Abstract
The Class I MAGE proteins are normally expressed only in developing germ cells but are often aberrantly expressed in malignancies, particularly melanoma, making them good therapeutic targets. MAGE proteins promote tumor survival by binding to the RBCC region of KAP-1 and suppressing p53. Although, suppression of MAGE expression, by RNA interference, relieves p53 suppression and inhibits tumor growth, its therapeutic uses are limited by lack of methods for systemic delivery of small interfering RNA. To overcome this barrier, we sought to discover chemical compounds that inhibit binding between MAGE and KAP-1 proteins. Based on previously published effects of MAGE suppression, we developed a strategy for screening a small molecule library based on selective death of MAGE positive cells, activation of p53 and lack of caspase activity. We screened the Maybridge HitFinder library of compounds and eight compounds fulfilled these criteria. Seven of these compounds interfered with co-precipitation of MAGE and KAP-1, and three interfered with binding of MAGE and KAP-1 in a mammalian two hybrid assay. We now report identification of three potential compounds that interfere with MAGE/KAP-1 binding and can be developed as novel chemo-therapeutic agents for treatment of advanced melanoma and other cancers.
Keywords: MAGE, KAP-1, MHD-MAGE, RBCC-KAP-1, Small molecule compounds
Introduction
Melanoma is not the most common but it is the most deadly form of skin cancer. The American Cancer Society estimates that about 120,000 new cases of melanoma are diagnosed every year in the USA (http://www.skincancer.org/Melanoma/). Early stages of melanoma can be surgically removed but advanced melanoma is associated with mortality and requires comprehensive treatments often associated with toxicity. Melanoma Associated Anti-GEns (MAGE) were first discovered in melanoma and are also known as Cancer Testes Antigens because they are expressed in cancers and male germ cells, but not in normal tissue. In addition to melanoma, MAGE proteins are expressed in many other types of tumors, such as head and neck squamous cell carcinoma, lung carcinoma and breast carcinoma.
The human MAGE genes are classified into two categories based on their chromosome location [1]. The Class I MAGE genes include 28 members of the human A, B, and C families and two murine Class I families, mMage-a and mMage-b [2–9]. The conserved MAGE homology domain (MHD) represents about 70% of the protein. Functions of the MHD MAGE have not been well studied. Expression of Class I MAGE genes is normally suppressed in all somatic tissues by hypermethylation of critical promoter region CpG dinucleotide islands [7,10]. As a result, MAGE proteins are normally expressed only in developing sperm and fetal ovarian germ cells [11–17]. Global hypomethylation, which occurs during epigenetic reprogramming in many cancers, frequently causes hypomethylation of Class I MAGE promoters and aberrant MAGE protein expression in a wide variety of cancers [8]. We have previously shown that suppression of MAGE proteins, by RNA interference, decreases tumor cell survival in vitro [18,19] and prolongs survival in a syngenic mouse model of melanoma [19]. However, effective systems for suppression of MAGE by systemically administered small interfering RNA in humans are not currently available although it may be possible to achieve the same result by interfering with MAGE function.
Expression of Class I MAGE genes results in suppression of p53 expression and function. p53 dependent apoptosis is induced by MAGE knockdown in MAGE positive cancer cells [19,20]. MAGE proteins cause p53 suppression by binding to KRAB domain-associated protein 1 (KAP-1) [19,20]. KAP-1, also known as TRIM28, Tif1β, or Krip1, is a universal co-repressor protein, causes transcriptional repression and acts as a molecular scaffold [19]. KAP-1 can suppress p53 by forming complexes with HDM2 and stabilizing HDM2 interactions with p53 [21]. It may also target p53 by acting as E3 ubiquitin ligase, whose activity is enhanced by MAGE proteins [19,20]. MAGE dependent activation of KAP-1 E3 ubiquitin ligase and suppression of p53 requires its binding to the RBCC region of KAP-1 [19].
Unlike ubiquitously expressed KAP-1 and p53, MAGE proteins are selectively expressed in cancer cells and can be specific therapeutic targets. As binding of MAGE to KAP-1 is required for p53 suppression, interference with MAGE and KAP-1 binding offers exquisitely tumor specific target [19,20]. To test this possibility and to develop new approaches for cancer therapy, we have developed a system for identifying small molecule inhibitors of MAGE-KAP-1 binding and function. In this study, we focused on MAGE-A3 and MAGE-C2 as they are the MAGE family members that are commonly expressed in cancers. Here we demonstrate high throughput screening of Maybridge HitFinder library, a collection of 14,400 compounds, based on cytotoxicity, p53 activation, and negligible caspase activity in MAGE positive cells. We, hereby, report three potential compounds that disrupt association of MAGE and KAP-1 and can be further developed as therapeutic agents for advanced melanoma and other cancers.
