Abstract
RNA interference-based screening is a powerful new genomic technology which addresses gene function en masse. To evaluate factors influencing hit list composition and reproducibility, we performed two identically designed small interfering RNA (siRNA)-based, whole genome screens for host factors supporting yellow fever virus infection. These screens represent two separate experiments completed five months apart and allow the direct assessment of the reproducibility of a given siRNA technology when performed in the same environment. Candidate hit lists generated by sum rank, median absolute deviation, z-score, and strictly standardized mean difference were compared within and between whole genome screens. Application of these analysis methodologies within a single screening dataset using a fixed threshold equivalent to a p-value ≤ 0.001 resulted in hit lists ranging from 82 to 1,140 members and highlighted the tremendous impact analysis methodology has on hit list composition. Intra- and inter-screen reproducibility was significantly influenced by the analysis methodology and ranged from 32% to 99%. This study also highlighted the power of testing at least two independent siRNAs for each gene product in primary screens. To facilitate validation we conclude by suggesting methods to reduce false discovery at the primary screening stage.
In this study we present the first comprehensive comparison of multiple analysis strategies, and demonstrate the impact of the analysis methodology on the composition of the “hit list”. Therefore, we propose that the entire dataset derived from functional genome-scale screens, especially if publicly funded, should be made available as is done with data derived from gene expression and genome-wide association studies.
Keywords: RNA interference, analysis, RNAi screen analysis, siRNA, RNAi, siRNA screening, sum rank, median absolute deviation, strictly standardized mean difference, genome-wide, whole-genome, comparison, overlap, hit list
Introduction
The development of high-throughput systems for the large scale application of RNA interference-based assays swiftly followed the discovery of RNA interference (RNAi) in eukaryotic systems.8,9 Dissemination of genome-scale libraries utilizing RNAi throughout the research community has driven the interrogation of the myriad of biological pathways resulting in many salient discoveries previously impossible. The progression of RNAi technology necessitates continual evaluation of the methodology to identify valuable targets.
The human genome can be interrogated by expressed short hairpin RNAs (shRNA) or transfected small interfering RNAs (siRNA).5,19 In this study, we focus on synthetic siRNAs arrayed in micro-well format. The evolution of RNAi screening to the genome-scale parallels the emergence of high-density microarray technology. As RNAi technology joins the “omics” echelons, so does the need for analysis methodology to make sense of the enormous datasets generated from genome-scale loss-of-function studies. These datasets can range from the simple output of luminescence or fluorescence of a well, to the generation of high-content cell-based data containing as many as a hundred separate parameters for each of the thousands of cells in the well. Multiple analysis methods have been used to generate the all important “hit list”, though none is considered standard procedure.1
One specific field benefiting from genome-scale siRNA screening technology is the field of host-pathogen interactions. To date, multiple groups have pursued the identification of factors influencing viral propagation using genome-scale RNAi technology. Recently, three groups identified host factors supporting the HIV lifecycle in human cells. However, upon comparison of the results, the significant dissimilarity between the proposed host factors posited more questions than any one project answered.2,15,28 The low level of overlap among hit lists can be explained by the dissimilar methodologies employed, however it also raises a question that can be explored experimentally, “To what degree would one expect RNAi-based genome-wide screens to agree?”
Our study explores the effect of screening strategy and analysis methodology on the results of two genome-scale siRNA screens. These two screens utilized high-content cell-based imaging and analysis to score for siRNAs that inhibited yellow fever virus propagation in human cells. Both were performed using the same siRNA library, cell line, viral stock, equipment and procedure, but were separated by five months. We use these data to illustrate the advantages of testing independent siRNAs during the primary screen. Additionally, we compare the performance of four accepted analysis methods with respect to the variability and overlap of intra- and inter-screen hit lists. Our work defines multiple factors contributing to the variability between genome-scale siRNA screens.
Materials and Methods
siRNA Screening
Both genomic screens to identify human host factors of yellow fever virus propagation were performed using the Qiagen Human Genome siRNA Library v1.0 at the Duke RNAi Screening Facility. HuH-7 cells were reverse transfected with 1.0 pmol siRNA in a total volume of 65 μL media. Briefly, assay plates were pre-arrayed from stock library plates using a Velocity11/Agilent Bravo precision liquid handling robot. 5 μL of dH2O containing 1.0 pmol of siRNA was dispensed into 384-well microplates (Corning 3712) using the 384 channel ST head of the Bravo. 10 μL of Opti-MEM I (Gibco 11058) containing 0.5% RNAimax (Invitrogen 13778-150) was dispensed to each well using a Matrix WellMate liquid dispenser and incubated for 30 minutes at room temperature in the presence of 1pmol siRNA in 5 μL dH2O. After incubation, 1,200 HuH-7 cells in 50 μL of DMEM (Gibco 11995) supplemented with 5% FBS (Gibco 16140) and 1% antibiotics (Gibco 15140) were dispensed into each well using a Matrix WellMate.
Approximately, fifty-one hours after transfection, the HuH-7 cells were infected with yellow fever virus vaccine strain 17D at an M.O.I. of 0.1 by addition of 20 μL of viral containing media to each assay well using the 384 channel ST head of the Bravo followed by mixing by triteration. The virus infection was allowed to proceed for forty-two hours at 37°C. The cells were then immuno-stained for the viral envelope protein using the primary antibody 4G2 followed by Alexa488-conjugated secondary antibodies.11 Reagents were added using a Matrix automated 12-channel pipette, and wash steps were performed using a BioTek ELx405 automated plate washer. Both genomic screens strictly adhered to a common protocol in which 11 batches of 14 plates per day were assayed within 27 days.
High-content imaging and cell-based assay
After fixation and staining, 2 of the 4 available fields in each assay well were imaged with a 10x objective using the Cellomics Array Scan VTI system. The acquired images were analyzed using the vHCS Scan Version 5.1.2 (Build 268). The Compartmental Analysis bio-application was optimized to determine the percentage of HuH-7 cells that were infected by yellow fever virus. The vHCS View software package reports the number of cells identified in channel one as the “Valid Object Count” (VOC) and the percent of the analyzed cell population infected by the virus is reported as the “% Selected” and henceforward referred to by the moniker “percent infection.”
Statistical Analysis
Valid Object Count
Only genes in which both assay wells in a single genomic screen had VOC ≥ 1.9 standard deviations less than the mean cell density of the population.
