Abstract
Sporisorium reilianum is a biotrophic maize (Zea mays) pathogen of increasing economic importance. Symptoms become obvious at flowering time, when the fungus causes spore formation and phyllody in the inflorescences. To understand how S. reilianum changes the inflorescence and floral developmental program of its host plant, we investigated the induced morphological and transcriptional alterations. S. reilianum infection promoted the outgrowth of subapical ears, suggesting that fungal presence suppressed apical dominance. Female inflorescences showed two distinct morphologies, here termed “leafy ear” and “eary ear.” In leafy ears, all floral organs were replaced by vegetative organs. In eary ears, modified carpels enclosed a new female inflorescence harboring additional female inflorescences at every spikelet position. Similar changes in meristem fate and organ identity were observed in the tassel of infected plants, which formed male inflorescences at spikelet positions. Thus, S. reilianum triggered a loss of organ and meristem identity and a loss of meristem determinacy in male and female inflorescences and flowers. Microarray analysis showed that these developmental changes were accompanied by transcriptional regulation of genes proposed to regulate floral organ and meristem identity as well as meristem determinacy in maize. S. reilianum colonization also led to a 30% increase in the total auxin content of the inflorescence as well as a dramatic accumulation of reactive oxygen species. We propose a model describing the architectural changes of infected inflorescence as a consequence of transcriptional, hormonal, and redox modulation, which will be the basis for further molecular investigation of the underlying mechanism of S. reilianum-induced alteration of floral development.
Fungal plant pathogens can severely impact plant development. While necrotrophic fungal pathogens hinder plant development and seed set by killing the plant or parts of the plants, biotrophic fungal plant pathogens are more subtle, since they depend on the living host for survival and reproduction. For example, the rust fungus Puccinia monoica induces pseudoflowers on host plants of the genus Arabis that are made up of infected leaves but resemble flowers and attract pollinators, helping fungal propagation (Roy, 1993). The hemibiotrophic fungus Moniliophthora perniciosa causes witches broom disease on cacao (Theobroma cacao), leading to hyperplasia, loss of apical dominance, and proliferation of axillary shoots, which results in the formation of a broom-like structure on the host plant (Meinhardt et al., 2008).
While the fungi of the latter examples affect only vegetative plant development, other fungi interfere also with floral development of the host. The smut fungus Ustilago maydis affects vegetative and reproductive development of maize (Zea mays) by inducing tumors on leaves and stems and in inflorescences (Schirawski et al., 2006; Brefort et al., 2009). An interesting case is the biotrophic pistil smut fungus (Salmacisia buchloëana), which induces hermaphroditism (i.e. development of pistils) in florets of male buffalograss (Buchloe dactyloides; Chandra and Huff, 2010). Some fungi can completely revert the floral program, leading instead to the development of vegetative organs (Meyer, 1966; Raghavendra and Safeeulla, 1979; Semisi and Ball, 1989). Floral reversion leading to phyllody formation is induced by the downy mildew fungus Sclerospora graminicola on its host plant pearl millet (Pennisetum americanum; Meyer, 1966; Raghavendra and Safeeulla, 1979; Semisi and Ball, 1989). Phyllody in inflorescences is also induced by the smut fungus Sporisorium reilianum on its host plants maize and sorghum (Sorghum bicolor; Reed et al., 1927; Matheussen et al., 1991).
For phyllody formation to occur, the fungus must alter the floral developmental program. In maize, the male inflorescence (tassel) develops at the apex of the plant, whereas the female inflorescences (ears) develop on the main shoot from axillary meristems (Veit et al., 1993). During inflorescence development of tassel and ear, several types of meristems develop that follow a very similar order. A first step toward development of the male inflorescence is the conversion of the shoot apical meristem into tassel inflorescence meristem. In the developing tassel, the inflorescence meristem develops branch meristems, which leads to the formation of branches. At the side of each branch, spikelet pair meristems develop that differentiate into spikelet meristems, which develop to a spikelet. Eventually, each spikelet develops into two floral meristems (Veit et al., 1993). The floral meristems differentiate into upper and lower florets. The two florets are surrounded by the spikelet remnants called outer and inner glumes. Together, the axillary meristems (branch meristems, spikelet pair meristems, spikelet meristems, and floral meristems) give the maize inflorescence its architecture. Similar meristem differentiations take place in the developing ear, except that there is no formation of branch meristems (Bennetzen and Hake, 2009). The maize floret initially develops five floral whorls that form lemma, palea, two lodicules, three stamens, and a pistil. The pistil in the most inner whorl is formed from three carpels; one remains rudimentary and two are fused to form the silk that surrounds a single ovule (Nickerson, 1954). The upper floret matures in advance of the lower floret, which aborts in the ear. Another difference between tassel and ear is obvious after initiating floral organ primordia, where stamens are aborted in ear florets and pistils are aborted in tassel florets, giving rise to monoecious flowers (Veit et al., 1993; Bennetzen and Hake, 2009).
The mechanistic causes of the change in developmental program have only in the rarest cases been elucidated. In the case of the hermaphroditism occurring in S. buchloëana-infected male buffalograss, down-regulation of a putative female-suppressor gene homologous to the maize TASSELSEED2 has been identified as a potential cause (Chandra and Huff, 2010). The ability of U. maydis to induce tumors on different floral organs in the maize tassel is dependent on the developmental stage of the floret at the time of infection (Skibbe et al., 2010; Walbot and Skibbe, 2010). Tassel infection has been suggested to be dependent on GA signaling, since the dwarf8 mutant failed to induce tumors (Walbot and Skibbe, 2010). GA has been proposed to play an important role for symptom formation of S. reilianum on sorghum. The presence of the fungus has been shown to lead to a decreased GA concentration, which was suggested to be the reason for increased tillering of infected plants (Matheussen et al., 1991). However, the addition of GA to in vitro-grown sorghum inflorescences has been used to mimic the phyllody caused by S. reilianum infection (Bhaskaran et al., 1990). While these reports imply a (albeit contrasting) role of GA in symptom formation of S. reilianum, GA addition did not lead to phyllody formation when applied to in vitro-grown maize inflorescences (Touraud et al., 1997), throwing doubt on GA concentration as the sole explanation for floral changes caused by S. reilianum. Thus, the question of how S. reilianum redirects the development of its hosts remains elusive.
S. reilianum is currently being established as a model organism to investigate fungus-host interaction on the molecular level. The genome sequence has recently been published, and targeted gene deletion strains can readily be generated (Schirawski et al., 2010). Since also the genomes of its host plants maize and sorghum are available (Paterson et al., 2009; Schnable et al., 2009), the S. reilianum-host interaction offers a unique opportunity to study the molecular basis of symptom formation on both the fungus and the plant side.
In this work, we provide a detailed morphologic analysis of the effects of S. reilianum on maize inflorescence and flower development. In addition, we describe the transcriptional changes induced in infected inflorescences at an early stage of symptom development and propose a mechanistic model of how S. reilianum might impact floral development.
RESULTS
S. reilianum Infection of Maize
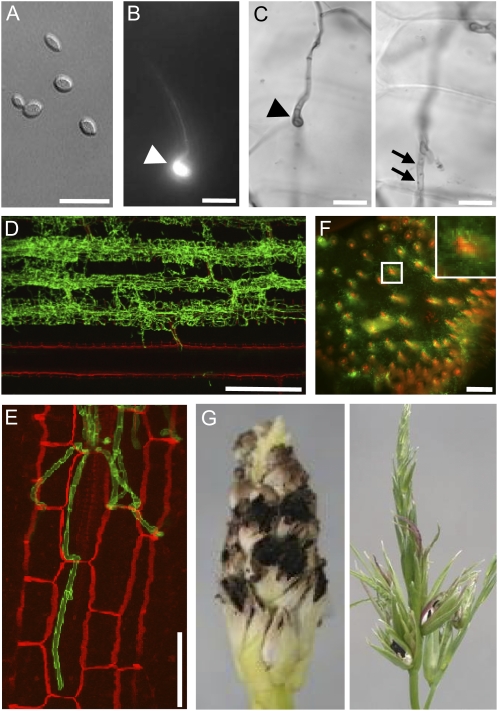
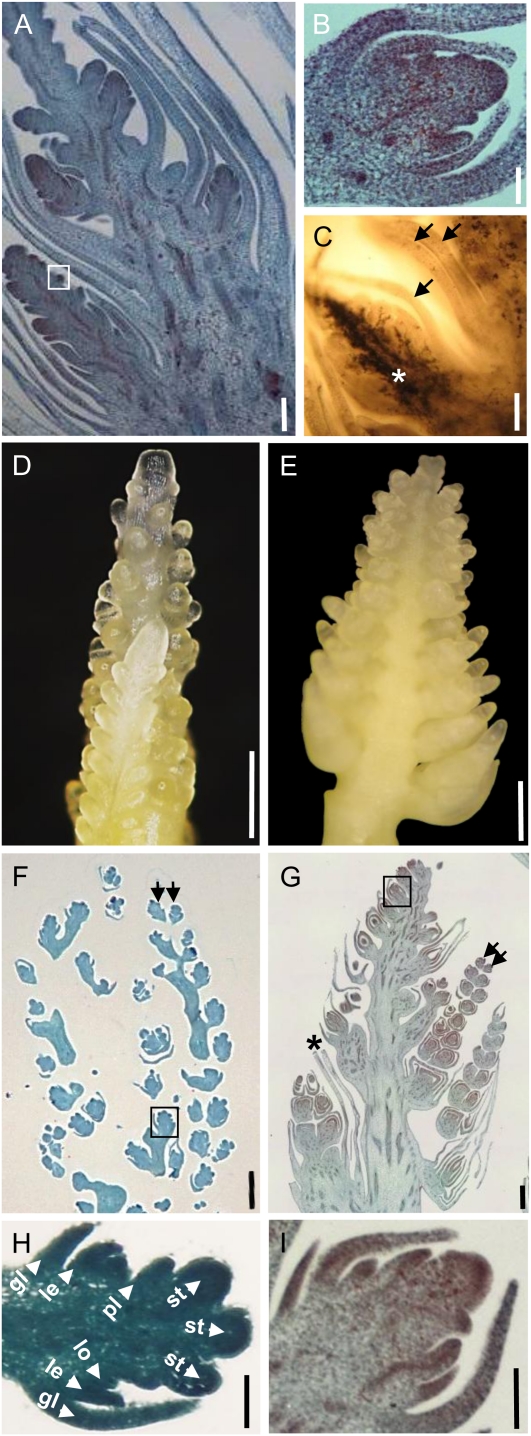
To understand the symptoms caused by S. reilianum on maize, we inoculated a mixture of compatible sporidia of S. reilianum strains (Fig. 1A) into the leaf whorl of 7-d-old maize seedlings. At 16 to 18 h post inoculation, Calcofluor White-stained leaf surfaces revealed the presence of appressoria that had developed at the tip of fungal hyphae (Fig. 1B). Appressoria marked the entry point of fungal hyphae into the leaf epidermal cells (Fig. 1C). Hyphae traversed epidermal cells and colonized bundle sheath cells (Fig. 1, D and E). From there, S. reilianum progressed toward the leaf sheath and could be detected in the nodes at 15 d post inoculation. In the node, the fungus mainly proliferated around the vascular bundles (Fig. 1F). Although hyphae of S. reilianum could be observed in all these tissues, sporulation occurred only in the male (tassel) or female (ears) inflorescences (Fig. 1G). In addition to spore formation, we observed morphological changes in the inflorescences that are described in detail below.
