Abstract
Three alternatively spliced forms of the human NOL7 gene coding for relatively small proteins were identified. The two shorter forms were generated by intron retention events, and each isoform was differently localized within the cell. The NOL7-SP1 long form (29 kD) localized to the nucleolus, SP2 was nucleoplasmic, while SP3 was distributed throughout the whole cell. NOL7-SP1 was confined to the nucleolar granular component, and during cell division disassociated from the nucleolus. Knockdown of NOL7-SP1 levels abrogated nucleolar architecture, in particular the internal regions, and reduced cell proliferation. Analysis of the nucleolar dynamics of the SP1 protein during interphase showed nucleolar high binding affinity. Dissection of protein domains showed that nucleolar targeting was mediated by a unique C-terminal nucleolar localization sequence (NoLS). However, this sequence was not sufficient for conferring high binding affinity, which required additional regions of the protein. Our analysis shows that NOL7 is important for maintaining internal nucleolar structure and cell growth rates, and that while specific protein localization can be obtained by specific short localization motifs, nucleolar residency through binding must be mediated by a synergistic combination of protein modules.
Key words: nucleolus, nuclear dynamics, alternative splicing, NOL7, nucleolar localization sequence
Introduction
The nucleolus plays a major role in ribosome biogenesis, namely in the transcription and processing of rRNAs, and the assembly of pre-ribosomal subunits.1,2 Other non-ribosomal nucleolar functions have been characterized in recent years and have brought to light the multi-functionality of the nucleolus in normal and diseased states.3,4 Although the nucleolus is not membrane-surrounded like other cellular organelles, it is possible to define specific nucleolar activities within three sub-structures.5 These have been termed morphologically, based on electron microscopy, as the fibrillar center (FC), dense fibrillar component (DFC) and granular component (GC).6 Specific functions are assigned to each region thereby generating a factory-like structure. Tandemly repeated rRNA genes are clustered mostly in the FCs which are enriched in RNA polymerase I, topoisomerase I and upstream binding factor (UBF). The FC-DFC boundary is the predominant site of pre-rRNA transcription by RNA polymerase I. Pre-rRNA undergoes processing, modifications and maturation within the DFC by proteins such as fibrillarin, nucleophosmin (B23) and nucleolin (C23). Some processing steps may also occur in the GC, together with the assembly of the mature rRNAs (18S, 5.8S, 28S from Pol I transcription and 5S from Pol III transcription) and ribosomal proteins into the two pre-ribosomal subunits.7–9 Altogether, the main body of the nucleolus is made up of the GC, and embedded in this granular mass are the FCs, each surrounded by a compact layer of the DFC. Although nucleoli are usually considered to be nuclear bodies that hold rather fixed positions in the interphase nucleus, most nucleolar protein components are in constant flux with the nucleoplasm, as has been measured using a variety of photobleaching techniques10,11 and demonstrated by proteomic analysis.12
The separation between the cytoplasmic and nuclear compartments requires a molecular mechanism that enables the recognition of protein cargo in the cytoplasm, translocation through the NPC, and release in the nucleoplasm. Nuclear localization sequences (NLS) facilitate NPC mediated import of nuclear proteins into the nucleus. The first identified NLS was found in the SV40 T antigen (PKKKRKV).13,14 Nucleolar localization sequences (NoLS) are less well-defined,15 although some sequence-specific information is available.16 For instance, the NoLS of NFκB inducing kinase (NIK) is mediated by the basic RKKRKKK sequence.17 Viral proteins can contain arginine-rich NoLS motifs,18–20 and a common R/K-R/K-X-R/K motif has been designated on the basis of several proteins such as nucleophosmin, ribosomal protein S6 and the ARF protein.21 The effects of the NoLS are not well understood. For instance, does the NoLS serve as a physical nucleolar targeting sequence only, or can this sequence also confer high binding affinity and prolonged residence times within the nucleolus.
We examined the nuclear and nucleolar dynamics of three alternatively spliced products of the NOL7 gene (chr. 6p23). A previous study showed that the NOL7 gene can undergo allelic loss in certain cervical carcinoma (CC) cell lines and tumor samples.22 Loss of heterozygosity (LOH) is a common molecular genetic alteration found in human cancers, which may result in the loss of a normal tumor suppressor gene allele, thereby promoting cell growth and conferring survival advantages during tumorigenesis. Therefore, NOL7 is considered a tumor suppressor gene candidate. An important role of LOH in the development of CC has been suggested, and indeed a number of allelotyping studies of CC tumor DNA have shown that chromosome regions 6p, along with 3p and 11q, are most often frequently affected in CC.23 Detailed LOH analysis with a larger set of markers has revealed allelic losses at chromosome band 6p23.24 Expression of NOL7 inhibited the growth of CC cells in mouse xenografts and its effect might be related to the modulation of the angiogenicity of the tumor. The originally published GFP-NOL7 gene-product exhibited nucleolar localization,22 as expected from proteomic analysis of isolated nucleoli,12,25 and recently several of its NLS and NoLS sequences were characterized.26
We characterized 3 alternative transcripts generated from the NOL7 gene and detected the endogenous proteins of the two larger forms. Even though these proteins have low molecular weights, and therefore could disperse throughout the whole cell by diffusion, we found that the proteins have distinct distribution patterns. We determined the amino acid sequences conveying the nuclear localization and nucleolar localizations of the NOL7 products. Using truncations and dynamic analysis we demonstrated how targeting sequences can define the sub-cellular targeting and location of a protein, but were not sufficient for generating a long-lasting high affinity interaction within the structure. Finally, we found that some nucleolar processing functions were maintained even when defects in nucleolar architecture and cell proliferation rates were induced under conditions of NOL7 depletion.
Results
Alternative spliced forms of the NOL7 gene.
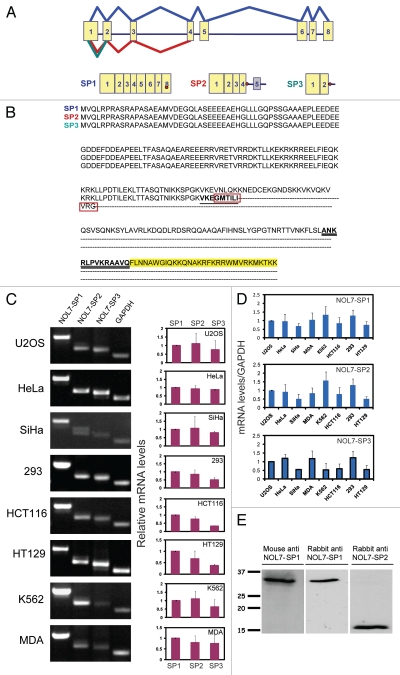
In order to examine the kinetic properties of the NOL7 protein in living cells we cloned the full length NOL7 coding sequence. The NOL7 gene is situated on chromosome 6 in humans and consists of 8 exons. The gene and coding region are highly conserved in chimpanzee, dog, cow, mouse, rat, chicken and zebrafish. The human coding region (874 bp, 257 aa) encodes a 29 kD protein, termed NOL7-SP1 (Fig. 1A). Since partial sequence information in EST sequence databases indicated the possibility of additional transcripts transcribed from this gene, we used different primer combinations to identify two alternatively spliced forms of the gene, both formed by an intron retention event. Intron 4 retention was found in NOL7-SP2, while in NOL7-SP3 intron 2 was retained (Fig. 1A). The span of intron 4 retention in NOL7-SP2 extended into intron 5, as further examined by RT-PCR with a sense primer to exon 4 and an antisense primer to intron 5 (not shown). We did not identify other forms with retained introns 3 or 5. Both of the alternatively spliced mRNAs encode for significantly shorter proteins (145 aa, 16 kD and 112 aa, 12 kD) in which a stop codon appears immediately at the 5′ end of the retained intron, thereby only slightly modifying the C-termini of these proteins in comparison to the NOL7-SP1 sequence (red boxes in Fig. 1B).
Figure 1.
Alternatively spliced products of the NOL7 gene. (A) Scheme of the genomic layout of the human NOL7 gene and the splicing pattern of the three gene products. Exons = yellow boxes; stop codons = red dots; grey box = retained exon. (B) Amino acid sequence alignment of the three NOL7 protein products. Differences are seen at the C-terminal ends (red boxes). Peptide sequences used for immunizing rabbits are underlined. The C-terminal NoLS is highlighted in yellow. (C) Expression of the alternatively spliced mRNAs in a variety of cell lines quantified by semi-quantitative RT-PCR. The gels show one representative experiment and the histograms show the average of at least 3 repeats. (D) Expression levels per transcript were normalized to GAPDH mRNA levels and presented as histograms. Standard deviation is produced from three independent experiments. (E) Western blotting of NOL7 in U2OS cells with: mouse anti-NOL7, rabbit anti-SP1, rabbit anti-SP2.
