Abstract
Phage-display technology has been widely used for developing tumor-targeting agents. Laser capture microdissection (LCM) has proven to be an accurate method to select specific cells from histological sections. Our goal was to develop a method to combine phage-display with LCM to obtain phage-displayed ligands that bind to selected cells in human solid tumors. Two panning strategies were evaluated and optimized. The first strategy was to pan on patient tissue mounted to LCM slides before LCM occurred. The poor panning output showed that phage did not tolerate the drying conditions during LCM. The second strategy was to pan on tumor cells from the patient tumor tissue that were isolated by LCM. The catapulted tumor cells were transferred to a filter unit which retained cells but allowed rinsing of unbound phage. Six commercially available filter units were evaluated and the one with the lowest nonspecific binding to phage was selected for the panning steps. The smallest number of cells (500) in which panning could be successfully accomplished was also determined. A micropipette system was developed to further decrease background by removing catapulted cells from the filter unit after panning was complete. This left behind nearly all background binding phage in the filter unit. This strategy led to the selection of individual phage antibody clones (5 out of 79 tested) specific for tumor cells of the patient’s cancer tissue. Immunofluorescence staining on tumor tissues from the same patient showed that these clones have selective signals on tumor island cells, while the scFv library only showed low nonspecific signals on tumor tissues. We established a method of panning on a small number of LCM-captured solid tumor specimens. The quick identification of specific phage-displayed antibodies in the cancer tissue of human patients will greatly enhance the therapy and diagnosis of cancer.
Introduction
Phage display is a powerful and widely used method to generate ligands for potential detection and therapy of cancer, Alzheimer’s disease, atherosclerosis, diabetes, and other autoimmune diseases[1]. A phage-display antibody library has billions of antibody candidates generated from human immune cells. After incubation of a phage-display library with any interested target, binding antibodies can be recovered while non-binding antibodies are rinsed off. In the biopanning step, the selection of ligands to clinically relevant targets is important. Intravenous infusion of a phage library in cancer patients with recovery of phage from surgically removed tumors is the most direct method of panning. We have successfully applied this method with identification of tumor selective ligands[2]. This method is challenging and can only be used for a small subset of patients. An alternative strategy, which largely retains the tumor in the native state, is to pan on tumor tissue that has been surgically harvested and immediately preserved. This strategy has been used to pan on human tissue such as thymic stroma[3], skeletal muscle[4], and breast cancer[5]. However, without guide of morphology, ligands may be selected against undesirable elements such as nonmalignant tumor components. Laser capture microdissection (LCM) is a method that allows accurate selection of specific cell types from histological specimens. Our goal is to develop methods of panning on clinical tumor histological specimens combined with accurate target selection by LCM. These methods should be applicable to any histological specimen and may speed up the process of identifying tumor selective ligands or ligands to any desired subset of cells in a tissue.
Material and Methods
Phage-display library
Two filamentous phage libraries were used. The first was a peptide library we previously constructed in the fUSE5 vector which displays X4CX10CX4 on the pIII protein. EC-1 is a clone from this peptide library that binds to extracellular domain of ErbB2[6]. The second library was Tomlinson I single-chain variable fragment (scFv) library cloned in ampicillin resistant phagemid vector pIT2 that was obtained from MRC, HGMP Resource Centre (Hinxton, Cambridge, UK). The library size is about 1.47 × 108 different scFv fragments. Clone 799 which binds to SK-BR-3 breast cancer cells was isolated by our group by panning of the scFv library on a breast tumor specimen[7].
Cell culture
Human breast carcinoma SK-BR-3 and MCF7 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). SK-BR-3 cells were cultured in McCoy5A medium supplemented with 10%(v/v) fetal bovine serum and 2mM L-glutamine. MCF7 cells were cultured in DMEM medium with 10%(v/v) fetal bovine serum, 2mM L-glutamine and 10μg/ml Bovine Insulin (Sigma).