Figure 1.
Morphological stages of S. reilianum outside, on, and in maize cv Gaspe Flint. A, Axenically grown haploid sporidia of S. reilianum. Bar = 15 μm. B, On the plant surface, dikaryotic hyphae of S. reilianum form appressoria for plant penetration at 1 d after infection. An appressorium (arrowhead) is visualized here by fluorescence microscopy after Calcofluor staining. Bar = 15 μm. C, Appressoria of S. reilianum penetrate the leaf surface. From the appressorium (arrowhead; left panel), a hypha penetrates the underlying epidermal tissue (arrows; right panel). Images in the left and right panels are of different focal planes visualized by bright-field microscopy after staining with Chlorazole Black E. Bars = 10 μm. D, In the plant leaf, fungal hyphae proliferate along leaf vascular bundles. The image shows a Z-stack of WGA-Alexa Fluor-stained (green; fungal hyphae) and propidium iodide-stained (red; plant cells) samples visualized by confocal microscopy. Bar = 300 μm. E, Closeup of fungal hyphae colonizing bundle sheath cells. Bar = 50 μm. F, In the nodes of the plant, fungal hyphae (green) surround the plant vascular bundles (red). A cross-section stained with WGA-Alexa Fluor and propidium iodide was visualized by confocal microscopy. Bar = 500 μm. G, Spores of S. reilianum forming on maize ear (left) and tassel (right).
S. reilianum Alters Ear-Branching Architecture
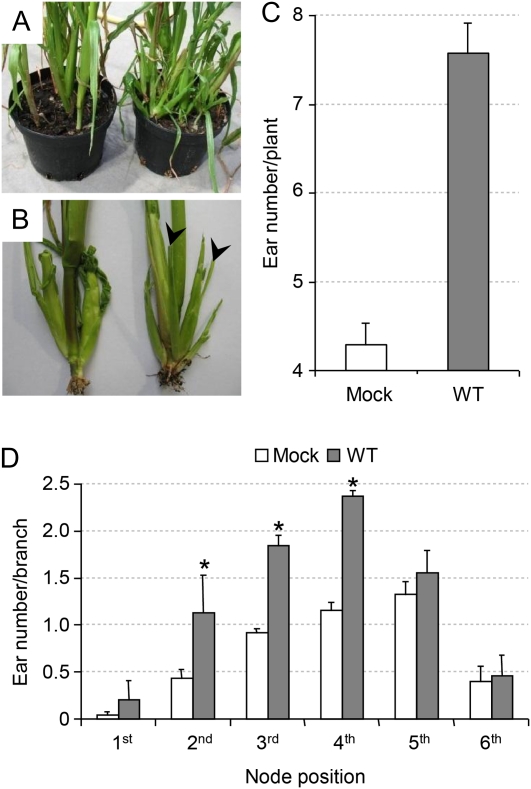
We noticed that S. reilianum-infected plants showed more ear branches per plant than mock-infected ones (Fig. 2A). Ear branches developed from axillary meristems born at the axils of husk leaves (Fig. 2B), indicating that S. reilianum infection resulted in a loss of apical dominance in ear-bearing branches. To quantify the formation of female inflorescences, we removed the husk leaves around the primary ear and counted all visible inflorescences. At 8 weeks post infection, S. reilianum-infected plants showed on average 7.6 ears per plant, whereas mock-infected plants had only 4.3 ears per plant (Fig. 2C). To find out whether the increase in the number of ears is restricted to a branch appearing at a specific node, we determined the ear number per branch at each node. For this purpose, the second node was defined as the top-most node with brace roots, and counting was toward the top. Secondary ear branches appearing from axils of husk leaves of the primary ear could be observed on any ear branches. In infected plants, subapical ears appeared at a significantly higher frequency at the second to fourth nodes (Fig. 2D). However, the relative increase in average ear number per branch was highest at the lowest node considered and ranged from 2.8-fold at the second node to 1.1-fold at the sixth node (Fig. 2D). This indicates that the increase in the total number of ears is derived from a loss of apical dominance at ear branches appearing at lower nodes.
Figure 2.
Effect of S. reilianum infection on ear number of maize cv Gaspe Flint. A, S. reilianum-infected plants (right) show a higher ear number then mock-infected plants (left). Each pot contains four plants. B, In mock-infected plants (left), ears form at apical points of a side branch. In infected plants (right), additional secondary ears (arrowheads) develop on subapical side branches of an ear-bearing side branch. C, Quantification of the ear number per plant. Error bars represent se of three independent experiments with 23 ± 2 plants per infection and replicate. The difference is significant (P = 0.01). D, S. reilianum-infected plants significantly (* P = 0.05) develop more ears per branch at the second, third, and fourth nodes. The second node was defined as the top-most node with brace roots, and counting was toward the top. Error bars represent se of three independent experiments with more than 25 plants per infection and replicate. WT, Wild type. [See online article for color version of this figure.]
S. reilianum Changes the Morphology of Maize Flowers
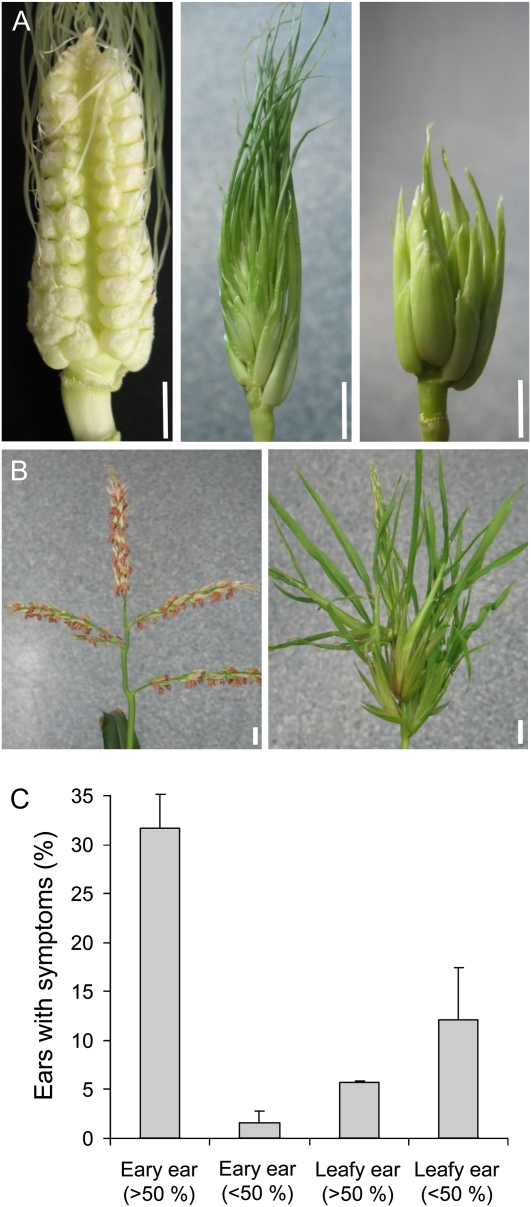
An inoculation of maize seedlings with S. reilianum did not lead to any plant developmental defects until flowering time. When inflorescences appeared, a range of morphological changes in both male and female inflorescences could be observed in infected plants. In addition to the formation of white sori harboring dark-brown fungal spores (Fig. 1G), we observed the formation of phyllody in tassels and ears of infected plants (Fig. 3, A and B). Phyllody occurred to different extents, affecting a range of spikelet numbers, from one individual spikelet to all spikelets of an inflorescence. In female inflorescences, we could distinguish two morphologic forms of phyllody, which we named “leafy” and “eary,” because they either seemed to be replaced by leaf-like structures or by ear-like structures, respectively (Fig. 3A). Interestingly, while leafy spikelets could cover different portions of the inflorescence, eary spikelets mostly covered the whole inflorescence (Fig. 3C). Ears carrying both leafy and eary spikelets were only rarely observed. In male inflorescences, we observed only one morphologic form, which we called “phyllodied tassel.” In phyllodied tassels, the morphologic change could affect one or more spikelets, and spikelets seemed to be replaced by ear-like structures (Fig. 3B). S. reilianum infection of cv Gaspe Flint led to phyllody at a higher frequency (91%; 114 of 126 plants) in female inflorescences emerging at basal nodes than in male inflorescences (5%; six of 126 plants) that form at the apex.
Figure 3.