We examined the expression levels of the three alternatively spliced transcripts (using DNase-treated RNA) in different human cell lines using specific primers for each variant and found varying levels of expression. In most cell lines the expression of the SP1 variant was predominant (Fig. 1C and D). A commercial mouse antibody generated against the full NOL7 protein showed a 29 kD band only, indicating that it is specific to the C-terminus of NOL7-SP1 (Fig. 1E and Sup. Fig. 1A–C). Two rabbit antibodies were generated against peptides that are unique to either the SP1 or SP2 forms. The SP1-specific peptide was from the unique C-terminus of SP1 (exon 7), while the SP2-specific peptide consisted of the unique C-terminal tail of SP2 (underlined in Fig. 1B). Western blots showed the specific detection of a 29 kD protein by the anti-SP1 antibody and a 16 kD protein by the anti-SP2 antibody (Fig. 1E and Sup. Fig. 1A–C).
Cellular distribution of the NOL7 proteins.
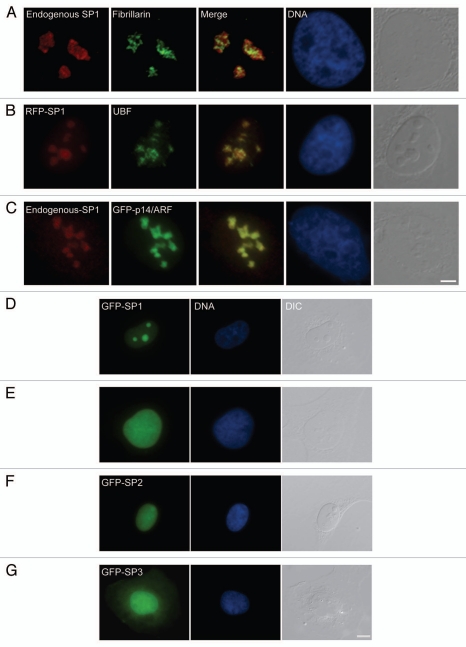
The three NOL7 proteins are small proteins (29, 16, 12 kD, respectively) and theoretically could easily diffuse in and out of the nucleus, since the cutoff for diffusion through the nuclear pore is ∼40 kD. We examined the sub-cellular distribution of the protein products. The rabbit antibodies did not perform in immunofluorescence, but the mouse antibody did and showed distinct nucleolar staining as described for GFP-NOL7 (Fig. 2A).22 In order to identify the nucleolar compartment in which NOL7-SP1 resides we performed co-localization experiments with known nucleolar markers. NOL7-SP1 exhibited a rounded nucleolar pattern reminiscent of GC proteins. Indeed, NOL7-SP1 surrounded the DFC and FC nucleolar compartments, as seen with co-staining to fibrillarin (DFC, Fig. 2A; and not shown—Nopp140, Nhp2), or UBF (FC, Fig. 2B; and not shown—Gar1). NOL7-SP1 co-localized with proteins in the GC, for instance p14/ARF (Fig. 2C; and not shown—B23). In addition, we constructed fluorescently tagged NOL7 proteins by C-terminal fusion to GFP or RFP and generated human U2OS cells stably expressing these proteins. NOL7-SP1 was predominantly nucleolar with some nucleoplasmic distribution (Fig. 2D). NOL7-SP2 was nuclear showing both nucleoplasmic as well as moderate nucleolar distribution (Fig. 2E and F). NOL7-SP3 was mostly nucleoplasmic with some cytoplasmic distribution (Fig. 2G).
Figure 2.
NOL7-SP1 is found in the GC region of the nucleolus. (A) Co-staining of endogenous NOL7-SP1 and fibrillarin (DFC). (B) RFP-NOL7-SP1 and endogenous UBF (FC). (C) Endogenous NOL7-SP1 and GFP-p14/ARF (GC). Bar, 5 µm. (D) Stably integrated GFP-NOL7-SP1 (green) localized to the nucleolus, while GFP-NOL7-SP2 was (E) predominantly nucleoplasmic with (F) some nucleolar representation. (G) GFP-NOL7-SP3 was detected throughout the cell. DNA was counterstained with Hoechst (blue) and the whole cell in observed in DIC (grey). Bar, 10 µm.
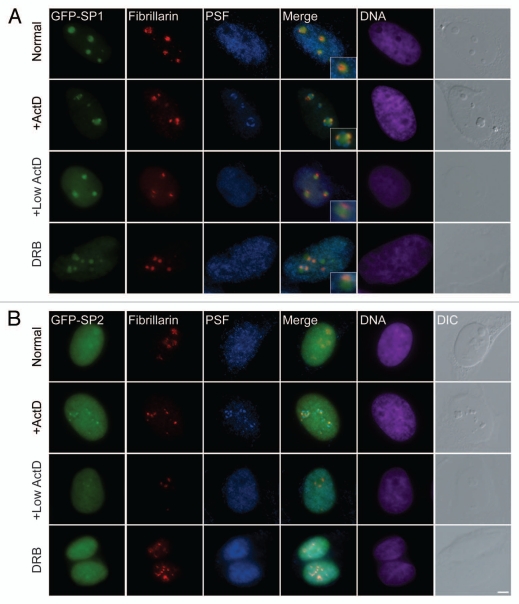
Experiments in which nucleolar segregation and nucleolar capping are induced, assist in examining the association of nucleolar proteins with their particular nucleolar compartment.27 For example, following nucleolar segregation, GC proteins like B23 are released into the nucleoplasm while the only GC protein known to be retained within the large central body is p14/ARF.27 On the other hand, DFC and FC proteins can usually be found in what has been termed light nucleolar caps (LNCs). In addition, some nucleoplasmic RNA binding proteins, like the PTB associated splicing factor PSF, localize in another set of cap structures, termed dark nucleolar caps (DNCs).27 We induced nucleolar segregation with actinomycin D (high 5 µg/ml or low 0.05 µg/ml levels), and found NOL7-SP1 in fibrillar caps (LNCs) which co-localized with fibrillarin and Nopp140, and were situated in between the PSF-positive caps (DNCs), in addition to some NOL7 protein that remained in the central body (Fig. 3A; and Sup. Fig. 1D).27 Under these conditions, GFP-NOL7-SP2 was mostly nucleoplasmic, but a portion of the protein was also observed together with fibrillarin and Nopp140 in nucleolar caps, in agreement with the moderate nucleolar localization of SP2 under regular conditions (Fig. 3B and Sup. Fig. 1E). Similarly, during Pol II inhibition by DRB, NOL7-SP1 remained in the central cloud of the disrupted nucleolar structure, with some protein localizing with the fibrillarin pool (Fig. 3B and Sup. Fig. 1D). A series of other treatments such as kinase inhibition by staurosporin, translation inhibition with cycloheximide or puromycin, proteosome inhibition by MG132, hypoxic stress by arsenite, heat shock, cold shock, osmotic shock with sorbitol, DNA damage with cisplatin, doxorubicin and neocarzinostatin (NCS), or inhibition of ATP synthesis by sodium azide, did not abrogate the nucleolar localization of NOL7-SP1 (data not shown). This implies that NOL7-SP1 has a high affinity to the GC compartment of the nucleolus.
Figure 3.
NOL7-SP1 is found in nucleolar caps during nucleolar segregation. (A) GFP-NOL7-SP1 (green), endogenous fibrillarin (red) and PSF (blue) show different nuclear localization. During nucleolar segregation by actinomycin D treatment (high 5 µg/ml or low 0.05 µg/ml), NOL7-SP1 is found in nucleolar caps containing fibrillarin, while some of the protein remains in the central body. PSF is localized to a second and larger subset of nucleolar caps (dark nucleolar caps). Inhibition of RNA Pol II only by DRB shows segregation of NOL7 and fibrillarin signals. (B) GFP-NOL7-SP2 is mostly nucleoplasmic with some nucleolar fraction. Upon nucleolar segregation (low and high ActD or DRB treatment) some of the protein is found in fibrillarin containing nucleolar caps. DNA is pseudo-colored in purple, DIC is grey. Insets show enlarged nucleolar areas. Bar, 5 µm.
Nuclear and nucleolar targeting of NOL7 proteins.
