Abstract
A-type lamins are a major component of the nuclear lamina. Mutations in the LMNA gene, which encodes the A-type lamins A and C, cause a set of phenotypically diverse diseases collectively called laminopathies. While adult LMNA null mice show various symptoms typically associated with laminopathies, the effect of loss of lamin A/C on early post-natal development is poorly understood. Here we developed a novel LMNA null mouse (LMNAGT−/−) based on genetrap technology and analyzed its early post-natal development. We detect LMNA transcripts in heart, the outflow tract, dorsal aorta, liver and somites during early embryonic development. Loss of A-type lamins results in severe growth retardation and developmental defects of the heart, including impaired myocyte hypertrophy, skeletal muscle hypotrophy, decreased amounts of subcutaneous adipose tissue and impaired ex vivo adipogenic differentiation. These defects cause death at 2 to 3 weeks post partum associated with muscle weakness and metabolic complications, but without the occurrence of dilated cardiomyopathy or an obvious progeroid phenotype. Our results indicate that defective early post-natal development critically contributes to the disease phenotypes in adult laminopathies.
Key words: laminopathies, lamin A, LMNA, knock-out mouse, cardiac hypertrophy, muscular dystrophy, differentiation
Introduction
Nuclear lamins are intermediate filament proteins mainly localized in the nuclear lamina, a protein meshwork lining the nucleoplasmic face of the inner nuclear membrane. Lamins are classified as A-type or B-type. The A-type lamins consist of the three LMNA splicing isoforms lamin A, AΔ10 and C. B-type lamins are encoded by the LMNB1 and B2 genes. Whereas B-type lamins are expressed in all mammalian cells throughout development, A-type lamin expression is developmentally regulated. Lamin A/C expression commences in the zygote but is lost at the morula stage.1–3 Lamin A/C expression is then first detectable in embryonic endoderm, yolk sac and trophoblast preceeding expression in myoblasts at the beginning of tissue differentiation, in mouse at day E12.0.4 In many other tissues, lamin A/C was reported to be absent until well after birth.4,5 The notion of lamin A/C being expressed upon tissue differentiation is further strengthened by its absence in embryonic stem cells and its re-expression after loss of pluripotency markers upon neuronal or cardiac myocyte differentiation.6 Similarly, embryonic carcinoma cells are devoid of lamin A/C in contrast to differentiated derivatives.3,7 Loss of lamin A/C results in increased proliferation due to hyperphosphorylation of the retinoblastoma protein and correlates with poor histological differentiation and prognosis in primary gastric carcinomas.8,9
Mutations in A-type lamins cause a group of phenotypically diverse diseases, collectively called laminopathies. They include several types of muscular dystrophies, lipodystrophies, cardiomyopathies, neurological disorders and premature aging syndromes.10 Several of the LMNA mutations affect cell differentiation. Mutations that lead to Dunnigan familial partial lipodystrophy (FPLD) interfere with the binding of lamin A to the adipocyte differentiation factor sterol element binding protein 1 (SREBP1) and impair adipocyte differentiation.11 In the premature aging disease Hutchinson-Gilford Progeria Syndrome (HGPS), the disease-causing lamin A mutant isoform progerin inhibits adipogenic differentiation and favors osteogenic differentiation of human mesenchymal stem cells via increased Notch signaling.12 To study the role of lamin A/C in skeletal, cardiac and adipose tissue and to probe how LMNA mutations affect the functioning of these tissues in laminopathies, several lamin A/C mouse models have been created.13 Tissue defects in these models mostly become manifest in young adult mice and are therefore mainly studied in weaned mice. As a consequence, little is known about the effect of loss of lamin A/C on early post-natal development or the contribution of defective development to laminopathies.
In this study we set out to probe the effects of loss of lamin A/C on early post-natal development as this is the period in which most tissue differentiation and maturation occurs. To this end, we created a novel LMNAGT−/− mouse model which combines loss of LMNA function with LMNA-controlled reporter gene expression. We demonstrate early activity of the LMNA promoter at day E11 during embryonic development in heart, liver and somites. Loss of lamin A/C results in growth retardation at 2 weeks post partum, with impaired post-natal hypertrophy of cardiac myocytes, skeletal muscle hypotrophy, decreased subcutaneous adipose tissue deposits, decreased adipogenic differentiation of MEFs and metabolic derangements. Ultimately these tissue differentiation and maturation defects are lethal before mice are weaned. These results demonstrate that lamin A/C is crucial in the early post-natal period and they provide novel insights into the physiological function of A-type lamins in the context of the developing animal.
Results
Generation of the laminGT−/− mouse.
We created LMNA null mice by interruption of the endogenous lamin A/C locus by a promoter-trap construct, which introduces an in-frame LMNA-βgeo fusion allele. The construct was inserted into LMNA intron 2 (Fig. 1A). Lamin A and C transcripts were expressed at an expected 50% reduced level in heterozygote PP15 LMNAGT+/− animals and undetectable by RT-PCR in LMNAGT−/− mice (Fig. 1B). RT-PCR analysis ruled out the formation of truncated, in-frame LMNA transcripts by alternative splicing of exon 1 and 1C to exons 2 or 7 in LMNAGT−/− mice (see material and methods for primers). Western blot analysis and immunofluoresence staining confirmed the absence of full length or truncated lamin A and C protein variants in LMNAGT−/− mice (Fig. 1C and D). The absence of lamin A and C impaired retention of emerin at the nuclear envelope and resulted in interrupted lamin B1 and nuclear pore complexes formation as previously reported in LMNAKO−/− mice (Fig. 1D).20 Combined these findings confirm the absence of functional or dominant-interfering LMNA/C protein in our gene-trap model.
Figure 1.
General phenotypical characterization of the LMNAGT−/− model. (A) Overview of the ROSAFARY genetrap (GT) sequence insertion in the LMNA gene. The GT consists of a splice acceptor (SA) containing βgeo-reporter with a polyA signal (bpA) and a PGK promoter driven hygromycin antibiotics resistance cassette surrounded by FRT sites and harbouring a splice donor (SD) site. 5′ and 3′ ends of the GT sequence are demarcated by long terminal repeats (LTR). FRT sites were not used for recombination in ES cells and the LMNAGT mice. Arrows indicate PCR primers used for detection of full length lamin A, lamin C and the LMNA-βgeo splicing product (See Fig. 1B). (B) Relative mRNA expression levels for full length lamin A, C and the LMNA-βgeo splicing product (n = 3). Levels are normalized to HPRT mRNA. (C) Western blot using polyclonal antibodies against LacZ (Abcam, Ab616), Lamin A/C (SantaCruz, Sc-6215), and a monoclonal antibody against α-Actin antibodies (Sigma, A781) on WT, LMNAGT+/− and LMNAGT−/− mouse embryonic fibroblasts protein extracts. (D) Immonofluorescence microscopy on WT, LMNAGT+/− and LMNAGT−/− mouse embryonic fibroblasts with antibodies against lamin A/C (SantaCruz, Sc-6215), emerin (Leica, cl.4G5), Lamin B1 (SantaCruz, Sc6217) and various NPC proteins (Abcam, mAB 414). DNA is stained with DAPI. Arrows indicate a local loss of lamin B1 and NPC proteins.