Phyllody caused by S. reilianum infection in female and male inflorescences of maize cv Gaspe Flint. A, Morphology of female inflorescences of S. reilianum-infected (leafy ear, middle; eary ear, right) and healthy (left) plants. Bars = 1 cm. B, Morphology of male inflorescences of S. reilianum-infected (phyllodied tassel; right) and healthy (left) plants. Bars = 1 cm. C, Symptom distribution of eary and leafy ears. Eary or leafy morphology could cover more than 50% or less than 50% of the inflorescence. Error bars represent sd of three independent experiments with more than 25 plants each.
S. reilianum Transforms Reproductive into Vegetative Organs
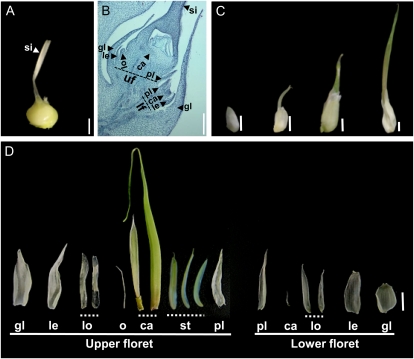
To understand the floral modifications caused by S. reilianum infection of maize, we analyzed different spikelet developmental stages. Spikelets of female maize inflorescences (Fig. 4A) are enclosed by two glumes that surround two florets, a lower one that aborts and an upper one that develops floral organs. The flower primordium produces floral organs in whorls, first forming the palea/lemma, then lodicules, and finally carpels that enclose the ovule, while stamen development is arrested at an early stage (Fig. 4B). Fused carpels generate the silk that is required for pollen tube guidance to ensure fertilization. In male flowers, stamens develop, while female reproductive organs (carpels and ovule) are aborted (Veit et al., 1993).
Figure 4.
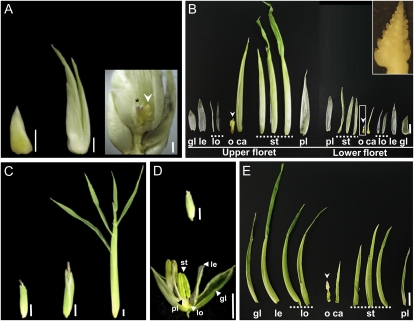
Spikelet development of healthy and S. reilianum-colonized leafy ears of maize cv Gaspe Flint. A, A healthy ear spikelet. Bar = 2 mm. B, Longitudinal section of a young healthy ear spikelet showing the upper floret (uf) with developing floral organs and the aborted lower floret (lf). Bar = 1 mm. C, Developmental stages of the leafy spikelet. The leafy spikelet first elongates, then a silk-like protrusion emerges. The protrusion elongates and thickens and gives rise to an onion leaf-like structure. Bars = 5 mm. D, Manually dissected floral structures of a leafy spikelet. Vegetative structures were formed at positions of all floral organs, except at stamen positions in the lower floret, indicating loss of organ identity. Tubular structures were formed at the position of the fused carpels, which infrequently enclosed a leafy structure and a needle-like structure. Bar = 5 mm. Floral organs forming in the healthy florets (A and B) are as follows: ca, carpel; gl, glume; le, lemma; lo, lodicules; o, ovule; pl, palea; si, silk; st, stamen. In phyllodied spikelets (D), these abbreviations indicate the positions at which vegetative structures appeared.
In comparison with healthy spikelets, leafy spikelets started to elongate before a silk-like protrusion emerged from the tip (Fig. 4C). This protrusion grew longitudinally and radially and gave rise to a green tubular structure (Fig. 4C, right). None of the floral whorls developed reproductive organs, which are readily formed in the healthy spikelet (Fig. 4, B and D). Instead, leaf-like organs formed in all floral whorls. In the upper floret, glume, lemma, lodicules, and palea appeared as thin, elongated, and translucent organs. In the innermost whorl, an elongated green tubular structure replaced the carpel. Within this tube-like organ, a smaller leaf-like structure was present infrequently that was folded in half to cover a transparent needle-like protrusion emerging from the center of the floret (Fig. 4D). The needle like-protrusion and the folded leaf-like structure might correspond to the ovule and the rudimentary carpel, respectively or might be the result of the formation of indeterminate organs at the central whorl.
Interestingly, leaf-like organs with a yellowish to green color could be found in upper florets of infected inflorescences at the place where stamens normally abort (Fig. 4D). In the lower floret, thin elongated translucent membranous organs corresponding to glume, lemma, lodicule, and palea were formed. In the center of the lower floret, a small closed tubular structure appeared in place of the carpel (Fig. 4D). In summary, vegetative structures replaced all reproductive organs, indicating that S. reilianum infection of maize led to floral reversion. In addition, meristem termination is disturbed, leading to the formation of leaf-like organs instead of aborted stamens.
S. reilianum Modifies Floral Meristem Fate
In eary ears, spikelet development also started with spikelet elongation, but, unlike in leafy ears, there was no development of a silk-like protrusion. Spikelets eventually developed two ear-like structures that contained new developing inflorescences (Fig. 5A). As in leafy spikelets, the floral whorls of eary spikelets formed vegetative structures at almost every floral whorl. While glume, lemma, lodicule, and palea appeared as thin, elongated translucent membranous organs, stamens appeared as thicker, leaf-like organs of yellowish to green color that resemble husk-like leaves (Fig. 5B). In the center of the floret, we found a husk-like leaf that covered a newly formed inflorescence replacing the carpels (Fig. 5B, inset).
Figure 5.
Loss of meristem and organ identity in spikelets of eary ears and phyllodied tassels caused by S. reilianum infection of maize cv Gaspe Flint. A, Developmental stages of eary spikelets. Eary spikelet development starts with spikelet elongation. Elongation continues but no silk emerges, and finally, two ear-like structures appear that correspond to the upper and lower florets. On the right, the surrounding husk-like leaves have been partially removed to show a new developing inflorescence (arrowhead) that also bears fungal sori (star). Bars = 5 mm. B, Manually dissected floral whorls of the eary spikelet. All the floral organs in the upper and lower florets, including the two carpels that normally form the silk, were replaced by husk leaf-like structures. The most inner whorl that normally gives rise to the ovule was transformed into a new inflorescence (arrowheads and inset). Bar = 5 mm. C, Spikelet developmental stages of phyllodied tassels. Bars = 5 mm. D, Closed (top) and open (bottom) healthy spikelets of a tassel. Bars = 5 mm. E, Manually dissected floral whorls of the upper floret of a spikelet of a phyllodied tassel spikelet. All floral organs were elongated and transformed into husk leaf-like structures, except the most inner whorl, which developed a new inflorescence (arrowhead). Bar = 5 mm. Floral organs forming in the healthy florets (D) are as follows: ca, carpel; gl, glume; le, lemma; lo, lodicules; o, ovule; pl, palea; st, stamen. In phyllodied spikelets (B and E), these abbreviations indicate the positions at which vegetative structures or new inflorescences appeared.
Similar morphological changes to those observed in the eary ear could also be observed in the phyllodied tassel. Spikelets of phyllodied tassels had glumes that were elongated compared with healthy spikelets (Fig. 5, C and D) and developed into husk-like leaves (Fig. 5C). Manual dissection of the floral whorls of the tassel spikelet revealed that most of the whorls developed husk-like leaves (Fig. 5E). However, while in spikelets of healthy tassels carpel and ovule development was aborted at an early time point, the innermost whorl in spikelets of the phyllodied tassel was altered to a husk-like leaf that covered a newly developed inflorescence (Figs. 5E and 6E). This suggests that in spikelets of both eary ears and phyllodied tassels, most of the floral whorls developed into vegetative organs, with the exception of the most inner whorl, which developed a new inflorescence instead of ceasing meristematic activity for carpel abortion. Thus, the remnant of the floral meristem in the inner whorl has changed identity into an inflorescence meristem.
Figure 6.
Loss of meristem determinacy in spikelets of eary ears and phyllodied tassels of S. reilianum-infected maize cv Gaspe Flint. A, Longitudinal section of an eary spikelet showing the development of new inflorescences in the upper and lower florets. Bar = 1 mm. B, Magnification of the part boxed in A showing the development of an inflorescence meristem instead of a spikelet meristem. Bar = 200 μm. C, Longitudinal section of an eary spikelet showing massive hyphal growth of S. reilianum (black area; white star) in the newly formed inflorescence. Arrows highlight noncolonized vegetative tissues. Fungal hyphae were stained with Chlorazole Black E. Bar = 500 μm. D, Tassel inflorescence of a healthy plant showing the development of a branched inflorescence. Bar = 500 μm. E, Tassel-like inflorescence formed in the center of an infected phyllodied tassel floret. Bar = 500 μm. F, Longitudinal section of a healthy tassel inflorescence showing the development of spikelet pairs (arrows). Bar = 500 μm. G, Longitudinal section of a newly formed tassel-like inflorescence from a phyllodied tassel floret of an infected plant showing the development of spikelet pairs (arrows). Branches of the newly formed tassel-like inflorescence are enclosed within leafy structures (star). Bar = 500 μm. H, Magnification of the part boxed in F showing the development of floral organs. Bar = 200 μm. gl, Glume; le, lemma; lo, lodicules; pl, palea; st, stamen. I, Magnification of the part boxed in G showing the development of an inflorescence meristem instead of a floret meristem. Bar = 200 μm. Sections were stained with O-Safranin-Fast Green.
Sections of eary spikelets showed the development of two inflorescences at the lower and the upper florets, respectively (Fig. 6A). In contrast to spikelets of healthy inflorescences that developed floral organs (Fig. 4B), spikelets of the newly developed inflorescence in eary spikelets developed new inflorescence meristems (Fig. 6B). The newly formed inflorescences were heavily colonized by hyphae of S. reilianum (Fig. 6C, star), whereas the floral organ-derived vegetative tissues were not (Fig. 6C, arrows). Phyllodied tassels also showed the development of new inflorescences, which appeared to be more highly branched than those of eary inflorescences (Fig. 6, D and E). The branches of the tassel-like inflorescence had spikelet pairs similar to those of the healthy tassel but were enclosed by leafy structures (Fig. 6, F and G). In contrast to spikelets of healthy tassels that developed floral organs (Fig. 6H), spikelets of the newly developed inflorescence in phyllodied tassel spikelets developed new inflorescence meristems (Fig. 6I). Thus, the meristem remnants at the inner whorl have not only changed identity from floral to inflorescence meristem, but the newly formed inflorescence meristems have in addition lost their determinacy: instead of terminating in floral meristems, they continue to form new inflorescence meristems.