The following analysis methodologies were performed treating the genomic data as a single dataset of 21,853 pairs for GS1 and 21,843 pairs for GS2.
Sum Rank (SR)
The SR p-value was calculated for each genomic screen separately.20 The percent infection values for each set of siRNAs (AB, CD; Figure 1) are ordered lowest to highest and given a rank from 1 to n (n= number of assay wells), respectively. The summation of the ranks for corresponding pairs between each set is calculated. Using the equation presented in Supplemental Table 1, the p-value for each pair is determined. The p-values are ranked. The genes with the lowest 200 p-values and the lowest 500 p-values comprise the top 200 and top 500 hit lists, respectively.
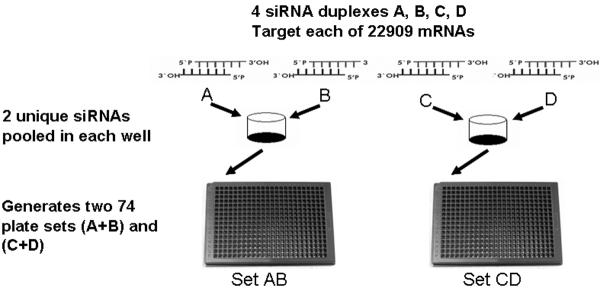
Figure 1. The human whole genome siRNA library design.
The target gene is assayed by four unique siRNAs (A, B, C, D) which are pooled as two siRNAs per well (AB, CD) and arrayed in corresponding wells within 74 pairs (set AB, set CD) of 384 well assay plates.
Median Absolute Deviation (MAD)
The MAD was calculated as reported in the supplemental table 1 separately for each set within each genomic screen.6 The median percent infection value for each set within each genomic screen was used in the calculations. In order to expedite the identification of the MAD limits for the top 200 and top 500 gene sets, the number of MAD units away from the median was calculated for each percent infection value by:
| 1) |
The thresholds for the top 200 or top 500 populations were identified as the point in which the MADn limit for set AB equaled the MADn limit for set CD and the population within the limits contained 200 or 500 hits, respectively.
z-score
The calculation for z-score is reported in Supplemental Table 1. As applied to these genomic screens, the negative control population consisted of all percent infection values from wells with siGFP for both sets within each genomic screen. The thresholds for the top 200 or top 500 populations were identified as the point in which the z-score for set AB equaled the z-score for set CD and the population within the limits contained 200 or 500 hits, respectively.
Strictly Standardized Mean Difference (SSMD)
The calculations for SSMD were reported in the Supplemental Table 1.25,27 The negative control population consisted of all percent infection values from wells with siGFP for both sets within each genomic screen. The genes with the lowest 200 and lowest 500 SSMD values within each genomic screen comprised the top 200 and top 500 hit lists.
Results
Duplicate siRNA-based genomic screens
Two genome-scale siRNA screens were performed to identify host factors supporting yellow fever virus propagation in HuH-7 cells. Approximately five months separated the first genomic screen (GS1) and the second genomic screen (GS2). A high-content imaging and analysis platform quantified infection as described in Materials and Methods.
In order to eliminate unreliable data, assayed pairs in which cells failed to proliferate after plating were removed from further analysis. Out of the 22,909 assayed pairs in each genomic screen, 95% of the pairs retained sufficient cell density to be further analyzed for percent infection (Table 1).
Table 1.
Genomic data set
| Genomic Screen | ||
|---|---|---|
| GS1 | GS2 | |
| Total Pairs | 22,909 | 22,909 |
| Pairs removed | 1,056 | 1,066 |
| Pairs analyzed | 21,853 | 21,843 |
Library design and screening procedure to reduce False Discovery Rate
Four distinct siRNA duplexes denoted A, B, C and D targeted each of the 22,909 predicted mRNAs assayed by a human whole genome siRNA library. We chose a 2×2 pooled siRNA screening format which consists of two distinct 74 plate sets, each containing two unique siRNA duplexes per well (SetAB, SetCD). This format results in 148 paired 384-well microplates (Figure 1). The library design allowed each gene to be assayed by two independent tests. Genes that scored strongly for a particular phenotype by both siRNA pools were considered a candidate.
We employed a total effective siRNA concentration of 15.4nM for screening, resulting in each siRNA being present at 7.7nM. This concentration of siRNA is significantly lower than that commonly used (Supplemental Table 2), and was chosen to reduce off target effects (OTE).12
The low siRNA concentration and the 2×2 screening strategy are designed to limit the false discovery rate in the primary screen. These criteria were selected to provide a high-confidence primary hit list, with minimal false positives for academic screeners who may face both personnel and financial constraints during assay validation.
Analysis of GS1 and GS2 control wells to assess behavior of the two screens
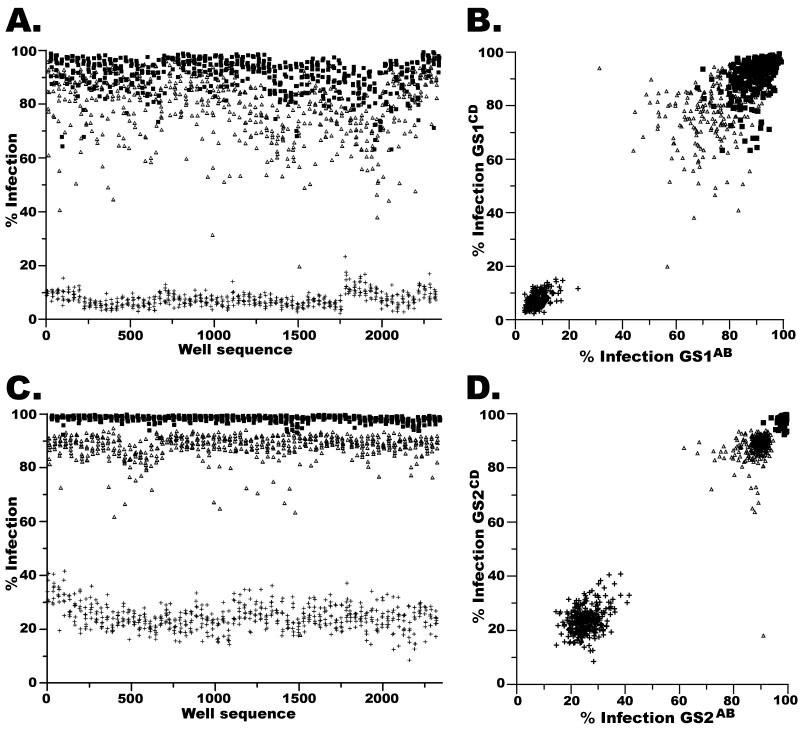
Twelve siRNA controls were arrayed on each assay plate. Four negative control wells targeted GFP (siGFP), a protein not present in our system. Four wells contained a non-targeting siRNA (siNSC) as a negative control (Qiagen). The remaining four wells contained a siRNA duplex that targeted a subunit of the vATPase (si-vATPase), which is required for flaviviral infection and served as a positive control.16,17,20 The mean and standard deviation of the percent infection in GS1 for the negative controls, siGFP and siNSC, was 78.6 ± 11.4 and 90.8 ± 6.3, respectively, while the mean and standard deviation of the percent infection for the positive controls was 7.5 ± 2.7 (Figure 2A). The mean and standard deviation of the percent infection in GS2 for the negative controls, siGFP and siNSC, was 87.9 ± 5.2 and 98.3 ± 1.1, respectively, while the mean and standard deviation of the percent infection for the positive controls was 24.6 ± 4.7 (Figure 2C). Despite our efforts to reproduce the screens in an identical fashion, these data begin to reveal a significant difference in the overall behavior of GS1 and GS2. We attribute this difference to slight changes in the infectivity of the yellow fever virus stocks, and minor variability of the HuH-7 cell line after storage in the freezer for five months, however we have no definitive explanation for the change in behavior.