Since the NOL7-SP1 and SP2 proteins are small enough to diffuse through the nuclear pore and shuttle in between the nucleoplasm and cytoplasm, yet still show specific nuclear/nucleolar distribution, together with the fact that many different treatments did not release NOL-SP1 from the nucleolus, suggested that NOL7-SP1 can form high affinity interactions with the nucleolar compartment. We first examined whether the NOL7 proteins are shuttling proteins. Using the heterokaryon assay in which proteins can be detected when they shuttle from a human to a mouse nucleus, we found that NOL7-SP1 did not shuttle, while NOL7-SP2 did shuttle in and out of the nucleus (Sup. Fig. 2). Raver1 was used as a positive control.28 This indicated that NOL7 contains specific nucleolar, in addition to nuclear, targeting sequences.
Recently, 4 NLS sequences were identified in NOL7-SP1, located at amino acids 1–10, 88–112, 144–162 and 242–257.26 However, the NOL7-SP3 protein (aa 1–112) is distributed throughout the whole cell (Sup. Fig. 3A), whereas NOL7-SP2 (aa 1–145) is completely nuclear (Sup. Fig. 3C). This would mean that another sequence affecting nuclear localization should lie within the C-terminal portion unique to NOL7-SP2 (after aa 112 and before aa 144). We therefore generated a NOL7 version slightly larger than SP3 (aa 1–121), and found that this protein was completely nucleoplasmic (Sup. Fig. 3B). This means that the leucine-rich sequence LLPDTILEK, aa 114–121 contributes to the nuclear localization of NOL7-SP2.
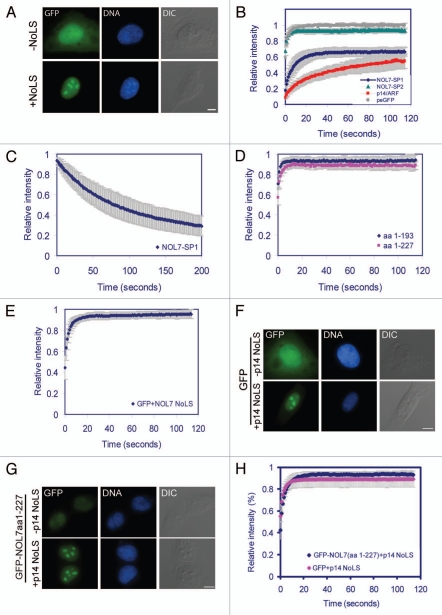
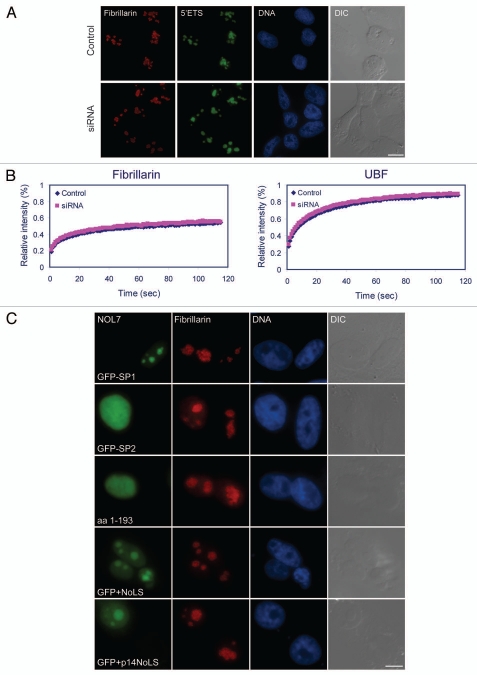
Additional constructs that were generated: aa 1–160, 1–193 and 1–227 (Sup. Fig. 3D–F) showed that the general nucleoplasmic distribution did not change, although in some cells faint nucleolar localization could be identified. This implied that a strong nucleolar targeting sequence might be contained within the very C-terminus of NOL7-SP1 (Sup. Fig. 3G). Therefore, we fused the C-terminal portion of NOL7-SP1 (aa 228–257) to a GFP protein that is normally distributed throughout the cell (Fig. 4A) and to a YFP-NLS protein that is completely nuclear (Sup. Fig. 4A), and found that this sequence (FLNNAWGIQ KKQNAKRFKRRWMVRKMKTKK) specifically targeted these fusion proteins to the GC region of the nucleolus. This region is lysine- and arginine-rich with amino acid stretches that concur with the R/K-R/K-X-R/K NoLS motif, such as KRFK and RKMK. Shorter C-terminal portions of the 228–257 region (aa 228–247, 235–257 and 235–247) fused to GFP showed only a faint nucleolar localization (Sup. Fig. 4B), indicating that the complete C-terminal tail of NOL7-SP1 is required for efficient nucleolar targeting.
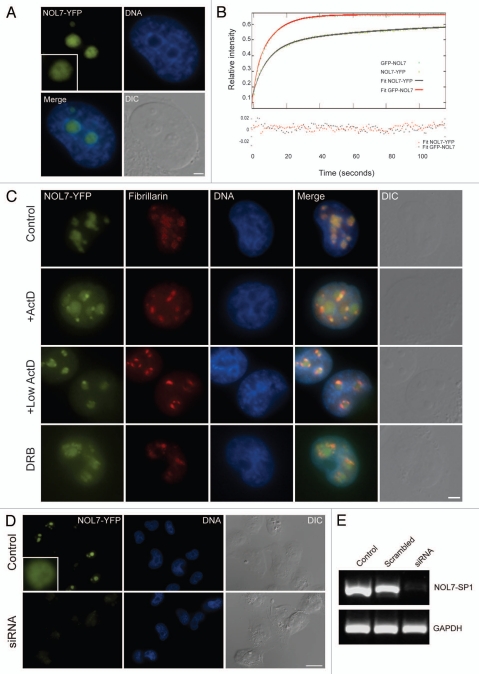
Figure 4.
Nucleolar targeting and dynamics of NOL7. (A) A peGFP protein is dispersed throughout the whole cell, while the addition of the NOL7 C-terminus conveys specific and efficient nucleolar targeting. (B) The recovery dynamics of NOL7-SP1 (blue), p14/ARF (red), NOL7-SP2 (green) and diffuse peGFP (grey). (C) FLIP of NOL7-SP1. (D) FRAP recovery of the truncated versions of NOL7 (aa 1-193, blue; aa 1-227, pink). (E) FRAP recovery of peGFP fused to the NOL7 NoLS. (F) The p14/ARF NoLS confers nucleoar targeting but not nucleolar residency. The C-terminal NoLS of the GC protein p14/ARF was fused to peGFP, or to (G) NOL7-SP1 (aa 1-227) lacking its normal NoLS. Bar, 10 mm(H) FRAP recovery of NOL7-SP1 aa 1-127 + p14-NoLS compared to GFP + p14-NoLS.
The intra-nuclear dynamics of NOL7 proteins.
To measure the intra-nuclear dynamics of the different NOL7 proteins we used fluorescence recovery after photobleaching (FRAP). The recovery curves of NOL7-SP1 showed a half-time of recovery of t1/2 = 5.3 sec, in comparison to the GC protein p14/ARF (t1/2 = 16.4 sec) (Fig. 4B) or nucleolin (DFC and GC) (t1/2 = 9 sec),29 that exhibited slower recovery kinetics. The 65% recovery suggested that a substantially fixed and highly interacting fraction (∼35%) of NOL7-SP1 exists within the nucleolus. On the other hand, FRAP measurements of NOL7-SP2 showed rapid and full recovery in the nucleoplasm (t1/2 = 1.2 sec) very similar to a free GFP protein, implying that NOL7-SP2 is highly mobile in the nucleoplasm (Fig. 4B). Fluorescence loss after photobleaching (FLIP) that measures the exit rates of a protein from a region of interest showed t1/2 = 53.4 sec for NOL7-SP1 and a similar fixed fraction as measured by FRAP (Fig. 4C). FRAP on the NOL7 constructs lacking the C-terminal NoLS targeting sequence (aa 1–193, 1–227) showed rapid recoveries as well as loss of the fixed fraction (Fig. 4D). Interestingly, FRAP on the GFP-NoLS fusion that was specifically targeted to the nucleolus also showed rapid and full recovery with low nucleolar residency times (Fig. 4E).
These data suggested that the high residency times and binding affinity of the NOL7-SP1 protein to the GC compartment in the nucleolus cannot be mediated solely via its NoLS, and that other regions of the protein are required for prolonged nucleolar residency times. Therefore, we examined whether the specific NOL7 NoLS only conveys the correct localization of the protein to the GC compartment of the nucleolus, while the binding per se is mediated by the remainder of the protein. To this end, the NoLS sequence of the GC protein p14/ARF (RGAQLRRPRHSHPTR ARRCP),21 was fused to the GFP protein and also to the NOL7-SP1 version lacking its original NoLS (aa 1–227; Sup. Fig. 3F). Both proteins harboring the p14/ARF NoLS showed specific nucleolar localization to the GC compartment (Fig. 4F and G). However, FRAP measurements of NOL7-SP1 containing the p14/ARF-NoLS showed rapid recovery kinetics similar to GFP-NoLS meaning that no long-term interactions with the nucleolus were occurring (Fig. 4H). These results mean that both the interaction and the high binding affinity of NOL7 with the nucleolus are mediated by the NOL7-specific NoLS region, and that the NoLS region per se does not confer the high affinity binding characteristics to the protein, thus the N-terminal part of the protein is also required for these interactions.