Human tissue and slides
Human tumor tissues were obtained from cancer patients immediately following therapeutic surgical resection using a protocol approved by the University of Vermont Committees on Human Research. After resection, the tissues were embedded in Optimal Cutting Temperature (OCT) compound (Tissue-Tek, Sakura Finetek U.S.A., Inc., Torrance, CA), and frozen at −80°C. Frozen sections (6μm thick) were cut on a cryostat, mounted onto clean uncoated glass slides and PALM membrane slides, then stored at −80°C until further processing. For paraffin slides, tissues were fixed in 10% paraformaldehyde overnight and processed using standard protocol.
Laser capture microdissection
After or before phage incubation with tissues, LCM was performed using a PALM MicroBeam System (P.A.L.M. Microlaser Technologies, Germany). Tissues to be microdissected were viewed through a video microscope and groups of the target cells were cut by UV laser. The cut tissue was catapulted by the laser beam to a collection tube containing 25μl PBS. About 500 cells were catapulted from frozen or paraffin tissue sections and placed in PBS buffer.
Phage biopanning
Strategy 1: Phage biopanning before LCM
Paraffin sections of SK-BR-3 and MCF7 cells were prepared on PALM membrane slides, xylene-treated to remove paraffin, re-hydrated in serially diluted (100%, 95%, and 75%) ethanol. The tissue sections were placed in PBS containing 0.1% casein blocking solution at room temperature (RT) for 1 hour, washed, and incubated with 1×1012 TU (transducing unit) peptide library or EC-1 clone (as positive control) for 2 hours at RT. The slides were washed three times in PBS to remove unbound phage. Bound phages were recovered directly from specimens by LCM. Preliminary experiments indicated that phage viability was diminished with dry conditions. Attempts to optimize the conditions were done by panning and capturing the specimens in the following condition: 1) ambient humidity, 2) high humidity in an enclosure, and 3) sections covered with varying concentrations of glycerol after rinsing unbound phage. Collected samples were placed in PBS buffer and phages were recovered by infection into the K91-Kan host strain. The infected cells were selected for on a LB-tetracycline plate. The colonies were counted to determine the number of phage recovered after LCM.
Strategy 2: Phage biopanning after LCM
Biopanning directly on the slide followed by LCM resulted in compromised phage viability (see Results), so a strategy was developed to pan on samples after LCM. Several conditions were evaluated in this strategy.
Determination of smallest number of cells in which target binding clones could be enriched with serial panning
We previously established a method to pan on breast cancer cell lines in an ultrafree-MC centrifugal filter unit (Millipore Corporation Bedford, MA) [7]. This method allows panning on as few as 10,000 tumor cells. However, when working on LCM-captured tumor cells, this number is still high. We evaluated the smallest number of cells in which target binding clones could be enriched by panning on SK-BR-3 cells with a mixture of phage library and a positive control clone (799). SK-BR-3 cells, 500 or 10,000 cells were incubated in the Millipore centrifuge filter units with a mixture of 1×10 8 TU Tomlinson I scFv library and 1×104 TU phage clone 799. The incubation, rinsing, and phage recovery steps were performed as described earlier[7]. Five rounds of panning were performed and individual phage colonies were randomly selected from the 5th panning output for DNA sequence analyses.
Evaluation of background phage binding to filter cups
Six commercially available filter units were evaluated for the lowest level of nonspecific (background) phage binding (see Table 1). The wells used for panning in 96 filter units were also called as filter cups in the following text. The Tomlinson scFv library (1×1012 TU) was incubated with 1% casein block in each filter unit with no specimen for 2 hours at RT. The filter units were rinsed 10 times with 0.6ml TBST ( TBS, 50 mM Tris-HCl buffer, pH 7.4, 150 mM NaCl, containing 0.1% Tween-20 ) and once with TBS. Phages were eluted with 100μl elution buffer (1mg/ml Trypsin in 50mM Tris-HCl pH 7.4, 1mM CaCl2) by incubation for 15 minutes at RT. The eluted samples were used for infecting TG1 bacteria (OD=0.4). Plating and amplification of phage were done based on MRC protocol (http://www.geneservice.co.uk/products/proteomic/datasheets/tomlinsonIJ.pdf), using KM13 helper phage[8] and TG1 E.coli bacteria.
Table 1.
