to 70% over 30 min and 70 to 100% over the final 3 min was used. For both gradient conditions, water comprised the balance of the solvent, and a total of 0.1% acetic acid was present. The LC effluent was directly fed into the mass spectrometer with similar ESI source conditions as described above. Tandem mass spectra were acquired using collision induced dissociation (CID) with isolation widths of 1.0 Da and excitation voltages between 0.6 and 1.0 V. Peptide sequences were determined from the MS/MS data via de novo sequencing or with the help of BioTools (Bruker Daltonics, Billerica, MA).
To monitor the formation of oligomers, the incubated solutions of β2m were separated by size-exclusion chromatography (SEC) using a Superdex 75 PC 3.2/30 column (Amersham Biosciences) installed on an Agilent HP 1100 series HPLC system. Before analysis of the sample, the SEC column was first equilibrated with a 20 mM ammonium acetate mobile phase (pH 7.4) at a 0.06 mL/min flow rate for 1 h. During the analysis, 5 µL of an incubated sample solution was injected into the sample loop. Either a variable wavelength detector set to 214 nm or a Bruker Esquire-LC quadrupole ion trap mass spectrometer, equipped with an ESI source (Billerica, MA), was used for detection. The identity of the separated oligomers was confirmed by comparing to a molecular weight (MW) calibration curve or from the m/z ratios measured by the mass spectrometer. For the MW calibration, a solution containing a mixture of the following proteins and peptides was used: 1.5 µM bovine serum albumin (MW 66,000 Da), 3 µM carbonic anhydrase (MW 29,040 Da), 3 µM myoglobin (MW 16,951 Da), and 3 µM β2m (MW 11,731 Da).
Determination of Modification Percentages
The percent modification of each labeled amino acid was determined by comparing the LC-MS abundances of modified and unmodified proteolytic peptide fragments containing the amino acid of interest. Because the ionization efficiencies of the modified and unmodified residues are not identical, our evaluation of the modified residues relies on relative, rather than absolute changes, in the extents of modification. All peptide fragments containing the modified residue were accounted for, including overlapping peptides that are commonly generated during chymotryptic digestions. For each peptide fragment, the modified form eluted after the unmodified form with the difference in retention times ranging from 1–12 minutes. The ion abundances of both modified and unmodified peptides were determined from extracted ion chromatograms; as examples, see Figures S1, S2, and S3 in the Supporting Information. The percent modification was obtained by dividing the total ion abundance of the modified fragment (Imodified) by the sum of the total ion abundances for the modified (Imodified) and unmodified (Iunmodified) fragments as shown in equation 1. All the charge states for a given peptide fragment were considered in the determination of the total ion abundances. The errors are reported as the standard deviation of the mean using the data from three separate modification reactions. Because ion intensity ratios were used, low modification levels could be accurately determined. Indeed, reliable covalent labeling approaches typically yield relatively low modification levels to avoid label-induced structural changes (25, 26 and references therein). Even so, these methods have been successfully used to obtain accurate protein structural information.
| Equation (1) |
Molecular Dynamics Simulations
All simulations were initiated using a dimer, the B and C chains, from the H13F mutant hexamer crystal structure (PDB ID: 3CIQ) (21), which contains 98 amino acid residues. Met0 was eliminated from the original set of coordinates because this residue is not present in the wild-type protein. Two dimers were added together before the simulations began in order to produce a tetramer structure that was consistent with the covalent labeling data. The two dimers were added together using the docking simulation program Zdock (version 3.0.1) [27,28]. Three descriptors were taken into account for dimer-dimer docking: i) the electrostatic potential of the dimer surface; ii) the shape complementarity of the dimer-dimer interface; and iii) the desolvation energy associated with the attachment process. The resulting tetramer structure was then energy-minimized, using an explicit model of the solvent and with the AMBER94 force field (29,30) implemented in GROMACS (31–33).
During the minimization, hydrogen atoms were explicitly added to the tetramer model and His residues were considered to be neutral, as shown in nuclear magnetic resonance (NMR) studies by Esposito and co-workers (34). Water molecules were simulated using a TIP3P model (35), and molecules were added until a cubic simulation cell of side length 9.6 nm was achieved.
RESULTS
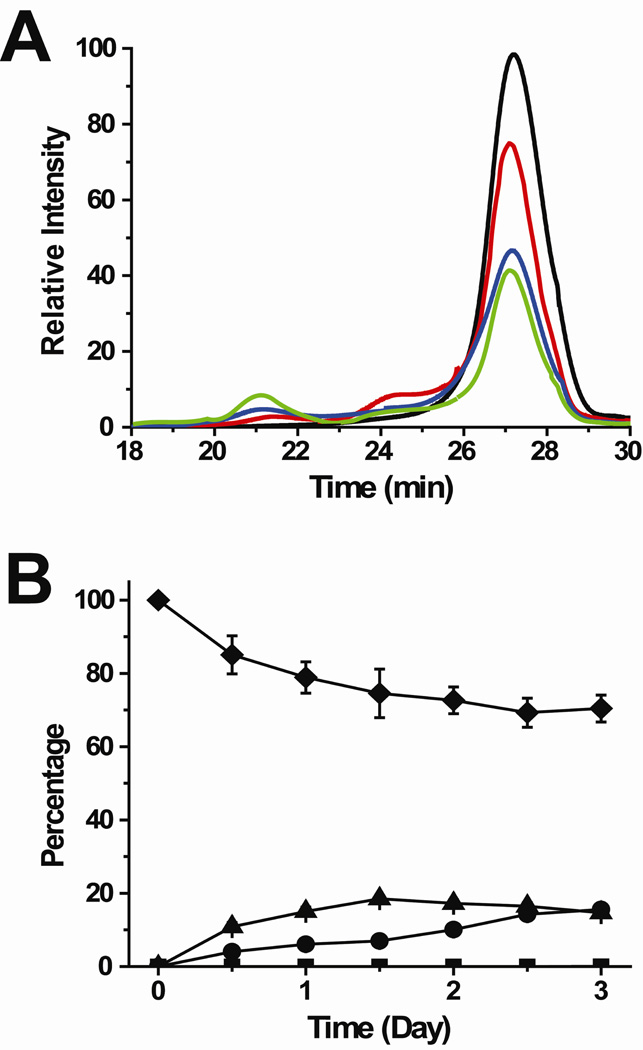
Structural studies of the oligomeric intermediates that precede β2m amyloid formation are challenging because these oligomers are transient, present as a mixture, and can dissociate back into the monomer during isolation. Using SEC, dynamic light scattering, and ESI-MS, we found previously that β2m oligomers are formed via the discrete addition of dimer units (13). The dimer first appears within about 1 hour after adding Cu(II), a first form of the tetramer appears within 12–24 hours, a second Cu(II)-free form of the tetramer appears after approximately 3 days, and the hexamer appears soon after the Cu(II)-free tetramer. Since complete isolation of the tetramer is very difficult without some dissociation back to the dimer and monomer, covalent labeling experiments were performed at different times before (t = 0 min) and after (t = 2 min, 2 hrs, 0.5 day, 1 day, 1.5 days, 2 days, and 2.5 days) adding Cu(II) to initiate the amyloid fibril-forming reaction. At time points between 0.5 and 2.5 days, the mixture contains monomer, dimer, and tetramer as indicated by SEC experiments (Figure 2). The percentage of tetramer at time points after the addition of Cu(II) increases from 0 to 15% based on ESI-MS measurements of the desalted sample under amyloid-forming conditions, as described previously (13), and these results are consistent with the SEC data. Also, the percentage of dimer increases from 0 to about 30% from 0 to 2.5 days, as indicated by ESI-MS measurements; the ESI-MS and SEC data are also roughly consistent with regard to the dimer as the SEC data indicates a dimer increase from 0 to about 20% (Figure 2).
Figure 2.
Formation of monomers, dimers, and tetramers as studied by size-exclusion chromatography (SEC). (A) SEC profiles before (black) and 1 day (red), 2 days (blue), and 3 days (green) after the addition of Cu(II). (B) Plot showing the percentage of each oligomer as a function of time: monomer (diamond), dimer (triangle), tetramer (circle), and hexamer (square). The oligomer identities in the size-exclusion chromatogram were confirmed by comparison to a molecular weight calibration curve and by on-line SEC-ESI-MS.