NOL7 dynamics during cell division.
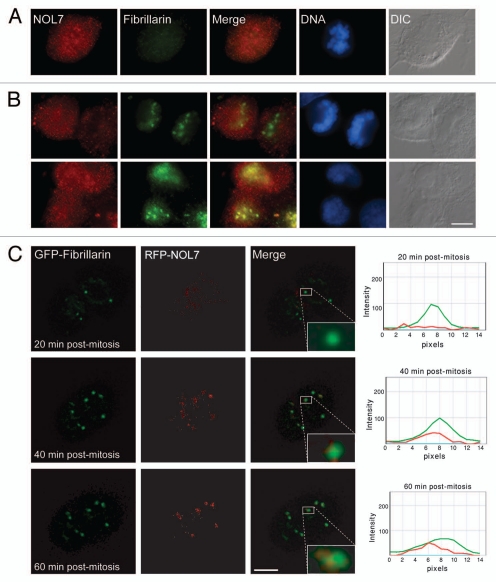
While we detected high affinity of NOL7-SP1 to the nucleolus during interphase and nucleolar segregation, many nucleolar proteins disperse during mitosis. During telophase fibrillarin concentrates in five NORs, with GC proteins arriving later on.30 Nucleolar assembly occurs through pre-nucleolar bodies (PNBs) that are known to form during telophase.31 We followed the cellular distribution of NOL7 during mitosis using time-lapse imaging. NOL7-SP1 was diffusely distributed in the nucleoplasm after nucleolar and nuclear disruption. Towards the end of mitosis, NOL7-SP1 localized to small bodies, later to form larger nucleoli (Sup. Fig. 5A and Sup. Movie 1). NOL7-SP2 remained diffusely distributed throughout cell division (Sup. Fig. 5B and Sup. Movie 2). Immuonofluorescence also showed that NOL7 dispersed in the cell during mitosis (Fig. 5A). NOL7 entry to the forming nucleolus occurred only after the formation of fibrillarin containing foci (Fig. 5B and C; and Sup. Movie 3). Many proteins become hyper-phosphorylated during mitosis, however, NOL7 protein from untreated cells compared to mitotically synchronized cells (nocodazole) or G1/S blocked cells (thymidine), did not show any changes in molecular weight that would be indicative of a high level of post-translational modifications (Sup. Fig. 5C).
Figure 5.
Subcellular localization of NOL7 during mitosis. (A) Immunofluorescence of endogenous NOL7 and fibrillarin during mitosis and (B) during cell division. (C) Frames from a live-cell movie following RFP-NOL7 (red) and GFP-fibrillarin (green) during the last stages of cell division. A box around one of the forming nucleolar bodies shows the appearance of fibrillarin followed by NOL7. The plots on the right measure the intensity of the two signals at different time points through the center of the box. Bar, 10 µm.
Nucleolar architecture is abrogated after NOL7 knockdown.
During the course of this study, a dynamic study of 1,000 endogenous YFP-tagged proteins expressed in H1299 human lung carcinoma cells was published, among them NOL7.32 Therefore, we examined the above cell line with regard to intra-nuclear dynamics of NOL7 proteins. The YFP coding region was inserted into exon 6 of one of the NOL7 alleles in human H1299 cells, thereby yielding endogenously expressed NOL-YFP. The NOL7-YFP protein was nucleolar and localized to the GC region (Fig. 6A). Since FRAP measurements are usually performed on exogenously overexpressed fusion proteins, we were interested to compare the above measurements to the dynamics of endogenous NOL7-YFP. The nucleolar dynamics of the endogenous protein as measured by FRAP were very similar to the overexpressed exogenous GFP-NOL7 protein, resulting in the same range of fixed nucleolar fraction (∼40%) (Fig. 6B). However, the initial recovery dynamics of GFP-NOL7 were faster than NOL7-YFP, (t1/2 = 10.1 sec) implying that there might be a substantial free population of the GFP fusion protein that is not bound to the nucleolus, due to the nature of overexpression. The fast kinetics can be seen from the best fit of the two plots to two exponentials, corresponding to a fast free fraction and a slow, most probably bound, fraction. NOL7-YFP re-localized similarly to GFP-NOL7 under different transcriptional inhibition treatments (Fig. 6C).
Figure 6.
Endogenous NOL7-YFP. (A) Endogenous NOL7-YFP is nucleolar. Bar, 2 µm. (B) NOL7-YFP nucleolar dynamics compared to overexpressed GFP-NOL7 measured by FRAP and fit to a two-exponent equation. Goodness of fit evaluated by checking for a random distribution of residuals around 0 is plotted at the bottom. (C) Nucleolar segregation of endogenous NOL7-YFP during high and low ActD, and DRB treatments. Bar, 5 µm. (D) NOL7-YFP expression levels before and after siRNA. Bar 10 µm. (E) NOL7-YFP mRNA levels after siRNA (RT-PCR).
The NOL7-YFP cells served as a robust assay for RNAi effects, since the NOL7-YFP protein was expressed at endogenous levels and knockdown could be assessed visually. We screened for siRNAs that reduced NOL7-YFP expression and identified the conditions under which a dramatic decrease in NOL7-YFP was observed both on the RNA and protein levels (Fig. 6D and E). We then used these conditions to reduce the protein levels of endogenous NOL7 in U2OS cells. The decrease in NOL7 was monitored by immunofluorescence with the commercial antibody. RNAi was highly efficient and a total disappearance of NOL7 staining was observed in over 95% of the cells (Fig. 7A). Cells were viable and nucleoli continued to be observed using DIC microscopy. Immunofluorescence to fibrillarin and Nopp140 showed a change in the appearance of the DFC compartment, namely loss of DFC structural definition and the dispersal of the DFC throughout the whole nucleolar volume (Fig. 7A; and Sup. Fig. 6A). Moreover, UBF puncta of the FC region were also dispersed, and instead, UBF staining was detected in the whole nucleolus, as well as the nucleolar periphery (Fig. 7B). (Since the antibodies to NOL7, UBF and B23 are all of mouse origin, the co-immunofluorescence experiments were performed with rabbit anti-fibrillarin instead, as a marker of the change in nucleolar structure due to NOL7 knockdown, as in Fig. 7A). On the other hand, B23 localization was not disrupted, indicating the relative integrity of the GC region (Fig. 7C). C23 was detected mostly in the GC (Sup. Fig. 6B). All of the above findings were detected also in HeLa cells and in the H1299 NOL7-YFP cells (not shown).
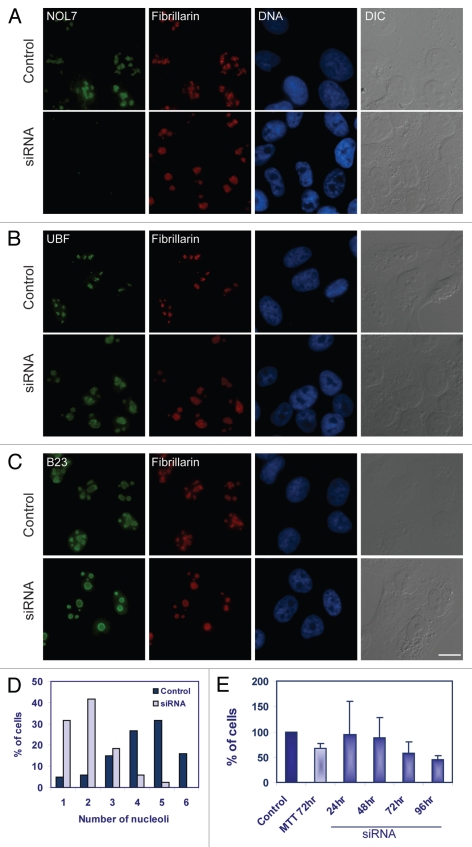
Figure 7.
Nucleolar substructure and cell proliferation change under NOL7 RNAi conditions. NOL7 siRNA treatment was performed and the following endogenous proteins were stained: (A) NOL7 and fibrillarin. (B) Fibrillarin and UBF. (C) Fibrillarin and B23. (D) Counting of number of nucleoli per cell before and after siRNA. (E) Proliferation of cells is reduced in siRNA treated cells as measured by the MTT assay for measuring cell viability (at 72 hrs) or cell counting (24, 48, 72 and 96 hrs). Standard deviation from n = 6 different plates. Bar, 10 µm.
