Abstract
Alternative splicing is a widespread means of increasing protein diversity and regulating gene expression in eukaryotes. Much progress has been made in understanding the proteins involved in regulating alternative splicing, the sequences they bind to, and how these interactions lead to changes in splicing patterns. However, several recent studies have identified other players involved in regulating alternative splicing. A major theme emerging from these studies is that RNA secondary structures play an under appreciated role in the regulation of alternative splicing. This review provides and overview of the basic aspects of splicing regulation and highlights recent progress in understanding the role of RNA secondary structure in this process.
Introduction
The removal of introns and joining together of exons through pre-mRNA splicing is an essential part of eukaryotic gene expression. Shortly after the discovery of introns and split genes [1,2], it was recognized that splicing can be regulated to produce multiple mRNA molecules from a single gene. Although it was initially expected to be rare, alternative splicing has since been found to occur in plants, animals, and fungi. Because many alternative splicing events are tissue-specific, this process plays an important role in cellular differentiation and organismal development.
By allowing single genes to encode mRNA for multiple proteins, alternative splicing has a dramatic impact on the amount of information encoded in the transcriptome. There is a general correlation between organismal complexity and the proportion of genes which are alternatively spliced. For example, alternative splicing has been found in ~25% of C. elegans genes [3-5], ~60% of D. melanogaster genes [6], and ~95% of Human genes [7,8]. Similar studies have found alternative splicing affecting a large fraction of many plant transcriptomes, with ~42% and 56% of genes in A. thaliana [9] and Z. mays [10]. Thus organisms with more tissue and cell types tend to have more alternative splicing.
The great impact of alternative splicing on the proteome suggests a very complex regulatory regime. Over the last three decades, much has been learned regarding the biochemical mechanisms that regulate alternative splicing. In this review, we will give a brief overview of the most common splicing regulatory mechanisms, and highlight recent discoveries which suggest that pre-mRNA structures may play a more important role in regulating splicing than previously appreciated.
The basics of splicing regulation
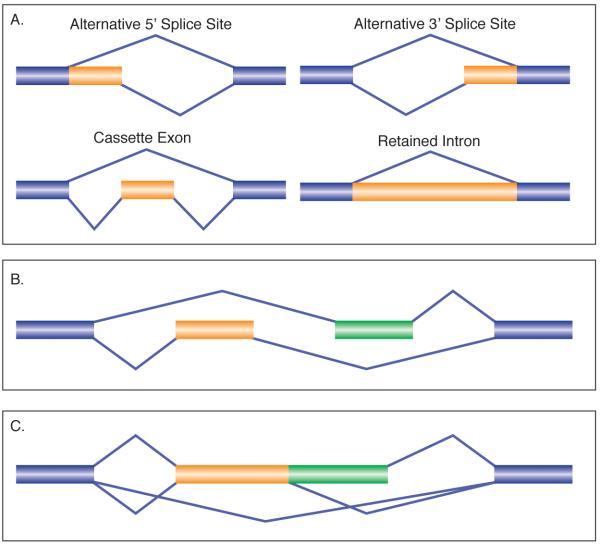
All alternative splicing events can be grouped into one of four categories: alternative 5′ splice sites, alternative 3′ splice sites, cassette exons, and retained introns (Figure 1A, types of alternative splicing). More complex splicing events can be made by combining two or more of these events together. For example, a mutually exclusive splicing event (Fig. 1B) is composed of two cassette exons that are included in a mutually exclusive manner. Similarly, an event such as that shown in Fig. 1C, involves a cassette exon and an alternative 5′ splice site and can therefore result in one of two different exons included in the transcript. Along these same lines, genes can evolve complex splicing patterns that can give rise to many isoforms by using these splicing events at multiple locations in the gene.
Figure 1.
Types of alternative splicing. A. The basic building blocks of all types of splicing events are depicted. Alternative 5′ splice sites; alternative 3′ splice sites; Cassette exons; Retained introns. B. An example of a mutually exclusive splicing event which is built from two cassette exons. C. An example of a complex splicing event containing a cassette exon and an alternative 5′ splice site.