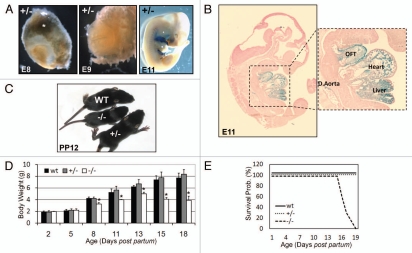
The LMNA-β-geo fusion protein is encoded by the first LMNA exon followed by β-galactosidase-neomycin cDNA (Fig. 1A–C), which allows for direct visualization of LMNA promoter activity in LMNAGT+/− and LMNAGT−/− whole mounts and tissue sections. At E8.0 and E9.0, only LMNAGT+/− placental tissue showed LMNA promoter activity (Fig. 2A). At E11.0 the LMNA promoter was active in heart, the outflow tract, dorsal aorta, liver and embryonic somites (Fig. 2A and B). Specificity of β-galactosidase signals were confirmed by staining adult LMNAGT+/− mice, which showed absence of LMNA promoter activity in various cell types, as reported previously (data not shown).4,5
Figure 2.
Embryonic and postnatal phenotypical characterization of the LMNAGT−/− mouse. (A) LMNA promoter activity is visualized by β-galactosidase staining in LMNAGT+/− embryo's (E8.0, E9.0, E11.0). LMNAGT+/− placental tissue is indicated by an asterisk. (B) β-galactosidase stained LMNAGT+/− embryo E11.0 tissue section (7 µm) counterstained with Azo Phloxine (magnification 2.5×), including a close-up image (magnification 5.0×) of the heart, the heart's outflow tract (OFT), liver and dorsal aorta (D. Aorta). (C) Macroscopical view of WT, LMNAGT+/− and LMNAGT−/− siblings 12 days post partum (PP12). (D) Body weight over time graph (PP2–PP18). Asterisks indicate a significant difference for LMNAGT−/− to LMNAGT+/− and WT littermates (N = 10, p < 0.05). (E) Survival curves for all three genotypes (N = 10) during the first 3 weeks post partum.
At birth, genotypes were present in normal Mendelian ratios and no obvious macroscopic differences were detected between WT, LMNAGT+/− and LMNAGT−/− mice. At PP7, onset of initial growth retardation was observed in LMNAGT−/− mice, which was confirmed by decreased body weight (BW) (Fig. 2C and D). LMNAGT−/− body weight peaked at PP13 and decreased slightly thereafter, leveling off at approximately half the BW of that of WT and LMNAGT+/− at PP16 (Fig. 2D). Complete loss of lamin A/C due to insertion of the GT sequence was invariably lethal between PP16 and PP18 (Fig. 2E). BW and survival curves of LMNAGT+/− mice were comparable to those of WT siblings. LMNAGT−/− mice showed normal grooming behavior until day PP16, when mice appeared scruffy and hunch-backed. Foraging, assessed by the ability to suckle, was diminished only at days PP16-PP18 in LMNAGT−/− mice compared to LMNAGT+/− and WT mice. Autopsy of LMNAGT−/− animal at PP17 revealed increased intestinal gas, but did not reveal obvious differences in food content or GI tract morphology of the glandular mucosa, muscularis mucosa, submucosa and muscularis externa on H&E stained tissue sections. Pathological examination at PP15 revealed heart, skeletal muscle myocytes and adipose tissue abnormalities (see below). LMNAGT−/− mice did not show overt gross morphological and histological (H&E) abnormalities in stomach, intestines, liver, kidneys, spleen, thymus, thyroid, lung, bladder, pancreas or brain between WT and LMNAGT−/− mice. No obvious indications of abnormal bone or dentition were observed (data not shown), in contrast to reports on other LMNA mouse models.13 Hence, we conclude that loss of lamin A/C leads to defects in early post-natal development and ultimately death.
Transcriptome analysis.
To assess how loss of lamin A/C affects tissue differentiation and maturation, microarray expression analysis was performed on left ventricle (LV) cardiac tissue at PP5 and PP13. At PP5 expression of 687 genes changed more than 1.2 fold (184 up, 464 down) in LMNAGT−/− mice compared to WT siblings (Fig. 3A, B and Sup. Table 1). 39 of those genes were still misregulated at PP13, 7 of which in similar and 32 in opposite direction. In addition, 334 genes were more than 1.2 fold deregulated at PP13 (159 up, 175 down) while normally expressed at PP5 (Fig. 3A, B and Sup. Table 1). Genmapp analysis indicates that the deregulated genes are significantly (p < 0.05) over-represented in fatty acid beta oxidation (PP5), striated muscle contraction (PP5), electron transport chain (PP13), adipogenesis (PP13) and hypertrophy (PP13). The most highly deregulated genes included Mybcp2, My6, Tnni1 (striated muscle contraction); Hdac9, Gata4, Atf3 (hypertrophy); Dci, Slc25a20 (fatty acid beta oxidation); Cebpd, Nrip, Dlk (adipogenesis) and ATP5g2, Cox8a, Cox8b (electron transport chain) (Fig. 3C). These results indicate that loss of lamin A/C resulted in myogenic, adipogenic and metabolic defects during early post-natal stages of development.
Figure 3.
Transcriptome analysis of the LMNAGT−/− mouse. (A) mRNA expression levels of left ventricle cardiac tissue were analyzed by microarrays at PP5 and PP13. Scatter plots indicate logarithmic expression levels of mRNA of LMNAGT−/− mice (N = 2) vs. WT littermates (N = 2) at both time points. Twofold up- and downregulation borders are indicated by blue lines within the scatter plots. (B) A Venn diagram of >1.2 fold up or down regulated genes in LMNAGT−/− mice compared (N = 2) to WT siblings (N = 2) at PP5 and PP13. The number of genes and direction of change are indicated for all deregulated genes at PP5, PP13 and genes which are deregulated at both time points. (C) Heatmaps of mRNA expression levels of the most deregulated genes at PP5 and PP13 within the biological pathways of striated muscle contraction, hypertrophy, fatty acid beta oxidation, adipogenesis and electron transport chain. WT expression levels for each gene were set at 1.0 (black color code). Relative differences in expression levels for LMNAGT−/− mice are indicated as fold change values and color coded.
Cardiac morphology phenotype.
In keeping with transcriptome analysis, histological examination revealed that cardiac myocytes were smaller in the LV of LMNAGT−/− mice compared to WT controls (PP15; Fig. 4A). We examined cardiac hypertrophy by weighing LV as a function of time post partum. LV weight (LVW; normalized to tibia length) was decreased by 14% at PP11 and up to 47% at PP15 in LMNAGT−/− mice compared to WT age-matched controls (PP11 2.2 ± 0.2 mg/mm LMNAGT−/− vs. 2.6 ± 0.2 mg/mm WT p < 0.05; PP15 1.9 ± 0.3 mg/mm LMNAGT−/− vs. 3.6 ± 0.3 mg/mm WT p < 0.01) (Fig. 4B). Post-natal hypertrophy was significantly impaired at PP11 in homozygous GT animals: cross-sectional area (CSA) counts were nearly 2-fold reduced (116 ± 17 µm2 LMNAGT−/− vs. 220 ± 34 µm2 WT; PP11; Fig. 4C p < 0.01), and reached a substantially lower maximum of 153 ± 17 µm2 in LMNAGT−/− mice vs. 380 ± 8 µm2 in WT siblings at PP15 (p < 0.001). Real-time quantitative PCR analysis showed comparable expression levels of the proliferative markers Ki67 and PCNA in the LV of all genotypes in early post-natal development and a general decrease in levels after birth (p > 0.05; Fig. 4D). The cardiac hypertrophy markers BNP and ANF showed 10- to 100-fold upregulation in LMNAGT−/− mice, at PP11 and PP13 and onwards, respectively (p < 0.05; Fig. 4D). No cardiac myocyte degenerative areas, fibrosis or obvious abnormalities in interstitial spaces and Z-disc organization were observed (data not shown). These data show that A-type lamins are essential for normal post-natal cardiac hypertrophy.