Materials and methods
Cell lines
A375 melanoma cells and CHO cells were obtained from ATCC (Manassas, VA) and propagated as recommended. The CellSensor® p53RE-bla HCT-116 cell line was purchased from Invitrogen (Carlsbad, CA) and cultured under continuous selection with Blasticidin. Cells were maintained at 37 °C, with 5% CO2 and 90–95% humidity in a tissue culture incubator.
The Maybridge HitFinder library
The Maybridge HitFinder library was purchased from Thermo-Fisher Scientific (Waltham, MA). It consists of 14,400 premier compounds representing drug-like diversity. All screening compounds fit Lipinski guidelines for “drug-likeness”, and all have purity greater than 90%. The library is arrayed in 45 384-well microtiter plates and is available for screening at 1 mM and 0.1 mM stock concentrations. These are individually designed compounds, produced by innovative synthetic techniques and have the potential for structural alterations to increase in efficacy and decrease potential toxicity.
High throughput screening
We screened Maybridge HitFinder library, a collection of 14,400 compounds, for their ability to (1) cause selective death of MAGE positive cells, (2) activate p53 in MAGE positive cells, and (3) cause death with low or no activation of caspase. High throughput screening was performed at the Keck Small Molecule screening Facility at the University of Wisconsin Comprehensive Carbone Center (UWCCC), Madison, WI.
p53 activation screen
We used a p53 responsive assay with a quantitative β-lactamase read out that employs CellSensor® p53RE-bla HCT-116 cell line (Invitrogen, Carlsbad, CA). CellSensor® p53RE-bla HCT-116 cell line has been modified from parental HCT-116 human colon cancer cell line by stable integration of a beta-lactamase reporter gene under control of the p53 Response Element (p53RE). Modulation of p53 pathway can be measured selectively and quantitatively as β-lactamase reporter activity. Cells were plated in 384 well plates and next day were treated with compounds at 10 μM concentration. 24 h later, β-lactamase assay was performed according to manufacturer’s directions. Briefly, cells were loaded with Live-BLAzer™-FRET B/G Substrate (Invitrogen, Carlsbad, CA) for 2.5 h and fluorescence emission values at 460 nm and 530 nm were obtained using a fluorescence plate reader. Live-BLAzer™-FRET B/G Substrate is an engineered fluorescent substrate containing two fluorophores, coumarin and fluorescein. In the absence of β-lactamase activity, the substrate molecule remains intact and excitation of coumarin results in fluorescence resonance energy transfer to the fluorescein moiety and emission of green light. However, in presence of β-lactamase activity, the substrate is cleaved, separating the fluorophores and disrupting energy transfer. Excitation of the coumarin in the presence of beta-lactamase enzyme activity results in a blue fluorescence signal. The resulting coumarin-to-fluorescein ratio provides a normalized reporter response represented as Relative Fluorescence Units (RFU).
Cytotoxicity assay
Cytotoxicity of the compounds was estimated by CytoTox-Glo Cytotoxicity Assay (Promega Corporation, Madison, WI), a luminescent assay that estimates dead cells ina population. The assay isbased on the activity of intra-cellular proteases that are released when cell membranes become porous in dying or dead cells. In this assay, the peptide substrate used for these proteases, AAF-aminoluciferin (alanyl-alanylphenylalanyl-aminoluciferin), is impermeable and hence can be utilized only by extracellular proteases. The lysed aminoluciferin is luminescent and can be estimated by a luminometer. Cytotoxicity results are represented as Relative Luciferase Units (RLU).
Caspase assay
We have previously shown that apoptosis induced by MAGE knockdown is caspase independent. In order to eliminate compounds that cause cell death by caspase dependent mechanisms, therefore, probably not acting via MAGE-KAP-1 interactions, we measured caspase activity by Caspase-Glo® 3/7 Assay (Promega Corporation, Madison, WI). We chose caspase 3/7 assay because caspase 3 is a key mediator of caspase dependent apoptosis in mammalian cells. This is a luminescent assay that utilizes the caspase-3/7 DEVD-aminoluciferin substrate. Degradation of this substrate by active caspase 3/7 decreases luminescence. Results are represented as Relative Luciferase Units (RLU).
