Summary
Cancer cells couple heightened lipogenesis with lipolysis to produce fatty acid networks that support malignancy. Monoacylglycerol lipase (MAGL) plays a principal role in this process by converting monoglycerides, including the endocannabinoid 2-arachidonoylglycerol (2-AG), to free fatty acids. Here, we show that MAGL is elevated in androgen-independent versus androgen-dependent human prostate cancer cell lines, and that pharmacological or RNA-interference disruption of this enzyme impairs prostate cancer aggressiveness. These effects were partially reversed by treatment with fatty acids or a cannabinoid receptor-1 (CB1) antagonist, and fully reversed by co-treatment with both agents. We further show that MAGL is part of a gene signature correlated with epithelial-to-mesenchymal transition and the stem-like properties of cancer cells, supporting a role for this enzyme in pro-tumorigenic metabolism that, for prostate cancer, involves the dual control of endocannabinoid and fatty acid pathways.
Cancer cells display alterations in metabolism that provide a biochemical foundation for tumors to progress in their etiology (DeBerardinis et al., 2008a; Kaelin and Thompson, 2010). These changes include aerobic glycolysis (Warburg, 1925), glutamine-dependent anaplerosis (Deberardinis et al., 2008b; Kovacevic and McGivan, 1983), and de novo lipid biosynthesis (Menendez, 2010). Each of these metabolic pathways appears to be important for the transformation of cells from a non-cancerous to a cancerous state; however, much less is understood about the metabolic pathways that confer the aggressive properties observed in malignant cancers, such as high migratory and invasive activity. Since most cancer deaths are related to cancer malignancy and metastasis (Jemal et al., 2010), understanding metabolic pathways that contribute to these pathogenic features is critical to both disease diagnosis and treatment. We recently discovered that monoacylglycerol lipase (MAGL) is elevated in aggressive cancer cells and primary tumors, where this metabolic enzyme regulates a fatty acid network that supports high migratory, invasive, and pro-tumorigenic activity (Nomura et al., 2010).
MAGL also plays an important role in regulating endocannabinoid signaling in the nervous system and some peripheral tissues, where it is responsible for degrading the endogenous cananbinoid receptor (CB1 and CB2) ligand, 2-arachidonoylglycerol (2-AG, C20:4 monoacylglycerol, C20:4 MAG) (Blankman et al., 2007; Chanda et al., 2010; Dinh et al., 2002; Long et al., 2009; Schlosburg et al., 2010). Since direct cannabinoid receptor agonists have been shown to impair cell growth, tumorigenicity, and metastasis (Caffarel et al., 2010; Guzman, 2003; Sarfaraz et al., 2005), it is possible that, at least for certain cancer types, MAGL could also support tumorigenicity by limiting the negative impact of endocannabinoids on cancer cell growth. However, in our studies to date with ovarian, breast, and melanoma cancer cells, we have yet to uncover evidence that endocannabinoid metabolism serves as a basis for the pro-tumorigenic function of MAGL (Nomura et al., 2010), which instead appears to be due principally to the fatty acid products of this enzyme. Prostate cancer cells has been shown to express significant levels of cannabinoid receptors (Sarfaraz et al., 2005), as well as endocannabinoids and their corresponding metabolic enzymes, including MAGL (Bifulco et al., 2008; Chung et al., 2009; Endsley et al., 2008b; Fowler et al., 2010; Mimeault et al., 2003; Nithipatikom et al., 2004; Nithipatikom et al., 2005; Thors et al., 2010; Wang et al., 2008). Cannabinoid receptor agonists have also been shown to impair prostate cancer cell malignancy (Guzman, 2003; Sarfaraz et al., 2005). Nonetheless, whether MAGL plays a pro-tumorigenic role in prostate cancer and, if so, by what mechanism are questions that remain unanswered.
Here we show that MAGL activity is elevated in androgen-independent human prostate cancer cell lines, where it supports migration and tumor growth through a mechanism that involves dual control over tumor-suppressing endocannabinoid and tumor-promoting fatty acid pathways. Furthermore, through global transcriptional profiling of aggressive and non-aggressive human cancer lines across several tumor types, we have found that MAGL is part of a gene expression signature that contains many markers of epithelial-to-mesenchymal transition (EMT) and cancer-stem cells (CSCs) (Mani et al., 2008; Polyak and Weinberg, 2009). Several additional metabolic enzymes are also found as part of this EMT/CSC signature, most of which, interestingly, have not yet been examined for functional roles in cancer.
Results
MAGL activity is elevated in aggressive human prostate cancer cell lines
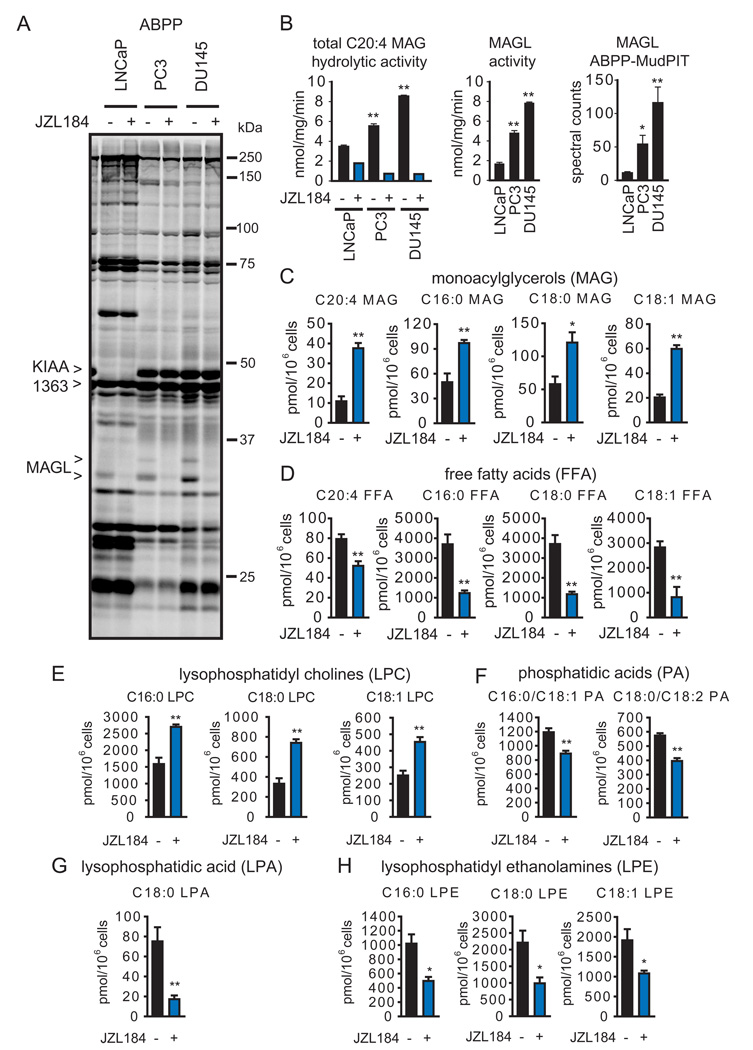
In a recent activity-based protein profiling (ABPP) analysis of a panel of human cancer cell lines (Nomura et al., 2010), we found that MAGL activity was higher in aggressive compared to nonaggressive breast, melanoma, and ovarian cancer cells. Here, we performed a similar ABPP experiment comparing the aggressive (highly migratory and invasive) androgen-independent human prostate cancer lines PC3 and DU145 to the less aggressive (poorly migratory and invasive) androgen-dependent prostate cancer line LNCaP (Hoosein et al., 1991{Chang, 2011 #960). Cell proteomes were treated with the activity-based probes fluorophosphonate (FP)-rhodamine (Patricelli et al., 2001) and FP-biotin (Liu et al., 1999) for gel- and mass spectrometry (MS)-based detection of serine hydrolase activities, respectively. FP-rhodamine-labeled enzymes were visualized by SDS-PAGE (Figure 1A), which identified MAGL and KIAA1363/AADACL1, both migrating as doublets likely due to alternative splicing (Karlsson et al., 2001) and glycosylation (Jessani et al., 2002), respectively, as being elevated in PC3 and DU145 cells compared to LNCaP cells. The FP-rhodamine labeling of both forms of MAGL was blocked by the selective small-molecule inhibitor JZL184 (Long et al., 2009) (1 µM), which showed no detectable cross reactivity with other serine hydrolase activities in PC3 or DU145 cells (Figure 1A). Substrate assays using C20:4 monoacylglycerol (MAG) confirmed that the total and JZL184-sensitive portions of MAGL hydrolytic activity were significantly higher in PC3 and DU145 compared to LNCaP cells (Figure 1B). Notably, a substantial proportion of C20:4 MAG hydrolytic activity in LNCaP cells (~50%) was not sensitive to JZL184, indicating that other serine hydrolases contribute to MAG hydrolysis in these cells (see below).