Figure 4.
Post-natal cardiac hypertropy in the LMNAGT−/− mouse. (A) Haematoxylin and eosin (H&E) staining on left ventricle sections of WT and LMNAGT−/− mice (PP15). (B) Bar graph indicating left ventricle weight (LVW) standardized to tibia length over time (PP5–PP18) for all three genotypes (N = 5 per time point). (C) Bar graph of cardiac myocytes cross sectional area over time (PP2–PP18) for LMNAGT−/− and WT siblings (N = 5 per time point). (D) mRNA levels of proliferative markers Ki67 and PCNA and hypertrophic markers ANF and BNP, standardized for GAPDH. Asterisks in Figure 4 indicate significant differences for LMNAGT−/− vs. both LMNAGT+/− and WT values (N = 5, p < 0.05).
Cardiac function phenotype.
To determine whether decreased cardiac hypertrophy results in compromised cardiac function, LMNAGT mice were subjected to echocardiography and electrocardiography (ECG). Impaired post-natal hypertrophy was confirmed by lower relative LVW in LMNAGT−/− mice (PP17: 2.1 ± 0.3 mg/cm LMNAGT−/− vs. 3.7 ± 0.1 mg/cm WT p < 0.01; Table 1). Nevertheless, fractional shortening (FS), which measures the fraction of LV diastolic dimension lost in systole, as an indicator for LV contractility, was comparable in all 3 genotypes at days PP10, PP13 and PP17 (PP17: 31 ± 7% LMNAGT−/−, 32 ± 9% LMNAGT+/−, 32 ± 8% WT). No signs of LV dilatation nor thinning of the LV wall were observed as all echocardiographic parameters appeared comparable for all genotypes (Table 1). ECG analysis, which captures electrical activity of the heart over time, did not reveal any differences in P wave duration (reflects atrial depolarization), PR interval (reflects atrial-ventricular node functionality), QRS time (reflects ventricular depolarization), or QT time (prolonged QT time is a risk factor for ventricular tachy-arrythmias and sudden death). ECG analysis revealed a significantly lower heart rate in LMNAGT−/− mice from day PP15 onwards (PP17: 279 ± 34 bpm. LMNAGT−/− vs. 420 ± 57 bpm. WT p < 0.05). In line with this, the RR interval was significantly increased at day PP17 (232 ± 27 ms. LMNAGT−/− vs. 158 ± 24 ms. WT p < 0.05), but not earlier (PP10, PP13). Decreasing cardiac workload by lowering blood pressure and decreasing heart rate was reported to be beneficial in dilated cardiomyopathy, which is observed in various LMNA mouse models.13,21 We therefore assessed whether treatment with the α- and β-adrenergic antagonist labetalol had beneficial effects in LMNAGT−/− mice (Sup. Fig. 1A). Labetalol increased QRS time in all genotypes (p < 0.05). In contrast to WT and LMNAGT+/− mice, labetalol treatment significantly increased P time (from 23.7 ± 2.6 ms to 29.8 ± 3.2 ms; p < 0.05) and lowered heart rate (396 ± 51 bpm to 232 ± 20 bpm; p < 0.05) in LMNAGT−/− mice (Sup. Fig. 1B). However, treatment with labetalol lead to earlier post-natal death in LMNAGT−/− mice compared to LMNAGT+/− and WT siblings (Sup. Fig. S1C).
Table 1.
Cardiac functioning upon partial and complete loss of lamin A/C
 |
*p<0.05 LMNAGT−/− vs. both age-matched LMNAGT+/− and WT values; #p<0.05 within the same genotype compared to the start of angiotensin II treatment. Abbreviations: IVSs = Intraventricular Septal Wall Thickness in systole, LVIDs = Left Ventricular Internal Dimension in systole, LVPWs = Left Ventricle Posterior Wall Thickness in systole, IVSd = Intraventricular Septal Wall Thickness in diastole, LVIDd = Left Ventricular Internal Dimension in diastole, LVPWd = Left Ventricle Posterior Wall Thickness in diastole, FS = Fractional Shortening.
To assess whether age-related physiological cardiac hypertrophy is subject to gene dosage effects 60–70 weeks old WT and LMNAGT+/− mice were subjected to echocardiography. Although the average FS was slightly decreased in LMNAGT+/− mice, this difference was not significant (23 ± 7% LMNAGT+/− vs. 30 ± 8% WT p > 0.05; Table 1). Other cardiac parameters did not differ significantly. Similarly, angiotensin II treatment induced comparable levels of hypertrophy under acute pathological conditions in 16–18 weeks old LMNAGT+/− and WT mice, based on relative LVWs (LMNAGT+/− from 5.8 ± 0.4 to 7.3 ± 0.6 mg/cm p < 0.05; WT from 6.1 ± 0.5 to 7.9 ± 0.6 mg/cm p < 0.05). No signs of cardiac failure were observed as FS, other echocardiographic parameters and relative lung weight were all comparable (p > 0.05; Table 1). Therefore, we conclude that partial loss of lamin A/C only slightly decreased LV weight without impairing the ability to respond structurally or functionally to increased load. Complete loss of lamin A/C impairs post-natal cardiac hypertrophy, lowers the heart rate moderately but does not cause any other direct functional abnormalities.
Skeletal muscle phenotype.
Initial gross anatomical examination showed hunched posture and abnormal gait, characterized by splayed hind legs from day PP13 onwards in LMNAGT−/− mice. Histological examination revealed smaller quadriceps skeletal myocytes at day PP15 (Fig. 5A). Based on transcriptome analysis and these initial findings we further assessed post-natal skeletal muscle development. Pathological findings were confirmed by quantification of the CSA of skeletal myocytes and the total quadriceps muscle weight (normalized to tibia length). Quadriceps muscles in LMNAGT−/− mice showed hypotrophy at PP15 (relative quadriceps weight 5.0 ± 0.5 mg/mm LMNAGT−/− vs. 7.1 ± 0.6 mg/mm WT p < 0.05) whereas they did not at PP9 (3.8 ± 0.95 mg/mm LMNAGT−/− vs. 4.7 ± 0.35 mg/mm WT p > 0.05; Fig. 5B). Accordingly, myocyte CSA did not differ at PP9 (195 ± 100 µm2 LMNAGT−/− vs. 259 ± 59 µm2 WT p > 0.05), but was significantly lower at PP15 (270 ± 55 µm2 LMNAGT−/− vs. 500 ± 50 µm2 WT p < 0.01; Fig. 5C). Consistent with these findings, LMNAGT−/− mice displayed reduced muscular strength (lag time dropping from grid: 0.9 ± 0.5 sec LMNAGT−/− vs. 6.7 ± 1.4 sec WT PP15; p < 0.05; Fig. 5D). Histological analysis did not show any overt signs of immature muscle fibers or centrally located nuclei (based on H&E staining), fibrosis (Sirius Red staining) or disorganized Z-disc (TEM; data not shown). These observations suggest that lamin A/C is important for maintaining skeletal muscle mass and strength.
Figure 5.
Post-natal skeletal muscle morphology and functioning in the LMNAGT−/− mouse. (A) Typical example of a haematoxylin and eosin (H&E) staining on quadriceps skeletal muscle sections of LMNAGT−/− and WT sibling (PP15). (B) Total quadriceps muscle weight normalized to tibia length assessed at PP9 and PP15 in LMNAGT−/−, LMNAGT+/− and WT mice (N = 8 each time point). (C) Quantification of the cross sectional area of quadriceps skeletal muscle myocytes for all 3 genotypes (PP9, PP15) (N = 8 each time point). (D) Assessment of skeletal muscle strength: time (in seconds) indicates the lag time until a mouse, hanging upside down, releases the grip; Asterisks in Figure 5 indicate significant differences for LMNAGT−/− vs. both LMNAGT+/− and WT values (p < 0.05; N = 4).