S. reilianum Modulates the Floral Transcriptome
To understand the basic molecular mechanisms of S. reilianum-induced changes in the floral architecture of maize, we performed comparative RNA microarray analysis of healthy and S. reilianum-infected ears. Small ears (less than 2 cm) from 20 plants each were collected at 4 weeks post inoculation, when infected ears displayed elongation of at least one spikelet (Supplemental Fig. S1). Expression patterns using Affymetrix GeneChip Maize Genome Arrays were compiled after statistical analysis of three biological replicates. Significance analysis revealed 169 differentially regulated genes, 76 down-regulated and 93 up-regulated (Supplemental Table S1). To validate the data obtained by microarray analysis, gene expression of five randomly chosen differentially regulated genes was also analyzed by quantitative real-time (qRT)-PCR. qRT-PCR analysis supported expression-level changes observed by microarray analysis in all five cases (Supplemental Fig. S2).
Differentially regulated genes were manually annotated by comparison with sequence databases available at the National Center for Biotechnology Information (NCBI) using BLAST analysis (Altschul et al., 1990) and classified according to predicted biological functions. Of the 108 genes that could be classified, most showed involvement in biotic stress (27%) and transcriptional regulation (16%). Fewer genes were predicted to have a function in development (12%), metabolism (11%), hormone biosynthesis and response (9%), or signaling (6%). A small fraction of the genes were predicted to be involved in transport (3%), nuclear processes (3%), abiotic stress (3%), or energy (2%) or grouped to several biological functions (8%). In every group, both up- and down-regulated genes were represented in approximately equal parts. However, in the transcription factor group, members of the same family were regulated in the same manner. While MADS and NAC transcription factors were all down-regulated, the members of the APETALA2 (AP2), MYB, C2H2 zinc finger, and basic helix-loop-helix (bHLH) transcription factor families were up-regulated (Table I).
Table I. Functional classification and approximate tissue gene expression patterns of maize genes with altered expression in S. reilianum-infected ears.
| Accession | Fold Change | Annotation | Approximate Gene Expression Patterna |
| Biotic stress | |||
| AF244689.2 | 12.3 | GST 24 | Ovary |
| BM380003 | 10.4 | GST 31 | Root, ovary |
| U12679.1 | 6.0 | GST IV | Silk, root, ovary, ear |
| BM381077 | 4.9 | GSTU6 | Shoot |
| CF626259 | 4.7 | GST 25 | Endosperm |
| AF244684.1 | −3.8 | GST 19 | Ovary, ear, endosperm |
| BM332131 | 5.3 | Cytochrome P450 monooxygenase | Aerial organs, root, embryo |
| BM382553 | 3.9 | Similar to cytochrome P450 | Ovary, shoot |
| BU050860 | 4.5 | Similar to putative peroxidase | Ovary, ear |
| AY107230.1 | 3.2 | Similar to peroxidase | Silk, ovary, glume |
| BG874182 | −6.0 | Putative peroxidase | Root |
| BG842197 | −5.4 | Similar to putative peroxidase | Root |
| CF629008 | −5.1 | Peroxidase 1 | Root |
| AW424608 | −4.3 | Peroxidase 1 | Endosperm |
| BM381423 | −3.3 | Similar to putative peroxidase | Endosperm, tassel |
| CF014750 | 48.3 | Putative sesquiterpene cyclase | Silk, glume |
| BM379420 | 22.1 | Pathogenesis-related protein 10 | Endosperm, meristem |
| BM351351 | 15.3 | Pathogenesis related protein-1 | Leaf |
| BM379802 | 11.5 | Putative Ser-type endopeptidase inhibitor | Ovary, root |
| BM079805 | 5.2 | Ustilago maydis induced 11 | Silk |
| BM379606 | 4.5 | Polygalacturonase inhibitor | Sheath, ovary |
| BM348442 | 4.1 | Similar to glycosyl hydrolase family 17 protein | Ovary, endosperm |
| CF649483 | 4.0 | O-Methyltransferase ZRP4 | Silk, ear, meristem |
| BM331999 | 3.5 | Xylanase inhibitor protein 1 | Aleurone, ovary, root |
| AW330894 | −4.7 | Lichenase-2 | Glume, leaf, ovary |
| AI834666 | −3.9 | Protease inhibitor/seed storage/LTP family protein | Tassel, ear, pollen, endosperm, meristem |
| BM074215 | −3.9 | Protease inhibitor/seed storage/LTP family protein | Shoot, meristem |
| AW355997 | −3.2 | Elicitor-responsive protein 3 | Ovary, root |
| Transcription factors | |||
| U31522.1 | −5.5 | Zea mays MADS1 (ZMM1) | Pedicel, pericarp, ear, embryo, ovary, tassel |
| U31521.1 | −4.1 | Zea AGAMOUS2 (ZAG2) | Pedicel, pericarp, ovary, ear, endosperm |
| AW055920 | −3.7 | MADS box protein 29 (ZMM29) | Ear, silk, pedicel, pericarp, pollen, shoot |
| L46397.1b | −2.6a | Zea AGAMOUS3 (ZAG3) | Pedicel, ear, pericarp |
| L18924.1b | −2.3a | Zea AGAMOUS1 (ZAG1) | Pericarp, ear |
| CF348980 | 3.9 | AP2 domain-containing protein | Pedicel, ovary |
| CF637428 | 4.8 | Similar to AP2 domain-containing protein (AP28) | Unknown |
| AI629804 | 5.4 | MYB-related protein Hv33 | Aerial organs, meristem |
| BM075809 | 4.0 | MYB domain protein 43 | Mixed tissues |
| AF099413.1 | 3.7 | MYB domain protein | Unknown |
| AF099391.1 | 3.3 | MYB domain transcription factor family | Mixed tissues |
| CF045441 | −3.8 | NAC domain transcription factor family | Endosperm |
| AW352507 | −3.6 | Similar to vascular-related NAC domain protein 7 | Leaf |
| AI670293 | −3.3 | NAC domain-containing protein | Mixed tissues |
| CO533291 | 3.7 | C2H2 zinc finger family | Mixed tissues |
| BM380514 | 5.1 | bHLH family protein | Sheath, meristem |
| BG842397 | 4.5 | Nuclear transcription factor Y | Meristem, embryo, shoot |
| Development | |||
| CD999944 | 7.5 | 1-Cys peroxiredoxin antioxidant | Aleurone, embryo, endosperm |
| CA401976 | 5.2 | 1-Cys peroxiredoxin antioxidant | Endosperm |
| AY108635.1 | 9.3 | Seed maturation protein | Embryo, endosperm |
| BM080758 | 4.9 | Similar to nodulin MtN3 family protein | Root, aerial organs, meristem |
| BM338644 | 3.4 | Embryonic abundant protein | Pollen, shoot |
| BQ539573 | 6.3 | Meiosis 5 | Aerial organs, meristem |
| AF332177.1 | 3.6 | β-Expansin4 (EXPB4) | Aerial organs, meristem |
| AF332180.1 | −3.9 | β-Expansin7 (EXPB7) | Mixed tissues, root, silk |
| BM075217 | −9.9 | Vegetative cell wall protein gp1 | Root |
| CK370036 | −4.0 | Vegetative cell wall protein gp1 | Root, silk |
| CK369379 | −4.0 | l-Ascorbate oxidase precursor | Mixed tissues, root, silk |
| CK986211 | −4.1 | Late embryogenesis abundant protein | Root |
| AY108650.1 | −3.9 | Lower-specific γ-thionin | Mixed tissues |
| CD446261 | −3.3 | Phosphatidylethanolamine-binding proteins (ZCN1) | Endosperm |
| Hormones | |||
| AY254104.1 | 7.7 | Trp synthase α-subunit | Meristem, pericarp, tassel |
| CF650494 | 5.2 | Similar to Trp synthase α-subunit | Root |
| CO533600 | 3.8 | IAA-amino acid conjugate hydrolase ILR1-like 4 | Pollen |
| CK347988 | −3.3 | l-Trp:2-oxoglutarate aminotransferase | Endosperm |
| BM339000 | 4.1 | SAUR36, auxin-responsive SAUR family member | Meristem, aerial organs |
| BM379588 | 3.2 | SAUR55, auxin-responsive SAUR family member | Mixed tissues |
| AY562491.1 | 11.5 | Kaurene synthase2 | Leaf |
| CF028197 | 3.2 | GA 20-oxidase 2 | Silk, leaf |
| AY105651.1 | −3.4 | GA 20-oxidase (GAO20) | Meristem, pericarp, pollen |
| CN845512 | −3.8 | Cytokinin-O-glucosyltransferase 2 | Ovary, root, meristem |
| Signaling | |||
| CF650678 | −4.7 | Transducin family protein | Ovary, root, meristem, endosperm, shoot |
| BG319707 | −4.3 | Protein phosphatase 2C | Pericarp, ear, ovary, endosperm, meristem |
| AY105086.1 | −3.8 | Phosphatase | Ovary, tassel, shoot |
| D87045.1 | −3.4 | Protein kinase | Ear |
| CF028241 | 6.2 | Brassinosteriod-insensitive 1-associated receptor kinase 1 | Leaf, root |
| BQ528747 | 4.1 | Protein Ser/Thr phosphatase | Pollen |
Estimated gene expression patterns obtained from the UniGene database at NCBI, which were inferred from EST counts and the cDNA library sources (as reported by sequence submitters at NCBI). Libraries known to be normalized, subtracted, or otherwise biased had been removed. Tissues or organs are ordered according to their potential to express the gene.