Mammalian two hybrid assay
We used a modified mammalian two hybrid assay to directly measure binding of MAGE-A3 or C2 to KAP-1 and if the compounds interfere with this interaction. We modified a commercially available mammalian two hybrid assay (Matchmaker™ Mammalian Two Hybrid Assay Kit, Clontech, Mountain View, CA) by replacing the promoter of the bait plasmids with a stronger CMV promoter, to drive expression of the bait fusion proteins, resulting in increased sensitivity and reproducibility. pM plasmid (bait), empty or expressing MAGE-C2, MAGE-A3, or the MHD regions of MAGEC2 and MAGE-A3, were cloned under CMV promoter in p3xflag-CMV plasmid (Sigma–Aldrich, St Louis, MO). Similarly, KAP-1 or RBCC region of KAP-1 were cloned in the pVP-16 plasmid (prey). To quantify the interaction of MAGE-A3 or C2 with KAP-1, and to determine if the compounds interfere with this interaction, MHD region of MAGE-A3 or C2 was used as bait and the RBCC region of KAP-1 was used as prey. The interaction was measured by the secreted alkaline phosphatase (SEAP) assay using the Phospha-Light™ Secreted Alkaline Phosphatase Reporter Gene Assay System (Applied Biosystems/Life Technologies Corporation, Carlsbad, CA) performed according to manufacturer’s directions. Briefly, CHO cells were co-transfected with pM-3XFLAG-CMV, pVP16 and pG5SEAP vectors with Lipofectamine reagent (Invitrogen, Carlsbad, CA). The next day, cells were treated with various concentrations of compounds and 48 h later, SEAP activity was estimated. The output was measured as secreted alkaline phosphatase (SEAP) and analyzed by a luminescent substrate.
Western immunoblotting
A375 melanoma cells were treated with compounds at 10 μM concentration and collected after 24 h. Cells were lysed in RIPA buffer and subjected to SDS–PAGE. Membranes were probed with MAGE-C2 and Actin antibodies (SantaCruz Biotechnology, Santa Cruz, CA).
Co-immunoprecipitation
Protein lysates (500 μg) were immunoprecipitated with MAGE-C2 antibody (SantaCruz Biotechnology, Santa Cruz, CA). Immunoprecipitates were subjected to SDS–PAGE and immunoblotted with antibodies for KAP-1 (Novus Biologicals, Littleton, CO).
Results
Eight compounds were selected based on Primary High Throughput Screening
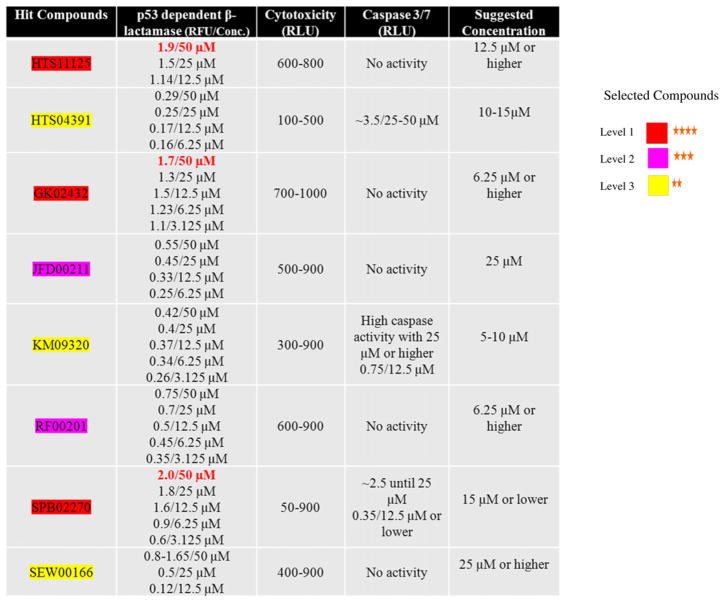
Fourteen thousand and four hundred compounds of the May-bridge HitFinder library were screened in HCT116bla cells by high throughput testing of the library. Indiscriminant cytotoxic and auto-fluorescent compounds were eliminated. Based on our previous studies and other published data, we selected the criteria to be (1) selective cell death, (2) p53 activation and (3) negligible caspase activity in MAGE positive cancer cells. Based on these criteria, 22 compounds that induced cell death independent of caspase activation and induced p53 activity as measured by β-lactamase read out were selected. After confirmation of these results, eight compounds were further selected, based on highest p53 activity (1.0 RFU or higher) and lowest caspase activity (2.5 RLU or higher), as compared to controls (Fig. 1).