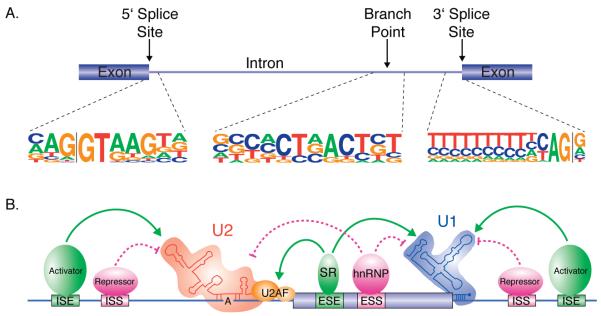
Many cis-acting sequences are involved in pre-mRNA splicing, some of which direct the splicing reaction, and others that regulate alternative splicing (Fig. 2). The most basic of these sequences are those that direct splicing (Fig. 2A). For example, the 5′ splice site has the sequence AG/GURAGU (where “/” designates the splice site), and the 3′ splice site contains a polypyrimidine tract followed by an AG dinucleotide at the actual 3′ splice site. An additional sequence element upstream of the 3′ splice site, called the branchpoint sequence, encompases the nucleophile for the first step of splicing.
Figure 2.
Basics of the mechanisms of alternative splicing. A. The architecture of a pre-mRNA and the important cis-acting sequence elements that direct the splicing reaction. The consensus sequences for the 5′ splice site, branchpoint and 3′ splice site for human introns is shown. B. Schematic diagram of the sequences and proteins involved in regulating alternative splicing. Four types of regulatory sequences are known: intronic splicing enhancers (ISE), intronic splicing silencers (ISS), exonic splicing enhancers (ESE) and exonic splicing silencers (ESS). The enhancer elements are recognized by activator proteins. Within exons, these activators are most commonly members of the SR protein family. The silencer elements are bound by repressor proteins. Within exons, these repressors tend to be members of the hnRNP protein family. Regardless of their binding location, activators tend to enhance the binding of spliceosomal components to the regulated splice site while repressors tend to inhibit binding or function of the spliceosomal components.
The splicing reaction is catalyzed by the spliceosome, a large multicomponent and highly dynamic complex [11]. The spliceosome consists of 5 small nuclear ribonucleoprotein particles (snRNPs) each of which contains one small nuclear RNA (snRNA) and several proteins. In addition, the spliceosome contains 100-200 non-snRNP protein components. The cis-acting sequences that direct splicing are recognized by various components of the spliceosome. For example, U1 snRNP binds to the 5′ splice site, U2 snRNP binds to the branchpoint, and U2 Auxiliary Factor (U2AF) recognizes the polypyrimidine tract and AG at the 3′ splice site. Most of the splicing regulatory mechanisms that have been described to date act by enhancing or preventing the binding of these factors to the pre-mRNA. However, recent studies also indicate that the components of the spliceosome themselves are able to regulate splicing [12-14].
In addition to the general sequences described above that direct the splicing reaction, several classes of auxiliary regulatory signals have been defined that function to regulate alternative splicing [15]. These are broadly categorized based on their location (in exons or introns) and their effects on splicing (enhancers or silencers), resulting in exon splicing enhancers (ESEs) and silencers (ESSs), and intron splicing enhancers (ISEs) and silencers (ISSs). To add to the complexity, the functions of these regulatory elements can depend on their locations in a pre-mRNA. Thus an ESE can also act as an ISS, depending mainly on whether it is located in an exon or and intron, respectively.
There are also a large number of RNA binding proteins that function in splicing regulation. The two largest families are SR proteins and hnRNPs [15]. SR proteins usually promote splicing, while hnRNPs are usually inhibitors. These proteins can be regulated both at their expression level and post-translationally through signaling networks to modulate their activity in specific tissues and cell-types. The mechanisms of splicing involving these classes of regulatory factors have been the subject of many excellent reviews. We therefore devote the remainder of this review to a less appreciated aspect of splicing regulatory mechanisms – RNA structure.