In recent covalent labeling studies of the β2m dimer, we identified 20 amino acids that undergo changes in labeling reactivity, relative to the monomer, after Cu(II) binding and dimer formation (Figure 1) (22). In the current study, we have sought to identify residues that only undergo changes in labeling reactivity as the tetramer is formed. To do this, the covalent labeling data that is obtained when the tetramer is present (0.5 – 2.5 days) is compared to the data acquired when only dimer and monomer are present in solution (0.5 – 2 hours). Specifically, we examined the modification trend (i.e. increase, decrease, or leveling off) from the covalent labeling data in the 0.5 – 2.5 day time period with the modification trend from the covalent labeling data in the 0.5 – 2 hour time period (22). During the 0.5 – 2.5 day time period, the rate of tetramer formation is almost twice that of dimer formation and similar to the rate of monomer decrease (Figure 2). Indeed, the amount of dimer levels off after 1.5 days. Hence, any residues that undergo a new modification trend (i.e. increase, decrease, or leveling off) after 0.5 days, as compared to the 0.5 – 2 hour time period, are then residues that likely undergo changes in solvent accessibility important for the formation of the tetramer. Furthermore, we have baseline modification data for the monomer (i.e. no Cu(II) added) to ensure that any decreases in the amount of modification are due to tetramer formation. We also performed control experiments in which covalent labeling reactions were carried out on a solution containing only monomeric β2m (i.e. no Cu(II) added) that was incubated under identical conditions for time periods up to 2 days. No significant changes in the labeling of any residue were observed, indicating that the protein undergoes no significant conformational changes during this time period (Table S1 in the Supporting Information). In addition, both SEC and ESI-MS measurements indicate no oligomer formation over this time period without the addition of Cu(II).
Covalent labeling with NHSA
Amino groups such as the ε-NH2 of lysine residues and the N-terminal α-NH2 can react with NHSA, but information from such surface mapping experiments is reliable only if the structural integrity of a protein is preserved during the reaction. Our group recently demonstrated that monitoring the reaction kinetics for individual modification sites is powerful way to ensure a protein’s structural integrity upon modification [36]. This can be done by monitoring the ion abundance of peptide(s) that contain the residue of interest as a function of labeling reagent concentration. Considering these reactions are second order, such dose-response plots will be linear over the range of reagent concentrations where the protein’s structural integrity is maintained and will deviate from linearity when reagent concentrations are reached that perturb the protein’s structure (see Figure S4 as an example). By generating such plots for all modified peptides, we identify the maximum reagent concentration that can be used to ensure the protein’s structure is not perturbed. In the studies here, we found that reaction of β2m with a 3-fold molar excess of NHSA for 1 min at 37 °C does not induce any protein structural changes, which is consistent with our previous work on this protein (22). The reaction time was kept short (1 min) to minimize tetramer dissociation.
Proteolytic digestion and LC-MS analyses are necessary to identify the amino acids that change in reactivity as the tetramer concentration increases. Under solution conditions in which β2m forms amyloid fibrils, LC-MS/MS analyses show that the N-terminus, almost all of the lysines (Lys6, Lys19, Lys41, Lys58, Lys75, Lys91, and Lys94), and Asn83 are labeled to different degrees. The unmodified and modified fragments containing these amino acids are detectable at all time points; however, the level of modification for some residues changes over time (vide infra). In our previous study, definitive MS/MS data could not be obtained for the fragment Lys41-Leu54 (22). Here, the better sensitivity of the quadrupole ion trap used in these experiments allowed identification of Lys41 as the modification site.
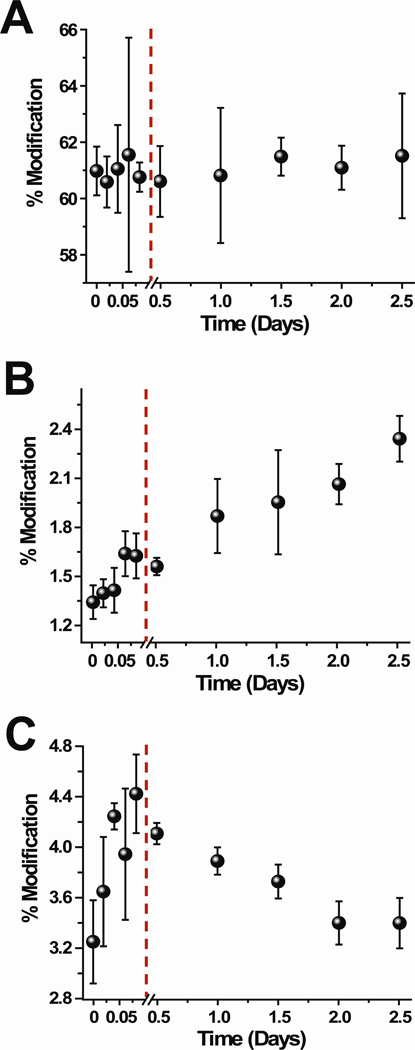
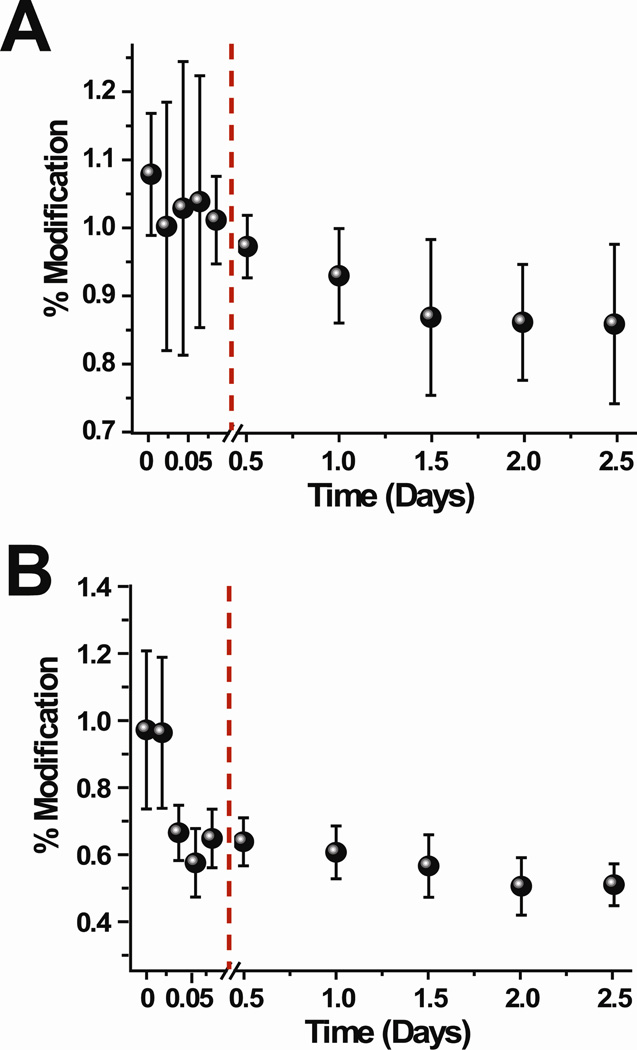
The reactions with NHSA at different time points after formation of the tetramer reveal that the modification extents of some residues change as the concentration of the tetramer increases in solution (Figure 3 and Figure S5 in the Supporting Information). The percentage modification for the peptide fragments containing the indicated residues were determined from the LC-MS intensities of the modified and unmodified fragments, as described in the Materials and Methods section. For each time point, the modification reaction was repeated three times. Because ion intensity ratios of the unmodified and modified peptide fragments are used to determine the extent of modification, small changes in modification levels can be accurately and precisely determined, as we demonstrated in our previous work (22). Also, it should be noted that the low levels of modification for some residues reflect the need to minimize the overall degree of labeling to prevent the labels from perturbing protein structure (25).
Figure 3.