Figure 1. MAGL is elevated in androgen-independent prostate cancer cells where it regulates monoacylglycerol and free fatty acid metabolism.
(A) ABPP of serine hydrolase activities in the androgen-dependent LNCaP and androgen-independent PC3 and DU145 cells lines. Serine hydrolase activities were labeled in whole cell protoemes with the activity-based probe FP-rhodamine and detected by SDS-PAGE and in-gel fluorescence scanning (fluorescent gel shown in greyscale). MAGL and KIAA1363 are elevated in PC3 and DU145 cells compared to LNCaP cells. Proteomes were also prepared from cancer cells pretreated with DMSO or the selective MAGL inhibitor JZL184 (1 µM, 4 h in situ) to confirm the identities of the 33 and 35 kDa bands as MAGL. (B) The left panel shows C20:4 MAG hydrolytic activity of cancer cells in the presence or absence of JZL184 (1 µM, 4 h in situ). The middle panel shows MAGL-specific activity derived from subtracting the JZL184-insensitive portion from total MAG hydrolytic activity. The right panel shows spectral counts for MAGL in proteomes treated with FP-biotin and subjected to ABPP-MudPIT. (C,D) Inhibition of MAGL by JZL184 (1 µM, 4 h, in situ) raises MAG (C) and lowers FFA (D) levels in PC3 cells. (E–H) JZL184-treated PC3 cells also show elevations in lysophosphatidyl cholines (LPCs) (E), and reductions in phosphatidic acids (PAs) (F), lysophosphatidic acid (LPA) (G), and lysophosphatidyl ethanolamines (LPEs) (H). *p<0.05, **p<0.01 PC3 or DU145 versus LNCaP cells for (B) and JZL184-treated versus DMSO-treated control groups for (C, D). Data are presented as means ± standard error of the mean (SEM); n=4–5/group. See also Table S1, Table S2, and Figure S1.
Prostate cancer cell proteomes were next treated with FP-biotin and probe-labeled enzymes were enriched by avidin chromatography, digested with trypsin, and the resulting peptide mixture analyzed by multidimensional liquid chromatography (LC)-MS. This method, referred to as ABPP-MudPIT (Jessani et al., 2005), identified more than 50 serine hydrolases in prostate cancer cells (Table 1, Table S1). Several of these enzymes, including MAGL (Figure 1B) and KIAA1363, were elevated in activity in PC3 and DU145 cells relative to LNCaP cells as measured by the semi-quantitative method of spectral counting (Liu et al., 2004a). Additional serine hydrolase activities that were higher in PC3 and DU145 included CPVL, DPP9, LYPLA2, PNPLA6, and PPT2 (Table 1). In contrast, the serine hydrolase activities ABHD11, FAAH, LYPLA1, DPP7, and SIAE were enriched in LNCaP. FAAH has previously been shown to possess C20:4 MAG hydrolytic activity and could account for much of the JZL184-insensitive portion of this activity LNCaP cells (Endsley et al., 2008a; Goparaju et al., 1998).
Table 1. ABPP-MudPIT analysis of serine hydrolase activities in human prostate cancer cell lines.
Serine hydrolase activities were filtered for enzymes that displayed: 1) > 10-fold higher spectral counts in FP-biotin-treated proteomes compared to "no-probe" control proteomes for at least one of the cancer cell lines examined, and 2) an average of ≥ 6 spectral counts for at least one of the cancer cell lines examined. Data represent average values ± SEM for 3–4 independent experiments per cell line. See also Table S1.
| LNCaP |
PC3 |
DU145 |
|||||
|---|---|---|---|---|---|---|---|
| Protein | abbreviation | av | sem | av | Sem | av | sem |
| prolyl endopeptidase | PREP | 455 | 64 | 423 | 144 | 132 | 11 |
| FASN Fatty acid synthase | FASN | 1850 | 135 | 1324 | 111 | 1385 | 170 |
| Acyl-protein thioesterase 1 | LYPLA1* | 81 | 5 | 21 | 8 | 55 | 1 |
| Abhydrolase domain-containing protein 10 | ABHD10 | 622 | 132 | 534 | 111 | 252 | 29 |
| Acylamino-acid-releasing enzyme | APEH | 571 | 45 | 642 | 88 | 196 | 32 |
| Probable serine carboxypeptidase CPVL | CPVL* | 0 | 0 | 27 | 4 | 6 | 1 |
| Platelet-activating factor acetylhydrolase 2 | PAFAH2 | 19 | 6 | 22 | 4 | 25 | 4 |
| prolyl endopeptidase-like isoform C | PREPL | 63 | 13 | 98 | 12 | 145 | 25 |
| Acyl-protein thioesterase 2 | LYPLA2* | 31 | 15 | 138 | 7 | 106 | 13 |
| platelet-activating factor acetylhydrolase IB subunit gamma | PAFAH1B3 | 72 | 13 | 0 | 0 | 43 | 17 |
| Dipeptidyl peptidase 9 | DPP9* | 50 | 8 | 445 | 45 | 244 | 24 |
| Patatin-like phospholipase domain-containing protein 4 | PNPLA4 | 49 | 14 | 42 | 3 | 15 | 3 |
| Protein phosphatase methylesterase 1 | PPME1 | 63 | 5 | 48 | 12 | 14 | 3 |
| palmitoyl-protein thioesterase 2 | PPT2* | 3 | 2 | 22 | 8 | 42 | 5 |
| Uncharacterized protein ABHD12 | ABHD12 | 109 | 23 | 107 | 19 | 107 | 19 |
| Platelet-activating factor acetylhydrolase IB | PAFAH1B2 | 60 | 23 | 101 | 14 | 66 | 31 |
| Abhydrolase domain-containing protein 11 | ABHD11* | 258 | 26 | 136 | 17 | 89 | 17 |
| Retinoid-inducible serine carboxypeptidase | SCPEP1 | 121 | 33 | 47 | 14 | 63 | 5 |
| arylacetamide deacetylase-like 1 (KIAA1363) | AADACL1* | 58 | 15 | 373 | 59 | 593 | 107 |
| Abhydrolase domain-containing protein 6 | ABHD6 | 55 | 11 | 62 | 9 | 81 | 11 |
| Tripeptidyl peptidase II | TPP2 | 4 | 2 | 10 | 1 | 3 | 1 |
| prolylcarboxypeptidase | PRCP | 77 | 13 | 49 | 27 | 33 | 9 |
| Retinoblastoma-binding protein 9 | RBB9 | 9 | 5 | 10 | 3 | 1 | 0 |
| Dipeptidyl peptidase 8 | DPP8 | 9 | 3 | 16 | 3 | 23 | 11 |
| cathepsin A | CTSA | 12 | 5 | 8 | 0 | 17 | 2 |
| Abhydrolase domain-containing protein FAM108A1 | FAM108A1 | 7 | 3 | 8 | 2 | 31 | 9 |
| Neuropathy target esterase | PNPLA6* | 46 | 11 | 211 | 27 | 106 | 22 |
| Lysophospholipase-like protein 1 | LYPLAL1 | 73 | 21 | 25 | 11 | 27 | 7 |
| Presenilins-associated rhomboid-like protein | PARL | 8 | 3 | 8 | 1 | 9 | 2 |
| Abhydrolase domain-containing protein FAM108B1 | FAM108B1 | 12 | 6 | 31 | 10 | 51 | 9 |
| BAT5 | BAT5 | 28 | 7 | 20 | 3 | 20 | 3 |
| Abhydrolase domain-containing protein 3 | ABHD3 | 2 | 1 | 7 | 2 | 9 | 4 |
| Sialate O-acetylesterase | SIAE* | 94 | 20 | 10 | 1 | 7 | 0 |
| Isoamyl acetate-hydrolyzing esterase 1 | IAH1 | 2 | 1 | 5 | 1 | 1 | 1 |
| Sn1-specific diacylglycerol lipase beta | DAGLB | 6 | 2 | 20 | 1 | 10 | 1 |
| monoglyceride lipase (MAGL) | MGLL* | 11 | 2 | 54 | 14 | 116 | 24 |
| carboxylesterase 2 | CES2 | 18 | 4 | 9 | 2 | 10 | 3 |
| PNPLA8 89 kDa protein | PNPLA8 | 15 | 3 | 11 | 2 | 2 | 0 |
| Abhydrolase domain-containing protein 4 | ABHD4 | 27 | 7 | 5 | 1 | 6 | 1 |
| btyrylcholinesterase | BCHE | 10 | 4 | 0 | 0 | 0 | 0 |
| Uncharacterized protein LACTB | LACTB | 2 | 1 | 2 | 1 | 5 | 1 |
| Lon protease | LONP1 | 48 | 4 | 34 | 9 | 14 | 2 |
| Fatty-acid amide hydrolase 1 | FAAH* | 181 | 56 | 0 | 0 | 9 | 2 |
| urokinase plasminogen activator | PLAU | 0 | 0 | 1 | 0 | 16 | 4 |
| similar to Dipeptidyl-peptidase 2 precursor | DPP7* | 170 | 48 | 14 | 1 | 8 | 2 |
| Dipeptidyl peptidase 4 | DPP4 | 38 | 7 | 89 | 9 | 1 | 0 |
| acetylcholinesterase | ACHE | 0 | 0 | 4 | 1 | 1 | 1 |
| Abhydrolase domain-containing protein 2 | ABHD2 | 22 | 7 | 0 | 0 | 0 | 0 |
| Fatty-acid amide hydrolase 2 | FAAH2 | 6 | 2 | 0 | 0 | 0 | 0 |
| Isoform 1 of Tissue-type plasminogen activator | PLA | 0 | 0 | 9 | 2 | 0 | 0 |
denotes serine hydrolase activities that were significantly higher (highlighted in red) or lower (highlighted in blue) in both PC3 and DU145 cells versus LNCaP cells (i.e. p<0.01 for PC3 versus LNCaP and DU145 versus LNCaP)
These data, taken together, identified several hydrolytic enzymes with altered activities in aggressive versus non-aggressive prostate cancer cells. Recent studies have uncovered a pro-tumorigenic role for KIAA1363 in PC3 and DU145 cells (Chang et al., 2011). Here, we asked whether MAGL also supports the aggressiveness of these prostate cancer cell lines and, if so, by what mechanism.