A structured environment
Diagrams similar to Figure 2B are helpful in thinking about the regulatory mechanisms of specific alternative splicing events. However, these representations are an oversimplification, as pre-mRNAs adopt complex secondary and tertiary structures in vivo [16]. These structures can modulate alternative splicing by altering the function of splicing regulatory elements and proteins [17,18]. The study of these RNA structures has mostly occurred on a case-by-case basis, thus the total impact of pre-mRNA structures on splicing regulation remains to be determined. However, several intriguing examples have been recently reported.
Because initial folding of pre-mRNA occurs co-transcriptionally, much in vivo RNA structure is expected to be local. This expectation, combined with the availability of many local RNA structure prediction programs such as mfold and evoFold [19,20], has led to much research on the effects of local structure on alternative splicing. There are many examples of local pre-mRNA structures that regulate alternative splicing, often by preventing spliceosomal recognition of the 5′ splice site, 3′ splice site, and branch point sequence elements [17]. More recently, several groups have performed genome-wide studies of pre-mRNA structure potential near these core elements. By combining structure predictions with conservation information, Shepard and Hertel [21] found that conserved secondary structures are enriched at alternative splice sites. The genes harboring these elements were functionally enriched for splicing factors, suggesting that the presence of these structures may in turn affect the regulation of genes further downstream in splicing regulatory networks. Another recent study showed that alternative cassette exons are enriched for high GC content that likely promotes stable RNA structures obscuring splice sites [22].
Local RNA structures can also affect the availability of other splicing cis-regulatory elements. One example comes from the EDA exon of the fibronectin gene. This cassette exon contains both ESE and ESS elements. However, Baralle and colleagues found that the function of these elements in recruiting SR proteins is dependent on their structural context [23,24]. These types of effects do not appear to be restricted to specific genes. For example, Hiller et al [25] used folding prediction programs to determine the genome-wide likelihood of known human splicing regulatory elements being single-stranded or in a base-paired state. There was a clear tendency of these elements to remain single stranded, and further experiments showed that sequestration of splicing regulatory elements in RNA structures reduced their regulatory activities. Thus the structural contexts of splicing regulatory elements affect their regulatory activities.
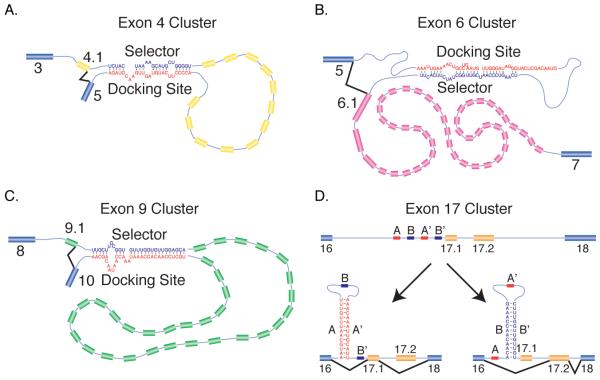
While the above examples highlight the splicing regulatory effects of local RNA structure, long-range interactions can also affect alternative splicing. Perhaps the most striking example of RNA structure in alternative splicing comes from the Drosophila Dscam gene. Dscam encodes a cell adhesion protein important for fly neuronal wiring and immune responses. An incredibly large number of Dscam isoforms, 38,016, can be generated through mutually exclusive splicing of 4 cassette exon clusters [26]. The exon 4, 6, 9, and 17 clusters encode 12, 48, 33, and 2 alternative cassette exons, respectively. The introns within the exon 6 cluster contains highly conserved sequences with secondary structure potential which could span several thousand nucleotides [27,28]. The first intron contains a “docking” sequence and subsequent introns contain selector sequences with varying base-pairing potentials to the docking site (Fig. 3B). This base-pairing is required for inclusion of exon 6 variants, and the strength of this pairing interaction contributes to the frequency of exon 6 variant inclusion [29]. However pairing strength alone does not determine exon 6 variant inclusion frequency, suggesting that other factors have a large effect in Dscam exon 6 inclusion frequencies.