Extent of NHSA modification for selected residues, (A) N-terminus, (B) Lys75, and (C) Lys94 throughout the course of the tetramer formation reaction. The changes in modification during the dimer formation are shown (points before red line) as reference points.
Of all the residues acetylated by NHSA, the N-terminus (Figure 3A) and Asn83 do not significantly change in the extent of modification from 0.5 to 2.5 days after addition of Cu(II). The level of modification of Lys75 (Figure 3B) increases from 0.5 days to 2.5 days after Cu(II) is added. In contrast, the reactivity of Lys6, Lys19, Lys41, Lys58, Lys91 (Figures S5A–D, F), and Lys94 (Figure 3C) decrease as the tetramer’s concentration increases in solution. The extent of modification of Lys6 decreases slightly within 2 hours after adding Cu(II), and the reactivity of this residue continues to decrease up to 2.5 days. In our previous covalent labeling studies on the dimer, we concluded that Lys6 was part of the dimer interface, and so its continual decrease in reactivity is consistent with the dimer’s concentration continuing to increase in solution over time.
The rest of the lysine residues, Lys19, Lys41, Lys58, Lys91, and Lys94, behave differently. The reactivity of these five lysine side chains also decrease as the tetramer concentration increases, but their reactivities either do not change or increase prior to tetramer formation. For example, Lys94 increases in reactivity as the dimer’s concentration increases in solution, but as soon as the tetramer begins to significantly populate the solution, Lys94 decreases in reactivity. On the other hand, Lys41 undergoes no change in reactivity as the dimer is formed, whereas it decreases in reactivity as the tetramer’s concentration increases in solution.
In summary, our results indicate that the reactivity of NHSA with the N-terminus and Asn83 do not change as the tetramer is formed in solution. In contrast, the reactivity of Lys75 increases as the tetramer is formed, whereas the reactivities of Lys6, Lys19, Lys41, Lys58, Lys91, and Lys94 decrease as the tetramer’s concentration increases in solution. Importantly, the modification extent for Lys6 decreases within the first 2 h after adding Cu(II), which suggests that its continuing drop in modification extent is due to the formation of more dimer in solution rather than the formation of tetramer. Tetramer formation is then likely responsible for the decreased reactivity of the remaining lysine residues (i.e. Lys19, Lys41, Lys58, Lys91, and Lys94).
Covalent labeling with DEPC
DEPC reacts readily with histidine residues but can also react with amine and hydroxyl groups at neutral pH. Using dose-response plots like that shown in Figure S4, we found that a 2.5-fold molar excess of DEPC is sufficient to label several amino acids in β2m while minimizing any DEPC-induced structural changes to the protein. As was the case with the NHSA reactions, the reaction time was kept short (1 min) to minimize tetramer dissociation but also to minimize DEPC hydrolysis by water.
Proteolytic digestion of the protein and LC-MS/MS analyses indicate that DEPC reacts with the N-terminus, Thr4, Lys6, His13, Lys19, Tyr26, Ser28, His31, Ser33, Lys41, His51, Ser57/Lys58, Tyr63, Tyr67, Lys75, Ser88, and Lys94. These modifications are observed at all time points, but the level of modification for some residues changes over time (vide infra). In all cases but one (Ser57/Lys58), the specific amino acids that are modified could be determined unambiguously from the MS/MS data.
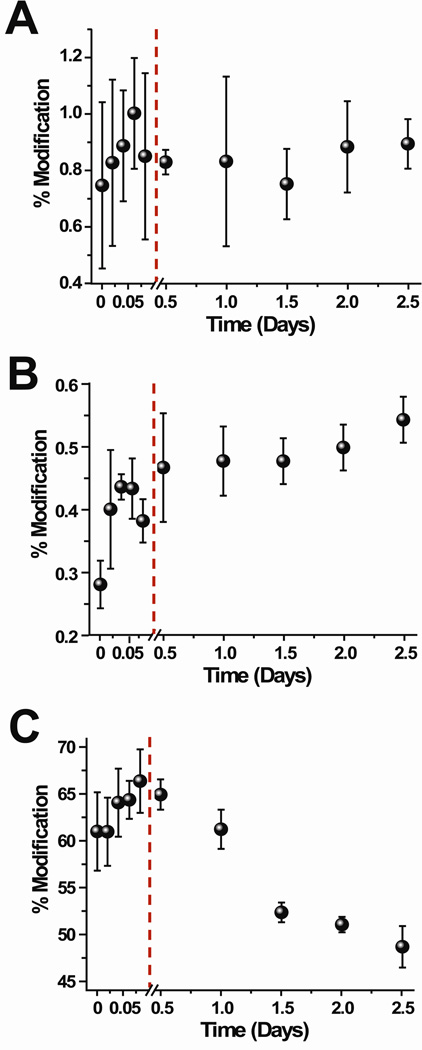
When the covalent labeling reactions with DEPC are performed at different time points after the tetramer begins to form, the modification levels of 13 of the 17 labeled residues change as the tetramer’s concentration increases in solution (Figure 4 and Figure S6 in the Supporting Information). The labeling data reveal that only Lys75 (Figure 4B) shows an increase in modification extent upon tetramer formation. The level of modification of six residues, Lys6, His13, Lys19, Ser28, Tyr63, and Tyr67, initially decrease within 2 hours after adding Cu(II) and continue to decrease up to 2.5 days. Five residues, Thr4, Lys41, His51, Ser57/Lys58, and Lys94, increase within 2 hours after adding Cu(II) but then decrease as the tetramer concentration increases in solution. Five residues, the N-terminus, Tyr26, His31, Ser33, and Ser88, undergo no change in reactivity as the tetramer is formed. In comparing the DEPC and NHSA reactivity for those residues that react with both reagents, we find complete consistency for the N-terminus, Lys6, Lys41, Lys58, Lys75, and Lys94. Only the reactivity of Lys19 is slightly different.
Figure 4.
Extent of DEPC modification for selected residues, (A) His31, (B) Lys75, and (C) His51 throughout the course of the tetramer formation reaction. The changes in modification during the dimer formation are shown (points before red line) as reference points.
In summary, the reactions with DEPC indicate that 12 residues undergo a notable change in reactivity as the tetramer is formed in solution. The modification extent of Lys75 increases whereas the reactivity of the other 11 amino acids decreases. The decrease in modification extents for Lys6, His13, Lys19, Ser28, Tyr63, and Tyr67 within 2 h after adding Cu(II) suggests that the continual drop in the extents of modification for these residues is due to formation of more dimer in solution rather than the formation of tetramer. Consequently, tetramer formation is then likely responsible for the reactivity changes of Thr4, Lys41, His51, Ser57/Lys58, Lys75, and Lys94.
Covalent Labeling with Butanedione
Unlike DEPC, which reacts with many nucleophilic groups, BD reacts specifically with arginine. The reaction is reversible at pH < 9 and is less efficient than the NHSA or DEPC reactions, so higher reagent doses are necessary to improve the product yield. Using dose-response plots like that shown in Figure S4, we determined that a 350-fold molar excess of BD is suitable to avoid any modification-induced structural changes as well as to obtain readily detectable modifications. The LC-MS data indicate that BD reacts with Arg3, Arg12, Arg45, and Arg97 but not Arg81. The reactions of BD with β2m reveal that Arg3 and Arg12 undergo a very slight decrease in reactivity upon formation of the tetramer (Figure 5 and Figure S7 in the Supporting Information). The modification levels of Arg3 and Arg12 decrease slightly as the tetramer’s concentration increases in solution, but because the reactivities of these residues also decrease slightly before 0.5 days their continual decreases in reactivity are likely due to formation of more dimer in solution rather than the formation of tetramer. The unchanged reactivity of Arg45 and Arg97 is curious since both of these residues increase in reactivity up to 2 h as the dimer’s concentration increases in solution. It is likely that these residues slightly decrease in reactivity as the tetramer is formed, but this decrease is counterbalanced by the increase in reactivity as the dimer’s concentration continues to increase in solution up to day 2.5. Overall, the BD reactivity suggests that Arg3 and Arg12 undergo no significant changes in reactivity as the tetramer is formed but Arg45 and Arg97 might.
Figure 5.