To learn whether the members of transcription factor families were expressed in a tissue-specific manner, we compiled data on tissue-specific expression from the UniGene EST database at NCBI for the differentially regulated genes. In this database, EST counts from different cDNA libraries were used to calculate approximate gene expression patterns (number of gene ESTs per total number of ESTs in the pool; Wheeler et al., 2007). Interestingly, all down-regulated MADS box transcription factors were estimated to be specifically expressed in reproductive organs of uninfected plants according to the calculated EST profile (Table I). Floral organ-specific expression was also shown experimentally by in situ hybridization for the Zea AGAMOUS (ZAG) homologs ZAG1 and ZAG2, the AGL6-like gene ZAG3 (BEARDED EAR), and the GLOBOSA homolog ZMM29 (Schmidt et al., 1993; Münster et al., 2001; Thompson et al., 2009). In Arabidopsis (Arabidopsis thaliana) and maize, AGAMOUS (AG) and AP2 mutually repress each other (Weigel and Meyerowitz, 1994; Chuck et al., 2008). Interestingly, the two AP2 domain-containing transcription factors with unknown function identified in this study are up-regulated (Table I). However, whether they are regulated by AG is unclear.
The group of regulated genes belonging to hormone biosynthesis and response included genes potentially involved in auxin and GA biosynthesis as well as auxin and cytokinin mobilization. Two GA biosynthesis genes were up-regulated, kaurene synthase 2 and GA 20-oxidase 2, estimated to be expressed in the leaf according to the calculated EST profile of the UniGene database (Table I). In contrast, one GA 20-oxidase gene that was meristem and floral organ-specifically expressed was down-regulated in S. reilianum-infected ears relative to mock-infected ears. The up-regulation of leaf-specific and down-regulation of meristem- and floral organ-specific GA biosynthesis genes could reflect the reversion of floral organs into leaf-tissue or could be the cause for the floral reversion process. Therefore, it is difficult to assess whether GA concentration actively influences the observed floral reversion process. In our experiments, concentrations of GAs (GA1, GA3, GA4, GA5, GA8, GA9, GA20, and GA34) in collected tissues were below the detection limit (see “Materials and Methods”).
Three genes involved in auxin biosynthesis as well as two auxin-responsive genes were up-regulated in infected ears (Table I). To know whether gene regulation resulted in increased auxin levels, we measured the auxin concentration of mock-infected and S. reilianum-infected ears. Mock-infected ears had an auxin concentration of 20 ± 0.5 pmol g−1 fresh weight, while that of S. reilianum-infected ears showed an increase of 30% (26 ± 0.7 pmol g−1 fresh weight; each the mean of three independent biological replicates), while auxin amino acid conjugates were again below the detection limit. This S. reilianum-induced increase in the total auxin concentration in ears with elongating spikelets could indicate a potential role of auxin in the floral reversion process.
S. reilianum-Colonized Inflorescences Show an Elevated Level of Reactive Oxygen Species
Most of the differentially regulated maize transcripts detected by microarray analysis had a predicted function in the response to biotic stress. Notable was the presence of 15 genes involved in the detoxification of oxidative stress. Of these, nine reproductive organ- or shoot-expressed genes (five glutathione S-transferase [GST], two cytochrome P450, and two peroxidase genes) were up-regulated, whereas five root- or endosperm-specific peroxidase genes were down-regulated (Table I). This suggests a need for detoxification of reactive oxygen species (ROS) in S. reilianum-colonized inflorescence tissue and implies that the level of ROS is higher in S. reilianum-colonized than in healthy inflorescences.
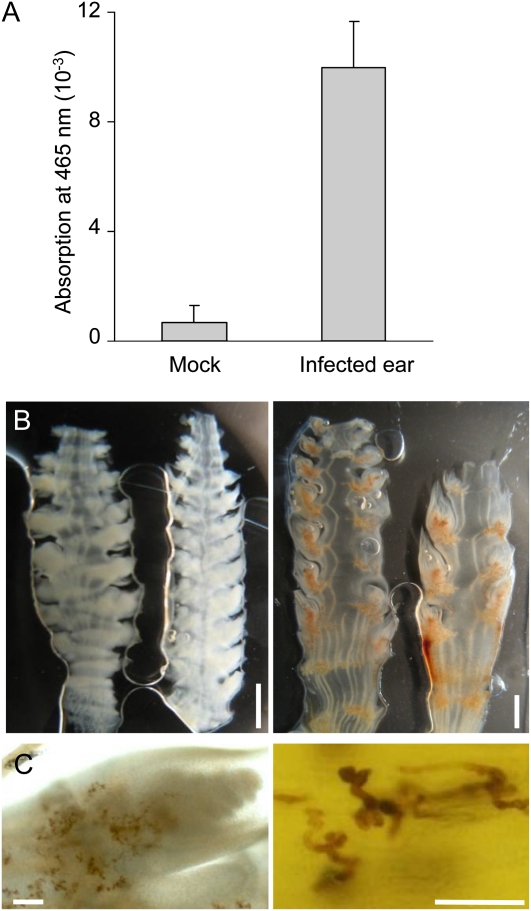
To verify whether S. reilianum-colonized inflorescences contained a higher level of ROS, we prepared manual sections of young ears of infected or healthy plants that were immersed for 2 h in a solution containing 3,3′-diaminobenzidine (DAB) that forms a brown oxidation product when exposed to ROS. When supernatants were collected and absorbance of oxidized DAB at 465 nm was measured, a 15-fold increase in absorbance was detected in DAB solutions that had contained sections of infected inflorescences compared with supernatants of the healthy inflorescence sections (Fig. 7A). When the DAB-treated sections were microscopically analyzed, brown oxidized DAB precipitates could readily be detected on inflorescence sections of S. reilianum-infected plants but not on those of mock-infected plants (Fig. 7B). DAB oxidation seemed to be specifically associated with fungal colonization and surrounded fungal hyphae that colonized leafy or eary spikelets (Fig. 7C). This indicates that the presence of S. reilianum causes the observed increase in the ROS levels in inflorescences of infected plants, which could explain the up-regulation of reproductive organ-specific ROS detoxification genes (Table I).
Figure 7.
Detection of ROS in healthy and S. reilianum-infected female inflorescences of maize cv Gaspe Flint. ROS were stained and visualized by exposure to DAB. A, DAB oxidization as a measure of ROS production in the supernatant of DAB-stained sections of female inflorescences. Error bars correspond to the sd of six independent measurements. B, Sections of healthy ears (left) and S. reilianum-colonized ears (right) after staining with DAB. Bar = 2 mm. C, ROS accumulation (brown area) in sections of an eary spikelet (left) and around fungal hyphae (right). Bars = 100 μm.
DISCUSSION
S. reilianum caused a number of developmental changes in the inflorescences of infected maize plants. In addition to loss of apical dominance, infected ears showed loss of meristem identity and determinacy as well as loss of organ identity. Loss of organ identity occurred at all floral organs (glume, lemma, lodicule, carpel, palea, and stamen) in leafy ears (Fig. 4). In eary ears and phyllodied tassels, floral organs showed reversion into vegetative structures, with the exception of the most inner whorl that developed an inflorescence (Fig. 5).
Different Morphologies of S. reilianum-Colonized Inflorescences
The developmental decision of leafy or eary ear formation might be made by the crucial timing of inflorescence invasion by S. reilianum. If the fungus invades the female inflorescence at an early time point of inflorescence development before the appearance of silk tissue, it can redirect the developmental program of the floral meristem to that of an inflorescence meristem and thus cause the development of ears instead of flowers. If the fungus reaches the female inflorescence after silk development and initiation of floral organs, it redirects the floral developmental program to a vegetative program giving rise to leafy ears.
In healthy plants, each of the spikelet meristems differentiates into two florets that both initiate all floral organs, including carpel and stamen. Then, floret development continues differently in male and female inflorescences: whereas in male florets the carpel primordia are aborted, in female inflorescences the complete lower floret and the stamen primordia of the upper floret are aborted (Veit et al., 1991). In the female inflorescence, the inner whorls develop three carpels, two that elongate and fuse to form the tubular silk and one that remains rudimentary (Nickerson, 1954; Dellaporta et al., 1991). Inside the carpels, the ovule develops (Dellaporta et al., 1991).
In eary ears, a husk-like leaf forms an open structure developed at the position of carpel primordia (Fig. 5B), suggesting that loss of organ identity occurred before the fusion of carpel primordia was complete. In contrast, in leafy ears, the impact of S. reilianum on carpel development must have occurred after carpel fusion was completed, since a closed tubular onion leaf-like structure developed. In some cases, two more vegetative structures within the tubular structure could be observed that might either be the result of indeterminate growth of carpel primordia preceding loss of organ identity or correspond to the rudimentary carpel and the ovule (Fig. 4). In such cases, the primordia of the rudimentary carpel and the ovule could have been initiated, but organ development was not yet terminated before S. reilianum exerted its effect. If in eary ears, S. reilianum colonized the inflorescence at an early stage of inflorescence development, all spikelets would be equally affected. Accordingly, we observed that most of the spikelets on eary ears were completely converted to eary spikelets (Fig. 3C). Thus, the different morphologies of the S. reilianum-colonized inflorescences are most likely a result of differences in the developmental states of the florets at the time of fungal colonization.
The spatial and temporal colonization pattern of S. reilianum might also be responsible for the observed loss of apical dominance at ear branches appearing at basal nodes (Fig. 2D). Ears develop first at the more apical nodes, and ears that appear at lower nodes are formed later. Since young seedling plants were used for infection, S. reilianum enters the plant via leaves developing from basal nodes. This enables the fungus to reach lower nodes first. Thus, it has time to colonize the submeristematic tissue before development of the inflorescence meristem. Any S. reilianum-induced hormonal or metabolic changes would therefore immediately affect inflorescence development. In contrast, S. reilianum might reach the primordia at higher nodes only after inflorescence development has been completed. In this case, any S. reilianum-induced hormonal or metabolic changes would have little impact on inflorescence morphology. Such a scenario would also explain why female inflorescences on infected plants show phyllody much more frequently (91%; see above) than male inflorescences (5%). Since the male inflorescence emerges from the plant apex, the fungus might only rarely be able to reach it before the completion of tassel development.