Figure 2. The behavior of the negative and positive controls across GS1 and GS2.
For a,b,c,d: si-nonsilencing negative controls (■), si-GFP negative controls (△) and si-vATPase positive controls (+). a, The % Infection for the negative and positive control wells from the 148 assay plates in GS1 are plotted left-to-right in the order in which each was assayed. b, The % Infection for the negative or positive control wells from the 74 assay plates in GS1AB are plotted as coordinate pairs with the corresponding negative or positive control well within the 74 assay plates from GS1CD on the coordinate plane. c, Similar to a, excepting that the negative and positive controls were assayed on the 148 plates in GS2. d, similar to b, excepting that pairs of controls wells belong to GS2AB and GS2CD
Although the behavior of the controls was different between screens, when assessed individually each screen performed very well. Figures 2A (GS1) and 2C (GS2), display all positive and negative control wells left-to-right in the order in which the screen was performed. Visualization of controls in this fashion allows intra- and inter-batch anomalies to be detected. While minor batch-specific affects are noticeable, we decided to analyze each genomic screen as a single dataset (Materials and Methods).
A modified Z’-factor (Z’n) illustrates the power of replicate tests
Z’-factor (Z’) was developed as a common metric to classify the strength of high-throughput chemical screens and has been similarly applied to siRNA based screens.24 We chose to use the values of negative control siGFP to calculate the most conservative Z’. Z’ was calculated between the siGFP and si-vATPase to be 0.40 and 0.53 for GS1 and GS2, respectively. These Z’-factors indicated we could consistently differentiate the positive control wells from the negative controls wells within both respective siRNA screens.
Additionally, the paired design of our screening format allowed us to improve the resolution of the assay by plotting the performance of the control wells from SetAB and SetCD as respective x and y coordinates. In Figure 2B and 2D, each point represents the percent infection for two corresponding controls for all paired assay plates within each screen. Plotting the controls in 2-dimensions illustrates the enhanced resolution afforded by paired tests and results in greater separation of the values. Indeed, by applying a modified equation one can arrive at a modified Z’-factor, Z’n-factor (Z’n).
| 2) |
The adjustment of Z’ to Z’n quantifies the increased distance between the mean of the negative and positive controls in 2-dimentional space without increasing the standard deviations. It assumes that the mean and standard deviation along the x and y axis are equal for the respective control set and that there is no covariance for which to account. When those assumptions are fair, as described in Supplemental Figure 1 and accompanying text, Z’n can be interpreted in a manner similar to how Zhang et al. 1999 proposed interpretation of Z’.
Calculation of the Z’n –factor for GS1 and GS2 result in 0.58 and 0.66, respectively. The increase of Z’n –factor relative to Z’-factor demonstrates the well known positive influence replicate tests have on data quality.
The 2×2 pooled library design improved data integrity by providing independent tests
The increased resolution afforded by replicate tests and illustrated by the Z’n-factor would apply to any screening format performed in duplicate. Our 2×2 pooled library format fulfills this criterion, while additional power is achieved by utilizing two unique pools of siRNAs and requiring the phenotype to reproduce with both pools. We demonstrated the affects of this format by comparing the VOC between screens in several ways. Initially, we limited the analysis to the first three plate pairs of each screen: GS11-3AB, GS11-3CD, GS21-3AB, and GS21-3CD.
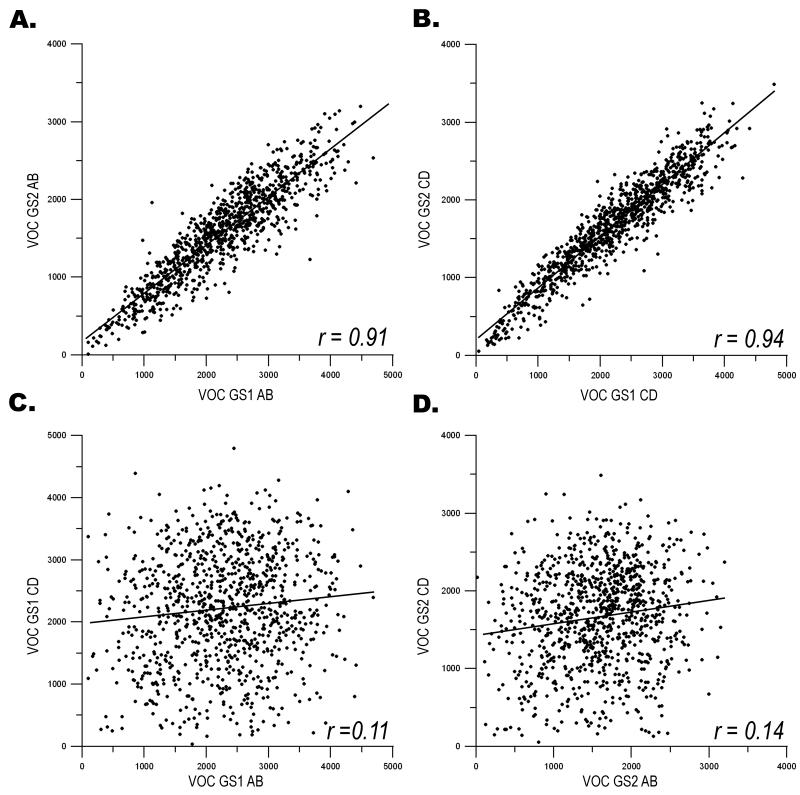
First, we examined the behavior of exactly the same plates between GS1 and GS2, by comparing the observed cell density as reported by VOC. A highly significant correlation was observed between the VOC for GS11-3AB and the VOC for GS21-3AB (r = 0.91, Figure 3A) demonstrating the reproducible affect on cell density of a single siRNA pool. Comparison of the VOC for GS11-3CD and GS21-3CD resulted in an equally strong correlation (r = 0.94, Figure 3B). These results clearly indicate that a specific siRNA pool will affect cell density in a way that is reproducible and predictive of its behavior in a replicate screen.