Metabolic Effects of Disrupting MAGL Activity in Prostate Cancer Cells
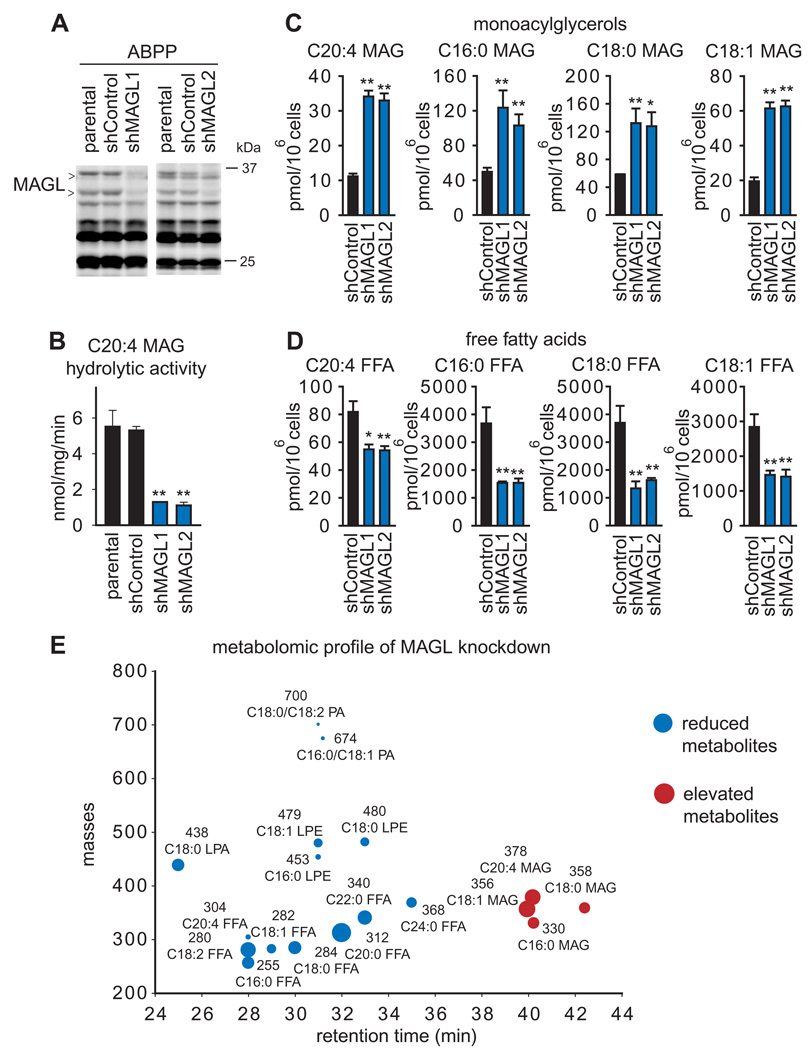
We previously found that MAGL regulates not only MAGs, but also free fatty acids (FFAs) in aggressive breast, melanoma, and ovarian cancer cells (Nomura et al., 2010). Consistent with these findings, acute pharmacological blockade of MAGL in PC3 and DU145 cells also led to elevations in several MAGs, including C20:4, C16:0, C18:0, and C18:1 MAG, and reductions in the corresponding FFAs (Figure 1C, 1D, Figure S1 and Table S2). With the exception of C20:4 FFA and MAG, the magnitude of the reductions of FFAs greatly exceeded the corresponding elevations in MAGs. We performed an untargeted lipidomic analysis of JZL184-treated versus DMSO-treated PC3 cells, and found that this discrepancy could partly be explained by JZL184–induced elevations in not only MAGs, but also lysophosphatidyl choline (LPC) levels (Figure 1E; Table S2). The cumulative magnitude of elevation of MAGs and LPC (5 nmoles) matched closely the reduction in the corresponding FFAs (7 nmoles) (Table S2), suggesting that MAGs are shunted to LPC in cancer cells where MAGL has been inactivated. These results are consistent our previous studies showing that metabolically labeled MAGs are directly converted to lysophosphatidyl cholines by cancer cells through a pathway that is greatly enhanced by MAGL inhibition (Nomura et al., 2010). Our lipidomic analysis also revealed not only reductions in FFAs, but also decreases in several additional metabolites, including the pro-tumorigenc signaling lipids, lysophosphatidic acid (LPA) (Mills and Moolenaar, 2003), phosphatidic acid (PA) (Foster, 2009), and lysophosphatidyl ethanolamines (LPE) (Park et al., 2007) (Figure 1F, G, H, Figure S1, Table S2). These bioactive lipids have been found to promote various aspects of cancer aggressiveness, including migration, invasion, and survival by signaling onto their cognate G-protein coupled receptors. (Foster, 2009; Mills and Moolenaar, 2003; Park et al., 2007). We next generated stable small hairpin (sh)RNA-mediated knocked down of MAGL using two independent probes (shMAGL1 and shMAGL2), both of which reduced MAGL activity by >75% in PC3 cells (Figure 2A and 2B), also caused significant elevations in MAGs (Figure 2C) and corresponding reductions in FFAs (Figure 2D). Lipidomic analysis of shControl versus shMAGL PC3 cells also showed not only elevations in MAGs and reductions in FFAs, but also decreases in LPA, PA, and LPE(Figure 2E, Figure S1, Table S2). We also found that, although acute JZL184 treatment elevated LPCs in PC3 cells, we did not observe this increase upon chronic JZL184 treatment (4 days, 1 µM JZL184), revealing metabolic differences between acute versus chronic blockade of MAGL and possible incorporation of transiently elevated LPCs into the more abundant phosphatidyl choline pools (Figure S1). We also observed similar decreases in LPA and PA in DU145 cells treated with JZL184 (1 µM, 4 h) (Figure S1, Table S2). These results mirror the metabolic effects of MAGL disruption in aggressive ovarian and melanoma cancer cells (Nomura et al., 2010). On the other hand, other changes observed in shMAGL ovarian and melanoma cancer cells, such as reductions in prostaglandins, lysophosphatidyl cholines, and monoalkylglycerol ethers were either unchanged or not detected in PC3. MAGL ablation in prostate cancer cells thus leads to reductions in cellular FFA levels and downstream FFA-derived pro-tumorigenic phospholipid signals. These results are consistent with our previous study showing that isotopically labeled free fatty acids in cancer cells are rapidly incorporated into phospholipid species such as LPA, PA, and LPEs (Nomura et al., 2010).
Figure 2. Stable shRNA-mediated knockdown of MAGL lowers FFA levels in PC3 cells.
(A) MAGL was stably knocked down using two independent short-hairpin RNA (shRNA) oligonucleotides (shMAGL1 and shMAGL2), resulting in >75 % reduction in MAGL activity in PC3 cells, as assessed by ABPP analysis of PC3 soluble proteomes, compared to shControl cells expressing an shRNA that targets a distinct serine hydrolase (DPP4). (B) Total C20:4 MAG hydrolytic activity of parental, shControl, and shMAGL1 and 2 PC3 whole cell lysate proteomes show significantly reduced MAGL activity in shMAGL cancer cells. (C, D) shMAGL cells show elevations in MAGs (C) and reductions in FFAs (D). The MAGL activity and MAG and FFA levels of shControl cells did not differ significantly from those of parental cancer cell lines. (E) Lipidomic analysis of PC3 shMAGL versus shControl cells shows not only elevations in MAGs and reductions in FFAs, but also lower levels of lysophosphatidyl ethanolamines (LPEs), phosphatidic acids (PAs), and lysophosphatidic acid (LPA). *p<0.05, **p<0.01 for shMAGL versus shControl groups. Data are presented as means ± SEM; n=4–5/group. See also Figure S1.