Characteristics and background phage binding to six blank commercially available filter units
| Name | ULTRAFREE- Centrifugal MC Filter Units Millipore |
MultiScreenHT S-BV Plate Millipore |
MultiScreenHTS 384 FC opaque, MILLIPORE |
HTS Transwell-96 system CORNING |
MultiScreen- MIC Plates96 well MILLIPORE |
HTS Transwell- 96 system CORNING |
|---|---|---|---|---|---|---|
| Cat# | UFC30DV00 | MSBVN1210 | MZFCN0W10 | 3380 | MAMI C5S 10 | 3388 |
|
Membrane
type |
Hydrophilic PVDF |
Hydrophilic PVDF |
Glass fiber/Polyester mesh |
Polyester | Polycarbonate | Polycarbonate |
|
Pore Size
(μm) |
0.65 | 1.2 | 1.2 | 1.0 | 5.0 | 5.0 |
|
Membrane
area (cm2) |
0.3 | 0.3 | 0.12 | 0.143 | 0.3 | 0.143 |
|
Background
level phage TU/ml |
(1.3±0.9)×106 | (2.2±0.8)×106 | (4.3±0.7)×105 | (1.1±0.1)×104 | (3.1±0.9)×105 | (1.0±0.3)×103 |
Biopanning SK-BR-3 cells after LCM
Starting with the naïve library, serial panning was done 5 times using the selected filter units with SK-BR-3 cells captured by LCM from frozen or paraffin specimens. About 500 cells were collected by LCM and transferred into filter units that had been previously blocked with 1% casein at 4°C overnight. Subtraction of filter binding phage was performed by incubating 1×1012 TU Tomlinson I scFv phage library in an empty filter unit at 4°C overnight. The subtracted library was collected by centrifugation and incubated with the SK-BR-3 cells for 2 hours at RT. The elution, plating, and amplification of phages were performed as described above. At each panning step, 1×1012 TU amplified panning output was used as the panning input for the next round of panning. Individual colonies were randomly selected from the 5th panning output for cell ELISA and DNA sequence analyses.
Using micropipette to transfer samples from filter unit after panning to decrease background phage
A method of removing the tissue specimens from the filter cup after completion of all panning and rinsing steps was established. A micromanipulator was mounted to an inverted microscope stage which allowed XYZ movement of the micropipette. A micropipette with a diameter of 250μm was custom manufactured by the Molecular Physiology and Biophysics department at the University of Vermont. Fluid movement in the pipette was controlled by a semiautomatic fluid controlled system (PV800 Pneumatic PicoPump, World Precision Instruments, Inc. FL). After panning, the micropipette was used to transfer specimens out of the filter cup to a second clean filter cup. This transfer step allowed the very small specimens to be removed from the filter cup used for panning and leave behind background phage bound to the filter cup. Elution, plating, and amplification of phage were done as described earlier.
Biopanning on colon cancer surgical specimen using optimized phage biopanning method
Colon cancer tissue frozen sections were mounted on PALM membrane slides and fixed in 2% paraformaldehyde in PBS at RT for 15 minutes. After rinsing 3 times in PBS, slides were stained with hematoxylin and prepared for LCM. Histologically the specimens consisted of tumor islands intermixed with noncancer stromal elements. Circular sections with diameter around 200μm were captured from tumor island areas of the slide. About 50 tumor cells were contained with each captured section. Ten captured sections with about 500 tumor cells were transferred into filter units. After blocking with 1% casein at 4°C overnight, 1×1012TU scFv library was incubated for 2 hours at RT with tumor island sections. After rinsing, tissue sections were transferred by micropipette to a new filter unit. Elution, plating, and phage recovery steps were done as described above. After one round of panning, randomly selected phage monoclones were amplified for binding assessment.
Immunofluorescence stain (IF) for binding assessment of selected phage clones
Frozen sections from the same tumor specimen that was used for panning were prepared for IF stain. Randomly selected phage monoclones from the panning output were normalized by chemititration[9]. Monoclones were incubated on slide specimens at 4°C overnight. After rinsing, mouse anti-M13 antibody (Amersham) and Goat anti-mouse IgG-AlexaFlour 488 (Molecular Probes) were used to label the phage. The slides were observed under fluorescence microscope. The naïve library was used as a negative control.