Extent of BD modification for selected residues, (A) Arg3 and (B) Arg12 throughout the course of the tetramer formation reaction. The changes in modification during the dimer formation are shown (points before red line) as reference points.
A summary of the reactivity changes for all of the modified amino acids monitored before Cu addition (t=0 h) and 2 h (t=2 h) and 2.5 days (t=2.5 days) after Cu(II) addition is shown in Table 1.
Table 1.
Summary of the modification percentages for the modified amino acids before the addition of Cu(II) (t = 0 min) and 2 hours (t = 2 hours) and 2.5 days (t = 2.5 days) after the addition of Cu(II).
| Residue | t = 0 min | t = 2 hours | t = 2.5 days | Statistically Significant Change?* |
|---|---|---|---|---|
| NHSA | ||||
| N-terminus | 84 ± 5 | 61 ± 3 | 61 ± 3 | No |
| Lys6 | 18.3 ± 0.9 | 18 ± 1 | 14.2 ± 0.7 | Yes |
| Lys19 | 7.1 ± 0.3 | 7.2 ± 0.3 | 6.42 ± 0.3 | Yes |
| Lys41 | 13 ± 1 | 13 ± 1 | 9.7 ± 0.5 | Yes |
| Lys58 | 16 ± 1 | 19 ± 1 | 13.9 ± 0.9 | Yes |
| Lys75 | 1.47 ± 0.1 | 1.6 ± 0.2 | 2.3 ± 0.2 | Yes |
| Asn83 | 1.48 ± 0.07 | 1.46 ± 0.05 | 1.6 ± 0.2 | No |
| Lys91 | 27 ± 1 | 24.1 ± 0.9 | 22.0 ± 0.8 | Yes |
| Lys94 | 3.2 ± 0.5 | 4.4 ± 0.5 | 3.4 ± 0.3 | Yes |
| DEPC | ||||
| N-terminus | 99 ± 5 | 68 ± 2 | 68 ± 3 | No |
| Thr4 | 87 ± 1 | 90 ± 4 | 61 ± 5 | Yes |
| Lys6 | 8.1 ± 0.1 | 7.64 ± 0.4 | 5.9 ± 0.6 | Yes |
| His13 | 45 ± 2 | 40 ± 2 | 34.2 ± 2 | Yes |
| Lys19 | 13.0 ± 0.9 | 12 ± 1 | 10.1 ± 0.5 | Yes |
| Tyr26 | 0.7 ± 0.1 | 0.4 ± 0.1 | 0.35 ± 0.04 | No |
| Ser28 | 0.24 ± 0.03 | 0.11 ± 0.01 | 0.078 ± 0.01 | Yes |
| His31 | 1.6 ± 0.3 | 0.8 ± 0.3 | 0.9 ± 0.1 | No |
| Ser33 | 1.7 ± 0.1 | 1.2 ± 0.2 | 1.3 ± 0.2 | No |
| Lys41 | 0.32 ± 0.07 | 0.34 ± 0.04 | 0.24 ± 0.02 | Yes |
| His51 | 60 ± 2 | 66 ± 3 | 49 ± 4 | Yes |
| Ser57/Lys58 | 39 ± 2 | 47 ± 5 | 33 ± 3 | Yes |
| Tyr63 | 6.3 ± 0.3 | 3.4 ± 0.3 | 2.7 ± 0.2 | Yes |
| Tyr67 | 2.2 ± 0.2 | 1.5 ± 0.1 | 1.2 ± 0.1 | Yes |
| Lys75 | 0.31 ± 0.04 | 0.38 ± 0.05 | 0.54 ± 0.06 | Yes |
| Ser88 | 66 ± 2 | 72 ± 4 | 69 ± 5 | No |
| Lys94 | 28 ± 3 | 36 ± 3 | 28 ± 2 | Yes |
| BD | ||||
| Arg3 | 0.5 ± 0.2 | 1.05 ± 0.06 | 0.8 ± 0.1 | Yes |
| Arg12 | 1.3 ± 0.2 | 0.64 ± 0.06 | 0.48 ± 0.07 | Yes |
| Arg45 | 3.8 ± 0.5 | 6.5 ± 0.8 | 6.6 ± 0.5 | No |
| Arg97 | 2.8 ± 0.5 | 4.9 ± 0.4 | 5.1 ± 0.3 | No |
A two-sample unpaired t-test was used to determine if the changes between 2 h and 2.5 days after the addition of Cu(II) are statistically significant. The calculated t-value was compared to the t-value at 95% confidence level (i.e. t=2.776). Residues with calculated t-values greater than 2.776 were considered to undergo a statistically significant change.
DISCUSSION
The goal of this study is to obtain structural insight into the pre-amyloid tetramer that is formed by β2m after adding Cu(II) at near physiological conditions. Our group has previously shown that Cu(II)-induced β2m amyloid fibril formation is preceded by the formation of dimers, tetramers, and hexamers (13). No odd-numbered oligomers are observed, indicating that the oligomeric intermediates form via the assembly of dimers. In addition, our previous work suggests that the first tetramer species that is formed has Cu(II) bound to it, but as the amyloid fibril-forming reaction proceeds, Cu(II) is lost from this first tetramer before forming Cu(II)-free tetramers, hexamers, and eventually amyloid fibrils. In a recent study, we showed that the pre-amyloid dimer is formed by the interaction of anti-parallel ABED β-sheets from two monomers (22). This dimer structure provides a starting point for gaining insight into the structure of the Cu(II)-bound tetramer.
Covalent modification with detection by MS was selected to probe the tetramer as this method has been effective in mapping protein surfaces, identifying ligand-binding sites, detecting ligand-induced conformational changes, and studying protein-protein complexes (26, 37–39), including the β2m dimer (23). As described above and in our previous studies (22), care was taken to select optimal reaction times and stoichiometries for each covalent labeling reagent to ensure that the amount of label added and the resulting modification do not disrupt the protein structure. A total of 24 residues in β2m are probed by NHSA, DEPC, and BD, and these residues are widely distributed along the polypeptide chain and on the surface of the protein, representing about 30% of the surface amino acids (see monomer in Figure 1A). Changes in the reactivity of amino acid side chains reflect changes in their solvent accessibility. Hence, patterns of modification during the course of the amyloid fibril-formation reaction permit us to build a low-resolution map of the protein and identify residues involved in mediating tetramer formation. Having baseline labeling data for the monomer and dimer allow us to readily identify those residues involved in tetramer formation, even though monomer, dimer, and tetramer are present simultaneously. In addition, being able to perform these reactions on wild-type β2m under amyloid fibril-forming conditions makes the resulting structural information relevant for understanding native β2m amyloid assembly.
Based on the data in Table 1, the amino acid side chains probed by the covalent labels can be grouped in three categories, namely, residues with (1) unchanged reactivity, (2) decreased reactivity, and (3) increased reactivity from 2 h to 2.5 days after adding Cu(II) when the first form of the tetramer is present. The covalent labeling experiments reveal that six of the 24 modified amino acids, the N-terminus, His31, Ser33, Arg45, Ser88, and Arg97, maintain the same reactivity with their respective labeling reagents at time points from 2 h to 2.5 days, although the unchanged reactivity of Arg45 and Arg97 is notably different (see below). Four of these residues, the N-terminus, His31, Ser33, Asn83 and Ser88 have the same reactivity in the dimer and tetramer, indicating that these residues retain similar microenvironments in both these oligomers. Therefore, these residues are not likely part of the tetramer interface. The unchanged reactivities of the N-terminus and His31 are consistent with these residues remaining Cu(II) binding sites in the monomer, dimer, and tetramer (40). The unchanged reactivity of Ser33 is also consistent with steric hindrance caused by nearby Cu(II) binding in the monomer and oligomers. Others have reported that Cu(II) binding induces a cis-to-trans backbone isomerization of Pro32. In the crystal structures of the P32A mutant dimer and the dimeric unit in the H13F mutant hexamer, which both show this cis-trans isomerization, Ser33 has low accessibility to solvent (20,21). Persistence of this structural feature in the tetramer due to Cu(II) binding would maintain the microenvironment around Ser33 in such a way that its reactivity does not change. The unchanged reactivity of Ser88 suggests that it remains solvent accessible in the monomer, dimer, and tetramer. The reactivity of Arg45 and Arg97 is different than the N-terminus, His31, Ser33, and Ser88. These arginine residues increase in reactivity as the dimer is formed, but then their reactivities level off as the tetramer is formed. This behavior suggests that they decrease in reactivity as the tetramer is formed, thereby counterbalancing their increased reactivity as the dimer continues to populate the solution. Thus, we consider Arg45 and Arg97 as residues that decrease in reactivity as the tetramer is formed.