Disruption of MAGL Activity Impairs Cancer Aggressiveness
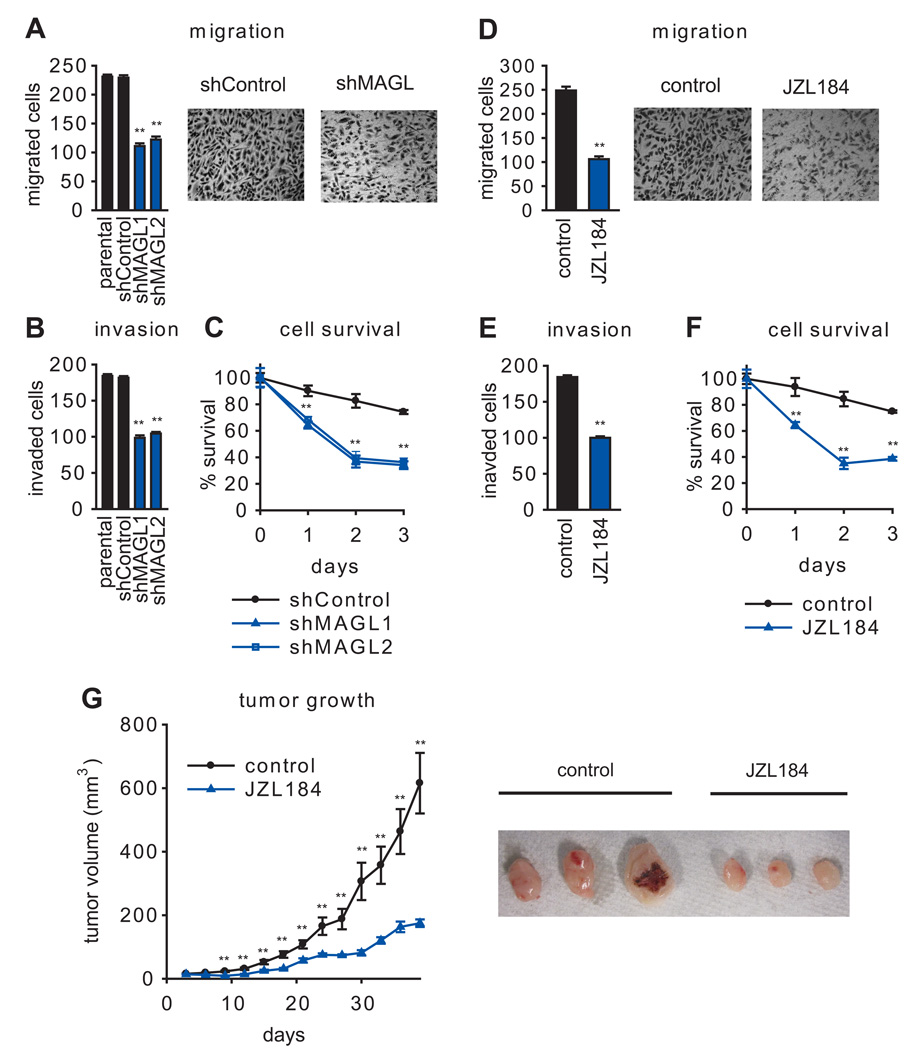
shMAGL PC3 cells showed significant reductions in migration (Figure 3A), invasion (Figure 3B), and survival (Figure 3C). Similar effects were observed for PC3 (Figure 3D–F) and DU145 (Figure S1) cells treated with JZL184 (1 µM, 4 hr). These data indicate that disrupting MAGL reduces prostate cancer cell aggressiveness in vitro. We next asked whether blocking MAGL would also impair the tumor growth of prostate cancer cells in vivo using a mouse xenograft model. shMAGL PC3 cells displayed significantly reduced tumor growth rates compared to control PC3 cell lines in immune-deficient SCID mice (Figure S2). Daily administration of JZL184 (40 mg/kg, oral gavage) was also found to slow PC3 tumor growth in this model (Figure 3G). Curiously, we did not observe an effect of JZL184 on tumor growth when PC3 cells were placed in a nude mouse xenograft model (Figure S3). Analysis of excised tumors revealed that, while JZL184 completely blocked MAGL activity in tumors grown in SCID mice, this compound only produced a partial inhibition of MAGL in tumors grown in nude mice (Figure S3). We also confirmed by assessing MAG hydrolytic activity that MAGL was more completely inhibited in brain tissue from SCID versus nude mice treated acutely with JZL184 (40 mg/kg, 4 and 24 h) (Figure S3). Thus, incomplete MAGL blockade could explain the lack of efficacy for JZL184 in tumor xenograft models performed in a nude mouse background. We also cannot exclude the possibility that the distinct effect on tumor growth in SCID versus nude mice could be due to inherent biological differences between the two mouse strains, such as immune-cell leakiness (Bankert et al., 2002; Sharkey and Fogh, 1984), which could also inform on possible mechanisms of JZL184 anticancer activity in vivo.
Figure 3. Pharmacological and shRNA blockade of MAGL impairs PC3 aggressiveness.
(A–C, D–F) Both shMAGL and JZL184 (1 µM) PC3 cells show impaired migration (A, D), invasion (B, E), and serum-free survival (C, F). Cancer cells were pretreated with JZL184 in serum-free media for 4 h before migration (5 h migration time) and invasion (24 h invasion time) assays and 24 h before cell survival (20 h in serum and 4 h in serum-free media with JZL184). For PC3 migration, representative fields of migrated cells are shown at 200 × magnification. (G) Pharmacological (40 mg/kg JZL184, daily oral gavage) inhibition of MAGL causes impairments in PC3 tumor xenograft growth in immune-deficient SCID mice. Representative tumors are shown on the right. **p<0.01 for shMAGL versus shControl or JZL184 versus vehicle treatment groups. Data are presented as means ± SEM. For (A,B,D,E), n=4–5/group and for (C,F,G), n=6–8/group. See Figure S2 and Figure S3.
We previously found that overexpressing MAGL in non-aggressive melanoma and ovarian cancer cells was sufficient to reduce MAGs, elevate FFAs, and confer heightened pathogenicity (Nomura et al., 2010). We therefore overexpressed MAGL in LNCaP cells (MAGL-OE cells) to determine whether we could mirror these effects in prostate cancer cells (Figure S4). Although MAG levels were significantly reduced in MAGL-OE LNCaP cells, FFA and phospholipid levels and migration were unaltered. While we do not yet understand why MAGL is incapable of controlling FFAs in LNCaP cells, we found that these cells express much higher levels of carnitine palmitoyltransferase 1 (CPT1), the rate-limiting step for fatty acid β-oxidation, compared to DU145 and PC3 cells (Figure S4; Table S3). It is possible that FFAs generated by LNCaP cells overexpressing MAGL, may be shunted into the mitochondria via CPT1 to oxidative pathways, thereby preventing their accumulation.