The affects of NOL7 knockdown on cell viability and nucleolar dynamics.
Larger and more pronounced nucleolar regions were observed with Hoechst DNA staining or in DIC imaging, when cells were treated with siRNA to NOL7 in comparison to control cells. In siRNA treated cells, nucleoli were larger, and in 73% of the cells there were only 1 or 2 nucleoli per cell, in comparison to untreated cells in which 4 to 6 nucleoli were typically detected (84%) (Fig. 7D). By fixing cells at different times after siRNA administration we could find the time frame during which the reduction in nucleolar numbers and increase in size occurred (from 48 hrs). We then imaged the GFP-fibrillarin expressing cells for long time-periods during this specific time-frame and indeed could detect events of nucleolar fusions in most of the cells (Sup. Fig. 7A and B and Sup. Movie 4). Efficient knockdown in these cells was verified by fixing the cells and staining with an anti-NOL7 antibody. Cells that were treated with a scrambled siRNA sequence under the same conditions did not show fusion events. In order to examine the effect of NOL7 knockdown on cellular growth rates the same number of U2OS cells were plated and transfected with siRNA to NOL7 or were mock transfected. Cells were counted over a time course of 96 hours, and a 46% ± 8 reduction in growth rates was observed (Fig. 7E). Similar reduction (72 hrs) was seen in the MTT cell viability assay. In this colorimetric assay, enzymes reduce the MTT compound to produce a purple color, and thereby the color product is a readout of viability levels (Fig. 7E). FACS analysis of the cell cycle (24–96 hrs) did not show any significant block in any of the stages (72 hrs; Sup. Fig. 7C). There was a small decrease in the G1 stage and very mild increases in S and G2/M phases (repeated 3 times). This analysis indicates that a prolonged lack of NOL7 protein does not have a dramatic effect on the cell cycle but might slow down cell division and altogether affect cell survival.
Using RNA FISH with fluorescent probes to nucleolar RNAs we examined whether nucleolar RNA processing functions could proceed under knockdown conditions. In the nucleolus, ribosomal genes transcribe a large precursor rRNA that is cleaved, from which the 5′ external transcribed sequence (5′ETS) is removed, and then degraded by the exosome. Using a specific probe to the 5′ETS, which specifically recognizes pre-rRNAs in the nucleolus, we found that the precursor rRNAs continued to build up in nucleoli during NOL7 depletion (Fig. 8A). Small nucleolar RNAs (snoRNAs) are required for the processes of ribosome assembly, and a probe to U3 snoRNA showed them present in nucleoli under knockdown conditions (Sup. Fig. 8A). The nucleolus also plays a role in the processing of RNA Pol III transcripts. One of these transcripts, the MRP RNA, which is the RNA component of the ribonucleoprotein enzyme RNase MRP that is involved in the formation of the 5′ end of 5.8S rRNA, was also found in nucleoli during NOL7 siRNA treatment (Sup. Fig. 8B). The fact that the processing of certain non-coding RNAs in the nucleolus continued, indicates that nucleolar activity, at least in part, can be maintained when NOL7 levels are reduced.
Figure 8.
Nucleolar RNA processing during NOL7 siRNA. (A) RNA FISH on NOL7 siRNA-treated cells with a probe against the 5′ETS region of pre-rRNA. (B) FRAP on GFP-fibrillarin and GFP-UBF before and after NOL7 siRNA. (C) The NOL7-SP1 protein can rescue the siRNA phenotype while small versions of the protein do not: NOL7-SP2, NOL7 lacking the NoLS (1-193), GFP containing the NOL7 NoLS or GFP containing the p14/ARF NoLS. Cells were stained with anti-fibrillarin to examine nucleolar structure. Bar, 10 µm.
Nucleolar dynamics as measured by FRAP were then examined after NOL7 knockdown using GFP fusions of nucleolar proteins. We first verified that the characteristic GFP-fibrillarin and GFP-UBF nucleolar structures were modified after NOL7 knockdown, as observed above with their endogenous counterparts. However, there was no change in the FRAP recovery dynamics of these proteins after NOL7 knockdown (Fig. 8B), which is a further indication that some nucleolar functions are retained even though NOL7 levels are low.
Finally, we examined whether nucleolar structure under NOL7 RNAi conditions could be rescued by the re-expression of one of the NOL7 isoforms. NOL7 knockdown was performed with fluorescently labeled siRNAs, thereby indicating which cells indeed received the siRNAs. The fluorescent tag does not interfere with the RNAi process. 48 hrs later the cells were transfected with different versions of GFP-NOL7. Only full length NOL7-SP1 rescued nucleolar structure in cells positive for cytoplasmic siRNAs, as seen by the return of a centralized and punctated DFC structure (Fig. 8C and Sup. Fig. 9). The smaller versions we generated such as NOL7-SP2, NOL7 without its NoLS (1-193), the NOL7-NoLS alone, or the p14-NoLS were unable to rescue the disrupted nucleolar phenotype (Fig. 8C and Sup. Fig. 9). Altogether, these data demonstrate the importance of the full length NOL7 protein variant in the maintenance of nucleolar structural integrity.
Discussion
The structure of the nucleolus during interphase is the sum of numerous protein-RNA, protein-DNA and protein-protein interactions. The majority of these interactions are dynamic, revealing a high rate of exchange between the nucleolus and the surrounding nucleoplasm, as demonstrated by live-cell studies of fluorescently tagged nucleolar proteins and RNAs.10,11,29,33–38 High-throughput proteomics provided a wide-scope quantitative analysis of hundreds of nucleolar protein constituents and their affinity to the nucleolus during a variety of drug treatments.12 The human nucleolar proteome now encompasses over 4,500 proteins based on the analysis of more than 50,000 peptides,39 hence portraying the dynamic complexity of this structure.
Our study focused on one of these proteins, namely on the protein products arising from alternative splicing events in the pre-mRNA transcribed from the NOL7 gene. Since EST sequences suggested the presence of additional non-confirmed spliced products of this gene, we searched for their presence and cloned two additional short alternatively spliced products generated through intron retention events. In both cases, a stop-codon was found immediately downstream of the alternatively spliced coding region thereby yielding short proteins. The mRNA expression levels of the NOL7 isoforms varied between various cell lines showing predominance of the NOL7-SP1 product. Using specific antibodies we could show the presence of the SP1 and SP2 proteins in human cells. The expected protein products have low molecular weights (29, 16, 12 kD, respectively). FRAP measurements showed that the NOL7-SP1 protein had a 56 sec residence time within the nucleolus, in agreement with FRAP and FLIP experiments that have demonstrated that the retention times of nucleolar proteins lie in the range of tens of seconds.29,40
Intron retention is probably the least studied type of alternative splicing, especially since the variants were believed for many years to be largely derived from unspliced or partially spliced pre-mRNAs. However, bioinformatics analyses have found that 15% of the genes have at least one intron retention event,41 and a recent study identified 16,288 intron retention events in 41% of human genes.42 These analyses suggest that a significant fraction of intron retention events is not spurious and probably reflects biological significance. To date, only several cases of intron retention events with biological consequences in mammals are known.42 Since intron retention involves avoiding nuclear surveillance mechanisms required to stop export of unspliced pre-mRNAs to the cytoplasm, it is expected to be regulated by proteins required for splicing and export processes. In our analysis two protein isoforms arising from the NOL7 gene and that co-exist in the same cells were identified and detected, in conjunction with differential levels of mRNA expression. This could signify the ability of the cell to regulate the alternative splicing pattern of this gene, and in turn the abundance of these proteins at different crucial points such as during the cell cycle or in response to stress. In light of our findings that the presence of the NOL7-SP1 protein is required for the integrity of nucleolar structure, we can speculate that it would make sense for the cell use the SP2 variant to interfere with SP1 function when necessary to downregulate SP1 function, rather than directly downregulating SP1 levels.