Including Arg45 and Arg97, 18 residues decrease in reactivity as the tetramer’s concentration increases. Nine of these residues, Arg3, Lys6, Arg12, His13, Lys19, Tyr26, Ser28, Tyr63, and Tyr67, were found previously to decrease in reactivity as the dimer is formed, and their continued decrease in reactivity is consistent with the dimer’s continued increase in concentration over time. The remaining nine residues, Thr4 on the A strand, Lys41 on the C strand, Arg45 on the CD loop, His51 on the D strand, Ser57/Lys58 on the DE loop, Lys91, Lys94, and Arg97 on the G strand, only decrease in reactivity once the tetramer is formed. Because solvent accessibility is the main factor that affects the reactivity of a given amino acid side chain, decreases in reactivity suggests that these latter nine amino acids may mediate interactions in the tetramer.
Even though no crystal structure of a wild-type or mutant β2m tetramer is available to which we can compare our covalent labeling data, several lines of evidence help us arrive at a model for the pre-amyloid tetramer of the wild-type protein. (1) Previous reports have shown that the β2m oligomers are native-like in structure (41), indicating that the tetramer will have many of the structural features present in the wild-type monomer for which several crystal structures exist (42,43). (2) Previous covalent labeling experiments from our group give insight into the structure of the wild-type β2m dimer, which has essentially the same structural features as the dimer unit formed by the B and C chains of the H13F mutant hexamer of β2m (22). This dimer structure (see Figure 1) serves as a convenient starting point for constructing a model of the tetramer since the tetramer very likely arises from the assembly of two dimers. (3) The H13F mutant hexamer contains another oligomeric interface (chains C and D) that could represent the interface of the wild-type tetramer and so our labeling data can be directly compared to the interface of these two chains. (4) The P32A mutant dimer, which was previously ruled out as a possible dimer interface for wild-type β2m, could represent the interface of the wild-type tetramer and our labeling data can be compared to this structure too.
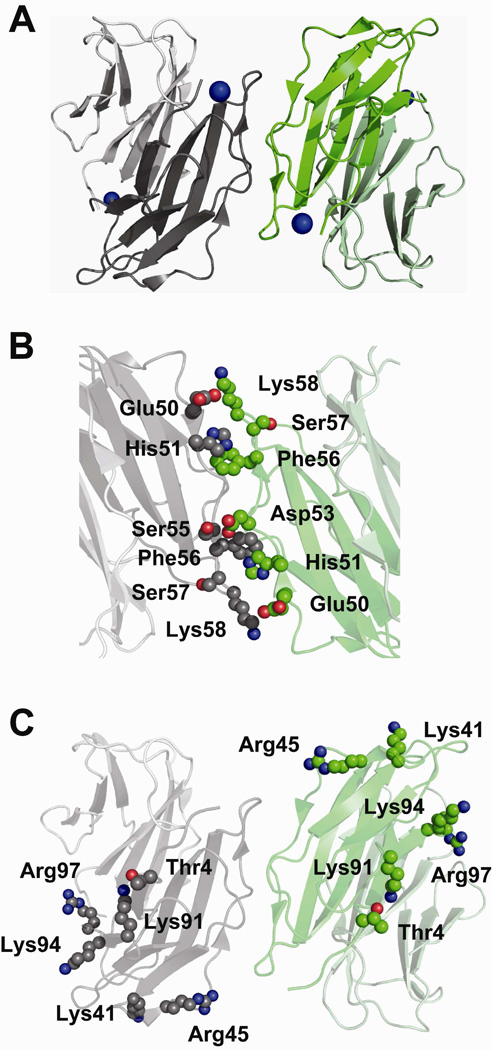
In comparing our labeling data with the oligomeric interface of chains C and D of the H13F hexamer (Figure 6), we find that only three of the nine residues that decrease in reactivity are found at the C–D interface. The interface of these two chains should result in reduced solvent accessibility for His51, Ser57, and Lys58, and we observe experimentally that these residues decrease in reactivity upon tetramer formation. In contrast, the other six residues (i.e. Thr4, Lys41, Arg45, Lys91, Lys94, and Arg97) that decrease in reactivity and thus are likely at the tetramer interface, are all solvent accessible when considering the C–D interface of the H13F hexamer (Figure 6C). This collective observation alone strongly suggests that the interface of the C and D chains of the H13F mutant does not reflect the tetramer interface in wild-type β2m. Indeed, no interface in the H13F hexamer is consistent with our covalent labeling data for the wild-type tetramer. This conclusion is not too troublesome, though, as the H13F hexameric mutant cannot form amyloid fibrils, whereas the wild-type protein can form tetramers, hexamers and eventually amyloid fibrils. Evidently, the wild-type protein forms a tetramer species capable of continuing on to form amyloid fibrils, whereas the H13F mutant cannot form such an interface and therefore stalls at the hexamer and does not progress to form amyloid fibrils.
Figure 6.
Possible tetramer formed by two dimer units with anti-parallel ABED interface as seen in the H13F hexamer (PDB ID: 3CIQ). The side chains involved in the interface are shown as spheres. (A) Possible tetramer formed by D strands and BC loops from two dimer units in the crystal structure of H13F hexamer (PDB ID: 3CIQ). One dimer unit (chains B–C) is shown in gray and the other (chains D–E) is shown in green. Chains C and D, which form a possible tetramer interface, are shown in dark gray and dark green, respectively. (B) Interaction of adjacent D strands. Inter-strand interactions of the side chains of Glu50 and His51 of one monomer with the side chain of Lys58 and the backbone of Phe56, respectively, of another monomer are shown. These interactions make Ser57 less exposed to solvent. (C) Six of the amino acids that decrease in reactivity upon tetramer formation, namely, Thr4, Lys41, Arg45, Lys91, Lys94, and Arg97, are solvent accessible in the H13F structure.