Adipogenic phenotype.
As pathological findings included reduced subcutaneous adipose tissue at PP15 in LMNAGT−/− mice, we further investigated the role of lamin A/C during early post-natal differentiation and maturation of adipose tissue. Subcutaneous adipose deposits at PP5 were comparable in weight for WT and LMNAGT−/− mice (weight corrected for tibia length: 0.38 ± 0.35 mg/mm LMNAGT−/− vs. 0.40 ± 0.17 mg/mm WT p > 0.05; Fig. 6A). During early post-natal development both WT and LMNAGT+/− mice increase their subcutaneous fat storage (PP14 1.13 ± 0.17 mg/mm WT p < 0.05 compared to PP5), while this storage was severely impaired in LMNAGT−/− mice (PP14 0.62 ± 0.16 mg/mm, p < 0.05 compared to age-matched WT). Gonadal fat patches and subscapular brown adipose tissue were present in all genotypes and macroscopically showed no obvious mass reduction.
Figure 6.
Adipogenic capacity of cells of the LMNAGT−/− mouse. (A) Quantification of 5 and 14 day old subcutaneous adipose tissue deposits normalized to tibia length (See Material & Methods; N = 5 per genotype for each timepoint). (B) Representative examples of Oil Red O staining at 16 days of differentiation for all genotypes. (C) Quantification of Oil Red O staining at 6, 10, and 16 days post induction. Asterisks in Figure 6 indicate significant differences for LMNAGT−/− vs. both LMNAGT+/− and WT values (p < 0.05; N = 3).
To directly investigate whether lack of subcutaneous adipose tissue could be caused by differentiation defects, E12.0 mouse embryonic fibroblasts (MEFs) were harvested from all 3 genotypes and adipogenic differentiation was induced ex vivo. Oil Red O lipid staining was substantially reduced upon complete loss of lamin A/C (0.284 ± 0.033 LMNAGT−/− vs. 0.615 ± 0.159 WT at 16 days post induction, p < 0.05; Fig. 6B and C). These findings suggest that that loss of lamin A/C results in impaired adipogenic differentiation and decreased post-natal subcutaneous fat deposition.
Metabolic parameters.
As loss of subcutaneous adipose tissue is expected to influence whole body metabolism, a metabolic fingerprint was taken at PP12 and PP16 by determining the status of carbohydrate, fat and protein metabolism. Carbohydrate metabolism was studied by measuring blood levels of glucose (the major carbohydrate energy source for the glycolytic pathway), liver glycogen levels (energy storage) and blood lactate (a by-product of anaerobic metabolism). Only lactate levels were reduced at PP12 (1.41 ± 0.37 mM LMNAGT−/− vs. 2.01 ± 0.54 mM WT p < 0.05). Metabolic complications were observed at PP16 (Table 2) as LMNAGT−/− mice were hypoglycemic (1.88 ± 0.97 mM LMNAGT−/− vs. 5.40 ± 1.54 WT p < 0.05) while blood lactate levels remained normal. In further agreement with lowered blood glucose levels, glycogen deposits in the liver were undetectable in LMNAGT−/− mice at PP18 in contrast to WT and LMNAGT+/− mice (Sup. Fig. 2).
Table 2.
Post-natal metabolic fingerprint of the LMNAGT−/− mouse
| Age | Parameter Measured | N= | wt | +/− | −/− | |||||||||
| PP9 | Body Core Temperature (°C) | 5 | 31.5 ± 2.0 | 32.1 ± 1.5 | 29.5 ± 0.9* | |||||||||
| Creatin Kinase (units/liter) | 4 | 646 ± 169 | 551 ± 231 | |||||||||||
| Creatinine (µM) | 4 | 8.0 ± 1.7 | 7.1 ± 1.2 | |||||||||||
| Urea (mM) | 4 | 8.7 ± 2.1 | 9.2 ± 1.2 | |||||||||||
| PP12 | Glucose (mM) | 6 | 3.20 ± 0.83 | 3.23 ± 0.80 | 3.08 ± 1.24 | |||||||||
| Lactate (mM) | 6 | 2.01 ± 0.54 | 2.52 ± 1.14 | 1.41 ± 0.37* | ||||||||||
| Betahydroxybutyrate (mM) | 6 | 0.97 ± 0.22 | 1.02 ± 0.20 | 0.90 ± 0.25 | ||||||||||
| C0, Free Carnitine (µM) | 6 | 31.88 ± 8.96 | 32.03 ± 2.85 | 29.07 ± 1.70 | ||||||||||
| C2, Non-Free Carnitine (µM) | 6 | 10.39 ± 3.60 | 11.52 ± 2.69 | 9.97 ± 1.33 | ||||||||||
| C0/C2, Free Camitine/Non-free Carnitine | 6 | 0.32 ± 0.06 | 0.31 ± 0.12 | 0.34 ± 0.05 | ||||||||||
| C5 carnitine (µM) | 6 | 0.38 ± 0.22 | 0.36 ± 0.21 | 0.38 ± 0.06 | ||||||||||
| C12 Carnitine (µM) | 6 | 0.13 ± 0.02 | 0.12 ± 0.02 | 0.08 ± 0.01* | ||||||||||
| C16, palmitoyi-carnitine (µM) | 6 | 0.75 ± 0.42 | 0.59 ± 0.22 | 0.40 ± 0.09 | ||||||||||
| C18:l, Oieoyl-carnitine (µM) | 6 | 0.26 ± 0.19 | 0.20 ± 0.11 | 0.12 ± 0.05 | ||||||||||
| PP15 | Body Core Temperature (°C) | 5 | 32.5 ± 1.0 | 32.5 ± 1.8 | 27.5 ± 0.8* | |||||||||
| Creatin Kinase (units/liter) | 4 | 667 ± 310 | 1561 ± 287* | |||||||||||
| Creatinin (µM) | 4 | 9.6 ± 2.3 | 8.8 ± 0.9 | |||||||||||
| Urea (mM) | 4 | 8.9 ± 3.3 | 10.2 ± 3.9 | |||||||||||
| PP16 | Glucose (mM) | 4 | 5.40 ± 1.54 | 4.94 ± 1.11 | 1.88 ± 0.97* | |||||||||
| Lactate (mM) | 4 | 3.28 ± 0.65 | 2.50 ± 0.62 | 2.81 ± 0.44 | ||||||||||
| Betahydroxybutyrate (mM) | 4 | 1.26 ± 0.21 | 1.26 ± 0.17 | 1.94 ± 0.18* | ||||||||||
| C0, Free Carnitine (µM) | 4 | 39.28 ± 9.06 | 33.01 ± 4.01 | 26.73 ± 2.6* | ||||||||||
| C2, Non-Free Carnitine (µM) | 4 | 14.76 ± 1.73 | 12.40 ± 2.06 | 18.86 ± 1.90* | ||||||||||
| C0/C2, Free Carnitine/Non-free Carnitine | 4 | 0.38 ± 0.05 | 0.38 ± 0.04 | 0.72 ± 0.17* | ||||||||||
| C5 carnitine (µM) | 4 | 0.41 ± 0.28 | 0.25 ± 0.13 | 0.40 ± 0.20 | ||||||||||
| C12 Carnitine (µM) | 4 | 0.15 ± 0.03 | 0.14 ± 0.05 | 0.23 ± 0.12 | ||||||||||
| C16, palmitoyi-carnitine (µM) | 4 | 0.57 ± 0.15 | 0.57 ± 0.08 | 0.86 ± 0.13* | ||||||||||
| C18:1, Oieoyl-carnitine (µM) | 4 | 0.14 ± 0.06 | 0.17 ± 0.06 | 0.25 ± 0.17 | ||||||||||
p<0.05 LMNAGT−/− vs. both LMNAGT+/− and WT values
Fat metabolism was monitored by determining the acylcarnitine profile (carnitine-coupled free fatty acid (FFA) substrates for β-oxidation), and by measuring blood levels of the ketone body betahydroxybutyrate (by-product of fatty acid catabolism; alternative energy source during hypoglycemia).17 At PP12 most of these parameters were normal, with the exception of lowered C12-carnitine levels (0.08 ± 0.01 pmol/mg LMNAGT−/− vs. 0.13 ± 0.02 pmol/mg WT p < 0.05). Metabolic complications became apparent in LMNAGT−/− mice at PP16 (Table 2), as betahydroxybutyrate levels were elevated (1.94 ± 0.18 mM LMNAGT−/− vs. 1.26 ± 0.21 mM WT p < 0.05) providing evidence for increased ketogenesis. Acylcarnitine analysis showed decreased free carnitine in combination with elevated acetylcarnitine and long chain acylcarnitines (Table 2), consistent with increased fatty acid catabolism. The combination of hypoglycemia with increased betahydroxybutyrate and acylcarnitine are highly suggestive of fasting.