The conversion to vegetative growth could have the potential advantage for the fungus to be provided with nutrients generated by photosynthetic activity of the green leaf-like tissue. If eary ears develop, the fungus can profit in two ways: first, the husk leaves of the newly developing inflorescence will provide nutrients by photosynthesis, while the additional inflorescences will enlarge the tissue suitable for fungal proliferation and spore formation.
Loss of Floral Organ Identity
A reversion of floral organs into vegetative structures implies a change in the identity of affected organs. Floral organ identity is regulated by a number of homeotic transcription factors, including those of the MADS and the AP2 families (Coen and Meyerowitz, 1991). According to the ABCDE model that was developed for dicots, transcription factors belong to five functional classes that determine floral organ identity in different combinations (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Ng and Yanofsky, 2001; Ditta et al., 2004). While genes that carry out B, C, D, and E functions were identified in rice (Oryza sativa) and maize, information about an A function outside the higher eudicots is ambiguous (Yanofsky et al., 1990; Ambrose et al., 2000; Dreni et al., 2007; Thompson et al., 2009). While AP2 carries out A function in Arabidopsis, diverse functions have been described for AP2-like genes in monocots and dicots (Tang et al., 2007; Chuck et al., 2008; Maeo et al., 2009). This suggests that monocots might follow different regulatory networks to govern floral development and that AP2-like genes regulate additional developmental processes in monocots.
It has been proposed that lemma/palea and lodicules of monocots correspond to sepals and petals of dicots, respectively (Ambrose et al., 2000). In dicots, the organ identity of sepals is governed by A class genes, while that of petals is defined by A and B class genes (Weigel and Meyerowitz, 1994). Active B and C class genes define organ identity of stamens, C class genes that of carpels, and D class genes that of ovules (Weigel and Meyerowitz, 1994; Colombo et al., 1995). E class genes work as scaffolds and are required for organ identity of all floral whorls (Ditta et al., 2004). In S. reilianum-infected leafy ears, floral organs corresponding to lemma, lodicule, stamen, and carpel have lost their organ identity and have reverted to vegetative leaf-like organs. This could indicate that S. reilianum affects the function of A, B, C, and D genes or affects E genes to cause the loss of floral organ identity.
Transcriptional analysis of young S. reilianum-infected ears showed that the E class-resembling gene ZAG3 (BDE; Thompson et al., 2009) was down-regulated (Table I). In addition, the C class gene ZAG1 (Mena et al., 1996) was down-regulated. Two AP2 family genes were up-regulated (Table I), indicating an A class-typical antagonistic regulation to the C class genes (Chuck et al., 2008), but whether the two AP2 family genes fulfill other A class functions is unknown. One gene likely belonging to the B class of floral transcription factors, ZMM29 (Münster et al., 2001), was down-regulated (Table I). ZAG2, a gene that is homologous to the rice D class gene OsMADS13 (Dreni et al., 2007) and whose expression is constrained to the ovule and extensions that form the silk (Schmidt et al., 1993; Colombo et al., 1998), was also down-regulated, as was its homolog ZMM1 (Theissen et al., 1995). According to the UniGene database, the B, C, D, and E class transcription factors, as well as one AP2-family protein, were found to be specifically expressed in floral organs (Table I). S. reilianum infection apparently greatly impacts the transcriptional profile of A, B, C, D, and E class genes, supporting the observation that in infected inflorescences, vegetative organs develop in place of floral organs. While S. reilianum infection might affect the regulation of floral organ identity genes directly, it is more likely that the infection has an impact on regulatory mechanisms acting upstream of flower development, such as meristem identity, local hormone concentration, or general metabolite levels, that all may affect gene expression.
In this respect, it is remarkable that ROS accumulation was observed around fungal hyphae in colonized inflorescences (Fig. 7). While it is unclear whether ROS generation stems from the fungus or the plant, it is obvious that ROS are detected by the plant, since we observed a dramatic increase in the expression level of plant genes involved in the detoxification of oxidative stress (Table I). In plants, ROS are known to have a role in signaling and development (Gapper and Dolan, 2006; Møller and Sweetlove, 2010). Therefore, it is possible that the morphological changes induced by S. reilianum colonization of the inflorescence are an indirect result of the increased ROS levels in the plant cells. Possibly, this indirect effect is exerted via redox regulation of glutaredoxins, which have recently been shown to play a crucial role in floral organ development (Xing and Zachgo, 2008; Li et al., 2009a, 2009b). On the other hand, the increased ROS levels in infected floral tissues might be a result of the down-regulation of a specific MADS box transcription factor. It has recently been shown in rice that a mutation in the C class gene MADS3 leads to ROS accumulation, which may be responsible for the observed decreased pollen viability (Hu et al., 2011).
Inflorescence Development in Eary Ears
In eary ears, the floral whorls corresponding to lemma/palea, lodicule, stamen, and carpel showed loss of organ identity leading to the formation of vegetative organs, similar to what happens in leafy ears. However, in eary ears, the whorl that corresponded to the ovule developed an inflorescence (Fig. 5). This indicates that the remnants of stem cells in the floral meristems did not terminate in forming ovules. Instead, the floral meristems changed identity to inflorescence meristems. These new inflorescence meristems showed a loss of meristem determinacy: transition of spikelet to floral meristems did not occur, but spikelets developed inflorescence meristems (Fig. 6). In addition, these newly formed inflorescences were heavily colonized by fungal hyphae, and fungal growth was prominent in the core of the inflorescence and did not extend to the surrounding vegetative organs (Fig. 7). Apparently, inflorescences are the preferred substrates for fungal growth. Thus, abolishing meristem determinacy may be advantageous for proliferation of the fungus, because more inflorescence tissue for colonization would be available.
How could fungal presence induce these described changes? Spikelet meristem determinacy was shown to be regulated by three AP2 family genes, INDETERMINATE SPIKELET1 (IDS1), SISTER OF INDETERMINATE SPIKELET1, and BRANCHED SILKLESS1 (Chuck et al., 2002, 2008). Of these, only IDS1 was represented on the arrays and did not show changes in transcript level. However, two uncharacterized AP2 family genes were up-regulated in S. reilianum-infected ears, and one of them was found to have a flower-specific expression pattern (Table I). This could hint at a role of these AP2 family members in regulating meristem determinacy.
Floral meristem identity has been shown to be additionally regulated by two redundantly acting genes, ZAG1 and ZAG3 (Thompson et al., 2009). The zag1 zag3 double mutants show a severe ear phenotype, in which the floral meristems fail to develop floral organs but instead produce branch-like structures that initiate ectopic meristems (Thompson et al., 2009). The indeterminate meristem phenotype in the zag1 zag3 double mutants is very similar to the occurrence of inflorescence meristems that continuously differentiate into additional meristems observed in S. reilianum-infected eary ears (Fig. 6). Since both ZAG1 and ZAG3 were down-regulated in young S. reilianum-infected ears (Table I), their down-regulation could at least partially explain the changes in meristem fate caused by S. reilianum, even though it is unknown how the infection influences the transcription levels of ZAG1 and ZAG3.
S. reilianum-Triggered Loss of Apical Dominance
S. reilianum infected plants show an increase in the number of ears per branch (Fig. 2). This observation can be explained by a loss of apical dominance leading to activation and outgrowth of the axillary meristems on the ear shank. Shanks of ears lacking silks show outgrowth of the subapical ear meristems in many maize inbred lines (V.A. Walbot, personal communication). Because loss of carpel identity precludes silk formation, the observed change of carpel identity induced by S. reilianum could explain the increased ear number per branch. How the outgrowth of axillary meristems in maize is regulated has not been deeply studied. However, two C2H2 zinc finger transcription factors with a role in axillary meristem fate, RAMOSA1 (RA1) and INDETERMINATE1 (ID1), have been identified. Whereas RA1 determines the fate of second-order meristems (Vollbrecht et al., 2005), ID1 controls the transition from vegetative to reproductive growth in maize, and id1 mutants fail to form ears (Colasanti et al., 1998). Neither RA1 nor ID1 was found to be regulated in the microarray experiments. While one C2H2 zinc finger family member was up-regulated in young S. reilianum-infected ears (Table I), it is unknown whether the gene has a function in the regulation of meristem outgrowth.
In addition to an increased number of ears per branch, S. reilianum-infected plants exhibited an increased ROS level in colonized ears (Fig. 7). A similar finding has been described for the Epichloe festucae-perennial ryegrass (Lolium perenne) interaction. Perennial ryegrass infected with E. festucae strains lacking the stress-activated mitogen-activated protein kinase gene sakA showed an elevated ROS level and increased tillering (Eaton et al., 2010), thereby linking ROS accumulation to loss of apical dominance.
The initiation of vegetative and reproductive development in maize has recently been shown to be mediated by the Trp aminotransferase VANISHING TASSEL2, an enzyme involved in auxin biosynthesis (Phillips et al., 2011). A gene encoding Trp aminotransferase was down-regulated upon maize infection with S. reilianum (Table I), possibly linking lack of auxin to the observed developmental defects in infected inflorescences. Although the Trp aminotransferase gene was clearly down-regulated, we did not observe an overall decrease in auxin concentration in the emerging ear. Instead, the auxin content of young S. reilianum-infected ears showed an increase of 30% relative to control ears. This increase was detectable even though complete inflorescences were collected at a stage where the inflorescences only partially showed symptoms. In addition, two auxin biosynthesis genes encoding Trp synthase α-subunits and one encoding an indole-3-acetic acid (IAA)-amino acid conjugate hydrolase for auxin mobilization were up-regulated, pointing to an overall increase in auxin production in the infected ears. How could an increase in auxin levels lead to a loss of apical dominance? The generation of local auxin maxima is known to be necessary for axillary meristem formation (Gallavotti et al., 2008). Local auxin maxima at axillary meristems could be responsible for the observed increase in the number of ears per branch in S. reilianum-infected plants. In addition, polar auxin transport is necessary to activate axillary meristems. In maize, the Ser/Thr protein kinase BARREN INFLORESCENCE2 (BIF2) and the bHLH protein BARREN STALK1 (BA1) have been implicated in polar auxin transport and the initiation of axillary meristems (McSteen and Hake, 2001; Gallavotti et al., 2004, 2008; McSteen et al., 2007; Wu and McSteen, 2007). BIF2 has been shown to interact with BA1 (Skirpan et al., 2008). It was suggested that BIF2 functions upstream of polar auxin transport and that polar auxin transport is required for BA1 expression (Bennetzen and Hake, 2009). The auxin canalization model predicts that, irrespective of absolute auxin concentration, polar auxin transport is necessary to activate axillary meristems (Domagalska and Leyser, 2011). The axillary bud is activated if it generates auxin and if this auxin is exported from the bud into the main polar auxin transport canal. This would suggest that there is auxin export from the axillary buds that developed on shanks of infected ears, leading to bud activation and the development of additional ears.