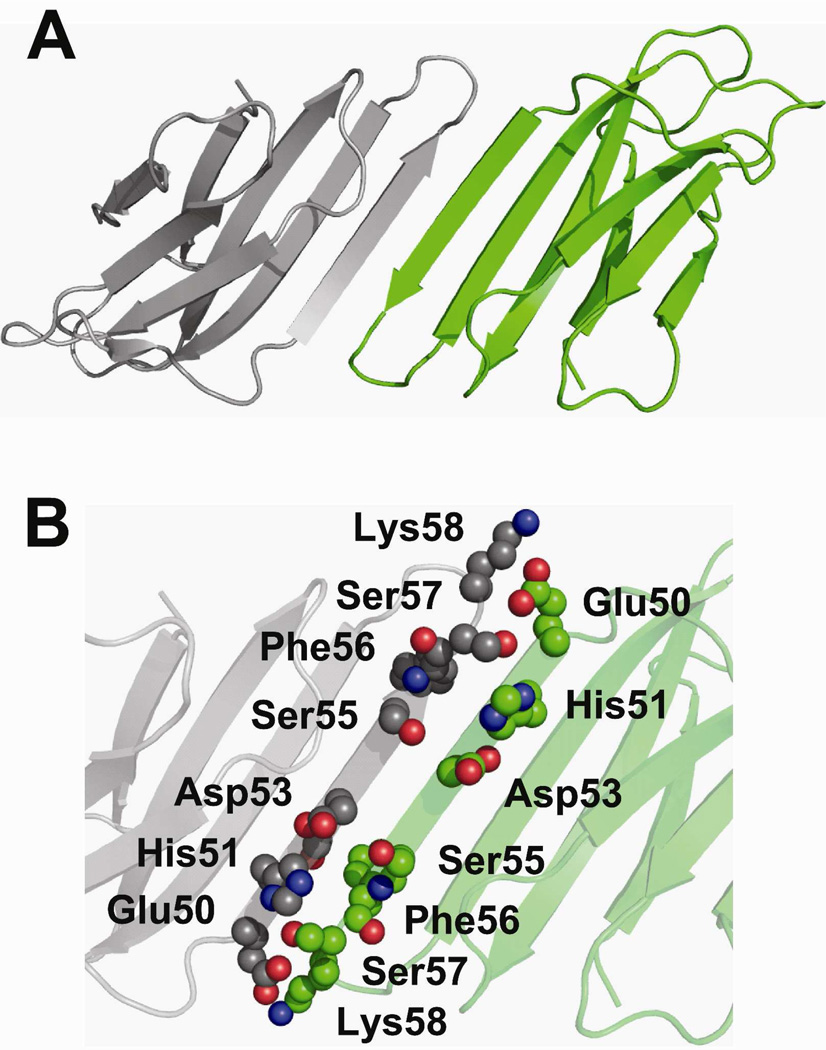
The P32A mutant also forms an oligomeric interface with which we can compare our labeling data (Figure 7). Like the interactions seen in the C–D chains of the H13F hexamer, the interaction of the anti-parallel D strands in P32A span primarily the amino acids Glu50-Lys58 (Figure 7B). These interfaces predict reduced solvent accessibility of His51 on the D strand and Ser57 and Lys58 on the DE loop. These interactions protect residues His51, Ser57, and Lys58 from solvent, which possibly explains why we see a decrease in the reactivity of these residues. Like the H13F mutant, though, Thr4, Lys41, Arg45, Lys91, Lys94, and Arg97 are solvent accessible in the P32A dimer, which is not consistent with the observed decrease in their reactivity upon tetramer formation. This result indicates that an interaction involving D strands from two protein sub-units does not represent the tetramer interface in wild-type β2m.
Figure 7.
Interface formed by D-D strand interactions in P32A. (A) Crystallographic dimer formed by P32A (PDB ID: 2F8O). (B) Amino acids Glu50-Lys58 on the D strands are shown as spheres. Inter-strand interactions of the side chains of Glu50 and His51 of one monomer with the side chain of Lys58 and the backbone of Phe56, respectively, of another monomer are shown.
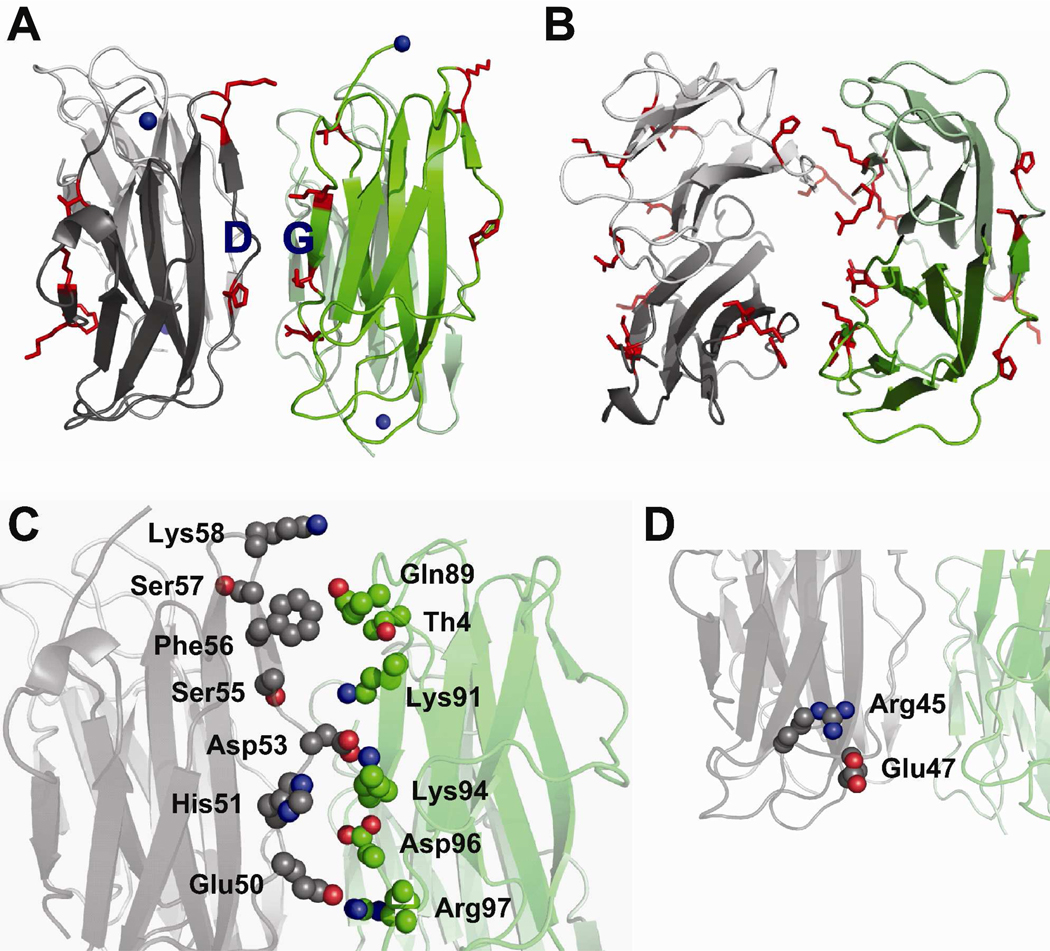
In arriving at a viable model for the tetramer, it is helpful to map the amino acids that decrease in reactivity upon tetramer formation. In doing so, it is clear that the residues that decrease in reactivity are localized on the edge strands A, D, C and G strands of β2m (Figure 8A). This observation suggests a tetramer interface that involves an anti-parallel arrangement of D and G strands from two dimers (Figure 8). In this tetramer model, two D strands from one dimer unit and two G strands from an adjacent dimer form the interface (Figure 8A and 8B). More specifically, a D strand from one monomer in a dimer and a D strand from the other monomer in the same dimer interact with the G strands from each of the monomers the other dimer. Analysis of such a model, which was computationally constructed and energy minimized as described in the experimental section, reveals that seven out of the nine residues that drop in reactivity as the tetramer is formed are found at the interface of the D and G strands of adjacent dimers. These seven residues are Thr4, His51, Ser57/Lys58, Lys91, Lys94, and Arg97 (Figure 8C). The amino acid that increases in reactivity upon tetramer formation, Lys75, is solvent accessible in the tetramer model. In this model, His51, Lys58, Lys91, Lys94, and Arg97 are involved in mediating the interactions at the tetramer interface, whereas Ser57 is protected from solvent by these interactions and Thr4 is enclosed by the D and G strands. Glu50 and His51 form salt bridges with Arg97 and Asp96, respectively. Such electrostatic interactions can substantially enhance the stability of protein-protein complexes (44,45), and the involvement of His51 in one of these interactions is consistent with a study by Blaho and Miranker in which they found from mutagenesis studies that His51 is a critical residue in mediating the interface (46). Asp53 is in close proximity to both Lys91 and Lys94, indicating that this residue can form a complex salt bridge with both of these lysine residues. Such complex salt bridges at protein-protein interfaces contribute significantly to protein complex stability (47–49). In addition, Lys58 is in close enough proximity to hydrogen bond with Gln89. Moreover, several hydrophobic residues are buried in this model structure, including Pro32, Leu54, Phe56, Ile92, and Val93 (residues not shown). In this model, Arg45 decreases in reactivity but is not buried at the interface. In this case, the orientation of the D strand is altered such that Arg45 can form a salt bridge with Glu47 (Figure 8D), which could explain its decreased reactivity with BD. These interactions might also alter the orientation of the D strand such that Lys41 becomes more protected from solvent than in the dimer, and thus decreases in reactivity upon tetramer formation.
Figure 8.