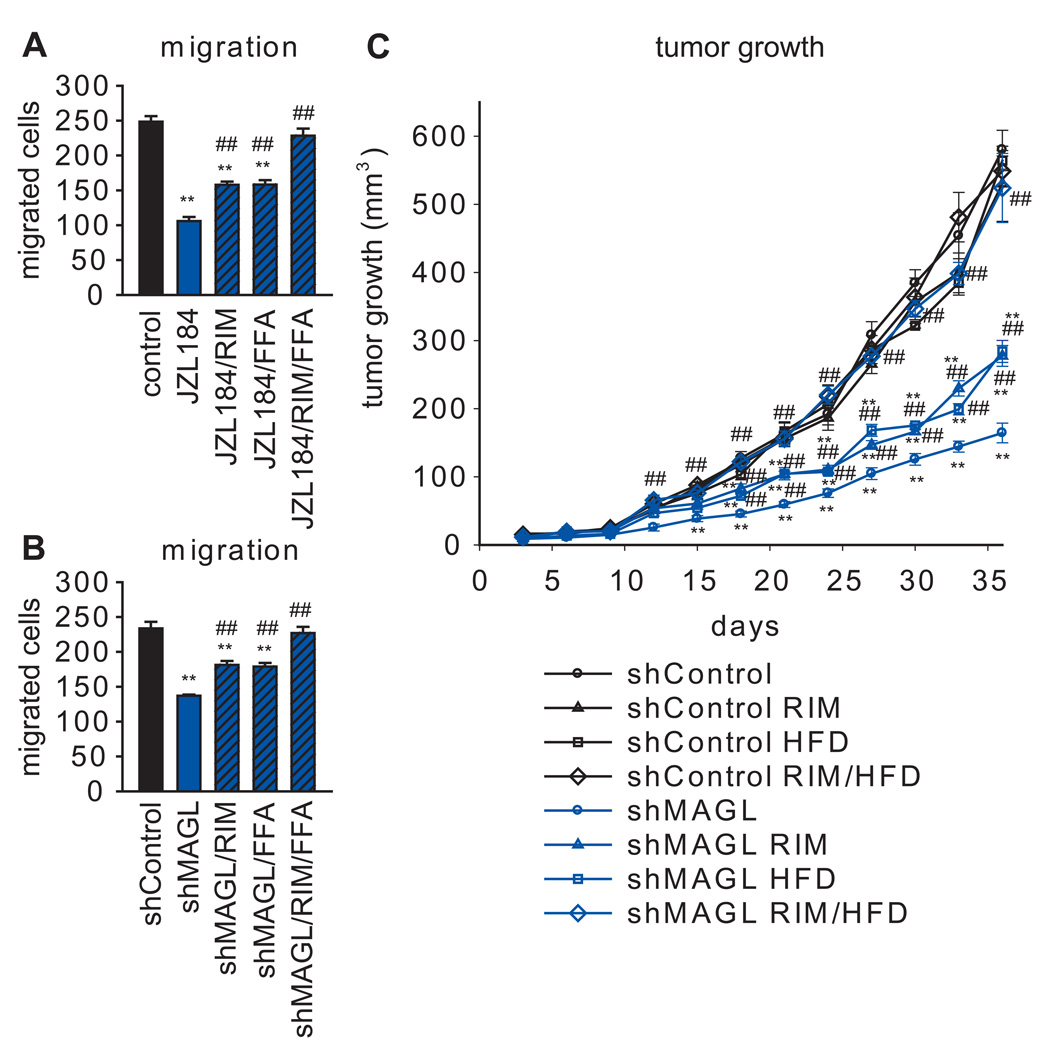
In our previous studies, we found that the reduced migratory activity of shMAGL breast, melanoma, and ovarian cancer cells could be fully rescued by fatty acids, but not cannabinoid receptor antagonists (Nomura et al., 2010), suggesting that the fatty acid products of MAGL, rather than its endocannabinoid substrates, were responsible for affecting migration in these cells. Here, we uncovered a distinct mechanism in human prostate cancer cells, where the migratory, invasion, and cell survival defects caused by JZL184 or an shMAGL probe were only partially reversed by treatment with palmitic acid (10 µM) (Figure 4A, B, Figure S4). Partial reversal was also observed with the CB1 receptor antagonist rimonabant (RIM) (1 µM), but not with the CB2 antagonist AM630 (1 µM) (Figure 4A, B, Figure S4), and a combination of palmitic acid and RIM fully restored migration in MAGL-disrupted prostate cancer cells (Figure 4A, B, Figure S4). We also found that PC3 migration was reduced by treatment with the endocannabinoid 2-AG (1 µM) (Figure S4). The partial rescue of the migratory activity of shMAGL cells produced by palmitic acid was blocked by the Gi/o-protein uncoupling agent pertussis toxin (PTX, 100 ng/mL) (Figure S4), suggesting the involvement of signaling lipids (such as LPA, PA, and LPE) that act on G-protein coupled receptors. Finally, the reduced tumor-growth of shMAGL cells in SCID mice was also partially reversed by RIM or a high-fat diet (HFD), and fully restored by co-treatment of RIM and a HFD (Figure 4C). These data indicate that MAGL supports prostate cancer aggressiveness by multiple mechanisms – 1) the production of pro-tumorigenic fatty acid products, and 2) the degradation of anti-tumorigenic endocannabinoids.
Figure 4. A combination of fatty acid and CB1 antagonist rescues the impaired migration and tumor growth of MAGL-disrupted PC3 cells.
(A, B) The migratory defects observed with JZL184 (A) and in shMAGL (B) PC3 cells are partially rescued upon co-treatment with the CB1 receptor antagonist rimonabant (1 µM) or palmitic acid (10 µM), denoted as FFA in the figure), and fully rescued upon addition of both. (C) Tumor growth defects observed with shMAGL PC3 cells are also partially recovered upon daily administration of rimonabant (3 mg/kg, oral gavage) or high-fat diet (60 kcal % diet) and fully rescued with both. **p<0.01 for shMAGL or JZL184 groups versus control or shControl groups; ## p<0.01 for JZL184 or shMAGL groups treated with RIM, FFA, and/or HFD versus shMAGL or JZL184 groups. Data are presented as means ± SEM. For (A, B), n=4–5/group and for (C), n=5–8 mice/group. See Figure S4.
MAGL is Part of a Gene Signature that Contains EMT and Cancer Stem Cell Markers
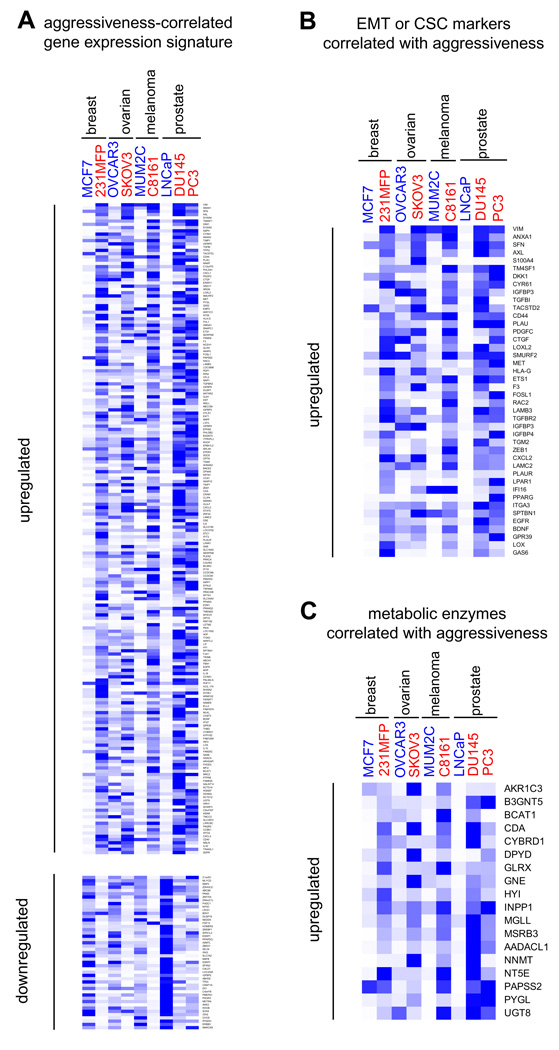
Although we and others have shown that MAGL expression and activity are elevated in aggressive human cancer cells and primary tumors (Gjerstorff et al., 2006; Nomura et al., 2010), the pathways responsible for regulating MAGL in cancer remain unknown. One strategy to map such pathways would be to identify gene expression signatures that correlate with MAGL activity in cancer cells. Toward this end, we transcriptionally profiled sets of aggressive and non-aggressive human cancer cell lines from four different tumor types (ovarian, melanoma, breast and prostate cancer) using Affymetrix HU133 Plus 2.0 microarrays (Table S3). Consistent with our ABPP results, MAGL expression was higher in all of the aggressive cancer cell lines tested (SKOV3, C8161, 231MFP, PC3 and DU145) compared to their non-aggressive counterparts (OVCAR3, MUM2C, MCF7, and LNCaP). To uncover genes that showed a similar expression pattern, we filtered the microarray datasets to include only genes that displayed > 3-fold higher signals in at least four out of five sets of aggressive versus non-aggressive cancer cells. This analysis identified 199 genes that were consistently overexpressed in the aggressive cancer cell line panel (Figure 5A and Table S3). Among these aggressiveness-correlated genes were numerous established markers of epithelial-to-mesenchymal transition (EMT) and cancer stem cells, including VIM, TGFB1, CD44, LOX, LOXL2, LAMB3, TGFBR2, ZEB1, LAMC2, EDN1, and EGFR (Polyak and Weinberg, 2009) (Figure 5B, Table S3, Table S4). We identified 28 additional metabolic enzymes that were also part of this aggressiveness gene signature, possibly pointing to other metabolic pathways important for sustaining cancer malignancy (Figure 5C and Table S3). A distinct set of metabolic enzymes showed consistently lower expression in aggressive cancer lines (Table S3), as did gene products known to be downregulated following EMT (e.g., E-cadherin) (Polyak and Weinberg, 2009) (Table S3). This large-scale transcriptional analysis suggests that the heightened expression of MAGL (and several other metabolic enzymes) in aggressive cells may be due, at least in part, to pro-tumorigenic pathways that support EMT and cancer ‘stemness’.
Figure 5. Gene signatures of aggressive human cancer cell lines.