As general markers for protein catabolism, creatin kinase, creatinin and urea were determined. Blood creatin kinase levels (marker for muscle damage or wasting), were elevated at PP15 (1,561 ± 287 U/L LMNAGT−/− vs. 667 ± 310 U/L WT p < 0.05), but not at PP9. However, urea and creatinin levels (indicators of protein breakdown and kidney functioning), did not differ significantly (urea: 10.2 ± 3.9 mmol/L LMNAGT−/− vs. 8.9 ± 3.3 mmol/L WT; creatinin: 8.8 ± 0.9 µmol/L LMNAGT−/− vs. 9.6 ± 2.3 µmol/L WT; PP15; p > 0.05). In addition to these metabolic parameters, core body temperature was lower both at PP9 (29.5 ± 0.9°C LMNAGT−/− vs. 31.5 ± 2.0°C WT p < 0.05) and PP15 (27.5 ± 0.8°C LMNAGT−/− vs. 32.5 ± 1.0°C WT p < 0.01).
As the metabolic fingerprint and the expression array analysis (Fig. 3B) supported an abnormal metabolic phenotype, morphology and function of mitochondria was assessed. Transmission electron microscopy revealed no differences in mitochondrial morphology (Sup. Fig. 3A). Mitochondrial DNA copy numbers were comparable in both heart and quadriceps skeletal muscle at PP15 (p > 0.05; Sup Fig. 3B). Electron transport chain subcomplex activities were determined in LV tissue at PP15 and were shown to be almost fully comparable between WT and LMNAGT−/− mice (Sup. Fig. 3B). A slight elevation in the amount of mitochondria was observed, however (CS 1,712 ± 307 LMNAGT−/− vs. 1,508 ± 118 WT p < 0.05), and complex II activity was slightly, but significantly lowered (0.114 ± 0.00024 LMNAGT−/− vs. 0.122 ± 0.007 WT p < 0.01). Together these observations show the LMNAGT−/− mice become severely catabolic in the 3rd week post partum, without apparent abnormal mitochondrial function.
Discussion
Lamin A/C expression is developmentally regulated and has been specifically linked to early mesenchymal commitment.4–6 Little is known, however, about the effect of A-type lamins in embryonic and early post-natal development. We have generated a novel LMNAGT−/− mouse model and demonstrate that loss of A-type lamins results in defective post-natal differentiation and defective maturation of adipose-, cardiac- and skeletal muscle tissue.
For many essential organs in mice, differentiation and maturation start during embryonic development and continues throughout early post-natal development.22–24 Immunohistochemical studies revealed a diffuse lamin A/C protein staining in connective tissue and myoblasts at E11.0.4,5 Lamin A/C protein levels increase until day E14.0 and lamin A/C protein was only detected in a few more tissues during embryonic development from this age onwards. Lamin A/C was reported to be absent in epithelia of lung, liver, kidney, intestine, heart and brain until well after birth. Using a β-galactosidase reporter gene in our LMNAGT−/− mouse model we here report LMNA promoter activation, coinciding with the onset of tissue differentiation (E11.0) in the heart, outflow tract, the dorsal aorta, liver and somites. In good agreement, promoter activity of the lamin A processing enzyme Zmpste24 in a GT mouse model partially mirrors this LMNA promoter activity.16 It is conceivable that previous immunohistochemical studies may have been limited in their sensitivity and specificity toward e.g., potential embryonic LMNA splicing variants. The detection of LMNA promoter activity from the earliest stages of tissue differentiation onward supports the importance of lamin A/C in tissue differentiation and maturation. Although we cannot formally exclude discordant mRNA and protein synthesis, e.g., as a result of pre-/posttranslational regulation, our findings suggest that lamin A/C is expressed earlier than previously reported.
Despite apparently morphologically normal overall fetal development, the consequences of loss of lamin A/C becomes apparent during early post-natal development. By and large, post partum development in the absence of LMNA reveals no gross phenotypical abnormalities from PP1 until PP6. The growth of LMNAGT−/− mice is retarded from PP7 to PP12 compared to WT and LMNAGT+/− siblings. We observe a prominent abnormal post-natal cardiac development in LMNAGT−/− mice: whereas normal LV myocytes undergo post-natal hypertrophy upon cell cycle exit in the first week after birth, cardiac hypertrophy is severely impaired in LMNAGT−/− mice.22,25 The comparable post partum decrease in cardiac proliferative activity (PCNA, Ki67) between WT and LMNAGT−/− siblings suggests altered hypertrophy is not caused by an inability of myocytes to exit the cell cycle. Instead, elevated expression of the hypertrophic markers ANF and BNP, suggests that pro-hypertrophic signaling is increased in the absence of productive physiological cardiac hypertrophy. Finally, overall body growth ceases between PP13 and PP18 and BW decreases in LMNAGT−/− mice. During this stage, skeletal muscle display hypotrophy and a prominent catabolic phenotype is manifested, with reduced blood glucose (PP16), absent hepatic glycogen storage (PP18) and increased utilization of alternative energy sources (ketone bodies, FFA and muscle tissue; PP15–16). Although the direct cause of death is currently not clear, it most likely relates to a combination of muscle weakness and metabolic complications. Cardiac function appears not sufficiently impaired to explain premature death at this stage, but we cannot exclude the possibility that the short life-span of these mice prevents further development of cardiac malfunction. Decreased heart rate and core body temperature are typical side effects of a catabolic state. Both parameters decline in human anorexia nervosa patients as does blood pressure.26 Progressive lowering of blood pressure and heart rate further deteriorates the condition during starvation.27 This notion also provides an explanation for the detrimental effects of labetalol in LMNAGT−/− mice, which furthermore demonstrates that in the end-stage increased sympathetic activity prolongs survival to some extent, possible by its effects on overall metabolism. Although it is unclear at this stage whether muscular wasting contributes to the abnormal metabolic state of LMNAGT−/− mice, the impaired adipogenic capacity upon loss of lamin A/C provides a direct etiological connection to the defective energy metabolism.