DNA sequencing
The vector DNA of each selected clone was purified using QIAprep spin miniprep kit (Qiagen, Inc., Chatsworth, CA) according to manufacturer’s instructions. The pIII sequencing primer, 5-CCC TCA TAG TTA GCG TAA CG-3, designed by Dr. George Smith was used for sequencing the DNA inserts. The sequence reactions were carried out using BigDye Ver.1 Dye Terminator kit (PE Biosystems) by the Vermont Cancer Center DNA Analysis Facility.
Results
Biopanning Strategy 1: Phage biopanning before LCM
LCM dissection on the biopanned slides failed to recover more than an occasional clone. Very few phage were recovered indicating that phages were intolerant of dry conditions beyond 15 minutes. Humidification of slides following phage incubation allowed better recovery of phage but still yielded only 1 phage per 30 to 100 cells.
Generation of 90% humidity around the LCM stage was impractical due to excessive condensation on the microscope parts. Placement of glycerol on the slide after incubation of phage maintained phage viability even up to 6 hours. However, glycerol resulted in many unsuccessful capturing events. In summary, panning first on the slide was unreliable under the experimental conditions evaluated.
Biopanning Strategy 2: Phage biopanning after LCM
Five rounds of panning were done on 0, 500, 10,000 SK-BR-3 cells with scFv library spiked with positive clone 799. For each round of panning, phage were recovered in all three conditions including the blank well. Sequencing of randomly selected monoclones from the final output demonstrated that in the group of 500 cells, 2 of 20 clones were clone 799, while in the 10,000 group, 1 of 20 clones was clone 799. These results demonstrated that there was enrichment of clone 799 in both the 500 and 10,000 cell panning group. This indicated that panning a target as small as 500 cells was feasible. We previously demonstrated that with 10,000 cells or more, background phage binding to a filter unit did not interfere with selection of cell target binding clones. However, background phage bound to the filter unit could become an important issue when the cell number decreases to around 500. Panning results of 6 commercially available empty filter units with no target present yielded phage from all the units (see Table 1). The variation in phage recovered from the different filter units was up to 1000-fold. Corning HTS Transwell-96 system had the lowest nonspecific binding to the phage library and was selected for the subsequent experiments.
Serial biopanning (5 rounds) with the scFv library was done on 500 SK-BR-3 cells captured by LCM from frozen or paraffin specimens, using the filter unit from Corning HTS Transwell-96 system. Cell ELISA assay of the output phage was conducted in the 96-well plates that were made of the same material as the panned filter units. The results showed an enrichment of phage clones with selective binding to the filter unit (data not shown). Pre-incubation of the library with an empty blocked filter unit to subtract filter-binding phage did not prevent subsequent selection of filter-binding phage. Although preceding experiments with 10,000 cells showed some enrichment in the spiked positive clone, the biopanning with naïve library on 500 cells recovered only filter-binding phage. It may be attributed to the very small size of the captured specimen, in present experiment, relative to the size of the filter unit.
In the next set of experiments, after the panning and rinsing steps, a micropipette was used to transfer the cells into a fresh filter cup for phage recovery, leaving behind the phage bound nonspecifically to the primary filter cup. The integrity of captured specimens was robust and maintained integrity during all rinsing steps and micropipette transfer steps. Figure 1A shows the on-slide specimen and Figure 1B shows the empty space after capturing. The diameter of the cut section was about 200 μm. Figure 1C shows the micropipette-transferred specimen in the second filter cup after panning in a primary filter cup. A single panning event on approximately 500 paraffin-embedded captured cells yielded recovery of about 1 phage per cell. This was a 30- to 100-fold increase in phage yield over panning prior to LCM. DNA sequencing of 24 randomly selected clones demonstrated all clones to be unique.
Figure 1.
The cutting and capturing steps of LCM and the appearance of samples in the filter unit after panning, rinsing and transferring steps. Figures (A) and (B) show selected tissue area with about 50 cells that was cut and captured, (C) shows that samples remained intact after panning, rinsing and the micropipette transfer steps. The multitudes of gray dots in C are filter membrane pores.