CONCLUSION
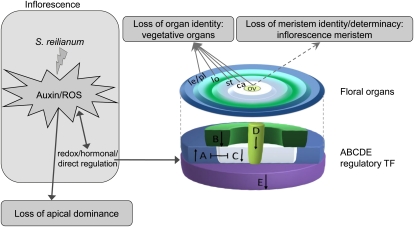
S. reilianum infection of maize cv Gaspe Flint induced a loss of apical dominance and led to two major modifications of the inflorescences. Loss of organ identity was evident in leafy ears, eary ears, and phyllodied tassels. Loss of meristem identity and determinacy was exclusive to eary ears and phyllodied tassels. The timing of S. reilianum colonization relative to the developmental state of the colonized inflorescence could be decisive for the outcome of the floral modification resulting in leafy or eary ears. S. reilianum-colonized inflorescences showed a higher level of ROS, a higher level of auxin, and misregulation of floral regulatory transcription factors. In accordance with the expectations of the ABCDE model, we observed up-regulation of A class and down-regulation of B, C, D, and E class genes. Together, these changes might explain how S. reilianum modulates the floral architecture of maize inflorescences (Fig. 8). While floral gene regulation might be a secondary consequence of increased ROS or auxin levels, it could also be that S. reilianum directly regulates key players controlling floral gene expression (e.g. via the secretion of small effector proteins that are translocated into the plant cells). Future experiments will clarify whether and how ROS, auxin, or fungus-secreted effectors contribute to symptom development of S. reilianum.
Figure 8.
Hypothetical model of changes in inflorescence and branching architectures of maize induced by S. reilianum. Fungal colonization leads to ROS and auxin accumulation in the inflorescence. Possibly, elevated ROS or auxin levels in the S. reilianum-infected ears promote outgrowth of the subapical inflorescence meristems, leading to a loss of apical dominance and consequently a change in branching architecture. Additionally, loss of organ identity triggered by S. reilianum could be the result of an altered expression of the A, B, C, D, and E class genes. Altering the expression of the ABCDE regulators could be the result of a direct regulation by fungal secreted proteins or of an indirect regulation by ROS and/or auxin. Alternatively, increased accumulation of auxin and ROS could be the result of a modulation of the ABCDE regulators. S. reilianum could trigger a loss of meristem identity and determinacy (dashed arrow) if it reached the floral meristem before stem cells terminate into an ovule. Black arrows indicate up- or down-regulation of ABCDE transcription factors (TF). ca, Carpel; le, lemma; lo, lodicules; ov, ovule; pl, palea; st, stamen.
MATERIALS AND METHODS
Maize Growth Conditions
The maize (Zea mays ‘Gaspe Flint’) plants were obtained from Regine Kahmann. Plants were grown in T-type soil (Frühstorfer Pikiererde) under 15-h-day conditions at 28°C, 50% relative humidity, and illumination at a minimum of 28,000 lux, with additional occasional sun radiation up to 90,000 lux, and 9-h-night conditions at 20°C and 60% relative humidity. Between day and night shifts, 2.5 and 3.5 h of light ramping was included to simulate sunset and sunrise, respectively. For transcriptome analysis, plants were transferred after 6 d to a phytochamber (Vötsch) with the same growth conditions as the greenhouse, with the exception of 1-h ramping between day and night shifts and 50% relative humidity during the day and not sun radiation.
Plant Inoculation with Sporisorium reilianum
The S. reilianum-compatible mating-type strains SRZ1 and SRZ2 (Schirawski et al., 2005) were inoculated in 2 mL of YEPSL medium (1% tryptone, 1% yeast extract, and 1% Suc) modified from Tsukuda et al. (1988) and incubated at 28°C with 200 rpm shaking for 8 to 12 h. Each culture was used to inoculate 50 mL of potato dextrose (2.4%) broth (Difco) overnight until an optical density at 600 nm of 0.5 to 1.0 was reached. The cultures were centrifuged at 3,500 rpm for 5 min. Cell pellets were resuspended in water to an optical density at 600 nm of 2.0. Suspension cultures of SRZ1 and SRZ2 were mixed and used to inoculate 7-d-old maize seedlings as described (Gillissen et al., 1992).
RNA Extraction
Ears (1-2 cm in length) from 20 plants at 4 weeks post inoculation were collected from healthy or S. reilianum-infected plants. Of infected ears, those displaying clear vegetative structures were excluded, and only ears showing elongation of spikelets were collected. RNA from three independent experiments was extracted with Trizol (Invitrogen). DNase treatment was performed using the RNase-Free DNase Set (Qiagen), and RNA was purified with the RNeasy Plant Mini Kit (Qiagen). The RNA quality was verified using a Bioanalyzer 2100 (Agilent), and the RNA purity and concentration were assessed with NanoDrop ND-1000 (Thermo Fisher Scientific).
Gene Expression Analyses
The Affymetrix GeneChip Maize Genome arrays were performed for three independent experiments. RNA amplification was performed according to the manual of the One-Cycle cDNA Synthesis Kit (Affymetrix). Copy RNA labeling and copy RNA fragmentation and labeling were performed according to the manual of the GeneChip IVT Labeling Kit (Affymetrix). Array hybridization, washing, and staining were performed according to the Midi_Euk2V3 protocols using GeneChip Fluidics Station 400, and arrays were scanned with the Affymetrix GSC3000 scanner (Affymetrix).
The microarray fluorescence signals were processed using the GCOS version 1.4 software package (Affymetrix). The software was used to produce presence-absence calls and to normalize the signal intensities from each array with scaling the target signal to 300 and using default parameters. The files produced were used to analyze statistically significant differences using the Bioconductor version 2.3 package. Genes with adjusted values of P < 0.05 were considered to be significantly misregulated. Microarray data were verified for five randomly selected genes by qRT-PCR using actin as an internal control (Ghareeb et al., 2011) with the primers listed in Supplemental Table S2.
Staining and Microscopy
To visualize the modifications in inflorescence structures, ears and tassels showing different developmental stages were fixed overnight at 4°C in 2.5% formaldehyde, 5% glacial acetic acid, and 60% ethanol and dehydrated in a graded ethanol series. Following substitution with Histo-clear (National Diagnostics), we embedded the samples in Paraplast Plus (McCormick Scientific) and sectioned them at 15-μm thickness using a rotary microtome. Sections were stained with O-Safranin-Fast Green staining according to Orashakova et al. (2009).
To stain fungal appressoria or fungal structures in planta, infected leaves at 1 d post inoculation were dipped in Calcofluor White (100 ng mL−1; Fluorescent Brightener 28) for 30 s and then washed once with water. The samples were immediately scanned by a Zeiss Axioplan II microscope using the 4′,6-diamino-phenylindole filter set (EX BP 365, BS FT 395, EM BP 445/50).
For visualization of fungal structures in planta, leaves, nodes, and ear sections were soaked in ethanol overnight. The samples were rinsed once with water and incubated in 10% KOH overnight. After washing with water, the leaves and nodes were incubated in WGA-Alexa Fluor 488 and propidium iodide staining solution (10 μg mL−1 WGA-Alexa Fluor 488, 20 μg mL−1 propidium iodide, and 0.02% Tween 20 in phosphate-buffered saline, pH 7.4) for 30 min with three 2-min intervals of vacuum infiltration. The samples were scanned with a TCS-SP5 confocal microscope (Leica). WGA-Alexa Fluor 488 was detected with excitation at 488 nm and emission at 500 to 540 nm, and propidium iodide was detected with excitation at 561 nm and emission at 580 to 660 nm. For staining the fungal hyphae in leaves or in inflorescences, leaves or ear sections were incubated in Chlorazole Black E staining solution (0.03% [w/v] Chlorazole Black E in a 1:1:1 mixture of water, lactic acid [80%], and glycerol [97%]) at 60°C overnight. The sections were destained in 50% glycerol overnight and then scanned using light microscopy.
ROS Quantification and Localization
To test ROS production in ears in response to S. reilianum proliferation, ROS were quantified using DAB obtained from Sigma (Herzog and Fahimi, 1973). Manual sections of 4-week-old female inflorescences were prepared and weighed (50–80 mg) prior to adding a 10-fold volume excess of freshly prepared DAB staining solution (1 mg of DAB per 1 mL of water). The samples were incubated in the dark for 5 h at room temperature before removing the sections and photometrically determining the absorption of the DAB solution at 465 nm.
To localize ROS production in infected ears, ears sections were stained with DAB-HCl solution for 3 h at room temperature. The samples were destained in ethanol:chloroform (4:1) and then kept in the dark in 60% glycerol until analysis by light microscopy.
Hormone Analysis
In order to measure auxin and GA concentrations, plant material was extracted as described previously for lipids, with some modifications (Matyash et al., 2008). Plant material (100 mg) was extracted with 0.75 mL of methanol containing 20 ng of D5-IAA (Eurisotop) and 10 ng of D3-GA3 (OlChemIm) each as an internal standard. After vortexing, 2.5 mL of methyl-tert-butyl ether was added, and the extract was shaken for 1 h at 4°C. For phase separation, 0.6 mL of water was added. The mixture was incubated for 10 min at room temperature and centrifuged at 450g for 15 min. The upper phase was collected, and the lower phase was reextracted with 0.7 mL of methanol:water (3:2.5, v/v) and 1.3 mL of methyl-tert-butyl ether as described above. The combined upper phases were dried under streaming nitrogen and resuspended in 100 μL of acetonitrile:water:acetic acid (20:80:0.1, v/v/v).