The LMNAGT−/− mouse complements previously established LMNA murine models. Using a conventional gene targeting strategy, Stewart and colleagues created a functional LMNA knock-out mouse (LMNAKO−/−) which displays growth retardation starting 2–3 weeks post partum, followed by skeletal abnormalities, decreased subcutaneous adipose deposits, cardiac dilatation and, ultimately at 6–8 weeks post partum, life-threatening impaired cardiac function.20,28,29 Considering the onset and progression of these symptoms over time, the overall phenotype of the LMNAKO−/− animals is comparatively milder than that of LMNAGT−/− mice. A possible explanation for this phenotypic difference is the targeting strategy used in the two models. We consider it unlikely that the β-geo fusion protein itself causes dominant negative effects as LMNAGT+/− mice are indistinguishable from WT mice in all assays of our study. In contrast to the LMNAGT−/− mouse, where a GT sequence is introduced, in the LMNAKO−/− mouse exons 8 to 10 of LMNA are replaced by a PgKNeo cassette.20 Differences in phenotype due to varying targeting are not uncommon. For instance, the lamin A processing enzyme Zmpste24 loss-of function mouse model shows a more dramatic phenotype when several exons were replaced by a GT sequence compared to a conventional gene targeting strategy.16,30 In addition, phenotypes may be affected by genetic background.31 While LMNAGT−/− mice were generated in a mixed C57Bl/6 and 129S4/SvJae background LMNAKO−/− mice were in a mixed C57Bl/6 and 129Sv1/Sv genotype.20 Compared to other LMNA mouse models, lethality in the LMNAGT−/− model occurs much earlier, and notably, in the absence of cardiac functional defects and HGPS related bone abnormalities and hair loss.13 Furthermore, in contrast to LMNAKO+/− mice, which develop progressive electrophysiological abnormalities and cardiac left ventricle dilation from the onset of 4 weeks after birth, we did not detect any such defects in LMNAGT+/− mice up to one year in age, further highlighting differences in the models.32
The combined observations presented here show that the LMNAGT−/− mouse model is the most severe lamin A/C mouse model to date and, relevantly, our results show that loss of lamin A/C induces severe and widespread defects in post-natal tissue maturation. This leads to an early post-natal and fully penetrant lethal phenotype, which is conceivably due to combined muscle weakness and metabolic complications, and occurs in the absence of a dilated cardiomyopathy or obvious progeroid phenotype. This novel LMNA mouse provides new opportunities to dissect the molecular and cellular mechanisms underlying the role of lamin A/C in early post-natal development.
Materials and Methods
Targeting of the LMNA gene.
LMNAGt(FHCRC-GT-S7-1F1)Sor AK7.1 ES cells (strain of origin 129S4/SvJaeSor) containing a ROSAFARY Genetrap (GT) vector in the LMNA gene were obtained from the International Genetrap Consortium (www.genetrap.org). Chimeric mice were created by ES cell injection into C57Bl/6 recipient blastocysts at the Transgenesis and gene Targeting Unit Maastricht (Maastricht, The Netherlands). Resulting chimeric males were bred to wild-type (WT) C57Bl/6 mice. Resulting heterozygous LMNAGT (LMNAGT+/−) mice were interbred to achieve homozygocity (LMNAGT−/−). For genotyping, PCR-primer pairs Fw (forward) 5′-GAC GGG TTG TTA CTC GCT CAC-3′ and Rev (reverse) 5′-CAG GTC AAA TTC AGA CGG CAA A-3′ were used to selectively amplify GT; Fw 5′-ACC ATC TCC CCA GCC CTT AG-3′ and Rev 5′-CAA CAT TCC TGA TTC TTT CTG C-3′ to detect the endogenous LMNA gene. All studies involving animal experiments were approved by the Animal Care and Usage Committee of Maastricht University (Maastricht, The Netherlands) or the National Institutes of Health (NIH, Bethesda, USA) and were performed in accordance with regulations formulated in Dutch or US law on care and use of experimental animals.
Histology, immunohistochemisty and transmission electron microscopy (TEM).
LMNAGT mice were sacrificed by decapitation or cervical dislocation. Blood was removed from the vascular system by perfusing 5 ml physiological buffered saline via the left ventricle (LV) of the heart; muscle relaxation was induced by a consecutive injection with 100 µl of a 0.1 N cadmium chloride solution. Tissues for histological analysis and immunohistochemistry were fixed for 4–6 h in 3.7% paraformaldehyde, washed three times 5 min in 70% ethanol, embedded in paraffin and sectioned at 5 microns. Tissues sections were stained with hematoxylin eosin (H&E) or Picro Sirius Red (SR) as described before in reference 16. H&E staining was used for quantification of cardiac- and skeletal-myocyte cross sectional areas. Impaired post-natal hypertrophy was quantified by cardiac myocyte cross-sectional area (CSA) counts in LV myocytes adjacent to the septum. Tissues for RNA isolation were snap-frozen in liquid nitrogen. Tissues for TEM analysis were fixed in 2.5% glutaraldehyde and further analysis was performed as previously described in reference 15. Whole-mount embryos were β-galactosidase stained as described in reference 16. For Azo Phloxine counterstaining tissue sections (7 µm) were deparrafinized in xylene (3 × 7 min), and rehydrated in a series of graded ethanol (3× 100, 96, 90, 80, 70 and 50%). Next, the sections were rinsed in running bi-distilled water for 5 min, after which the sections were incubated with Azo Phloxine (0.1% in bi-distilled water, filtered and addition of 0.05% acetic acid shortly before use) for 3 min. Sections were then rinsed in running tap water for 15 min, and in running bi-distilled water for 2 min. Next, sections were dehydrated quickly in a graded series of ethanol (50, 70, 80, 90, 96 and 3× 100%). Sections were then treated with xylene for 3 × 7 min and mounted with Entellan (Merck 7961).
RNA isolation and microarray analysis.
RNA was isolated from mouse embryonic fibroblasts (MEF) and LV of LMNAGT mice using the RNAeasy minikit (Qiagen, Hilden, Germany). For whole transcriptome analyses, RNA from LV's of 5 and 13 d old WT and LMNAGT−/− mice (N = 2 for each genotype and age) was isolated and hybridized to Nugo Mouse Affymetrix expression arrays. Intensity values after hybridization were normalized to the median signal intensity of the array, and further analyzed by using Microarray Analysis Suite Software (version 5.0).15 Enrichment of deregulated genes in specified biological pathways was tested using GENMAPP 2.1 software. For expression analysis by PCR cDNA was synthesized with the iScript™ cDNA synthesis kit (BioRad, Hercules, USA). SYBR Green real-time quantitative PCR analysis was performed with primers for ANF (Fw 5′-ATT GAC AGG ATT GGA GCC CAG AGT-3′, Rev 5′-TGA CAC ACC ACA AGG GCT TAG GAT-3′), BNP (Fw 5′-GTT TGG GCT GTA ACG CAC TGA-3′, Rev 5′-GAA AGA GAC CCA GGC AGA GTC A-3′), Ki67 (Fw 5′-TCA ACA GCT GGT ATG CCT AAC AG-3′, Rev 5′-TTC CAG TGG TCA AAG AGT CAT TAG C-3′), PCNA (Fw 5′-AGG GTT GGT AGT TGT CGC TGT AG-3′, Rev 5′-GGT CCC CCG ATT CAC GAT-3′), GAPDH (Fw 5′-GGT GGA CCT CAT GGC CTA CA-3′, Rev 5′-CTC TCT TGC TCA GTG TCC TTG CT-3′) HPRT (Fw 5′-GCG TCG TGA TTA GCG ATG ATG AAC-3′, Rev 5′-CCT CCC ATC TCC TTC ATG ACA TCT-3′), Lamin A specific (Fw 5′-GGA TCC ATC TCC TCT GGC TCT T-3′; exon 10, Rev 5′-AGG TAG GAG CGG GTG ACT AGG T-3′; exon 12), Lamin C specific (Fw 5′-GTT GAG GAC AAT GAG GAT GAC G-3′; exon 10, Rev 5′-AAA AGA CTT TGG CAT GGA GGT G-3′; exon 10), ?Geo (Fw 5′-ATT CTG TGG CCA AGG AGC G-3′; exon 1, Rev 5′-CAG GTC AAA TTC AGA CGG CAA A-3′; βGeo), Lamin RNA containing exon 1 spliced to exon 2 (Fw 5′-ATT CTG TGG CCA AGG AGC G-3′; exon 1, Rev 5′-TGC GCA GCC AAC AAG TCC-3′; exon 2), Lamin RNA containing exon 1 spliced to exon 7 (Fw 5′-ATT CTG TGG CCA AGG AGC G-3′; exon 1, Rev 5′-CTC GTT GGA CTT GTT GCG-3′; exon 7), Lamin RNA containing exon 1c spliced to exon 2 (Fw 5′-ATG GGG AAC GCG GAG GG-3′; exon 1c, Rev 5′-TGC GCA GCC AAC AAG TCC-3′; exon 2), Lamin RNA containing exon 1c spliced to exon 7 (Fw 5′-ATG GGG AAC GCG GAG GG-3′; exon 1c, Rev 5′-CTC GTT GGA CTT GTT GCG-3′; exon 7).