Panning on colon cancer surgical specimen identified specific phage binders to tumor island
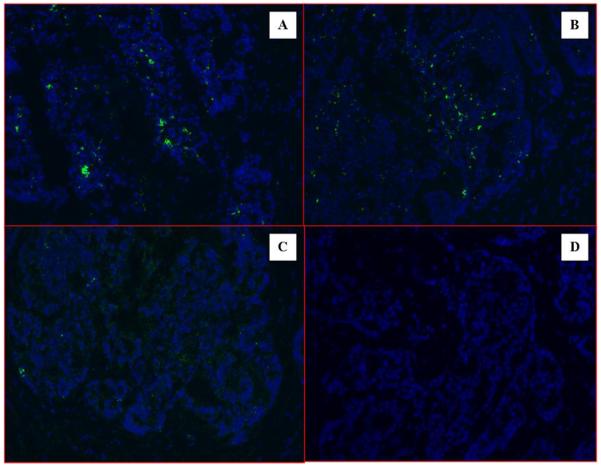
Cancer cells from a human colon cancer tissue slide were preferentially captured avoiding surrounding stroma and necrotic areas. The specimen with about 500 colon cancer cells was panned with the naïve scFv library as described above. Following one round of panning, the specimen was transferred with the help of a micropipette to a fresh filter cup for phage recovery. Seventy-nine clones were selected and DNA sequence data showed they all were unique. The clones were evaluated for binding using IF staining on a histological sample prepared from the same colon cancer tissue. Preferential binding of the selected clones to the tumor island cells and not the stroma was clearly identified (Figure 2). Five unique clones had preferential cancer cell-binding pattern with stronger signals and 9 unique clones had this pattern of binding with weaker signals. A number of clones showed positive binding to both tumor and stroma.
Figure 2.
Immunofluorescence stain of selected scFv phage clones on human patient cancer tissues on frozen slides. Clone T-146 (A) and T-25 (B) show selective signals (green dots) located on tumor islands. The scFv phage library stain (C) shows low nonspecific signals. No stain of tumor tissue was observed on no-phage, only antibody control group (D).
Discussion
Biopanning in a diseased organism may be the ideal situation for panning since the target tissue is in the native state. Cancer tissues that are preserved immediately upon surgical resection are an ideal target, as tissue sections keep the tumor features but avoid exposing the patient to intravenous phage. However, a tumor has a complex array of malignant and nonmalignant cells and panning directly on the specimen will be unselective relative to the various cell types in the tumor. LCM is a method of accurately selecting pure populations of cells from histological specimens[10]. Combining phage panning on tumor tissues in combination with LCM is attractive because of the following: 1) a pure population of desired cells can be selected, 2) the tissue is little changed from the native state, and 3) it is widely applicable since tumor tissue in commonly available for most cancer patients.
The most important technical difficulty in the effort to combine phage display and LCM is the loss of phage viability after LCM. The Liu group (2006) has used phage display combined with laser microdissection system to pan on human prostate cancer tissues[11]. They found that phages were not infectious after micro-dissection and they recovered specimen-bound phage by PCR amplification. Freeze drying was also used to process LCM specimens and this method appeared to maintain phage viability[12]. A third strategy was to use LCM to collect large number of cells (more than ten thousand) and then pan on the LCM recovered cells in immunotubes[13].
We compared two strategies to combine phage display and LCM. Phages were initially incubated directly on the slide-mounted specimen and then selected populations of cells were captured off the slide. Our results showed that phage did not tolerate this process and it appeared to be primarily related to the duration of low moisture during capturing episodes.
Humidification increased the yield of phage but this level of humidification was not compatible with the microscope system. Glycerol maintained phage viability but was incompatible with reliable capturing. Panning after capturing target cells avoided the problems of phage viability. A filter cup allows panning and rinsing with retention of target cells. With a small number of target cells we successfully demonstrated enrichment of a positive clone spiked into the library. However, subsequent serial panning experiments with a naïve library resulted in preferential selection of filter cup binding clones even though subtraction steps were used. The captured specimens had too few phages relative to the number of phages bound to the filter cup.