(A) Transcriptional profiling of aggressive (noted in red, PC3, DU145, C8161, SKOV, and 231MFP) versus non-aggressive (noted in blue, LNCaP, MUM2C, OVCAR3, and MCF7) cancer cell lines yields commonly dysregulated genes (left) (>3-fold changes in 4 out of 5 pairs of cell lines). (B) Several EMT and cancer stem cell markers are found among genes consistently elevated in aggressive cancer lines. (C) Metabolic enzymes consistently elevated (upper panel) or reduced (lower panel) in aggressive cancer lines. Heat maps were generated using Gene Tree View obtained from http://rana.lbl.gov/EisenSoftware.htm. Blue versus white denotes high versus low relative mRNA expression of each gene, respectively. Relative mRNA expression, as expressed by blue color, was normalized to the highest gene chip signal for each gene across the 9 cancer cell lines profiled. See also Table S3.
Discussion
While MAGL has been most extensively studied for its role in terminating endocannabinoid signaling in the nervous system (Blankman et al., 2007; Dinh et al., 2002; Long et al., 2009; Schlosburg et al., 2010), recent investigations have also pointed to a function for this enzyme in cancer, where it regulates a set of pro-tumorigenic fatty acid products (Nomura et al., 2010). Other components of the endocannabinoid system, including the CB1 receptor and FAAH, have also been implicated in cancer (Bifulco et al., 2008; Chung et al., 2009; Endsley et al., 2008b; Fowler et al., 2010; Guzman, 2003; Mimeault et al., 2003; Nithipatikom et al., 2004; Nithipatikom et al., 2005; Sarfaraz et al., 2005; Thors et al., 2010; Wang et al., 2008). Here, we have used ABPP to determine that MAGL and FAAH are highly expressed in androgen-independent (PC3 and DU145) and androgen-dependent (LNCaP) prostate cancer cells, respectively. Since androgen-independent prostate cancer is the more malignant and life-threatening form of this disease (Damber and Aus, 2008; So et al., 2005), we sought to characterize the role that MAGL plays in PC3 and DU145 cells.
Using a combination of selective inhibitors and shRNA probes, we determined that MAGL exerts dual control over both endocannabinoid and fatty acid pathways to support prostate cancer aggressiveness. Through expressing high levels of MAGL activity, androgen-independent prostate cancer cells limit the inhibitory impact of endocannabinoid signaling and promote the stimulatory effects of fatty acid pathways on pro-tumorigenic properties such as migration, invasion, and cell survival. This mechanism for MAGL action in prostate cancer cells contrast with other types of cancer (e.g. ovarian, breast, and melanoma), where endocannabinoid pathways were not found to play a significant role in regulating aggressiveness (Nomura et al., 2010). Previous studies have, however, shown that direct cannabinoid receptor agonists impair the proliferation and metastatic activity of cancer cells, as well as induce apoptosis in cancer cells (Alexander et al., 2009; Guzman, 2003). These results indicate that both exogenous and endogenous cannabinoids can impede the progression of certain forms of cancer.
In addition to their direct effects on tumor cells, cannabinoids have been used in cancer treatment to alleviate many of the side-effects from chemotherapy, including nausea, vomiting, weight loss, lack of appetite, and pain (Guzman, 2003). Although we do not know yet whether MAGL inhibitors will impact all of these processes, they have been shown to exhibit anti-emesis (Sticht et al., 2011) and anti-hyperalgesic activity (Kinsey et al., 2009; Long et al., 2009). Thus, MAGL inhibitors have the potential to not only impair cancer progression, but also remedy co-morbidity factors, such as pain and nausea, that accompany the treatment of this disease by more conventional chemotherapeutics. In this regard, we should also note that MAGL inhibitors do not produce the cataleptic effects of direct cannabinoid agonists and may therefore show an improved safety profile (Long et al., 2009). On the other hand, recent studies indicate that complete blockade of MAGL eventually leads to behavioral tolerance that is accompanied by downregulation of CB1 receptors in the nervous system (Schlosburg et al., 2010). It remains to be determined whether similar adaptations to MAGL inhibition occur in cancer cells, which could explain the blunted effect of chronic JZL184 on survival. It is also of future interest to determine whether the high FAAH expression in LNCaP cells may point to a dual role of 2-AG and anandamide in the control of androgen-dependent prostate cancer biology. Further studies are also required to determine whether MAGL is elevated in primary human prostate tumors, akin to its heightened expression in primary breast and ovarian cancers (Gjerstorff et al., 2006; Nomura et al., 2010).
We do not yet understand how MAGL is elevated in aggressive cancer cells, but our initial studies suggest that its heightened expression is transcriptional in origin. We therefore sought to identify an ‘aggressiveness’-associated gene signature that correlated with MAGL across a panel of nine human cancer cell lines originating from four different tumor types (breast, melanoma, ovarian, and prostate). This experiment revealed that aggressive cancer cells possessing high levels of MAGL also express numerous EMT and cancer stem cell markers, many of which have been found to correlate with, and contribute to cancer malignancy (see Table S3 and Table S4). Interestingly, other researchers have similarly observed that cancer cells showing the phenotypic hallmarks of EMT are enriched in stem cell markers (Polyak and Weinberg, 2009). Moreover, recent studies have found that MAGL is overexpressed in Ras-transformed cancer cells along with other markers of EMT (Chun et al., 2010; Joyce et al., 2009). These findings indicate that the MAGL-endocannabinoid/fatty acid pathway may constitute a key metabolic network that supports EMT and the stem-like properties of cancer cells. That several other metabolic enzymes are also part of the aggressiveness gene signature observed in this study points to fundamental differences in the biochemistry of cancer cells that have undergone EMT and stem-like conversion. Through our ABPP studies, we also identified other serine hydrolase activities that are dysregulated selectively in androgen-independent versus androgen-dependent prostate cancer cells that may regulate additional metabolic pathways that contribute to prostate cancer malignancy. Future studies on MAGL and these aggressiveness-related enzymes should increase our knowledge of the metabolic pathways that support cancer malignancy and identify new molecular targets for cancer treatment.
Significance
Characterizing the metabolic pathways that tumor cells use to support their malignant behavior is important for both understanding and ultimately treating cancer. We have previously shown that the lipolytic enzyme MAGL is elevated in aggressive breast, ovarian, and melanoma cancer cells, where it regulates a fatty acid network enriched in pro-tumorigenic signaling lipids. Here, we show that MAGL is also highly expressed in aggressive prostate cancer cells where it not only regulates pro-tumorigenic fatty acid products, but also suppresses anti-tumorigenic endocannabinoid signals. This dual control over fatty acid and endocannabinoid signaling pathways can be disrupted by inhibitors or shRNA probes that target MAGL, resulting in impairments in prostate cancer cell migration, invasion, survival, and tumor growth. We also show that MAGL expression and the expression of several other metabolic enzymes correlate with EMT and cancer stem cell markers across a broad panel of human cancer cell lines. These data thus point to a set of metabolic enzymes and pathways, such as the MAGL-endocannabinoid/fatty acid network, that may create key biochemical changes in cancer cells that support their progression to a high-malignancy state. Disrupting these pathways with small-molecule inhibitors, as we show with MAGL and its cognate inhibitor JZL184 in this study, could impair tumor cell aggressiveness and offer novel ways to treat the most aggressive forms of cancer.
Experimental Procedures
Materials
Prostate cancer cell lines LNCaP, PC3, and DU145 cell lines were obtained from ATCC. OVCAR3, SKOV3, and MCF7 cell lines were obtained from the National Cancer Institute's Developmental Therapeutics Program. The C8161 and MUM2C lines were provided by Mary Hendrix. The 231MFP cells were generated from explanted xenograft tumors of MDA-MB-231 cells, as described previously (Jessani et al., 2004). The SKOV3 cells used in this study were also derived from explanted xenograft tumors of SKOV3 cells. All lipid standards and internal standards were purchased from Sigma, Alexis Biochemicals, Cayman Chemicals, Nu-check Prep, or Avanti Lipids. FP-rhodamine (Patricelli et al., 2001) and FP-biotin (Liu et al., 1999) were synthesized by following previously described procedures. Rimonabant was purchased from AK Scientific. JZL184 was synthesized as previously detailed (Long et al., 2009).
Cell Culture
LNCaP, C8161, MUM2C, SKOV3, OVCAR3, and MCF7 cells were maintained in RPMI medium, PC3 cells in F12K medium, DU145 cells in DMEM high glucose medium, and 231MFP in L-15 medium. All media were supplemented with with 4 mM L-glutamine and 10 % (v/v) fetal calf serum and kept at 37°C in a humidified atmosphere of 5% CO2/95% air, except for 231MFP cells which were maintained in a CO2-free atmosphere.