We tried to disrupt the interaction of NOL7 with the nucleolus. Some granular component (GC) proteins are released from the nucleolus into the nucleoplasm upon transcriptional inhibition that leads to nucleolar segregation e.g., nucleophosmin, nucleolin, RNA helicase RH-II/Gu. NOL7 was not released from the nucleolus under conditions of actinomycin D treatment (low levels that inhibit RNA Pol I nor under high levels that inhibit all RNA polymerases), and was found to localize together with fibrillarin in structures termed nucleolar caps, while some of the protein was found in the central body.27,43,44 A similar redistribution was found when only Pol II transcription was inhibited with DRB. When examining NOL7 distribution during mitosis, NOL7 is released from the nucleolus and reappears shortly after fibrillarin puncta are observed. Although another study26 has shown that low ActD levels release NOL7 into the nucleoplasm, this might be due to the use of different cell types or by the expression of NOL7-GFP versus GFP-NOL7 that might be masking certain interaction motifs within the protein. Using a variety of treatments, mostly causing different types of cellular stress, we could not release NOL7 from the nucleolus. On the other hand, nucleolar architecture was affected under RNAi specific to NOL7-SP1, and the internal nucleolar structure was distorted. While B23 (nucleophosmin) continued to show a GC distribution under these conditions, fibrillarin, Nopp140 and UBF foci were not confined to the inner regions of the nucleolus as typically seen, but rather were dispersed throughout the whole nucleolar volume demonstrating a drastic modification in nucleolar sub-structure. Taken together, the transcriptional inhibition and mitosis findings can imply a functional role for NOL7 through interactions with other proteins of the DFC and the GC, while the knockdown data point to an architectural role in nucleolar integrity.
Other studies have examined the knockdown of nucleolar proteins that reside in the GC and a variety of phenotypes were observed. siRNA to nucleophosmin caused major distortion in nuclear and nucleolar structure.45,46 siRNA to Nop25 caused nucleolar fragmentation.47 Nucleostemin knockdown caused cell cycle arrest48,49 and modified the internal pattern of the DFC, causing the loosening of snoRNP clusters.46 USP36 depletion resulted in nucleophosmin redistribution in the nucleoplasm and smaller-sized nucleoli.50 Nucleolin (found in the DFC and GC),51 depletion resulted in the loss of nucleolar structures.52
The knockdown of GC proteins might cause loss of structural support of the nucleolus and therefore modify nucleolar structure. We suggest that although the GC region in NOL7-depleted cells is not physically disrupted, there is loss of internal nucleolar structure due to loss of NOL7 and possibly other NOL7-interacting proteins. Indeed, RNA FISH demonstrated that some RNA processing is maintained, since after NOL7 siRNA, events of pre-rRNA processing of Pol I and Pol III transcripts, as well as snoRNP presence in the nucleoli, were still detected. Yet while viable, a proliferation assay showed that cell growth rates were significantly reduced. Moreover, these nucleoli were very large due to the fusion of several nucleoli. Previous studies found a correlation between increased proliferation rates and nucleolar size.53 For instance, in cancer cells a phenotype of large nucleoli coincides with increased rRNA transcription and faster proliferation.54 It is possible that during NOL7 RNAi many nucleoli fuse to form one nucleolus. Still the general rate of nucleolar functions is not increased since proliferation does not increase.
The nucleolar targeting sequence was found in the C-terminus of the NOL7-SP1 protein. It has been suggested that NoLSs do not play a part in the nuclear import of the protein such as with NLSs, but rather mediate the interactions of nucleolar proteins with either other proteins or nucleic acids.5 Indeed, some nucleolar proteins do not contain a NoLS and instead interact with other nucleolar components that do contain a NoLS. For instance, nucleolin and protein phosphatase 1 (PP1) do not have a NoLS but are specifically targeted to the nucleolus by other binding partners.55,56 In the case of NOL7-SP1, the NoLS was determined to be the 30 C-terminal amino acids, since shorter versions did not confer the high specificity of nucleolar localization. NoLSs usually contain short Arg/Lys rich stretches, are externally oriented in the protein sequence, and can range from 8 to 30 amino acids.16 At least two such motifs are found in the NOL7-SP1, KRFKRR and RKMKTKK, with another upstream option of KRLPVKR. These regions also contain the R/K-R/K-X-R/K NoLS motif. This region of the protein is highly conserved across species. Only the combined presence of these sequences provided full nucleolar localization of the protein.
The NOL7-SP2 product did not contain the strong C-terminal NoLS and therefore was mostly nucleoplasmic, rapidly moving, and able to shuttle into the cytoplasm. Yet, some of the protein could still be identified within the nucleolus although with a very low retention time. A recent study has shown that the central region of the protein contains NLS sequences that function in nucleolar localization as well,26 thereby explaining the weak localization of NOL7-SP2 that lacks this region.
The location of the strong NoLS in the very C-terminus of the protein implies its possible functionality as an interaction domain with other nucleolar proteins. Indeed, when the full NoLS sequence was fused to eGFP or a nuclear YFP protein, predominant localization in the GC was detected. However, FRAP analysis showed that the high specificity of localization did not also mediate the protein-protein interactions required for the high residency times of the full-sized protein. Replacement of the NOL7 C-terminal NoLS with the GC protein p14/ARF C-terminal localization sequence did not contribute to prolonged residence times. The addition of the p14/ARF NoLS to NOL7 could change the folding of NOL7 and thereby affect its natural binding, or it could mean that the specific nucleolar residency of NOL7 can be obtained only through the complete protein structure. The interaction of NOL7 with the nucleolus could occur as part of a protein complex that functions within the nucleolus.
It has been shown for several nucleolar proteins that the functional portions of the protein can be distinct from the nucleolar localizing motifs.57 The interaction between ARF and mdm2 is necessary for the induction of a p53-dependent cell cycle arrest by ARF. The binding between mdm2 and ARF causes a conformational change in mdm2 that reveals a cryptic NoLS directing the complex to the nucleolus.21,58 The NoLS of nucleostemin mediates nucleolar targeting of the protein, but not the long residence times.59,60 The La protein (47 kD) can bind RNA through its RRM domain, but mutants lacking RRM2 were localized to the nucleolus through a 32 aa NoLS containing several R/K stretches.61 Finally, exchanging the NoLS on fibrillarin (DFC) with the NoLS of B23 (GC) causes the mis-localization of fibrillarin and its binding partner Nop56 to the GC region of the nucleolus.62
A common theme emerges from these examples. Thousands of proteins are known to pass through the nucleolus, many of them transiently passing by, and a minority staying around for a time period that is significant for functional activity. The cell has devised an amino-acid based coding mechanism for the specific targeting of proteins to sub-cellular compartments. Nevertheless, in the case of the nucleolus, initial targeting is merely one step—only synergistic interactions with other portions of the targeted protein will determine the full function and residency time. This might be regarded as a fail-safe mechanism ensuring that different levels of interactions and residency times of the mass of proteins or complexes traveling through or associating with the nucleolus and its variety of machineries and functions, are correctly maintained.
Materials and Methods
Plasmids.
NOL7 cDNAs were amplified and cloned into peGFP-C1 or RFP-C1 (Clontech) using a high fidelity Taq polymerase (Fermentas) and the following primers: Sense primer ATA CTC GAG CCA TGG TGC AGC TCC GAC CGC GAG and anti-sense: SP1-ATA GAA TTC GAT CTA TGA ATG ATT TAT TAG; SP2-ATA GAA TTC TCA CTT TAA GCA TCC ACT ACA; SP3-ATA GAA TTC ACT GTC AGA GGC AAT CTT GGT.
Intron 4 retention verification in SP2 was performed using a sense primer CAT CAA GAA ATC GCC AGG located in exon 4, and an antisense primer GAA ACA AAA GAC TCA TTA C located in intron 5.
The truncated versions of NOL7 were formed using:
1–121 aa, 365 bp, by HpaI restriction;
1–160 aa, 480 bp, anti-sense: ATA GAA TTC TTG TAC TTT AAC TTT CTT GGA;
1–193 aa, 581 bp, anti-sense: ATA GAA TTC ATG AAG GCT TGT GCT GCT TGT;
1–227 aa, 683 bp, anti-sense: ATA GAA TTC AAC TGG ACA GCA GCT CT.
The C-terminal NoLS of NOL7-SP1 was fused to peGFP-C1 or YFP-Nuc (Clontech) using primers: sense: ATA CTC GAG AGT TTT TGA ATA ATG CTT GG and anti-sense: ATA CTC GAG ACT TCT TAG TTT TCA TC.
Shorter C-terminal portions of the NoLS of NOL7-SP1 were generated using:
Sense: ATA CTC GAG CCA AAA AAA ACA AAA TGC C;
anti-sense: ATA CTC GAG ACC GTC TTT TAA ACC TCT TG.
The p14/ARF NoLS sequence was inserted to both GFP and the GFP-NOL7 1–227 aa constructs as an adaptor RGAQLRRPRHSHPTRARRCP using EcoRI/BamHI restriction sites. Ligations were performed with T4 ligase (New England Biolabs). GFP-p14/ARF, GFP-Nopp140 and GFP-NHP2 were previously described in reference 27 and 63. GFP-UBF was obtained from Miroslav Dundr (Rosalind Franklin University of Medicine and Science, Chicago).