Proposed model for the wild-type β2m tetramer. (A) Tetramer formed by the interaction of two D strands from one dimer unit (dark and light gray) and two G strands of another dimer unit (dark and light). The probed amino acids that decrease in modification upon tetramer formation (shown as red sticks) are located on the edge strands, A, C, D and G. (B) Top-view of the tetramer model. (C) Amino acids Thr4 on the N-terminal strand, Glu50-Lys58 on the D strand, and Gln89-Arg97 on the G strand are shown as spheres. Several interactions are shown: salt bridge between Glu50 and Arg97, salt bridge between His51 and Asp96, complex salt bridge between Asp53 and Lys94 and Lys91, and H-bonding between Gln89 and Lys58. (D) These interactions alter the orientation of the D strand such that Arg45 can form a salt bridge with Glu47.
An interesting feature of our proposed model for the pre-amyloid tetramer is the likely ability of this tetramer to further propagate into higher order oligomers. In our previous studies, we found that the tetramer can progress to form hexamers. As shown in Figure 8, interaction of D strands of one dimer and the G strands of a second dimer to form the tetramer leaves the G strands of the first dimer and the D strands of the second dimer available for further interaction with another dimer unit. Such unfulfilled interactions would presumably allow formation of a hexamer via another D–G interaction. Covalent labeling studies of the hexamer are beyond the scope of the present work. Such studies are on-going, though, and will be reported in the future.
Finally, it is tempting to compare our tetramer model with the structural information obtained from recent solid-state NMR data of β2m amyloid fibrils produced at low pH (50–52), especially since cryo-electron microscopy data indicate that the fibrils are assembled as a “dimer of dimers” (53). Such a comparison is challenging, however, because fibrils produced under different conditions can have structural variations at the molecular level (51,54). Furthermore, NMR data from one study suggest a β-sheet core that preserves β2m’s overall native structure (50), while more recent NMR studies indicate a core structure that is more non-native (51,52). If the latter is correct, then substantial structural reorganization may occur in going from the oligomeric states to the fibrils. Thus, a thorough comparison of our proposed tetramer with β2m fibril structure will have to await further characterization (e.g. solid-state NMR, labeling) of the fibrils produced by incubation with Cu(II).
SUMMARY AND CONCLUSIONS
Using a combination of selective covalent labeling and mass spectrometric detection, we identify structural features of the pre-amyloid tetramer of β2m, which is generated by incubation with Cu(II) under physiologically relevant conditions. The covalent labeling data allow us to develop a model for the tetramer in which the interface is mediated by interactions between D strands of one dimer unit and the G strands of another dimer unit. Our model, therefore, differs from previous suggestions that the tetramer is mediated by interactions between D strands on separate dimer units. Covalent labeling data that covers about 30% of the amino acids that are at the interface of the proposed tetramer structure provide strong support for our model. Future studies will further test this model, and more structural information about this pre-amyloid tetramer might be useful for the development of therapeutics against DRA. More broadly speaking, covalent labeling along with MS detection should have applicability to other amyloid-forming systems, especially those in which mixtures of pre-amyloid oligomers are formed. This approach could then take on greater importance since in several amyloid diseases the pre-amyloid oligomers may represent the species responsible for cellular toxicity rather than the amyloid fibrils themselves (55,56).
Supplementary Material
ABBREVIATIONS
- β2m
β-2-microglobulin
- MS
mass spectrometry
- DRA
dialysis-related amyloidosis
- MHC-I
class I major histocompatibility complex
- NHSA
sulfo N-hydroxysuccinimide acetate
- DEPC
diethylpyrocarbonate
- BD
2,3-butanedione
- SEC
size exclusion chromatography
- LC-MS
liquid chromatography-MS
- ESI-MS
electrospray ionization-MS
Footnotes
This work was supported by the National Institutes of Health (R01 GM075092).
SUPPORTING INFORMATION AVAILABLE
Plots illustrating the intensities of unmodified and modified forms of fragment Val27-Lys41 2 hours and 2.5 days after addition of Cu, intensities of unmodified and modified forms of fragment Tyr67–Tyr78 2 hours and 2.5 days after addition of Cu, formation of oligomers over time, extent of DEPC modification of a control β2m solution (i.e. no Cu(II)) in the absence and up to 2 days after addition of Cu(II), extent of NHSA modification of β2m residues throughout the course of the tetramer formation, plots showing the extent of DEPC modification of residues throughout the course of the tetramer formation, and plots illustrating the extent of BD modification of Arg residues throughout the course of the tetramer formation can be found in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Floege J, Ehlerding G. Beta-2-microglobulin associated amyloidosis. Nephron. 1996;72:9–26. doi: 10.1159/000188801. [DOI] [PubMed] [Google Scholar]
- 2.Okon M, Bray P, Vucelic D. H1-NMR assignments and secondary structure of human beta-2-microglobulin in solution. Biochemistry. 1992;31:8906–8915. doi: 10.1021/bi00152a030. [DOI] [PubMed] [Google Scholar]
- 3.Ayers DC, Athanasou NA, Woods CG, Duthie RB. Dialysis arthropathy of the hip. Clin. Orthop. Relat. Res. 1993;290:216–224. [PubMed] [Google Scholar]
- 4.Menaa C, Esser E, Sprague SM. β2-microglobulin stimulates osteoclast formation. Kidney Int. 2008;73:1275–1281. doi: 10.1038/ki.2008.100. [DOI] [PubMed] [Google Scholar]
- 5.Keating MJ. Chronic lymphocytic leukemia. Semin. Oncol. 1999;26:107–114. [PubMed] [Google Scholar]
- 6.Malaguarnera M, Restuccia S, Di Fazio I, Zoccolo AM, Trovato BA, Pistone G. Serum beta-2-microglobulin in chronic hepatitis C. Dig. Dis. Sci. 1997;42:762–766. doi: 10.1023/a:1018855928753. [DOI] [PubMed] [Google Scholar]
- 7.McParland VJ, Kad NM, Kalverda AP, Brown A, Kirwin-Jones P, Hunter MG, Sunde M, Radford SE. Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry. 2000;39:8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 8.Esposito G, Michelutti R, Verdone G, Viglino P, Hernandez H, Robinson CV, Amoresano A, Dal Piaz F, Monti M, Pucci P, Mangione P, Stoppini M, Merlini G, Ferri G, Bellotti V. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 2000;9:831–845. doi: 10.1110/ps.9.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Relini A, Canale C, De Stefano S, Rolandi R, Giorgetti S, Stoppini M, Rossi A, Fogolari F, Corazza A, Esposito G, Gliozzi A, Belloti V. Collagen plays an active role in the aggregation of β2-microglobulin under physiopathological conditions of dialysis-related amyloidosis. J. Biol. Chem. 2006;281:16521–16529. doi: 10.1074/jbc.M513827200. [DOI] [PubMed] [Google Scholar]
- 10.Ohhashi Y, Kihara M, Naiki H, Goto Y. Ultrasonication-induced amyloid fibril formation of β2-microglobulin. J. Biol. Chem. 2005;280:32843–32848. doi: 10.1074/jbc.M506501200. [DOI] [PubMed] [Google Scholar]
- 11.Morgan CJ, Gelfans M, Atreya C, Miranker AD. Kidney dialysis-associated amyloidosis: a molecular role for copper in fiber formation. J. Mol. Biol. 2001;309:339–345. doi: 10.1006/jmbi.2001.4661. [DOI] [PubMed] [Google Scholar]
- 12.Villanueva J, Hoshino M, Katou H, Kardos J, Hasegawa K, Naiki H, Goto Y. Increase in the conformational flexibility of β2-microglobulin upon copper binding: a possible role for copper in dialysis-related amyloidosis. Protein Sci. 2004;13:797–809. doi: 10.1110/ps.03445704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antwi K, Mahar M, Srikanth R, Olbris MR, Tyson JF, Vachet RW. Cu(II) organizes beta-2-microglobulin oligomers but is released upon amyloid formation. Protein Sci. 2008;17:748–759. doi: 10.1110/ps.073249008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bush AI, Tanzi RE. The galvanization of β-amyloid in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2002;99:7317–7319. doi: 10.1073/pnas.122249699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human α-synuclein. A possible molecular link between Parkinson’s disease and heavy metal exposure. J. Biol. Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 16.Jobling MF, Huang X, Stewart LR, Barnham KJ, Curtain C, Volitakis I, Perugini M, White AR, Cherny RA, Masters CL, Barrow CJ, Collins SJ, Bush AI, Cappai R. Copper and zinc binding modulates the aggregation and neurotoxic properties of the prion peptide PrP106-126. Biochemistry. 2001;40:8073–8084. doi: 10.1021/bi0029088. [DOI] [PubMed] [Google Scholar]