Fig. 1.
Compounds selected from the Maybridge HitFinder library: their p53 activity, cytotoxicity, and caspase 3/7 activity. Maybridge HitFinder library was screened by high throughput assays for p53 activity, caspase 3/7 and MAGE specific cytotoxicity in p53RE-bla HCT-116 cell line, as described in Materials and methods section. Twenty-two compounds scored high on these criteria and eight compounds were further selected based on the highest p53 activity (RFU of 1.0 or higher) and no or low caspase 3/7 activity (2.5 RLU or lower). p53 activity, cytotoxicity and caspase activation of these eight compounds is demonstrated in this figure 1. As shown in the figure, three of the compounds, HTS11125, GK02432 and SPB02270, show highest p53 activity (highlighted in red).
Seven compounds interfere with MAGE-C2 and KAP-1 binding
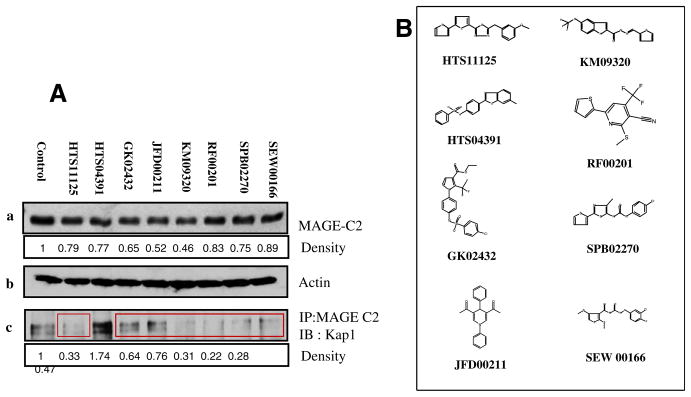
To determine effect of the selected compounds on MAGE positive melanoma cells, further testing was done on A375 cells. A375 cells were treated, for 24 h, with HTS11125 (25 μM), HTS04391 (15 μM), GK02432 (25 μM), JFD00211 (25 μM), KM09320 (10 μM), RF00201 (25 μM), SPB02270 (10 μM), SEW 00166 (25 μM). As shown in Fig. 2A, treatment of A375 cells with these compounds results in decrease in MAGE-C2 expression ranging from 9% to 54%. We hypothesize that decrease in MAGE-C2 expression might represent decreased stability or solubility of MAGE-C2 when it is not bound to KAP-1.
Fig. 2.
Six of the eight selected compounds interfere with MAGE-C2 and KAP-1 binding in A375 cells. (A) A375 cells were treated with the eight selected compounds, collected 24 h post-treatment and processed for SDS–PAGE and immuno-precipitation. (a) Immuno-blotting for MAGE-C2, (b) β-actin, and (c) immuno-precipitation with MAGE-C2 antibody and immuno-blotting for KAP-1. Note that six of the eight compounds decrease co-precipitation of MAGE-C2 and KAP-1 (highlighted in red boxes). (B) Chemical structures of the compounds.
We have previously reported that MAGE-C2 co-precipitates with KAP-1. To assess the effect of the selected compounds on binding of MAGE-C2 and KAP-1, protein lysates were immunoprecipitated with MAGE-C2 antibody and probed with KAP-1 antibody. As compared to control treatment, seven compounds HTS11125, GK02432, JFD00211, KM09320, RF00201, SPB02270 and SEW00166, decreased co-precipitation of MAGE-C2 and KAP-1, whereas, HTS04391 increased in MAGE-C2 and KAP-1 co-precipitation (Fig. 2B).