Total RNA purification.
Total RNA was isolated using Tri-Reagent (Sigma). DNA-free™ Kit (Ambion) was used to remove genomic DNA contamination. cDNA (1 µg RNA) was synthesized using the ReverseAid™ First Strand cDNA Synthesis Kit (Fermentas) with oligo-dT as a primer. Semi-quantitative RT-PCR was performed using Eppendorf Thermocycler amplification for 19–38 cycles (depending on the saturation level of the genes amplified) using 1 min denaturation at 94°C, 1 min annealing at 55°C, 1 min extension at 72°C; and 72°C for 10 min for final extension. Primers for GAPDH: sense: ACC ACA GTC CAT GCC ATC AC, anti-sense: TCC ACC ACC CTG TTG CTG TA.
Cell culture and transfections.
Human U2OS cells were maintained in low glucose DMEM (Biological Industries, Israel) containing 10% FBS (HyClone). H1299 cells were maintained in RPMI medium (Biological Industries) containing 10% FBS. U2OS cells were transfected by electroporation to form stable cell lines and selected for 2–3 weeks in 350 µg/ml G418. Drugs used: actinomycin D (Pol I & II-5 µg/ml, 2 hrs; Pol I 0.05 µg/ml, 4 hrs), DRB (100 µM, 4 hrs), α-amanitin (60 µg/ml, 6 hrs), cycloheximide (100 µg/ml, 4 hrs, Sigma), Na-azide (10 mM, 1 hr), sorbitol (200 mM, 30 min), doxorubicin (1 µg/ml, 2 hrs), NCS (100 ng/ml, 20 min), arsenite (1 mM, 30 min), MG132 (2–20 µM, 4–24 hrs, Sigma), puromycin (10 µg/ml, 4 hrs, Megapharm, Raanana, Israel), staurosporine (2 µM, 6 hrs, Almone, Jerusalem, Israel), cisplatin (3 µg/ml, ABIC, Fairfield, NJ). For the heterokaryon assay HeLa cells were transiently transfected with either of GFP-NOL7-SP1, GFP-NOL7-SP2 or Raver-1-YFP28 plasmids. Similar number of HeLa cells after transfection were mixed with untransfected NIH3T3 cells and seeded onto coverslips. After overnight incubation, medium was removed and cells were incubated for 1 hr in the presence of 100 µg/ml cycloheximide and fused with 50% polyethylene glycol (PEG 3350; Sigma) for 2 min, washed twice with PBS, and returned to medium supplemented with 100 µg/ml cycloheximide for another 4 hrs incubation. Cells were fixed with 4% paraformaldehyde (PFA).
Immunofluorescence and FISH.
Immunofluorescence was performed as previously described in reference 27. Cells were fixed in 4% PFA for 20 min. Primary antibodies: mouse-anti-Gar1,27 rabbit anti-Nopp140,64 rabbit anti-fibrillarin, mouse anti-B23, rabbit anti-C23 (Abcam), mouse anti-PSF (Sigma), mouse anti-UBF, mouse anti-NOL7 (Abnova). Secondary antibodies: anti-mouse Alexa488, anti-rabbit Alexa594 (Molecular probes), anti-mouse Cy3, anti-rabbit Cy5 (Jackson ImmunoResearch, West Grove, PA). DNA was stained with Hoechst (1 µg/ml, Sigma). Coverslips were mounted in mounting medium. RNA fluorescence in situ hybridization was performed as previously described in reference 65, and followed by immunofluorescence. Probes used have been described in reference 27.
Fluorescence microscopy, live-cell imaging and data analysis.
Wide-field fluorescence images were obtained using the CellR system based on an Olympus IX81 fully motorized inverted microscope (60X PlanApo objective, 1.42 NA) fitted with an Orca-AG CCD camera (Hamamatsu), rapid wavelength switching, active focus control (ZDC), and driven by the CellR software. For time-lapse imaging cells were plated on glass-bottomed tissue culture plates (MatTek, Ashland, MA) in medium containing 10% FCS at 37°C. The microscope is equipped with an on-scope incubator which includes temperature and CO2 control (Life Imaging Services, Reinach, Switzerland). For long-term imaging of cells during mitosis, several cell positions were chosen and recorded by a motorized stage (Scan IM, Märzhäuser, Wetzlar-Steindorf, Germany). Each position was imaged in a number of Z planes and in preset time points either using the internal Z-motor or a Z-piezo stage insert (Prior). For presentation of the movies, the 4D image sequences were transformed into a time sequence using the CellR software.
FRAP and FLIP.
For FRAP and FLIP, cells were maintained in Leibovitz's L-15 phenol red-free (Gibco) containing 10% FCS at 37°C. FRAP and FLIP image sequences were obtained on a Zeiss LSM 510 META inverted scanning confocal microscope equipped with a heated chamber and objective heater, and a Plan-Apochromat 63X, 1.4 NA oil objective (Jena, Germany). Cells were scanned using a 488 laser. FRAP data were normalized and calculated as described in reference 27. For FLIP, cells were scanned using a 488 laser. FLIP data were normalized and calculated as described in reference 29. The FRAP data were fit using MatLab and a best fit was found using a two exponent equation: This kind of kinetics can be simplified and presented as a two exponent equation:
Western blotting.
Peptide specific antibodies were produced in rabbits (Adar-Biotech, Ness Ziyona, Israel). For SP1 the immunizing peptide was ANKRLPVKRAAVQF (exon 7). For SP2 the immunizing peptide was KVKEGMTILI against the very C-terminus of SP2 that diverges from SP1. Antibodies were affinity purified on an anti-peptide column. Mouse anti-GFP (Roche) was used for fusion protein detection. SDS-PAGE and western blotting were performed as previously described in reference 66. Secondary antibodies: HRP-conjugated goat anti-mouse or anti-rabbit IgG (Sigma). Immunoreactive bands were detected by ECL (Pierce).
siRNA.
siRNA specific for NOL7 and negative control were purchased from Invitrogen. The siRNA sequence CCA GGA AAG GUG AAA GAA GUU AAU U against the coding region of NOL7, and a negative control siRNA sequence were introduced into U2OS cells using Lipofectamine 2000 following the manufacturer's instructions. Forty-eight hours after transfection, a second pulse of siRNA was transfected. Cells were immunostained for NOL7, fibrillarin, Nopp140, B23, C23 and UBF after a total of 72 hrs. We verified that Lipofectamine itself did not have any effects on NOL7 expression. In experiments using fluorescently labeled siRNAs, the siRNAs were first labeled with Cy5 using the Label IT nucleic acid labeling kit (Mirus, Madison, WI). The fluorescent tag does not interfere with the RNAi process. Cellular growth rates were measured by counting cells after the same number of U2OS cells were seeded and transfected with siRNA to NOL7 or with Lipofectamin only, for 72 hours (two independent experiments, n = 3 each). Nucleoli counting was performed on 6 different coverslips (n = 120 cells from each group). For imaging of nucleolar fusions, NOL7 siRNA-treated (48 hrs) GFP-fibrillarin expressing cells were imaged for 14 hrs. Movies from NOL7 siRNA-treated cells showed nucleolar fusions in most of the cells, while in cells treated with a scrambled siRNA no fusions were detected. After time-lapse imaging the plates were subjected to immunofluorescence with an anti-NOL7 Ab, and NOL7 knockdown was verified. For the rescue experiments, cells were treated with siRNA to NOL7 for 48 hrs and then transfected with different constructs in order to examine rescue of the knockdown nucleolar phenotype. These cells were stained with anti-fibrillarin to examine nucleolar structure.
For the MTT assay—a colorimetric assay that measures cell viability, U2OS cells were plated in triplicate in 24 well plates at a concentration of 1 × 105 cells per well. After 24 hrs cells were transfected with NOL7 siRNA or scrambled siRNA for negative control using Lipofectamine 2000. After 72 hrs of culture, cell proliferation was tested using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. 600 µl of 5 mg/ml of MTT (Sigma) solution was added per well. After 3 hrs of incubation at 37°C, 400 µl of dissolving solution colored crystals of formazan were added and the plate was incubated for an additional 6 hrs at 37°C. The plate was read on a multi-well scanning spectrophotometer (ELISA reader) at 570 nm.
Fluorescence-activated cell sorting (FACS) analysis.