Figure 3.
Secondary structures involved in alternative splicing of the Drosophila Dscam pre-mRNA. A. Mutually exclusive splicing of the exon 4 cluster. Schematic diagram of the basepairing interaction between the selector sequence downstream of exon 4.1 and the docking site upstream of exon 5. This interaction would facilitate splicing between exon 4.1 and exon 5. B. Mutually exclusive splicing of the exon 6 cluster. Schematic diagram of the basepairing interaction between the docking site downstream of exon 5 and the selector sequence upstream of exon 6.1. This interaction would facilitate splicing between exon 5 and exon 6.1. C. Mutually exclusive splicing of the exon 9 cluster. Schematic diagram of the basepairing interaction between the selector sequence downstream of exon 9.1 and the docking site upstream of exon 10. This interaction would facilitate splicing between exon 9.1 and exon 10. D. Mutually exclusive splicing of the exon 17 cluster. Schematic diagram indicating the location of the four sequences (A, A’, B, B’) predicted to be involved in basepairing. When sequences A and A’ pair (left) exon 17.1 would be included while when sequences B and B’ pair (right) exon 17.2 would be included.
Initially, the intricate competing RNA secondary structure mechanism present in the Dscam exon 6 cluster appeared to be an anomaly – similarly organized RNA structural elements were not found in the Dscam exon 4 or 9 clusters or in other genes containing large arrays of mutually exclusive exons. However, recently Yang et al. [30] found functionally analogous structures in several clusters of multiple mutually exclusive exons including the exon 4 and 9 clusters of Dscam (Fig. 3A and C). All of these new structural elements differed from the exon 6 structures in that the docking sites are located in the downstream intron of the cluster, while the exon 6 docking site is in the upstream intron. Nonetheless, in all cases, each variable exon contains a selector sequence in the adjacent intron that can basepair with the docking site in a mutually exclusive manner. Importantly, the sequences of the docking sites and selector sequences differ significantly between clusters indicating that the basepairing potential rather than the sequence of these elements is important. Moreover, the discovery of these elements in 6 other clusters from 3 genes indicates that the mechanism initially discovered to ensure mutually exclusive splicing of the Dscam exon 6 cluster is not an anomaly, but rather appears to be a common mutually exclusive splicing mechanism.
Dscam contains other RNA structures involved in alternative splicing. First, basepairing between two conserved structural elements called the iStem located in the first intron of the exon 4 cluster has been shown to be required for exon 4 inclusion [31]. Second, though the exon 17 cluster contains only two mutually exclusive exons, conserved sequence elements have been identified that could explain the mutually exclusive splicing of these exons [28]. The intron upstream of the first exon 17 variant contains four highly conserved sequences that can basepair in a mutually exclusive manner (Fig. 3D). In one configuration the 3′ splice site of exon 17.1 would be accessible and splicing could occur to this exon. However, in the other configuration, where sequence B pairs with sequence B’, the 3′ splice site of exon 17.1 would be sequestered and exon 17.2 would be included. These two structural elements, along with those previously described, highlight the important role of RNA structure in alternative splicing and pre-mRNA structure.
Long-range structures may form more frequently and play a larger role in regulating alternative splicing than currently appreciated. In support of this, Raker et al. [32] found evidence for 202 long range structures in Drosophila introns. Mutation of the sequences in these proposed structures led to changes in the alternative splicing of their respective genes. While many of these sequences were conserved across ~40 million years of evolution, other sequences are not so deeply conserved. For example, the RNA structures identified by Yang et al. [30] involved in mutually exclusive splicing were typically clade specific and not universally conserved among Drosophilids. Similarly, the iStem involved in Dscam exon 4 splicing is conserved at the sequence level in species closely related to D. melanogaster, but only at the structural level in more distantly related Drosophilids [31]. These results suggest that higher-resolution comparative genomics may be necessary to reveal structurally conserved intronic sequences.