Figure 3. The correlation of a phenotype between identical siRNA pools and independent siRNA pools targeting a subset of genes within Genomic Screen 1 (GS1) and Genomic Screen 2 (GS2).
a, The valid object count (VOC) from 984 siRNA pools (GS1AB1-3) within GS1 is compared across genomic screens to the VOC for 984 identical siRNA pools (GS2AB1-3) from GS2. The linear regression line is provided with the correlation coefficient (r). b, Similar to a, excepting that the VOC from 984 identical siRNA pools from GS1CD1-3 is compared to GS2CD1-3. Again, the linear regression line is identified along with the correlation coefficient (r). c, The VOC from 984 siRNA pools (GS1AB1-3) is compared to the corresponding 984 siRNA pools (GS1CD1-3) within GS1. Both the linear regression line and correlation coefficient (r) are provided. d, Similar to c, excepting that intra-screen comparison is made between the VOC from 984 wells (GS2AB1-3) to the 984 corresponding wells (GS2CD1-3) within GS2.
The power of our 2×2 pooled screening strategy is demonstrated when the independent siRNA sets are compared within screens: GSl1-3AB vs. GS11-3CD and GS21-3AB vs. GS21-3CD. The correlation coefficient of the VOC of GSl1-3AB vs. GS11-3CD was 0.11 (Figure 3C), and reflected the independence exhibited by the two sets of siRNAs. The second genomic screen also demonstrated independent behavior of the siRNA sets with a correlation coefficient of 0.14 for GS21-3AB vs. GS21-3CD (Figure 3D). We have also performed this analysis on seven plate pairs from the middle of the screen and seven pairs from the end of the screen, accounting for greater than 18% of the library, with similar results (data not shown). These results indicate that the effect of each AB siRNA pool on cell density is not predictive of the effect of the corresponding CD siRNA pool.
Our assay system used percent infection to identify hits therefore effects on cell number can be considered an OTE of siRNA treatment. When assaying cell number as a surrogate of OTEs, our results demonstrate that testing the same siRNA pool multiple times produced similar results (Figure 3A, 3B). However, our 2×2 pooled siRNA library format is intended to take fullest advantage of the power of two independent trials and supporting that strategy we clearly demonstrate the ability to alleviate this observed correlation by testing two unique siRNA pools (Figure 3C, 3D). It should be noted that these data are derived from a single siRNA library from one manufacturer and it remains possible that other algorithms for siRNA design may produce siRNA libraries which behave differently.
Four methods for hit selection resulted in significantly different hit lists
As an academic screening facility, we were interested in testing the reproducibility of siRNA screening on a genome-wide scale. As suggested by the behavior of the controls (Figure 2A, 2C), the distributions of the population of screening wells from GS1 were markedly different from those of GS2 (Figure 4A, 4B). In order to compare the screens, we first had to arrive at a collection of hit selection strategies that were appropriate for both data sets.
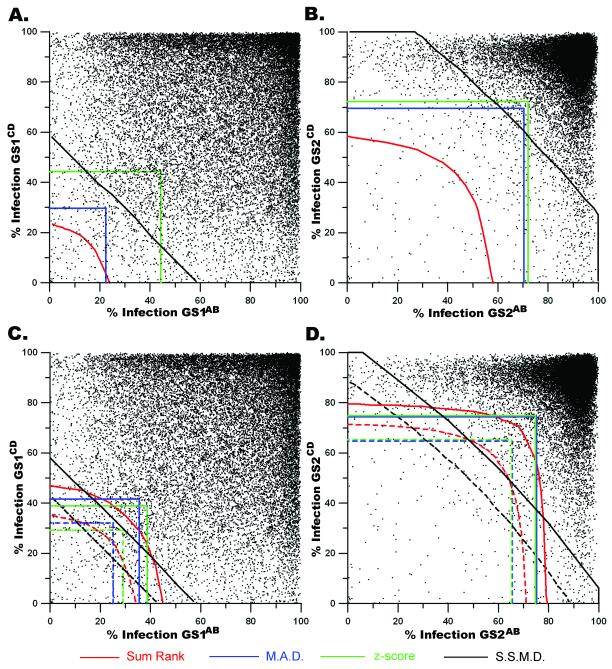
Figure 4. The illustrated thresholds from four statistical methodologies overlaid onto the associated genomic population.
For (a, b, c, d): genomic screen 1 (GS1), genomic screen 2 (GS2), sum rank (SR), median absolute deviation (MAD), z-score, and strictly standardized mean difference (SSMD). a, Each point (▪) represents the % Infection for the analyzed population in GS1AB plotted against the % Infection from the corresponding wells within GS1CD. The illustrated lines indicate the thresholds within which hits are declared if SR ≤ 0.001350, MAD ≤ −2.000, z-score ≤ −3.000 or SSMD ≤ −3.000. b, Similar to a, excepting that the % Infection from GS2AB is plotted against the % Infection for corresponding wells from GS2CD. The thresholds indicate the limits within which hits can be declared assuming SR ≤ 0.001350, MAD ≤ −3.000, z-score ≤ −3.000 or SSMD ≤ −3.000. c, The thresholds for the top 200 (dashed lines) and top 500 (solid lines) lists generated by SR, MAD, z-score and SSMD are overlaid onto the scatter plot of the % Infection (as described in a) for the paired siRNA pools in GS1AB and GS1CD. d, The % Infection for corresponding siRNA pools within GS2 are plotted as coordinates (GS2AB, GS2CD). The limits determining the top 200 (dashed lines) and top 500 (solid lines) lists are illustrated in the context of the GS2 population.