Compounds interfere with binding of MHD-MAGE C2/A3 to the RBCC region of KAP-1
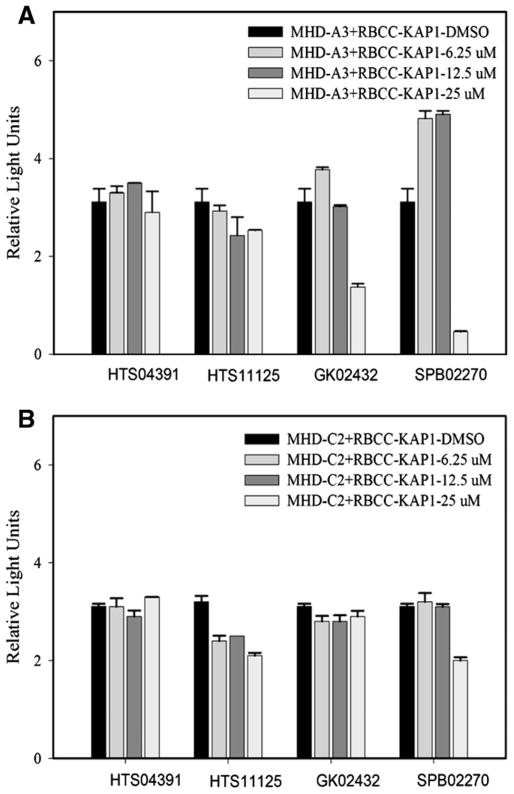
To more directly detect MAGE and KAP-1 binding, we developed a mammalian two hybrid assay employing modifications of the Matchmaker Mammalian Two Hybrid Assay (Clontech, Mountain View, CA). The modifications we made to the bait and prey plasmids, as described in Materials and methods section, increased the sensitivity and reproducibility and allowed us to quantify the interaction of MAGE-A3 or C2 with KAP-1, and to determine if the compounds interfere with this interaction. In our assays, we found that binding of the MHD region of MAGE to RBCC region of KAP-1 was stronger than binding of full length MAGE and KAP-1, which confirmed our hypothesis that a common function of the MAGE homology domain (MHD) is binding to KAP-1. Therefore, we used MHD MAGE and RBCC KAP-1 to test the ability of the compounds to interfere with this function of MHD MAGE. For our preliminary studies, we had focused only on MAGE-C2 but in these experiments we also investigated binding of MAGE-A3 with KAP-1 and effect of the compounds on it (Fig. 3). The bars, as value of SEAP activity, represent binding of MHD MAGE A3 and C2 to RBCC KAP-1, as shown in Fig. 3A and B, respectively. We found three promising candidates, HTS11125, GK02432 and SPB02270. As shown in Fig. 3, SPB02270 showed the strongest inhibition of binding of MHD-MAGE A3 and C2 with RBCC-KAP1, whereas, HTS11125 inhibited the binding to a lesser extent. GK02432 inhibits binding of MHD MAGE-A3 with RBCC-KAP-1, but no effect was observed on MHD-MAGE-C2 binding with RBCC-KAP1. HTS04391 was used as a negative control because it seems to increase MAGE-C2 and KAP-1 interaction in coimmunoprecipitation.
Fig. 3.
Three compounds interfere with MHD-MAGE C2/A3 binding to RBCC-KAP-1. We employed mammalian two-hybrid assay to investigate direct binding of MHD MAGE C2/A3 and RBCC-KAP-1 and the ability of the compounds to interfere with this binding. CHO cells were transfected with the indicated plasmids, treated with selected eight compounds at 6.25 μM, 12.5 μM and 25 μM concentrations for 48 h and SEAP (secreted alkaline phosphatase) activity was estimated. SEAP activity is demonstrated as RLUs. The bars represent binding of MHD MAGE to RBCC KAP-1. (A) MHD MAGE A3 binding to RBCC KAP-1. (B) MHD MAGE C2 binding to RBCC KAP-1. Based on the interference with MHD-MAGE and RBCC KAP-1 binding, three compounds, HTS11125, GK02432 and SPB02270, were identified with potential for drug development. HTS04391 was used as a negative control.