Some splicing regulatory mechanisms involve interactions between RNA structures and splicing regulatory proteins. For example, mutually exclusive splicing of Dscam exon 6 requires repression of all but one exon, such that repression is likely the default state. Olson et al. showed that the RNA binding protein hrp36 is required to repress the inclusion of multiple exon 6 variants, while several SR proteins likely promote inclusion [33]. Depletion of hrp36 allows multiple inclusion of exon 6 variants. Through an as of yet unknown mechanism, the docking site-selector sequence RNA structure are proposed to lead to the removal of hrp36 and activation of exon 6 splicing. Another example of dynamic interactions between RNA structure and splicing regulatory proteins comes from the human cardiac troponin T (cTNT) gene [34]. Splicing of cTNT exon 5 is regulated by the RNA binding protein MBNL1, and this activity requires MBNL1 binding at the 3′ end of the upstream intron. Intriguingly, MBNL1 binding stabilizes a local RNA hairpin structure, which in turn blocks the association of the splicing factor U2AF65. This dynamic interplay between splicing regulatory proteins and RNA structural elements may play a larger role in splicing regulation than currently known.
RNA structures can also change in response to binding small molecules, and this may be an important mechanism of splicing regulation. The recent discovery that riboswitches participate in alternative splicing is one of the most surprising examples of RNA structure-mediated splicing regulation. Riboswitches are metabolite-sensing small RNAs whose structure changes in response to binding specific small molecules. These structural changes then mediate changes in transcription and translation in response to the bound metabolite. Although nearly all identified riboswitches appear to be restricted to bacteria, recent studies have identified thiamine pyrophosphate (TPP) binding riboswitches in fungi [35], algae [36], and plants [37]. In contrast to their bacterial counterparts, eukaryotic TPP riboswitches affect gene expression by regulating alternative splicing. These changes in alternative splicing in response to TPP binding in turn alter translational regulation [38]. Thus alternative splicing can be regulated through RNA structural responses to both regulatory protein and small molecule binding.
The above examples underscore the importance of pre-mRNA secondary structure in specific alternative splicing events. However, the full affects of these structures is difficult to asses because their in vivo conformations are largely unknown. While RNA folding algorithms are helpful in identifying potential secondary structures, they often result in multiple structures of equivalent stability. In some cases comparative genomics can improve structure models, but the rapid evolution of intronic sequence limits the usefulness of this approach. Furthermore, there may be multiple conformers for a pre-mRNA in different tissues, dependent upon the presence of tissue-specific RNA binding proteins. Ultimately, higher resolution structures of pre-mRNAs would be needed to help understand the relationship between structure and function in splicing. PARS [39] and FragSeq [40] use enzymatic structure probing and high-throughput sequencing to study RNA structures. This approach and comparative genomics using more closely related species should reveal more about pre-mRNA structure, which may prove invaluable to understanding alternative splicing regulation.
Towards cracking the code
There are two main goals in alternative splicing research - predicting alternative splicing regulation from sequence alone (commonly called the “splicing code”), and determining the molecular mechanisms of alternative splicing. The former would be incredibly useful towards understanding the splicing regulatory effects of human polymorphisms, while the latter is necessary to attempt to design treatments for splicing related diseases. Barash et al. [41] took an integrative approach towards the first goal. By combining known and predicted sequence motifs with primary structural information, this group was able to make accurate predictions of cassette exon usage in four major mouse tissue types. The success of this approach suggests enormous promise for future studies of the splicing code. However, these predictions are currently limited to only one of the four basic types of alternative splicing, in very broadly categorized tissue types, and many alternative splicing events occur in other tissues or in specialized cell types. Given the multitude of splicing regulatory roles played by RNA structural elements, integration of more structural data may allow for substantial improvements in future iterations of the splicing code.