Analyses utilizing parametric statistics assume that the population in question can be accurately described by an established probability distribution. The consistent behavior of the siGFP negative control wells within each genomic screen (Figures 2A, 2C), moderately agrees with a normal distribution, so methods that utilized the siGFP population mean and variance were acceptable for analysis. In addition, non-parametric analysis can be applied when a population is not easily described by a normal distribution. The skewed distributions of the experimental populations GS1 and GS2 (Figure 4A and 4B) indicated that nonparametric statistics applied to the genomic population were also appropriate. We found no single batch of data behaving independently, therefore GS1AB, GS1CD, GS2AB and GS2CD were considered distinct populations and each was analyzed as a single batch. Supplemental Table 1 lists four statistical strategies: MAD, SR, z-score and SSMD. MAD and SR were representative nonparametric methods while z-score and SSMD were representative parametric methods. All four strategies had been previously described for genome-wide siRNA-based screen applications.6,20,25,27 Also described in the literature, both z-score and SSMD methods can be performed substituting the mean and standard deviation with the median and MAD, respectively, to generate non-parametric estimators for SSMD and z-score which are robust with respect to influences by strong outliers and/or skewed distributions, however these alternate calculations were not evaluated here.1,25
To arrive at comparable hit lists using the four methods, we initially sought to apply a consistent probability threshold to each. Three standard deviations greater or less than the mean of the negative control, or the population median or mean, is considered a robust threshold for hit detection.1 We chose a z-score ≤ −3.000 and its corresponding probability threshold of p ≤ 0.001350 to meet this convention. The SSMD and MAD values −3.000 were both defined by the respective authors as being equivalent to z-score of −3.000 while a p ≤ 0.001350 for SR could be used for analysis.6,25,27 In the specific instance of the application of MAD to the GS1 data set, a −3.000 was inappropriate because this threshold exceeded the limits of the assay (−4.3%) for the GS1AB set. As advised by the method’s authors’ we relaxed the MAD limit to k ≤ −2.000.6 Table 2 presents the number of hits each strategy identified and the overlap among the different lists (some fields intentionally left blank to reduce redundancy). It is immediately apparent that application of the four strategies resulted in significantly different hit list lengths even when the probability thresholds were held constant. For GS1, SR resulted in 75 hits while z-score generated the most extended hit list with 794 members. Analysis of GS2 produced lists ranging from 82 (SR) to 1,140 (SSMD) members.
Table 2.
The intra and inter screen comparisons of hit lists.
| Screen ID | Genomic Screen 1 | Genomic Screen 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Method & Threshold |
MAD k ≤ −2 |
z-score z ≤ −3 |
SSMD β≤ −3 |
SR p ≤ 0.001 |
MAD k ≤ −3 |
z-score z ≤ −3 |
SSMD β ≤ −3 |
||
|
Screen
ID |
Method
& Threshold |
List Length | 148 | 794 | 513 | 82 | 312 | 392 | 1140 |
|
Genomic
Screen 1 |
SR
p ≤ 0.001 |
75 | 75 | 75 | 75 | 43 | 55 | 56 | 59 |
|
MAD
k ≤ −2 |
148 | 148 | 148 | 53 | 81 | 86 | 105 | ||
|
z-score
z ≤ −3 |
794 | 445 | 68 | 179 | 199 | 290 | |||
|
SSMD
β ≤ −3 |
513 | 68 | 153 | 168 | 281 | ||||
|
Genomic
Screen 2 |
SR
p ≤ 0.001 |
82 | 82 | 82 | 82 | ||||
|
MAD
k ≤ −3 |
312 | 312 | 285 | ||||||
|
z-score
z ≤ −3 |
392 | 331 | |||||||
In order to better understand the wide range of hit list lengths generated using these analysis methods we illustrated the limits defined by each strategy upon the GS1 and GS2 population (Figure 4A and 4B). Visualization of the methods in this way allows one to quickly ascertain how lists can vary in both size and composition. Importantly, the assay design required that a phenotype be identified in both wells however the SSMD limit illustrated in Figure 4B indicates that for GS2, SSMD is not strictly adhering to the rule. These data clearly demonstrate that reproducibility of hit lists between, and even within, genome-scale siRNA screens can be greatly influenced by the analysis method. Although we present the overlap among and between these hit lists (Table 2), we believe that the considerable difference in list lengths confounded the interpretation of reproducibility between GS1 and GS2 and chose to pursue fixed length hit lists for comparison.
Generation of GS1 and GS2 hit lists using fixed list lengths of 200 and 500 members
As an alternative to determining the overlap of hit lists of such divergent lengths, we applied a consistent rule to all statistical strategies that resulted in a fixed number of hits and then studied reproducibility from that perspective. We scanned the literature for whole genome RNAi-based screens and considered the length of the primary hit lists. Primary hit lists for thirteen different genome wide screens ranged from 0.96% to 6.66% of the populations tested (Supplemental Table 2). The median of the thirteen lists identified 407 hits, or 2.09% of their genomic library.18 Our choice of bench mark lists included the top 200 and top 500 hits, representing 0.88% and 2.21% of the interrogated genome, respectively.
Table 3 presents the thresholds associated with the top 200 and top 500 hit lists generated when SR, MAD, z-score, and SSMD methods are utilized to rank hits for GS1. While the thresholds determining the top 200 hits, p ≤ 0.005735, k ≤ −1.900, z ≤ −4.326, and β ≤ −3.505, are no longer related in a statistical sense, they appeared to identify comparable regions of the genomic population (depicted in Figure 4C as dotted lines). Furthermore, for the top 500, the thresholds p ≤ 0.017673, k ≤ −1.510, z ≤ −3.480, and β ≤ −3.019 also appeared to identify comparable regions of the genomic population (as depicted in Figure 4C by solid lines). It should be noted that for the MAD-generated hit list, 201 hits are reported indicating that at the threshold k ≤ −1.900, two different genes performed in a statistically indistinguishable manner (a tie).