Echocardiography and electrocardiogram (ECG) analysis.
WT, LMNAGT+/− and LMNAGT−/− cardiac functioning was monitored by serial echocardiography and ECG analysis at days 10, 13 and 17 post partum (PP10, PP13 and PP17). ECG analysis was performed under 2% isoflurane sedation; RR interval, P wave, PR, QRS and QT time were assessed using IDEEQ v1.7 software (Instrument Development Engineering & Evaluation, Maastricht University, The Netherlands). Echocardiography was performed under similar sedative conditions, with a 30 MHz transducer on a Vevo770™ high-resolution in vivo micro-imaging system (VisualSonics BV, Amsterdam, The Netherlands). LV parameters were obtained from M-mode recordings in the short axis-view. Fractional shortening (FS) was calculated using the formula: FS = 100 × [(Left Ventricle Inner Diameter in diastole − Left Ventricle Inner Diameter in systole)/Left Ventricle Inner diameter in diastole]. To study the effects of α- and β-adrenergic receptors antagonists on LMNAGT−/− cardiac performance, labetalol was administered twice daily by subcutaneous interscapular injections at a dose of 10 mg/kg from day PP10 onward; ECG analysis was performed as indicated above at day PP13. To study the effects of LMNA heterozygosity on cardiac hypertrophy, echocardiography was performed as indicated above on 60–70 w old LMNAGT+/− mice and WT controls. Furthermore 16–18 w old WT and LMNAGT+/− mice were infused subcutaneously with 1.5 µg/g/day angiotensin II for 4 w by an osmotic minipump (Alzet osmotic minipumps, Cupertino, CA). Echocardiograms were made at 2 and 4 w after the start of the angiotensin II treatment.
Funtional assessment of skeletal muscles.
Total quadriceps muscle weight was normalized to tibia length for all genotypes. Skeletal muscle strength was assessed by quantifying the capacity to hang upside down on a grid with use of both fore- and hind limbs. Time in seconds indicates the time from placing the raster upside down until the mouse dropped from the grid. Measurements on each genotype were performed in triplicate (n = 4, PP15).
Adipocyte differentiation assay.
MEFs were isolated from E12.0 LMNAGT−/−, LMNAGT+/− and WT embryos by digesting skin isolates for 20 min in 3 ml 0.05% trypsin at 37°C during which the solution was resuspended every 10 minutes. This was followed by addition of an equal volume of fresh trypsin solution and a 20 min incubation step; this was repeated once and incubated for 5 additional min, after which a cell pellet was obtained by centrifugation and plated in Dulbecco's modified eagle medium (11360-070, Invitrogen, San Diego, USA) with 2 mM L-glutamine, 10 mM non-essential amino acids (NEAA), 110 mg/l pyruvate, 10% fetal bovine serum (FBS) and antibiotics. Passage 2 (P2) cells were seeded at 50% confluency in 6-wells plates for adipogenic differentiation and grown to confluency for 3 d. The medium was replaced and cells were left for 3 d on induction medium: hMSC basal medium (PT-3238, Lonza, Basel, Switzerland) with 2 mM L-glutamine, 10 mM NEAA, 110 mg/l pyruvate, 10% FBS, antibiotics, 10 µg/ml insulin, 0.5 mM 3-isobutyl-l-methylxantine (IBMX) and 1 µM dexamethasone. After 3 d the medium was replaced by maintenance medium (similar to induction medium with omission of IBMX and dexamethasone) and left for 2 d. This 5 d cycle of induction and maintenance was repeated three times. Oil Red O staining and quantification after each cycle (6, 10 and 16 d post induction) were performed in triplicate as previously described in reference 12.
Metabolic characterization.
To assess core body temperature, mice received an inter-intestinal temperature probe through a small abdominal incision directly after applying isoflurane anesthesia. Core body temperature was measured as soon as it reached a plateau phase (i.e., before ambient temperature decreased body temperatures). Subcutaneous adipose tissue deposits were determined by incising the skin along the saggital axis, collecting and weighing subcutaneous adipose tissue in between the left fore- and hind leg. Subcutaneous adipose tissue weights were standardized by tibia length. For blood chemistry measurements, WT, LMNAGT+/− and LMNAGT−/− mice were intraperitoneally injected with 50 µl of a 500 Units/ml heparin solution as an anticoagulant before decapitation and collection of blood. For glucose, lactate and betahydroxybutyrate analysis the collected blood was mixed with equal amounts of 1 M perchloric acid. After centrifugation the supernatant was neutralized using 2 M KOH, 0.5 M 2-(N-morpholino)ethanesulfonic acid. Glucose, lactate and β-hydroxybutyrate concentrations were measured in the neutralized supernatant after removal of KClO4 using established enzymatic procedures.17 The acylcarnitine spectrum was determined by tandem mass spectrometry on regular blood plasma as described in reference 17. Urea, creatinin and creatin kinase were determined on EDTA anticoagulated blood. For mitochondrial functioning LV samples were snap frozen in liquid nitrogen and used to determine catalytic activity of citrate synthase and individual oxidative phosphorylation complexes as described in reference 18. Mitochondrial DNA copy numbers were assessed in LV cardiac muscle and quadriceps skeletal muscle.18
Western blotting and immunofluorescence.
For immunofluorescence microscopy LMNAGT−/−, LMNAGT+/− and WT MEFs were grown on 0.1% gelatin-coated multi-wells glass slides (MP Biomedicals). After fixation (4% paraformaldehyde/PBS; 15 min), cells were permeabilized (0.5% Triton X-100/PBS; 5 min), blocked (1% BSA, 1% sucrose in milliQ; 1 h) and incubated for one hour with appropriate primary antibodies (See Fig. legend A). Further immunofluorescence detection was done as described in reference 19. Western blots were performed as described in reference 19.
Statistical analysis.
Data are represented as average ± SD. The data for each study group were compared using 2-way ANOVA or student's t-test where appropriate. Analyses were performed using the statistical package SPSS 11.0. p < 0.05 was considered to be statistically significant.