Discussion
MAGE proteins are expressed in many types of human tumors including at least 50% of advanced malignant melanomas, 75% of non-small cell carcinoma of the lung, 70% of squamous cell carcinomas of the head and neck, 91% of carcinomas of the breast, and smaller percentages of other cancers [5,22–27]. The fact that so many different malignancies express MAGE proteins suggests that there is selective pressure for MAGE expression, and several clinical correlations support this hypothesis. There is an association between MAGE-A expression and advanced or aggressive melanoma, and MAGE expression has been found in putative melanoma stem cells [5,22–28]. MAGE expression has also been associated with aggressive clinical behavior in myeloma and malignant gammopathies [22,25] and with acquisition of resistance to chemotherapeutic drugs by myeloma, ovarian carcinoma, and meduloblastoma cell lines [22,26,29,30]. In addition, overexpression of MAGE A3 promotes cell proliferation, increase in primary tumor size, and the number and size of metastatic foci in an orthotopic xenograft model of thyroid cancer [31]. We have previously shown that suppression of MAGE A and MAGE-C in melanoma and malignant mast cell lines increases apoptosis and decreases cell growth in vitro [18,19] and that knockdown of MAGE with systemically administered cholesterol conjugated siRNA prolongs survival in a syngenic mouse model of melanoma [19]. These observations suggest that interference with MAGE expression could have an impact on treatment of a wide range of human tumors. Furthermore, because MAGE proteins are not expressed in adult somatic tissues, target related side effects could be as limited as temporary infertility. Safe systems for suppression of MAGE or other proteins in humans by systemically administered RNAi are not currently available so we focused on identifying drug candidates that could interfere with MAGE function.
We have shown that Class I MAGE proteins bind to the KAP-1 RBCC region and suppress p53 [19]. As a multifunctional protein, KAP-1 is increasingly being recognized as a central molecule in the regulation of p53, cell cycle and mitosis, DNA damage response, KRAB domain zinc finger transcription factors (KZFTFs) functions and recently as a E3 ubiquitin ligase [20,21,32–39]. Therefore direct targeting of KAP-1 in tumors would be anticipated to have widespread side effects. However, because of selective expression of MAGE proteins, targeting MAGE protein expression or function can be exquisitely specific target for treatment of melanoma and other cancers. Herein, we show that MHD region of MAGE binds to the RBCC region of KAP-1 and that this interaction can be inhibited by small molecules. Here we report three compounds that inhibit MAGE and KAP-1 binding and may be used as potential drugs. Further studies are required to develop these compounds as potential therapeutic targets for cancer.
Acknowledgments
Funding
T.Z.X. is a recipient of NIH T32 training award (1T32AR055893-01A1).
References
- 1.Simpson AJG, Caballero OL, Jungbluth A, Chen YT, Old LJ. Nat Rev Cancer. 2005;5:615. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 2.Knuth A, Wolfel T, Klehmann E, Boon T, Meyer zum Buschenfelde KH. Proc Natl Acad Sci USA. 1989;86:2804–2808. doi: 10.1073/pnas.86.8.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Eynde B, Hainaut P, Herin M, Knuth A, Lemoine C, Weynants P, van der Bruggen P, Fauchet R, Boon T. Int J Cancer. 1989;44:634–640. doi: 10.1002/ijc.2910440413. [DOI] [PubMed] [Google Scholar]
- 4.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 5.Basarab T, Picard JK, Simpson E, Russell-Jones R. Br J Dermatol. 1999;140:106–108. doi: 10.1046/j.1365-2133.1999.02616.x. [DOI] [PubMed] [Google Scholar]
- 6.De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethe B, Lurquin C, et al. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 7.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas S, De Smet C, Arden KC, Viars CS, Lethe B, Lurquin C, Boon T. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]
- 9.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 10.Sigalotti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, Fonsatti E, Traversari C, Altomonte M, Maio M. Cancer Res. 2004;64:9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Shichijo S, Noguchi M, Hirohata M, Itoh K. Cancer Res. 1995;55:3478–3482. [PubMed] [Google Scholar]