Micropipette transfer to a new filter cup of the disc-shaped captured samples after panning and rinsing greatly minimized the problem of background phage. Using a 96-well plate with filter wells allowed simple transfer to a new cup in an adjacent well in the next row. Using a colon cancer specimen, malignant colon cancer cells were collected for panning. Panning on about 500 cancer cells yielded about 500 phage clones. Evaluation of individual clones randomly selected after only one panning demonstrated that 18% (14 out of 79) of the clones had some level of selective binding to the tumor island cells and not to the tumor stroma.
In conclusion, we present a new method of panning on a pure population of target cells isolated from a tumor specimen using LCM. The step of micropipette transfer allowed specimen-bound phage to be physically removed from a filter cup, leaving behind filter-bound phage. This allowed selection of phage ligands that bound preferentially to a subpopulation of cells from the colon cancer. The selection was performed using a single panning on only 500 target cells. This method should be broadly applicable to the selection of phage from any preserved tissue.
Acknowledgments
Financial support: This study was supported by the Department of Defense Congressionally Directed Medical Research Program U.S. Army grant W81XWH-05-1-0237, and in part by SD Ireland Professorship of Oncology for Research and Vermont Cancer Venter core grant P30CA022435.
Abbreviations
- LCM
Laser capture microdissection
- OCT
Optimal Cutting Temperature compound
- TU
transducing unit
- IF
Immunofluorescence stain
References
- 1.Smothers JF, Henikoff S, Carter P. Tech.Sight. Phage display. Affinity selection from biological libraries. Science. 2002;298(5593):621–2. doi: 10.1126/science.298.5593.621. [DOI] [PubMed] [Google Scholar]
- 2.Krag DN, et al. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006;66(15):7724–33. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 3.Van Ewijk W, et al. Subtractive isolation of phage-displayed single-chain antibodies to thymic stromal cells by using intact thymic fragments. Proc Natl Acad Sci U S A. 1997;94(8):3903–8. doi: 10.1073/pnas.94.8.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka T, et al. In situ phage screening. A method for identification of subnanogram tissue components in situ. J Biol Chem. 2002;277(33):30382–7. doi: 10.1074/jbc.M203547200. [DOI] [PubMed] [Google Scholar]
- 5.Jakobsen CG, et al. Phage display derived human monoclonal antibodies isolated by binding to the surface of live primary breast cancer cells recognize GRP78. Cancer Res. 2007;67(19):9507–17. doi: 10.1158/0008-5472.CAN-06-4686. [DOI] [PubMed] [Google Scholar]
- 6.Pero SC, et al. Identification of a small peptide that inhibits the phosphorylation of ErbB2 and proliferation of ErbB2 overexpressing breast cancer cells. Int J Cancer. 2004;111(6):951–60. doi: 10.1002/ijc.20306. [DOI] [PubMed] [Google Scholar]
- 7.Shukla GS, Krag DN. Phage display selection for cell-specific ligands: development of a screening procedure suitable for small tumor specimens. J Drug Target. 2005;13(1):7–18. doi: 10.1080/10611860400020464. [DOI] [PubMed] [Google Scholar]
- 8.Goletz S, et al. Selection of large diversities of antiidiotypic antibody fragments by phage display. J Mol Biol. 2002;315(5):1087–97. doi: 10.1006/jmbi.2001.5314. [DOI] [PubMed] [Google Scholar]
- 9.Shukla GS, Krag DN. A sensitive and rapid chemiluminescence ELISA for filamentous bacteriophages. J Immunoassay Immunochem. 2005;26(2):89–95. doi: 10.1081/ias-200051990. [DOI] [PubMed] [Google Scholar]
- 10.Bonner RF, et al. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278(5342):1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 11.Ruan W, et al. Identification of clinically significant tumor antigens by selecting phage antibody library on tumor cells in situ using laser capture microdissection. Mol Cell Proteomics. 2006;5(12):2364–73. doi: 10.1074/mcp.M600246-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Lu H, Jin D, Kapila YL. Application of laser capture microdissection to phage display peptide library screening. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;98(6):692–7. doi: 10.1016/j.tripleo.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Kubo N, et al. Identification of oligopeptide binding to colon cancer cells separated from patients using laser capture microdissection. J Drug Target. 2008;16(5):396–404. doi: 10.1080/10611860802088796. [DOI] [PubMed] [Google Scholar]