Acknowledgments
We thank S. Frints, M.W. Schellings, C. van het Hoofd and A. Voets for technical expertise and help. J.P. Cleutjens is thanked for help with quantification of myocyte cross-sectional areas, H. Duimel for electron microscopy imaging, and P. Scaffidi and S.G. Rane for adipogenic differentiation assays. Fast and reliable assistance in arranging animal experiments by members of the central animal facilities was instrumental for phenotypic analyses. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), NCI, Center for Cancer Research, grants from the Dutch Heart Foundation, ZonMW and the EU-KP7 grant ‘Inheritance’.
Abbreviations
- IBMX
3-isobutyl-l-methylxantine
- ANF
atrial natriuretic factor
- BW
body weight
- BNP
brain natriuretic peptide
- CS
citrate synthase
- CSA
cross-sectional area
- DCM
dilated cardiomyopathy
- DN
dominant negative
- FPLD
dunnigan-type familial partial lipodystrophy
- ECG
electrocardiogram
- FC
fold change
- Fw
forward
- FS
fractional shortening
- FFA
free fatty acids
- GT
genetrap
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- H&E
hematoxylin & eosin
- HE
heterozygous
- LMNAGT+/−
heterozygous LMNA genetrap
- LMNAGT−/−
homozygous LMNA genetrap
- HGPS
hutchison-gilford progeria syndrome
- HPRT
hypoxanthine-guanine phosphoribosyl transferase
- IVSd
intraventricular septal wall thickness in diastole
- IVSs
intraventricular septal wall thickness in systole
- KO
knock-out
- LV
left ventricle
- LVIDd
left ventricular internal dimension in diastole
- LVIDs
left ventricular internal dimension in dsystole
- LVPWd
left ventricle posterior wall thickness in diastole
- LVPWs
left ventricle posterior wall thickness in systole
- LVW
left ventricle weight
- MEF
mouse embryonic fibroblast
- PAS
periodic acid shiff
- SR
picro sirius red
- PP
post partum
- Rev
reverse
- SREBP1
sterol element binding protein 1
- TEM
transmission electron microscopy
- WT
wild-type
Supplementary Material
References
- 1.Prather RS, Sims MM, Maul GG, First NL, Schatten G. Nuclear lamin antigens are developmentally regulated during porcine and bovine embryogenesis. Biol Reprod. 1989;41:123–132. doi: 10.1095/biolreprod41.1.123. [DOI] [PubMed] [Google Scholar]
- 2.Schatten G, Maul GG, Schatten H, Chaly N, Simerly C, Balczon R, et al. Nuclear lamins and peripheral nuclear antigens during fertilization and embryogenesis in mice and sea urchins. Proc Natl Acad Sci USA. 1985;82:4727–4731. doi: 10.1073/pnas.82.14.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- 4.Röber RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 5.Röber RA, Sauter H, Weber K, Osborn M. Cells of the cellular immune and hemopoietic system of the mouse lack lamins A/C: distinction versus other somatic cells. J Cell Sci. 1990;95:587–598. doi: 10.1242/jcs.95.4.587. [DOI] [PubMed] [Google Scholar]
- 6.Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 7.Peter M, Nigg EA. Ectopic expression of an A-type lamin does not interfere with differentiation of lamin A-negative embryonal carcinoma cells. J Cell Sci. 1991;100:589–598. doi: 10.1242/jcs.100.3.589. [DOI] [PubMed] [Google Scholar]
- 8.Van Berlo JH, Voncken JW, Kubben N, Broers JL, Duisters R, van Leeuwen RE, et al. A-type lamins are essential for TGFbeta1 induced PP2A to dephosphorylate transcription factors. Hum Mol Genet. 2005;14:2839–2849. doi: 10.1093/hmg/ddi316. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z, Wu L, Weng D, Xu D, Geng J, Zhao F. Reduced expression of lamin A/C correlates with poor histological differentiation and prognosis in primary gastric carcinoma. J Exp Clin Cancer Res. 2009;28:8. doi: 10.1186/1756-9966-28-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 11.Lloyd DJ, Trembath RC, Shackleton S. A novel interaction between lamin A and SREBP1: implications for parial lipodystrophy and other laminopathies. Hum Mol Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- 12.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 14.Junqueira LC, Bignolas G, Brentani RR. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem J. 1979;11:447–455. doi: 10.1007/BF01002772. [DOI] [PubMed] [Google Scholar]
- 15.Schroen B, Leenders JJ, van Erk A, Bertrand AT, van Loon M, van Leeuwen RE, et al. Lysosomal integral membrane protein 2 is a novel component of the cardiac intercalated disc and vital for load-induced cardiac myocyte hypertrophy. J Exp Med. 2007;204:1227–1235. doi: 10.1084/jem.20070145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 17.Costa CC, de Almeida IT, Jakobs C, Poll-The BT, Duran M. Dynamic changes of plasma acylcarnitine levels induced by fasting and sunflower oil challenge test in children. Pediatr Res. 1999;46:440–444. doi: 10.1203/00006450-199910000-00013. [DOI] [PubMed] [Google Scholar]
- 18.van den Bosch BJ, van den Burg CM, Schoonderwoerd K, Lindsey PJ, Scholte HR, de Coo RF, et al. Regional absence of mitochondria causing energy depletion in the myocardium of muscle LIM protein knockout mice. Cardiovasc Res. 2005;65:411–418. doi: 10.1016/j.cardiores.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 19.Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szlachcic J, Hall WD, Tubau JF, Porter V, Vollmer C, Wollam G, et al. Left ventricular hypertrophy reversal with labetalol and propranolol: a prospective randomized, double-blind study. Cardiovasc Drugs Ther. 1990;4:427–433. doi: 10.1007/BF01857749. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293:1883–1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- 23.Hew KW, Keller KA. Postnatal anatomical and functional development of the heart: a species comparison. Birth Defects Res B Dev Reprod Toxicol. 2003;68:309–320. doi: 10.1002/bdrb.10034. [DOI] [PubMed] [Google Scholar]
- 24.Wu S, Platteau A, Chen S, McNamara G, Whitsett J, Bancalari E. Conditional overexpression of connective tissue growth factor disrupts postnatal lung development. Am J Respir Cell Mol Biol. 2009 doi: 10.1165/rcmb.2009-0068OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:2183–2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 26.Gottdiener JS, Gross HA, Henry WL, Borer JS, Ebert MH. Effects of self-induced starvation on cardiac size and function in anorexia nervosa. Circulation. 1978;58:425–433. doi: 10.1161/01.cir.58.3.425. [DOI] [PubMed] [Google Scholar]
- 27.Pirke KM. Central and peripheral noradrenalin regulation in eating disorders. Psychiatry Res. 1996;62:43–49. doi: 10.1016/0165-1781(96)02982-4. [DOI] [PubMed] [Google Scholar]
- 28.Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, Jogia D, et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cutler DA, Sullivan TS, Marcus-Samuels B, Stewart CL, Reitman ML. Characterization of adiposity and metabolism in Lmna-deficient mice. Biochem Biophys Res Commun. 2002;291:522–527. doi: 10.1006/bbrc.2002.6466. [DOI] [PubMed] [Google Scholar]
- 30.Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness and a prelamin A processing defect. Proc Natl Acad Sci USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rankin J, Ellard S. The laminopathies: a clinical review. Clin Genet. 2006;70:261–274. doi: 10.1111/j.1399-0004.2006.00677.x. [DOI] [PubMed] [Google Scholar]
- 32.Wolf CM, Wang L, Alcalai R, Pizard A, Burgon PG, Ahmad F, et al. Lamin A/C haploinsufficiency causes dilated cardiomyopathy and apoptosis-triggered cardiac conduction system disease. J Mol Cell Cardiol. 2008;44:293–303. doi: 10.1016/j.yjmcc.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.