- 12.Jungbluth AA, Chen YT, Busam KJ, Coplan K, Kolb D, Iversen K, Williamson B, Van Landeghem FK, Stockert E, Old LJ. Int J Cancer. 2002;99:839–845. doi: 10.1002/ijc.10416. [DOI] [PubMed] [Google Scholar]
- 13.Rajpert-De Meyts E, Jacobsen GK, Bartkova J, Aubry F, Samson M, Bartek J, Skakkebaek NE. Histopathology. 2003;42:217–226. doi: 10.1046/j.1365-2559.2003.01587.x. [DOI] [PubMed] [Google Scholar]
- 14.Yakirevich E, Sabo E, Dirnfeld M, Sova Y, Spagnoli GC, Resnick MB. Appl Immunohistochem Mol Morphol. 2003;11:37–44. doi: 10.1097/00129039-200303000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Gaskell TL, Esnal A, Robinson LL, Anderson RA, Saunders PT. Biol Reprod. 2004;71:2012–2021. doi: 10.1095/biolreprod.104.028381. [DOI] [PubMed] [Google Scholar]
- 16.Pauls K, Schorle H, Jeske W, Brehm R, Steger K, Wernert N, Buttner R, Zhou H. Hum Reprod. 2006;21:397–404. doi: 10.1093/humrep/dei325. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang R, Zhu Y, Fang L, Liu XS, Tian Y, Chen LH, Ouyang WM, Xu XG, Jian JL, Gure AO, Fortunato S, Ritter G, Old LJ, Simpson AJ, Chen YT, Jin B, Jungbluth AA. Cancer Immun. 2006;6:7. [PubMed] [Google Scholar]
- 18.Yang B, O’Herrin S, Wu J, Reagan-Shaw S, Ma Y, Nihal M, Longley BJ. J Invest Dermatol. 2006;127:267–275. doi: 10.1038/sj.jid.5700548. [DOI] [PubMed] [Google Scholar]
- 19.Yang B, O’Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, Peng H, Ivanov AV, Simpson AJ, Longley BJ. Cancer Res. 2007;67:9954–9962. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 20.Doyle JM, Gao J, Wang J, Yang M, Potts PR. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ, 3rd, Chen J. EMBO J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhodapkar MV, Osman K, Teruya-Feldstein J, Filippa D, Hedvat CV, Iversen K, Kolb D, Geller MD, Hassoun H, Kewalramani T, Comenzo RL, Coplan K, Chen YT, Jungbluth AA. Cancer Immun. 2003;3:9. [PubMed] [Google Scholar]
- 23.Hofbauer GF, Schaefer C, Noppen C, Boni R, Kamarashev J, Nestle FO, Spagnoli GC, Dummer R. Am J Pathol. 1997;151:1549–1553. [PMC free article] [PubMed] [Google Scholar]
- 24.Jungbluth AA, Busam KJ, Kolb D, Iversen K, Coplan K, Chen YT, Spagnoli GC, Old LJ. Int J Cancer. 2000;85:460–465. [PubMed] [Google Scholar]
- 25.Jungbluth AA, Ely S, Diliberto M, Niesvizky R, Williamson B, Frosina D, Chen YT, Bhardwaj N, Chen-Kiang S, Old LJ, Cho HJ. Blood. 2005;106:167–174. doi: 10.1182/blood-2004-12-4931. [DOI] [PubMed] [Google Scholar]
- 26.Park JW, Kwon TK, Kim IH, Sohn SS, Kim YS, Kim CI, Bae OS, Lee KS, Lee KD, Lee CS, Chang HK, Choe BK, Ahn SY, Jeon CH. J Immunol Methods. 2002;266:79–86. doi: 10.1016/s0022-1759(02)00105-9. [DOI] [PubMed] [Google Scholar]
- 27.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, Shevde LA, Li W, Eschrich S, Daud A, Ju J, Matta J. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sigalotti L, Covre A, Zabierowski S, Himes B, Colizzi F, Natali PG, Herlyn M, Maio M. J Cell Physiol. 2008;215:287–291. doi: 10.1002/jcp.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan Z, Duan Y, Lamendola DE, Yusuf RZ, Naeem R, Penson RT, Seiden MV. Clin Cancer Res. 2003;9:2778–2785. [PubMed] [Google Scholar]
- 30.Kasuga C, Nakahara Y, Ueda S, Hawkins C, Taylor MD, Smith CA, Rutka JT. J Neurosurg Pediatr. 2008;1:305–313. doi: 10.3171/PED/2008/1/4/305. [DOI] [PubMed] [Google Scholar]
- 31.Liu W, Cheng S, Asa SL, Ezzat S. Cancer Res. 2008;68:8104–8112. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- 32.Goodarzi AA, Noon AT, Jeggo PA. Biochem Soc Trans. 2009;37:569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 33.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 34.Kim YH, Jensen RA, Watanabe GL, Varghese A, Hoppe RT. Arch Dermatol. 1996;132:1309–1313. [PubMed] [Google Scholar]
- 35.Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- 37.White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ., 3rd Cancer Res. 2006;66:11594–11599. doi: 10.1158/0008-5472.CAN-06-4138. [DOI] [PubMed] [Google Scholar]
- 38.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 39.Rowe JJHM. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]