For ABPP, activity assay, or lipidomic analysis, cells were treated as described previously (Nomura et al., 2010). Briefly, 1 × 106 cells were plated in 6 cm dishes (subconfluent). A total of 20 hr after plating, cells were washed twice with PBS and inhibitors were incubated in the corresponding serum-free media. For pharmacological treatments, JZL184 (1 µM), rimonabant (1 µM), or FFA (10 µM), or vehicle (DMSO) at 0.1% were incubated with the cells for 4 h in serum-free media before cells were harvested by scraping and analyzed by ABPP or LC-MS. For chronic JZL184 treatments, cells were treated once daily with JZL184 (1 µM) for 4 days in serum-containing media. On the third day, DMSO or JZL184-treated cells (1 × 106 cells) were seeded into 6 cm dishes and treated with DMSO or JZL184. On the fourth day, cells were serum-starved for 4 h in the presence of DMSO or JZL184, after which cells were washed twice with PBS, and harvested by scraping.
ABPP of Cancer Cell Proteomes
ABPP analysis was performed as previously described (Nomura et al., 2010). Briefly, cells were washed twice and scraped in ice-cold PBS. Cell pellets were isolated by centrifugation at 1,400 x g for 3 min and sonicated in PBS. Either whole cell lysates or soluble proteomes were used for ABPP or MAG hydrolysis assays. To obtain soluble proteomes, cell lysates were centrifuged at 100,000 × g for 45 min and the supernatant was used for subsequence studies. Protein concentrations were adjusted to a final concentration of 1 mg/ml in PBS.
For gel-based ABPP experiments, cell lysate proteomes were treated with 2 µM FP-rhodamine for 30 min at room temperature before quenching reactions with one volume of standard 4x SDS/PAGE loading buffer (reducing), separated by SDS/PAGE (10% acrylamide), and visualized in-gel with a Hitachi FMBio IIe flatbed fluorescence scanner (MiraiBio). Integrated band intensities were calculated for the labeled proteins and averaged from three independent cell samples to determine the level of each enzyme activity.
For mass-spectrometry-based ABPP experiments, cell lysate proteomes were treated with 5 µM FP-biotin for 1 h at room temperature and processed as described previously (Nomura et al., 2010). After solubilization, avidin enrichment of FP-labeled proteins, and on-bead tryptic digestion, digested peptide mixtures were loaded on to a biphasic (strong cation exchange/reverse phase) capillary column and analyzed by two-dimensional liquid chromatography (2D-LC) separation in combination with tandem mass spectrometry as previously described (Liu et al., 2004b). Peptides were eluted in a 5-step MudPIT experiment (using 0, 10, 25, 80 and 100% salt bumps) and data was collected in an ion trap mass spectrometer, LTQ (Thermo Scientific) set in a data-dependent acquisition mode with dynamic exclusion turned on (60s). ms2 spectral data were searched using the SEQUEST algorithm (Version 3.0) (Eng et al., 1994) against a custom made database containing the longest entry from v3.26 of the human IPI database associated with each Ensembl gene identifier resulting in a total of 22935 unique entries. Additionally, each of these entries was reversed and appended to the database for assessment of false-discovery rates.
Hydrolytic Activity Assays
Activity assays were performed as previously described (Nomura et al., 2010). Briefly, cell lysates (20 µg) in PBS were incubated with 100 µM of C20:4 MAG at room temperature for 30 min in a volume of 200 µl before quenching with 600 µl 2:1 chloroform:methanol and subsequent addition of 10 nmol of C15:0 FFA internal standard. The products were extracted into the organic layer which was removed and directly injected into LC-MS. LC-MS settings were as previously described (Blankman et al., 2007). Product levels (e.g. C20:4 FFA for MAGL activity) were quantified in relation to internal standard levels.
RNA Interference Studies in Human Cancer Cell Lines
RNA interference studies were conducted as described previously (Chiang et al., 2006; Nomura et al., 2010). Briefly, short-hairpin RNA constructs were subcloned into the pLP-RetroQ acceptor system, and retrovirus was generated by using the AmphoPack-293 Cell Line (Clontech). Hairpin oligonucleotides utilized were: for MAGL (shMAGL1), 5’-CAACTTTCAAGGTCCTTGC-3’ and (shMAGL2), 5’-AGACTACCCTGGGCTTCCT-3’; for the shControl (shDPPIV), 5’-GATTCTTCTGGGACTGCTG-3’. Infected cells were expanded and tested for the loss of enzyme activity by ABPP and C20:4 MAG hydrolytic activity.
Lipidomic analysis of Prostate Cancer Cells
Lipidomic analyses were performed as previously described (Nomura et al., 2010). Lipid measurements were conducted in cancer cells grown in serum-free media for 4 hrs to minimize the contribution of serum-derived lipids to the cellular profiles. Cancer cells (1 × 106 cells/6 cm dish, 80 % confluency) were washed with twice with phosphate buffer saline (PBS), isolated by centrifugation at 1,400 × g, and metabolites were extracted and analyzed by mass spectrometry using previously described conditions (Nomura et al., 2010). Further details on lipidomic analyses are provided in Supplemental Materials.
Cell Migration, Cell Survival, and Invasion Studies
Migration, cell survival, and invasion studies of cancer cells were performed as described previously (Nomura et al., 2010). Briefly, migration assays were performed in Transwell chambers (Corning) with 8 µm pore-sized membranes coated with 10 µg/ml collagen for 5 h (for PC3) or 20 h (for DU145 and LNCaP) at 37°C in serum-free media. The cells that migrated were counted at a magnification of 400x, and 4 fields were independently counted from each migration chamber. An average of cells in 4 fields for one migration chamber represents n=1. Cell survival assays were performed using the Cell Proliferation Reagent WST-1 (Roche) as previously described (Roca et al., 2008; Siddiqui et al., 2005) in cells serum-starved 4 h before seeding. Invasion assays were conducted using the BD Matrigel Invasion Chambers per the manufacturer’s protocol.
Tumor Xenograft Studies
Human cancer xenografts were established by transplanting cancer cell lines ectopically into the flank of C.B17 SCID mice (Taconic Farms) or J/nu mice (The Jackson Laboratory). Briefly, cells were washed two times with PBS, trypsinized, and harvested in serum-containing medium. Next, the harvested cells were washed two times with serum-free medium and resuspended at a concentration of 2.0 × 104 cells/µl and 100 µl was injected. Growth of the tumors was measured every 3 days with calipers. For HFD studies, mice were placed on a 60 kcal % fat diet (Research Diets), two weeks prior to cancer cell injections. For chronic JZL184 or rimonabant treatment studies, mice were treated with JZL184 (40 mg/kg), rimonabant (3 mg/kg) or vehicle once daily (at approximately the same time everyday) by oral gavage in polyethylene glycol 300 (4 µL/g mouse). The treatments were initiated immediately after ectopic injection of cancer cells.
Gene chip comparisons of aggressive versus non-aggressive cancer cell lines
mRNA were extracted (Qiagen RNeasy kit) from aggressive (231MFP, SKOV3, C8161, PC3 and DU145) and non-aggressive (MCF7, OVCAR3, MUM2C, LNCaP) cells from breast, ovarian, melanoma, and prostate cancers, respectively, reverse transcribed, and hybridized to Affymetrix HU133 Plus 2.0 microarrays. Data were then filtered for genes that were commonly up- or down-regulated (>3-fold) in at least 4 out of the 5 pairs of aggressive versus non-aggressive cancer cell lines for further analysis.
Research Highlights.