Cells were trypsinized and fixed with 70% ethanol (4°C, overnight). After fixation, cells were centrifuged for 4 min at 200 g and incubated for 30 min at 4°C in 1 ml of PBS, centrifuged and resuspended in PBS containing 5 mg/ml propidium iodide and 50 µg/ml RNase A for 20 min at room temperature (25°C). Fluorescence intensity was analyzed using a BD Biosciences flow cytometer. Experiments were repeated 3 times for 24, 48, 72 and 96 hr time points.
Acknowledgments
We thank Uri Alon (Weizmann Institute) for the NOL-YFP cell line, Miroslav Dundr (Rosalind Franklin University of Medicine and Science) for GFP-UBF1, Tom Meier (Albert Einstein College of Medicine) for anti-Nopp140 Ab, Yehuda Brody for FRAP fitting, and Rakefet Ben-Yishay for graphic assistance. This work was supported by the German-Israeli Project Cooperation (DIP) Foundation, the Israel Science Foundation and the Alon Fellowship. Y.S.T. thanks the Israel Science Foundation for the fluorescence live-cell imaging microscope. Y.S.T. is the Jane Stern Lebell Family Fellow in Life Sciences at BIU.
Abbreviations
- DFC
dense fibrillar component
- FC
fibrillar center
- FRAP
fluorescence recovery after photobleaching
- GC
granular component
Supplementary Material
References
- 1.Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Busch H, Smetana K. The Nucleolus. Academic Press; 1970. [Google Scholar]
- 3.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 5.Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2:107–112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 6.Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 7.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 8.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Intl Rev Cytol. 2006;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- 10.Olson MO, Dundr M. The moving parts of the nucleolus. Histochem Cell Biol. 2005;123:203–216. doi: 10.1007/s00418-005-0754-9. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem Cell Biol. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 12.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 13.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 14.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 15.Leung AK, Andersen JS, Mann M, Lamond AI. Bioinformatic analysis of the nucleolus. Biochem J. 2003;376:553–569. doi: 10.1042/BJ20031169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birbach A, Bailey ST, Ghosh S, Schmid JA. Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NFkappaB inducing kinase NIK. J Cell Sci. 2004;117:3615–3624. doi: 10.1242/jcs.01224. [DOI] [PubMed] [Google Scholar]
- 18.Endo S, Kubota S, Siomi H, Adachi A, Oroszlan S, Maki M, et al. A region of basic amino-acid cluster in HIV-1 Tat protein is essential for trans-acting activity and nucleolar localization. Virus Genes. 1989;3:99–110. doi: 10.1007/BF00125123. [DOI] [PubMed] [Google Scholar]
- 19.Malim MH, Bohnlein S, Hauber J, Cullen BR. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 20.Siomi H, Shida H, Nam SH, Nosaka T, Maki M, Hatanaka M. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell. 1988;55:197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- 21.Weber JD, Kuo ML, Bothner B, DiGiammarino EL, Kriwacki RW, Roussel MF, et al. Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol Cell Biol. 2000;20:2517–2528. doi: 10.1128/mcb.20.7.2517-2528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasina R, Pontier AL, Fekete MJ, Martin LE, Qi XM, Brigaudeau C, et al. NOL7 is a nucleolar candidate tumor suppressor gene in cervical cancer that modulates the angiogenic phenotype. Oncogene. 2006;25:588–598. doi: 10.1038/sj.onc.1209070. [DOI] [PubMed] [Google Scholar]
- 23.Mullokandov MR, Kholodilov NG, Atkin NB, Burk RD, Johnson AB, Klinger HP. Genomic alterations in cervical carcinoma: losses of chromosome heterozygosity and human papilloma virus tumor status. Cancer Res. 1996;56:197–205. [PubMed] [Google Scholar]
- 24.Rader JS, Li Y, Huettner PC, Xu Z, Gerhard DS. Cervical cancer suppressor gene is within 1 cM on 6p23. Genes Chromosomes Cancer. 2000;27:373–379. [PubMed] [Google Scholar]
- 25.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 26.Zhou G, Doci CL, Lingen MW. Identification and functional analysis of NOL7 nuclear and nucleolar localization signals. BMC Cell Biol. 2010;11:74. doi: 10.1186/1471-2121-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, et al. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huttelmaier S, Illenberger S, Grosheva I, Rudiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savino TM, Gebrane-Younes J, De Mey J, Sibarita JB, Hernandez-Verdun D. Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol. 2001;153:1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelier N, Tramier M, Louvet E, Coppey-Moisan M, Savino TM, De Mey JR, et al. Tracking the interactions of rRNA processing proteins during nucleolar assembly in living cells. Mol Biol Cell. 2005;16:2862–2871. doi: 10.1091/mbc.E05-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 33.Shav-Tal Y, Darzacq X, Singer RH. Gene expression within a dynamic nuclear landscape. EMBO J. 2006;25:3469–3479. doi: 10.1038/sj.emboj.7601226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 35.Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Politz JC, Tuft RA, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell. 2003;14:4805–4812. doi: 10.1091/mbc.E03-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobson MR, Cao LG, Wang YL, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pederson T. Protein mobility within the nucleus—what are the right moves? Cell. 2001;104:635–638. doi: 10.1016/s0092-8674(01)00258-6. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res. 2009;37:181–184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 41.Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mollet IG, Ben-Dov C, Felicio-Silva D, Grosso AR, Eleuterio P, Alves R, et al. Unconstrained mining of transcript data reveals increased alternative splicing complexity in the human transcriptome. Nucleic Acids Res. 2010;38:4740–4754. doi: 10.1093/nar/gkq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smetana K, Busch H. The nucleolus and nucleolar DNA. In: Busch H, editor. The Cell Nucleus. New York: Academic Press; 1974. pp. 73–147. [Google Scholar]
- 44.Louvet E, Junera HR, Le Panse S, Hernandez-Verdun D. Dynamics and compartmentation of the nucleolar processing machinery. Exp Cell Res. 2005;304:457–470. doi: 10.1016/j.yexcr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 45.Amin MA, Matsunaga S, Uchiyama S, Fukui K. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J. 2008;415:345–351. doi: 10.1042/BJ20081411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romanova L, Kellner S, Katoku-Kikyo N, Kikyo N. Novel role of nucleostemin in the maintenance of nucleolar architecture and integrity of small nucleolar ribonucleoproteins and the telomerase complex. J Biol Chem. 2009;284:26685–26694. doi: 10.1074/jbc.M109.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki S, Fujiwara T, Kanno M. Nucleolar protein Nop25 is involved in nucleolar architecture. Biochem Biophys Res Commun. 2007;358:1114–1119. doi: 10.1016/j.bbrc.2007.05.069. [DOI] [PubMed] [Google Scholar]
- 48.Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;18:2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endo A, Matsumoto M, Inada T, Yamamoto A, Nakayama KI, Kitamura N, et al. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J Cell Sci. 2009;122:678–686. doi: 10.1242/jcs.044461. [DOI] [PubMed] [Google Scholar]
- 51.Biggiogera M, Burki K, Kaufmann SH, Shaper JH, Gas N, Amalric F, et al. Nucleolar distribution of proteins B23 and nucleolin in mouse preimplantation embryos as visualized by immunoelectron microscopy. Development. 1990;110:1263–1270. doi: 10.1242/dev.110.4.1263. [DOI] [PubMed] [Google Scholar]
- 52.Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci. 2007;120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- 53.Derenzini M, Trere D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol. 2000;191:181–186. doi: 10.1002/(SICI)1096-9896(200006)191:2<181::AID-PATH607>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 54.Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 55.Li YP, Busch RK, Valdez BC, Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur J Biochem. 1996;237:153–158. doi: 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]
- 56.Gunawardena SR, Ruis BL, Meyer JA, Kapoor M, Conklin KF. NOM1 targets protein phosphatase I to the nucleolus. J Biol Chem. 2008;283:398–404. doi: 10.1074/jbc.M706708200. [DOI] [PubMed] [Google Scholar]
- 57.Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 59.Meng L, Yasumoto H, Tsai RY. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J Cell Sci. 2006;119:5124–5136. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horke S, Reumann K, Schweizer M, Will H, Heise T. Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J Biol Chem. 2004;279:26563–26570. doi: 10.1074/jbc.M401017200. [DOI] [PubMed] [Google Scholar]
- 62.Lechertier T, Sirri V, Hernandez-Verdun D, Roussel P. A B23-interacting sequence as a tool to visualize protein interactions in a cellular context. J Cell Sci. 2007;120:265–275. doi: 10.1242/jcs.03345. [DOI] [PubMed] [Google Scholar]
- 63.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- 65.Chartrand P, Bertrand E, Singer RH, Long RM. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 2000;318:493–506. doi: 10.1016/s0076-6879(00)18072-3. [DOI] [PubMed] [Google Scholar]
- 66.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.