Table 3.
Intra-screen comparisons of the top 200 or 500 hits identified by SR, MAD, Z-score and SSMD for GS1
| Genomic Screen 1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Method & Threshold |
MAD k ≤ −1.900 |
z-score z ≤ − 4.326 |
SSMD β ≤ −3.505 |
MAD k ≤ −1.510 |
z-score z ≤ − 3.480 |
SSMD β ≤ − 3.019 |
||
| Genomic Screen 1 |
Method &
Threshold |
List
Length |
201 | 200 | 200 | 500 | 500 | 500 |
|
SR
p ≤ 0.00573 |
200 | 173 (87%) |
174 (87%) |
176 (88%) |
||||
|
MAD
k ≤ −1.900 |
201 | 178 (89%) |
151 (76%) |
|||||
|
z-score
z ≤ −4.326 |
200 | 153 (77%) |
||||||
|
SR
p ≤ 0.0176 |
500 | 423 (85%) |
428 (86%) |
436 (87%) |
||||
|
MAD
k ≤ −1.510 |
500 | 445 (89%) |
375 (75%) |
|||||
|
z-score
z ≤ −3.480 |
500 | 376 (75%) |
||||||
For GS2, the SR, MAD, z-score, and SSMD thresholds with their illustrated limits are presented in Table 4 and illustrated in Figure 4D. The thresholds for the top 200 hits were p ≤ 0.005653, k ≤ −3.726, z ≤ − 4.300 and β ≤ −5.211, while the thresholds for the top 500 hits were p ≤ 0.016596, k ≤ −2.281, z ≤ − 2.491 and β ≤ −4.142. In the instance of the SR hit list, 501 hits are identified due to two hits at the threshold having identical SR values (a tie).
Table 4.
Intra-screen comparisons of the top 200 or 500 hits identified by SR, MAD, Z-score and SSMD for GS2
| Genomic Screen 2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Method & Threshold |
MAD k ≤ −3.726 |
z-score z ≤ −4.300 |
SSMD β ≤ −5.211 |
MAD k ≤ −2.281 |
z- score z ≤ −2.491 |
SSMD β ≤ −4.142 |
||
| Genomic Screen 2 |
Method &
Threshold |
List
Length |
200 | 200 | 200 | 500 | 500 | 500 |
|
SR
p ≤ 0.005653 |
200 | 172 (86%) |
173 (87%) |
144 (72%) |
||||
|
MAD
k ≤ −3.726 |
200 | 198 (99%) |
129 (65%) |
|||||
|
z-score
z ≤ −4.300 |
200 | 129 (65%) |
||||||
|
SR
p ≤ 0.01659 |
501 | 422 (84%) |
428 (86%) |
284 (57%) |
||||
|
MAD
k ≤ −2.281 |
500 | 481 (96%) |
237 (47%) |
|||||
|
z-score
z ≤ −2.491 |
500 | 240 (48%) |
||||||
Intra-screen comparison of analysis methods using fixed length hit lists
To examine the impact of analysis methodology on the composition of hit lists, we determined the intra-screen overlap of identified targets using each method. Intra-screen overlap illustrates to what extent the results of differing analysis methods agree with respect to identifying the same factors. The thresholds determining the top 200 and top 500 hits lists for GS1 by the SR, MAD, z-score and SSMD methods are illustrated in Figure 4C, while the intra-screen overlap is tallied in Table 3. As presented in Table 3, the best intra-screen overlap for the top 200 and top 500 hit lists for GS1 were between hit lists generated by z-score and MAD methods in which 89% of the potential overlap was identified by both methods. Similarly, as depicted in Figure 4C, both z-score and MAD methods identify very similar regions of interest. The least reproducibility is consistently associated with comparisons between SSMD and either z-score or MAD methods for both the top 200 and top 500 lists in GS1, ranging from 75% to 77% of the population (Table 3). Again, Figure 4C depicts that the SSMD method identified two unique regions of interest in which one of the two assay pairs performed quite strongly, while the other had fairly weak signal, relative to the control population. However, z-score and MAD methods identified unique populations in which both assay wells performed moderately, yet similarly. Comparisons between MAD, z-score and SSMD reveals the extremes while SR identifies a region of interest depicted in Figure 4C sharing some of the populations independently identified by each, and as tabulated in Table 3, consistently identifies 85% to 88% of any other methods’ top 200 or top 500 hit list.
The intra-screen overlap for the SR, MAD, z-score and SSMD methods as applied to GS2 is tallied in Table 4, and the limits for each analysis method is depicted in Figure 4D. Similar to the performance of the MAD and z-score methods in GS1, applied to the GS2 population these have the greatest overlap at 99% for the top 200 list and 96% for the top 500 list, and their respective limits depicted in Figure 4D support the strong overlap as they identify nearly identical regions of interest. The SSMD method again behaves most differently from the other methods. Relative to the MAD and z-score methods, the SSMD methods shares 65% overlap for the top 200 datasets and 47-48% of the population for the top 500 datasets. Explaining the discrepancy, Figure 4D shows that the SSMD algorithm did not strictly adhere to the proposed goal that both siRNA pools had to perform similarly. In fact, some candidate genes identified by SSMD have a higher than average percent infection, relative to the negative siGFP control, for one assay well, while nearly completely inhibiting infection in the alternate well. The overlap between SR and MAD or z-score for the top 200 was 86% and 87% while for the top 500 list 84% and 86% respectively. SR overlaps with SSMD only 72% for the top 200 list and 57% for the top 500 list.
Overlap of the GS1 and GS2 hit lists
The inter-screen reproducibility was measured by comparing the overlap of the top 200 and/or top 500 hits generated for each analysis method, SR, MAD, z-score and SSMD from GS1to the top 200 and top 500 hit lists generated from GS2. Table 5 reports the thresholds associated with each hit list and the overlap for all paired comparisons. The least overlap (32%) was noted when comparing the SSMD generated top 500 list for GS2 to the z-score or MAD generated lists from GS1. The best overlap, 67%, is noted when comparing the SR generated top 500 list in GS1 to the GS2 SR top 200 generated list. The general trend indicates that while hit lists of exactly the same length from two screens may not have the best overlap, in the case in which a smaller hit list is first considered and then compared to a larger hit list, the best overlap can then be observed. As practically applied, the best reproducibility between hit lists can be observed when a threshold is set for one genomic screen, and then compared to list generated by a relaxed threshold in the second screen.