Conclusions
In summary, the regulation of alternative splicing is much more complex than was initially imagined. Although much progress has been made using gene-of-interest models to understand different alternative splicing mechanisms, there is still much that remains unknown. One of the biggest unknowns is how RNA structure affects alternative splicing globally. As the research community builds more sophisticated models of splicing regulation, we may find a much larger role for local and long-range RNA structures. Much remains to be done before we can truly understand how RNA sequences, structures, and regulatory proteins converge to direct splicing events in specific tissues and cell types. In the coming years, we anticipate more systems-level approaches towards understanding splicing regulation, especially in regards to the effects of RNA-structure.
Acknowledgements
The authors are grateful to members of the Graveley laboratory for fruitful and stimulating conversations. This work was supported in part by a grant from the National Institutes of Health to BRG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 3.Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 2011;21:342–348. doi: 10.1101/gr.114645.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- 11.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript Specificity in Yeast Pre-mRNA Splicing Revealed by Mutations in Core Spliceosomal Components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011;25:373–384. doi: 10.1101/gad.2004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••21.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–1469. doi: 10.1261/rna.1069408. In this study, the authors found that the potential for secondary structure near splice sites was higher at alternative splice sites compared to constitutive ones.
- 22.Zhang J, Kuo CJ, Chen L. GC content around splice sites affects splicing through pre-mRNA secondary structures. BMC Genomics. 2011;12:90. doi: 10.1186/1471-2164-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buratti E, Muro AF, Giombi M, Gherbassi D, Iaconcig A, Baralle FE. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol Cell Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muro AF, Caputi M, Pariyarath R, Pagani F, Buratti E, Baralle FE. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol Cell Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- ••27.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. This study first identified highly conserved sequences with the potential to base-pair across the Dscam exon 6 cluster. Pairing of these sequences could function to ensure mutually exclusive splicing.
- 28.Anastassiou D, Liu H, Varadan V. Variable window binding for mutually exclusive alternative splicing. Genome Biol. 2006;7:R2. doi: 10.1186/gb-2006-7-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.May GE, Olson S, McManus CJ, Graveley BR. Competing RNA secondary structures are required for mutually exclusive splicing of the Dscam exon 6 cluster. RNA. 2011;17:222–229. doi: 10.1261/rna.2521311. In this study, the function of the docking site and selector sequences in the Dscam exon 6 cluster was tested. The docking site was shown to be essential for exon 6 inclusion and the strength of base-pairing between the dock and each selector sequence was shown to play a role in exon 6 inclusion frequency.
- ••30.Yang Y, Zhan L, Zhang W, Sun F, Wang W, Tian N, Bi J, Wang H, Shi D, Jiang Y, et al. RNA secondary structure in mutually exclusive splicing. Nat Struct Mol Biol. 2011;18:159–168. doi: 10.1038/nsmb.1959. This study used high-resolution comparative genomics to identify potential RNA base-pairing interactions in pre-mRNAs from 6 mutually exclusive cassette exon clusters. The functions of these RNAs were confirmed using a mini-gene transfection system.
- 31.Kreahling JM, Graveley BR. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol. Cell. Biol. 2005;25:10251–10260. doi: 10.1128/MCB.25.23.10251-10260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••32.Raker VA, Mironov AA, Gelfand MS, Pervouchine DD. Modulation of alternative splicing by long-range RNA structures in Drosophila. Nucleic Acids Res. 2009;37:4533–4544. doi: 10.1093/nar/gkp407. This paper describes the identification of hundreds of potential long-distance RNA base-pairing structures in introns.
- 33.Olson S, Blanchette M, Park J, Savva Y, Yeo G, Yeakley JM, Rio DC, Graveley BR. A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol. 2007;14:1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •34.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci U S A. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. This study shows that the RNA-binding protein MBNL1 regulates splicing of cTNT by stabilizing an intronic RNA structure and blocking spliceosome assembly.
- 35.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 36.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci U S A. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- •39.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. In this study the authors show the utility of combining nuclease RNA structure probing with high-throughput sequencing to reveal secondary structure information in yeast mRNA.
- 40.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••41.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. This is the first study to provide an integrative model of the splicing code. In this case, the splicing code is presented as an algorithm that can assign probabilities to cassette exon inclusion in four tissue types using the pre-mRNA sequence alone.