Monoacylglycerol lipase (MAGL) is elevated in aggressive prostate cancer cells
MAGL controls both endocannabinoid and fatty acid pathways in prostate cancer cells
MAGL inhibition impairs prostate cancer pathogenicity
MAGL is part of an epithelial-to-mesenchymal transition and stem-like gene signature
Supplementary Material
Acknowledgements
We thank the members of the Cravatt laboratory for helpful discussion and critical reading of the manuscript. This work was supported by the National Institutes of Health (CA132630, K99DA030908, and UL1RR025773(KL2)) and the Skaggs Institute for Chemical Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander A, Smith PF, Rosengren RJ. Cannabinoids in the treatment of cancer. Cancer Lett. 2009;285:6–12. doi: 10.1016/j.canlet.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Bankert RB, Hess SD, Egilmez NK. SCID mouse models to study human cancer pathogenesis and approaches to therapy: potential, limitations, and future directions. Front Biosci. 2002;2:c44–c62. doi: 10.2741/A758. [DOI] [PubMed] [Google Scholar]
- Bifulco M, Malfitano AM, Pisanti S, Laezza C. Endocannabinoids in endocrine and related tumours. Endocr Relat Cancer. 2008;15:391–408. doi: 10.1677/ERC-07-0258. [DOI] [PubMed] [Google Scholar]
- Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarel MM, Andradas C, Mira E, Perez-Gomez E, Cerutti C, Moreno-Bueno G, Flores JM, Garcia-Real I, Palacios J, Manes S, et al. Cannabinoids reduce ErbB2-driven breast cancer progression through Akt inhibition. Mol Cancer. 2010;9:196. doi: 10.1186/1476-4598-9-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda P, Gao Y, Mark L, Btesh J, Strassle B, Lu P, Piesla M, Zhang MY, Bingham B, Uveges A, et al. Monoacylglycerol lipase activity is a critical modulator of the tone and integrity of the endocannabinoid system. Mol Pharmacol. 2010 doi: 10.1124/mol.110.068304. [DOI] [PubMed] [Google Scholar]
- Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chem Biol. 2011 doi: 10.1016/j.chembiol.2011.02.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Chun SY, Johnson C, Washburn JG, Cruz-Correa MR, Dang DT, Dang LH. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1alpha and HIF-2alpha target genes. Mol Cancer. 2010;9:293. doi: 10.1186/1476-4598-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SC, Hammarsten P, Josefsson A, Stattin P, Granfors T, Egevad L, Mancini G, Lutz B, Bergh A, Fowler CJ. A high cannabinoid CB(1) receptor immunoreactivity is associated with disease severity and outcome in prostate cancer. Eur J Cancer. 2009;45:174–182. doi: 10.1016/j.ejca.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Damber JE, Aus G. Prostate cancer. Lancet. 2008;371:1710–1721. doi: 10.1016/S0140-6736(08)60729-1. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008a;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008b;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci U S A. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endsley MP, Thill R, Choudhry I, Williams CL, Kajdacsy-Balla A, Campbell WB, Nithipatikom K. Expression and function of fatty acid amide hydrolase in prostate cancer. Int J Cancer. 2008a;123:1318–1326. doi: 10.1002/ijc.23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endsley MP, Thill R, Choudhry I, Williams CL, Kajdacsy-Balla A, Campbell WB, Nithipatikom K. Expression and function of fatty acid amide hydrolase in prostate cancer. International Journal of Cancer. 2008b;123:1318–1326. doi: 10.1002/ijc.23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Foster DA. Phosphatidic acid signaling to mTOR: signals for the survival of human cancer cells. Biochim Biophys Acta. 2009;1791:949–955. doi: 10.1016/j.bbalip.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CJ, Hammarsten P, Bergh A. Tumour Cannabinoid CB(1) receptor and phosphorylated epidermal growth factor receptor expression are additive prognostic markers for prostate cancer. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015205. e15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerstorff MF, Benoit VM, Laenkholm AV, Nielsen O, Johansen LE, Ditzel HJ. Identification of genes with altered expression in medullary breast cancer vs. ductal breast cancer and normal breast epithelia. Int J Oncol. 2006;28:1327–1335. [PubMed] [Google Scholar]
- Goparaju SK, Ueda N, Yamaguchi H, Yamamoto S. Anandamide amidohydrolase reacting with 2-arachidonoylglycerol, another cannabinoid receptor ligand. FEBS Lett. 1998;422:69–73. doi: 10.1016/s0014-5793(97)01603-7. [DOI] [PubMed] [Google Scholar]
- Guzman M. Cannabinoids: potential anticancer agents. Nat Rev Cancer. 2003;3:745–755. doi: 10.1038/nrc1188. [DOI] [PubMed] [Google Scholar]
- Hoosein NM, Boyd DD, Hollas WJ, Mazar A, Henkin J, Chung LW. Involvement of urokinase and its receptor in the invasiveness of human prostatic carcinoma cell lines. Cancer Commun. 1991;3:255–264. doi: 10.3727/095535491820873146. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010 doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, 3rd, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Joyce T, Cantarella D, Isella C, Medico E, Pintzas A. A molecular signature for Epithelial to Mesenchymal transition in a human colon cancer cell system is revealed by large-scale microarray analysis. Clin Exp Metastasis. 2009;26:569–587. doi: 10.1007/s10585-009-9256-9. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Thompson CB. Q&A: Cancer: clues from cell metabolism. Nature. 2010;465:562–564. doi: 10.1038/465562a. [DOI] [PubMed] [Google Scholar]
- Karlsson M, Reue K, Xia YR, Lusis AJ, Langin D, Tornqvist H, Holm C. Exon-intron organization and chromosomal localization of the mouse monoglyceride lipase gene. Gene. 2001;272:11–18. doi: 10.1016/s0378-1119(01)00559-5. [DOI] [PubMed] [Google Scholar]
- Kinsey SG, Long JZ, O'Neal ST, Abdullah RA, Poklis JL, Boger DL, Cravatt BF, Lichtman AH. Blockade of endocannabinoid-degrading enzymes attenuates neuropathic pain. J Pharmacol Exp Ther. 2009;330:902–910. doi: 10.1124/jpet.109.155465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic Z, McGivan JD. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol Rev. 1983;63:547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004a;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., III A Model for Random Sampling and Estimation of Relative Protein Abundance in Shotgun Proteomics. Anal Chem. 2004b;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez JA. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta. 2010;1801:381–391. doi: 10.1016/j.bbalip.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- Mimeault M, Pommery N, Wattez N, Bailly C, Henichart JP. Anti-proliferative and apoptotic effects of anandamide in human prostatic cancer cell lines: implication of epidermal growth factor receptor down-regulation and ceramide production. Prostate. 2003;56:1–12. doi: 10.1002/pros.10190. [DOI] [PubMed] [Google Scholar]
- Nithipatikom K, Endsley MP, Isbell MA, Falck JR, Iwamoto Y, Hillard CJ, Campbell WB. 2-arachidonoylglycerol: a novel inhibitor of androgen-independent prostate cancer cell invasion. Cancer Res. 2004;64:8826–8830. doi: 10.1158/0008-5472.CAN-04-3136. [DOI] [PubMed] [Google Scholar]
- Nithipatikom K, Endsley MP, Isbell MA, Wheelock CE, Hammock BD, Campbell WB. A new class of inhibitors of 2-arachidonoylglycerol hydrolysis and invasion of prostate cancer cells. Biochem Biophys Res Commun. 2005;332:1028–1033. doi: 10.1016/j.bbrc.2005.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Lee HY, Lee SY, Kim MK, Kim SD, Kim JM, Yun J, Im DS, Bae YS. Lysophosphatidylethanolamine stimulates chemotactic migration and cellular invasion in SK-OV3 human ovarian cancer cells: involvement of pertussis toxin-sensitive Gprotein coupled receptor. FEBS Lett. 2007;581:4411–4416. doi: 10.1016/j.febslet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Roca H, Varsos Z, Pienta KJ. CCL2 protects prostate cancer PC3 cells from autophagic death via phosphatidylinositol 3-kinase/AKT-dependent survivin up-regulation. J Biol Chem. 2008;283:25057–25073. doi: 10.1074/jbc.M801073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfaraz S, Afaq F, Adhami VM, Mukhtar H. Cannabinoid receptor as a novel target for the treatment of prostate cancer. Cancer Res. 2005;65:1635–1641. doi: 10.1158/0008-5472.CAN-04-3410. [DOI] [PubMed] [Google Scholar]
- Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen PT, Ramesh D, Booker L, Burston JJ, et al. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat Neurosci. 2010;13:1113–1119. doi: 10.1038/nn.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey FE, Fogh J. Considerations in the Use of Nude-Mice for Cancer-Research. Cancer Metast Rev. 1984;3:341–360. doi: 10.1007/BF00051459. [DOI] [PubMed] [Google Scholar]
- Siddiqui RA, Zerouga M, Wu M, Castillo A, Harvey K, Zaloga GP, Stillwell W. Anticancer properties of propofol-docosahexaenoate and propofol-eicosapentaenoate on breast cancer cells. Breast Cancer Res. 2005;7:R645–R654. doi: 10.1186/bcr1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A, Gleave M, Hurtado-Col A, Nelson C. Mechanisms of the development of androgen independence in prostate cancer. World J Urol. 2005;23:1–9. doi: 10.1007/s00345-004-0473-1. [DOI] [PubMed] [Google Scholar]
- Sticht MA, Long JZ, Rock EM, Limebeer CL, Mechoulam R, Cravatt BF, Parker LA. The MAGL inhibitor, JZL184, attenuates LiCl-induced emesis in the Suncus murinus and 2AG attenuates LiCl-induced nausea (assessed by conditioned gaping) in rats. Br J Pharmacol. 2011 in press. [Google Scholar]
- Thors L, Bergh A, Persson E, Hammarsten P, Stattin P, Egevad L, Granfors T, Fowler CJ. Fatty acid amide hydrolase in prostate cancer: association with disease severity and outcome, CB1 receptor expression and regulation by IL-4. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012275. e12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhao LY, Uyama T, Tsuboi K, Wu XX, Kakehi Y, Ueda N. Expression and secretion of N-acylethanolamine-hydrolysing acid amidase in human prostate cancer cells. J Biochem. 2008;144:685–690. doi: 10.1093/jb/mvn122. [DOI] [PubMed] [Google Scholar]
- Warburg O. Uber den Stoffwechsel der Carcinomzelle. Klin Wochenschr. 1925;4:534–536. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.