Table 5.
Inter-screen comparisons between lists identified by SR, MAD, Z, SSMD for either 200 or 500 hits.
| GS2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method & Threshold* |
SR p ≤ 0.005 |
MAD k ≤ −3.72 |
zscore z ≤ −4.3 |
SSMD β ≤ −5.2 |
SR p ≤ 0.016 |
MAD k ≤ −2.28 |
z- score z ≤ −2.5 |
SSMD β ≤ −4.1 |
||
| GS1 |
Method &
Threshold * |
List Size | 200 | 200 | 200 | 200 | 501 | 500 | 500 | 500 |
|
SR
p ≤ 0.005 |
200 | 98 (49%) |
91 (46%) |
91 (46%) |
88 (44%) |
130 (65%) |
127 (64%) |
126 (63%) |
120 (60%) |
|
|
MAD
k ≤ −1.9 |
201 | 90 (45%) |
83 (42%) |
83 (42%) |
77 (39%) |
117 (59%) |
119 (60%) |
118 (59%) |
108 (54%) |
|
|
z-score
z ≤ −4.3 |
200 | 91 (46%) |
82 (41%) |
82 (41%) |
78 (39%) |
116 (58%) |
117 (59%) |
116 (58%) |
108 (54%) |
|
|
SSMD
β ≤ −3.5 |
200 | 96 (48%) |
87 (44%) |
87 (44%) |
92 (46%) |
129 (65%) |
124 (62%) |
122 (61%) |
129 (65%) |
|
|
SR
p ≤ 0.017 |
500 | 133 (67%) |
127 (64%) |
127 (64%) |
118 (59%) |
201 (40%) |
195 (39%) |
194 (39%) |
181 (36%) |
|
|
MAD
k ≤ −1.5 |
500 | 127 (64%) |
121 (61%) |
120 (60%) |
105 (53%) |
184 (37%) |
184 (37%) |
183 (37%) |
161 (32%) |
|
|
z-score
z ≤ −3.48 |
500 | 125 (63%) |
122 (61%) |
121 (61%) |
108 (54%) |
188 (38%) |
186 (37%) |
184 (37%) |
161 (32%) |
|
|
SSMD
β ≤ −3.02 |
500 | 131 (66%) |
123 (62%) |
123 (62%) |
127 (64%) |
202 (40%) |
190 (38%) |
189 (38%) |
205 (41%) |
|
Thresholds have been truncated for space. Actual values are in the Results section and reflect four significant digits.
Discussion
The work presented here is the first published study to experimentally define the factors that influence reproducibility between genome-scale siRNA screens. The high throughput loss-of-function genomic screening community has reached a critical milestone in the application of this promising technology. Multiple teams have completed whole genome siRNA-based screens for factors involved in the same biological system, and the obvious first reaction was to compare the overlap between related screens.2,15,28 This seemingly straightforward task resulted in a marked lack of overlap among the published hit lists. A comprehensive meta-analysis attempted to reconcile the data from these substantially different assay systems, and did produce some additional overlap of gene families.4 Without a clear idea of the reproducibility expected, interpretation of the limited overlap was clouded. Our study attempts to shed light on the issue of reproducibility of genome-scale siRNA screening and provide a context for interpretation of published screening data.
Within six months, two human whole genome siRNA-based screens were completed. The genomic screens rigorously adhered to the same protocols, utilized the same instrumentation, used the same batch of virus and were performed by the same team. The only intentional difference was the four months separating the completion of GS1 and beginning of GS2. Variables that were not accounted for included changes in the batches of reagents and the passages of the cell line.
The 2 × 2 pooled library format described in Figure 1 ensured that hit identification would require that at least two of four siRNAs induce a phenotype.7 If both positive siRNAs resided in one well, the hit would be missed. The probability of such a distribution by random chance is 0.18, or 18% of the possible scored wells.
Off target effects are defined as changes in gene expression for genes not intentionally targeted by the siRNA design. Jackson et al. 2003 demonstrated that decreasing the effective concentration of siRNA targeting MAPK14 decreased OTEs while preserving the integrity of the target-specific knockdown. They also noted some OTEs could not be titrated away. Our assay conditions were designed to minimize OTE by utilizing a relatively low effective siRNA concentration of 15.4nM (7.7nM for each siRNA) when compared to other published screens (Supplemental Table 2).
We addressed the impact of screening format on OTE as well as reproducibility by examining the effect of pooled siRNA duplexes on cell density. In this case, effects on cell density can be considered an unintended or OTE. The scatter plots from Figure 3 demonstrate the remarkably reproducible effect a given pool of two siRNAs had on cell density, while independent pools had entirely independent effects on cell density. Although we are unable to rule out the observed effect could be due to influences such as relying on a single source (Qiagen) for the design and manufacturing of the siRNA library, these data demonstrate that a given pool of siRNAs will produce similar results when tested multiple times. The rationale that multiple tests of a single system better illustrates the true range of the system has sound statistical support. Unfortunately, studying the cell density results in which the same pool of siRNAs is tested twice (Figures 3A and 3B) and contrasting it to the population distributions generated by testing independent pools (Figures 3C and 3D) clearly shows that the experimental interpretation of the function of a target mRNA is not defined without independent tests. We understand that additional validation of our hit list is required, however by identifying those targets which display the appropriate phenotype with two independent pools, we fulfill the important criteria of redundancy in the primary screen.
Recently, Brass et al. 2009 published a screen which identified factors influencing H1N1 propagation in a human model cell line utilized three tests of the same pool of four siRNA duplexes. This study reported 334 targets from the primary screen, and a follow-up screen to de-convolute the pools resulted in 40% of the 334 putative hits being confirmed by at least two independent siRNAs. As a contrast, consider Zhang et al. 2009 screen for modifiers of circadian cycle. The authors here used the pooled 2 × 2 format and followed up on hits that scored well in both independent tests. 78% of the 343 putative hits reconfirmed with two or more siRNAs eliciting a strong effect in a validation screen. Despite screening the genome three times, representing a 50% increase in the workload relative to the Zhang et al. 2009, Brass et al. 2009 did not improve the resolution of the mRNA function.