- 17.Wadsworth JD, Hill AF, Joiner S, Jackson GS, Clarke AR, Collinge J. Strain-specific prion-protein conformation determined by metal ions. Nat. Cell. Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 18.Davis DP, Gallo G, Vogen SM, Dul JL, Sciarretta KL, Kumar A, Raffen R, Stevens FJ, Argon Y. Both the environment and somatic mutations govern the aggregation pathway of pathogenic immunoglobulin light chain. J. Mol. Biol. 2001;313:1021–1034. doi: 10.1006/jmbi.2001.5092. [DOI] [PubMed] [Google Scholar]
- 19.Calabrese MF, Miranker AD. Formation of a stable oligomer of β-2-microglobulin requires only a transient encounter with Cu(II) J. Mol. Biol. 2007;367:1–7. doi: 10.1016/j.jmb.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 20.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nature Struct. Mol. Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 21.Calabrese MF, Eakin CM, Wang JM, Miranker AD. A regulatable switch mediates self-association in an immunoglobulin fold. Nature Struct. Mol. Biol. 2008;15:965–971. doi: 10.1038/nsmb.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendoza VL, Antwi K, Baron-Rodriguez MA, Blanco C, Vachet RW. Structure of the preamyloid dimer of beta-2-microglobulin from covalent labeling and mass spectrometry. Biochemistry. 2009;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fliss H, Viswanatha T. 2,3-Butanedione as a photosensitizing agent – Application to alpha-amino acids and alpha-chymotrypsin. Can. J. Biochem. 1979;57:1267–1272. doi: 10.1139/o79-168. [DOI] [PubMed] [Google Scholar]
- 24.Riordan JF. Arginyl residues and anion binding sites in proteins. Mol. Cell Biochem. 1979;26:71–92. doi: 10.1007/BF00232886. [DOI] [PubMed] [Google Scholar]
- 25.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen R, Weng Z. Docking Unbound Proteins Using Shape Complementarity, Desolvation, and Electrostatics. Proteins: Struct., Funct., Genet. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 28.Chen R, Li L, Weng Z. ZDOCK: An Initial-Stage Protein-Docking Algorithm. Proteins: Struct., Funct., Genet. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 29.Cornell W, Cieplak P, Bayly C, Gould I, Merz K, Ferguson D, Spellmeyer D, Fox T, Caldwell J, Kollma P. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 30.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys. J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark A, Berendsen H. Gromacs: Fast, flexible, and free. J. Comp. Chem. 2005;26:1701–1719. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 32.Lindahl E, Hess B, van der Spoel D. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 33.Berendsen H, van der Spoel D, van Drunen R. Gromacs: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 1995;91:43–56. [Google Scholar]
- 34.Verdone G, Corazza A, Viglino P, Pettirossi F, Giorgetti S, Mangione P, Andreola A, Stoppini M, Bellotti V, Esposito G. The solution structure of human β2-microglobulin reveals the prodromes of its amyloid transition. Protein Sci. 2002;11:487–499. doi: 10.1110/ps.29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahoney M, Jorgensen A. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000;112:8911–8922. [Google Scholar]
- 36.Mendoza VL, Vachet RW. Protein surface mapping using diethylpyrocarbonate with mass spectrometric detection. Anal. Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biom. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 38.Guan JQ, Chance MR. Structural proteomics of macromolecular assemblies using oxidative footprinting and mass spectrometry. Trends Biochem. Sci. 2005;30:583–592. doi: 10.1016/j.tibs.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom. Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 40.Srikanth R, Mendoza VL, Bridgewater JD, Zhang G, Vachet RW. Copper binding to β-2-microglobulin and its pre-amyloid oligomers. Biochemistry. 2009;48:9871–9881. doi: 10.1021/bi901172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by β-2-microglobulin. Biochemistry. 2004;43:7808–7815. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 42.Trinh CH, Smith DP, Kalverda AP, Phillips SEV, Radford SE. Crystal structure of monomeric human β-2-microglobulin reveals clues to its amyloidogenic properties. Proc. Natl. Acad. Sci. USA. 2002;99:9771–9776. doi: 10.1073/pnas.152337399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kihara M, Chatani E, Iwata K, Yamamoto K, Matsuura T, Nakagawa A, Naiki H, Goto Y. Conformation of amyloid fibrils of β2-microglobulin probed by tryptophan mutagenesis. J. Biol. Chem. 2006;281:31061–31069. doi: 10.1074/jbc.M605358200. [DOI] [PubMed] [Google Scholar]
- 44.Kundrotas PJ, Alexov E. Electrostatic Properties of Protein-Protein Complexes. Biophys. J. 2006;91:1724–1736. doi: 10.1529/biophysj.106.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong F, Zhou H-X. Electrostatic Contribution to the Binding Stability of Protein–Protein Complexes. Proteins. 2006;65:87–102. doi: 10.1002/prot.21070. [DOI] [PubMed] [Google Scholar]
- 46.Blaho DV, Miranker AD. Delineating the conformational elements responsible for Cu2+induced oligomerization of beta-2 microglobulin. Biochemistry. 2009;48:6610–6617. doi: 10.1021/bi900540j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musafia B, Buchner V, Arad D. Complex salt bridges in proteins: Statistical analysis of structure and function. J Mol. Biol. 1995;254:761–770. doi: 10.1006/jmbi.1995.0653. [DOI] [PubMed] [Google Scholar]
- 48.Risal D, Gourinath S, Himmel DM, Szent-Gyorgyi AG, Cohen C. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc. Natl. Acad. Sci. USA. 2004;101:8930–8935. doi: 10.1073/pnas.0403002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu GZ, Liu RT, Zak O, Aisen P, Chance MR. Structural allostery and binding of the transferrin·receptor complex. Mol. Cell. Proteomics. 2005;4:1959–1967. doi: 10.1074/mcp.M500095-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Barbet-Massin E, Ricagno S, Lewandowski JR, Giorgetti S, Bellotti V, Bolognesi M, Emsley L, Pintacuda G. Fibrillar vs crystalline full-length beta-2-microglobulin studied by high-resolution solid-state NMR spectroscopy. J. Am. Chem. Soc. 2010;132:5556–5557. doi: 10.1021/ja1002839. [DOI] [PubMed] [Google Scholar]
- 51.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Magic angle spinning NMR analysis of beta(2)-microglobulin amyloid fibrils in two distinct morphologies. J. Am. Chem. Soc. 2010;132:10414–10423. doi: 10.1021/ja102775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Intermolecular alignment in beta(2)-microglobulin amyloid fibrils. J. Am. Chem. Soc. 2010;132:17077–17079. doi: 10.1021/ja107987f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White HE, Hodgkinson JL, Jahn TR, Cohen-Krausz S, Gosal WS, Muller S, Orlova EV, Radford SE, Saibil HR. Globular tetramers of beta(2)-microglobulin assemble into elaborate amyloid fibrils. J. Mol. Biol. 2009;389:48–57. doi: 10.1016/j.jmb.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiramatsu H, Lu M, Matsuo K, Gekko K, Goto Y, Kitagawa T. Differences in the molecular structure of β2-microglobulin between two morphologically different amyloid fibrils. Biochemistry. 2010;49:742–751. doi: 10.1021/bi901536j. [DOI] [PubMed] [Google Scholar]
- 55.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo JS, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding disease. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 56.Mendes Sousa M, Cardoso I, Fernandes R, Guimaraes A, Saraiva MJ. Deposition of transthyretin in early stages of familial amyloidotic polyneuropathy. Evidence for toxicity of nonfibrillar aggregates. Am. J. Pathol. 2001;159:1993–2000. doi: 10.1016/s0002-9440(10)63050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.