Our study used the data from each of the four 74 plate sets; GS1AB, GS1CD, GS2AB, and GS2CD as batches for analysis for several reasons. On each 384 well plate, four negative siGFP and four positive si-vATPase controls were arrayed. Both SSMD and z-score methods require a comparison to a negative control set, so the minimum dataset for analysis must be one assay plate. Zhang et al. 2008 demonstrated using SSMD how variability in control wells contributed strongly to the calculated assay performance. This indicated that opportunities to combine more plates into a single dataset could mask the impact normal variation in the negative control set had on the analysis. Further, Qiagen manufactured their library in a systematic fashion. Consequently, 65% of the siRNA duplexes targeting the GPCR family members were arrayed on a single plate, and the remaining ones were distributed on two adjacent plates. Other gene families are arrayed similarly. SR utilized the population to determine a hit, so any population with significant bias would inhibit the application of SR, thus we chose larger datasets to provide protection from plating bias. Figures 2A and 2C qualitatively indicated that data within each screen performed consistently, while the calculated Z’-factor quantitatively indicated any variability which was present was unlikely to negatively impact the resolution of hits as strong as the si-vATPase. Although we concluded that treating all wells in a single siRNA set as one dataset would be appropriate, one must carefully consider which approach best suits the behavior of the data sets.
Genomic screening is costly. Significant resources are invested during assay development to determine how best to pursue factors involved in a biological system. It seems contradictory that the investment in the cell-based assay has not been complemented by an investment in understanding the interpretation of screening results. We measured the reproducibility between two genomic screens by directly testing how much overlap there was between specific analysis methods. Applying the statistical methods SR, MAD, z-score and SSMD by following the recommendations of each methods’ authors produced a broad range of overlap.6,20,25,27 It would seem in some cases that there was little overlap between identical screens. Unfortunately, the length of the hit lists was dramatically different. For example, comparison between SSMD in GS1 and GS2, the potential overlap could include all 513 hits from GS1, but the potential overlap is only 45% of the 1,140 hits in GS2. Further, it was difficult to assess the reproducibility between hit lists within a single screen because the different analytical methods produced significantly different length lists.
In order to compare methodologies within and between genomic screens, we chose to compare lists composed of the same number of hits. The top 200 hits for any method produced between 39% to 49% overlap. Expanding the list to include the top 500 hits did not improve the apparent reproducibility as the range was 32% to 41% overlap. The best indication that genomic data is regularly reproducible was established by comparing the top 200 from any individual method to the top 500 from the alternate screen. This situation reflects the stochastic nature of biological assays. A subset of siRNA targets will always score particularly well because the siRNAs are robust, the gene is easily silenced, or the gene is simply at a critical juncture in a system or pathway. Other siRNAs that may score strongly in one assay but only moderately in a second assay demonstrate that some pathways may be more resistant or adaptable to change or some genes are not as efficiently silenced as others. Thus, a gene that strongly inhibited infection in one screen did not necessarily strongly inhibit infection in the other screen, but it was highly likely to perform well.
As a standard practice, when comparing genomic screens one must consider the behavior of assay wells which did not make the top tier hit lists. Unlike other –omic technologies such as microarray analysis of gene expression, proteomic studies of protein abundance and genotyping via deep sequencing, the results of siRNA loss-of-function studies are not directly quantitative. The relative strength of the “score” in the phenotypic assay may not be directly related to the abundance of a required target protein. Due to difference in effective concentration, and stoichiometry of reactions involved, the strength of a complex phenotypic score is very unlikely to scale proportionally with protein levels. A target that scores only moderately in a screen may be absolutely required, however due to factors such as siRNA efficiency, protein half-life and message abundance, the protein levels may only be reduced 30% in the course of the assay. This makes generation of “hit lists” a troubling facet of siRNA screening and certainly contributes to overlap and reproducibility. In order to alleviate these issues, the authors of screens should make available the performance of all the wells in a genomic screen at the time of publication, similar to what is currently done for microarrays.
The dual genomic screens identified reproducibility that exceeded 67%. This is the strongest published overlap for two complete screens. The 2×2 pooled siRNA library format and low siRNA concentration provided an efficient assay design to identify strong candidates without the costly validation screening. We demonstrated that each siRNA pool behaves quite reproducibility with respect to VOC, and posit that independent siRNA pools tested against the same system would provide more robust final data sets. The best practice we can promote at this time is that researchers use several analysis strategies, and that all relevant data from each control and each experimental well be provided to future researchers as is the case for microarray and genome-wide association studies.
Supplementary Material
Acknowledgements
The project described was supported in part by NIH grants to MGB; 1 R21 AI064925 and #U54 AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The Duke RNAi Screening Facility was supported by NIH S10 1SA0RR024572-01, North Carolina Biotechnology Center Grant the Institute for Genome Sciences & Policy, Duke Comprehensive Cancer Center, and Duke School of Medicine.
References
- 1.Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, Long A, Kelleher D, Smith Q, Beijersbergen RL, Ghazal P, Shamu CE. Statistical methods for analysis of high-throughput RNA interference screens. Nature methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science (New York, NY. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 3.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS pathogens. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen. 2008;13:149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- 7.Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nature methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 8.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user’s guide. Nature reviews. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 9.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 10.Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, Rao A. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- 11.Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. The American journal of tropical medicine and hygiene. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- 12.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nature biotechnology. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 13.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010 doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 14.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann H Heinrich, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. Human host factors required for influenza virus replication. Nature. 2009 doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan MN, Sukumaran B, Pal U, Agaisse H, Murray JL, Hodge TW, Fikrig E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. Journal of Virology. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & development. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell host & microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science (New York, NY. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, Su AI, Hogenesch JB, Kay SA. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89:552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XD, Espeseth AS, Johnson EN, Chin J, Gates A, Mitnaul LJ, Marine SD, Tian J, Stec EM, Kunapuli P, Holder DJ, Heyse JF, Strulovici B, Ferrer M. Integrating experimental and analytic approaches to improve data quality in genome-wide RNAi screens. J Biomol Screen. 2008;13:378–389. doi: 10.1177/1087057108317145. [DOI] [PubMed] [Google Scholar]
- 27.Zhang XD, Ferrer M, Espeseth AS, Marine SD, Stec EM, Crackower MA, Holder DJ, Heyse JF, Strulovici B. The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J Biomol Screen. 2007;12:497–509. doi: 10.1177/1087057107300646. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell host & microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.