The analysis of constituents was performed using an Agilent 1100 HPLC system coupled to an Applied Biosystems 3200 hybrid triple quadrupole/linear ion trap mass spectrometer (MDS Sciex). Nanoelectrospray (nanoESI) analysis was achieved using a chip ion source (TriVersa NanoMate; Advion BioSciences). Reverse-phase HPLC separation was performed on an EC 50/2 Nucleodure C18 gravity 1.8-μm column (50 × 2.1 mm, 1.8-μm particle size; Macherey and Nagel) applying a column temperature of 30°C. For analysis, 10 μL of extract was injected. The binary gradient system consisted of solvent A (water:acetic acid, 100:0.1, v/v) and solvent B (acetonitrile:acetic acid, 100:0.1, v/v) with the following gradient program: 5% solvent B for 1 min, followed by a linear increase of solvent B up to 95% within 10 min, and an isocratic run at 95% solvent B for 4 min. To reestablish starting conditions, a linear decrease to 5% B within 2 min was performed, followed by 10 min of isocratic equilibration at 5% B. The flow rate was 0.3 mL min−1. For stable nanoESI, 130 μL min−1 2-propanol:acetonitrile:water:acetic acid (70:20:10:0.1, v/v/v/v) delivered by a 2150 HPLC pump (LKB) was added just after the column via a mixing tee valve. By using another post column splitter, 790 nL min−1 eluent was directed to the nanoESI chip. Ionization voltage was set to −1.7 kV. Phytohormones were negatively ionized and detected in a scheduled multiple reaction monitoring mode. For the scheduled mode, the multiple reaction monitoring detection window was 72 s and a target scan time of 1.2 s was applied. Mass transitions were as follows: 179/135 (declustering potential [DP] −40 V, entrance potential [EP] −6.5 V, collision energy [CE] −22 V) for D5-IAA, 174/130 (DP −40 V, EP −6.5 V, CE −22 V) for IAA, 160/116 (DP −55 V, EP −10 V, CE −40 V) for D2-GA3, 345/143 (DP −85 V, EP −10 V, CE −38 V) for GA3, 347/273 (DP −115 V, EP −10 V, CE −30 V) for GA1, 331/213 (DP −105 V, EP −10 V, CE −40 V) for GA4, 329/145 (DP −120 V, EP −10 V, CE −34 V) for GA5, 363/275 (DP −160 V, EP −10 V, CE −24 V) for GA8, 315/271 (DP −95 V, EP −10 V, CE −28 V) for GA9, 331/287 (DP −95 V, EP −10 V, CE −30 V) for GA20, and 347/259 (DP −240 V, EP −10 V, CE −24 V) for GA34. The mass analyzers were adjusted to a resolution of 0.7 atomic mass units full width at half height. The ion source temperature was 40°C, and the curtain gas was set at 10 (given in arbitrary units). Quantification was carried out using a calibration curve of intensity (mass-to-charge) ratios of unlabeled/deuterium labeled versus molar amounts of unlabeled (0.3–1,000 pmol).
Microarray data included in this article were submitted to the Gene Expression Omnibus database under accession number GSE29747.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Representative developmental morphologies of mock-infected and S. reilianum-infected ears used for microarray experiments.
Supplemental Figure S2. Comparison of gene expression profiles determined by microarray or qRT-PCR.
Supplemental Table S1. Complete list of S. reilianum-regulated genes in maize inflorescences, and their correlated approximate gene expression patterns.
Supplemental Table S2. List of primers used for qRT-PCR analysis.
Acknowledgments
We thank Regine Kahmann, Alfred Batschauer, and Virginia Walbot for fruitful discussions and Cornelia Herrfurth for support with the establishment of the hormone measurements.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5: 569–579 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Hake SC. (2009) Handbook of Maize: Its Biology. Springer, New York [Google Scholar]
- Bhaskaran S, Smith RH, Frederiksen RA. (1990) Gibberellin A3 reverts floral primordia to vegetative growth in sorghum. Plant Sci 71: 113–118 [Google Scholar]
- Brefort T, Doehlemann G, Mendoza-Mendoza A, Reissmann S, Djamei A, Kahmann R. (2009) Ustilago maydis as a pathogen. Annu Rev Phytopathol 47: 423–445 [DOI] [PubMed] [Google Scholar]
- Chandra A, Huff DR. (2010) A fungal parasite regulates a putative female-suppressor gene homologous to maize tasselseed2 and causes induced hermaphroditism in male buffalograss. Mol Plant Microbe Interact 23: 239–250 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S. (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019 [DOI] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V. (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Colombo L, Franken J, Koetje E, van Went J, Dons HJM, Angenent GC, van Tunen AJ. (1995) The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo L, Marziani G, Masiero S, Wittich PE, Schmidt RJ, Gorla MS, Pè ME. (1998) BRANCHED SILKLESS mediates the transition from spikelet to floral meristem during Zea mays ear development. Plant J 16: 355–363 [Google Scholar]
- Dellaporta SL, Moreno MA, Delong A. (1991) Cell lineage analysis of the gynoecium of maize using the transposable element Ac. Dev Suppl 1: 141–147 [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PBF, An G, Colombo L, Kater MM. (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Eaton CJ, Cox MP, Ambrose B, Becker M, Hesse U, Schardl CL, Scott B. (2010) Disruption of signaling in a fungal-grass symbiosis leads to pathogenesis. Plant Physiol 153: 1780–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D. (2008) The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pè ME, Schmidt RJ. (2004) The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Gapper C, Dolan L. (2006) Control of plant development by reactive oxygen species. Plant Physiol 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghareeb H, Bozsó Z, Ott PG, Repenning C, Stahl F, Wydra K. (2011) Transcriptome of silicon-induced resistance against Ralstonia solanacearum in the silicon non-accumulator tomato implicates priming effect. Physiol Mol Plant Pathol 75: 83–89 [Google Scholar]
- Gillissen B, Bergemann J, Sandmann C, Schroeer B, Bölker M, Kahmann R. (1992) A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68: 647–657 [DOI] [PubMed] [Google Scholar]
- Herzog V, Fahimi HD. (1973) A new sensitive colorimetric assay for peroxidase using 3,3′-diaminobenzidine as hydrogen donor. Anal Biochem 55: 554–562 [DOI] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D. (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. (2009a) Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zachgo S, Jean-Pierre J. (2009b) Glutaredoxins in development and stress responses of plants. Adv Bot Res 52: 333–361 [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K. (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Matheussen AM, Morgan PW, Frederiksen RA. (1991) Implication of gibberellins in head smut (Sporisorium reilianum) of Sorghum bicolor. Plant Physiol 96: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. (2008) Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Hake S. (2001) barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S. (2007) barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt LW, Rincones J, Bailey BA, Aime MC, Griffith GW, Zhang D, Pereira GAG. (2008) Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao: what’s new from this old foe? Mol Plant Pathol 9: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. (1996) Diversification of C-function activity in maize flower development. Science 274: 1537–1540 [DOI] [PubMed] [Google Scholar]
- Meyer VG. (1966) Flower abnormalities. Bot Rev 32: 165–218 [Google Scholar]
- Møller IM, Sweetlove LJ. (2010) ROS signalling: specificity is required. Trends Plant Sci 15: 370–374 [DOI] [PubMed] [Google Scholar]
- Münster T, Wingen LU, Faigl W, Werth S, Saedler H, Theissen G. (2001) Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene 262: 1–13 [DOI] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Nickerson NH. (1954) Morphological analysis of the maize ear. Am J Bot 41: 87–92 [Google Scholar]
- Orashakova S, Lange M, Lange S, Wege S, Becker A. (2009) The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J 58: 682–693 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra S, Safeeulla KM. (1979) Histopathological studies on ragi (Eleusine coracana (L.) Gaertn.) infected by Sclerophthora macrospora (Sacc.) Thirum. Shaw and Naras. Proc Indian Natl Sci Acad (B Biol Sci) 88: 19–24 [Google Scholar]
- Reed GM, Swabey M, Kolk LA. (1927) Experimental studies on head smut of corn and sorghum. Bull Torrey Bot Club 54: 295–310 [Google Scholar]
- Roy BA. (1993) Floral mimicry by a plant pathogen. Nature 362: 56–58 [Google Scholar]
- Schirawski J, Brefort T, Molina L, Mendoza-Mendoza A, Müller O, Kahmann R. (2006) Ustilago maydis: new insights into the early infection phase. Sánchez F, Quinto C, López-Lara IM, Geiger O, , Biology of Plant-Microbe Interactions, Vol 5. International Society for Molecular Plant-Microbe Interactions, St. Paul, pp 559–564 [Google Scholar]
- Schirawski J, Heinze B, Wagenknecht M, Kahmann R. (2005) Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryot Cell 4: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J, Mannhaupt G, Münch K, Brefort T, Schipper K, Doehlemann G, Di Stasio M, Rössel N, Mendoza-Mendoza A, Pester D, et al. (2010) Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330: 1546–1548 [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Veit B, Mandel MA, Mena M, Hake S, Yanofsky MF. (1993) Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5: 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Semisi ST, Ball SFL. (1989) Infection of the pearl millet (Pennisetum americanum) inflorescence by the downy mildew fungus (Sclerospora graminicola). Plant Pathol 38: 571–576 [Google Scholar]
- Skibbe DS, Doehlemann G, Fernandes J, Walbot V. (2010) Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 328: 89–92 [DOI] [PubMed] [Google Scholar]
- Skirpan A, Wu X, McSteen P. (2008) Genetic and physical interaction suggest that BARREN STALK 1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J 55: 787–797 [DOI] [PubMed] [Google Scholar]
- Tang M, Li G, Chen M. (2007) The phylogeny and expression pattern of APETALA2-like genes in rice. J Genet Genomics 34: 930–938 [DOI] [PubMed] [Google Scholar]
- Theissen G, Strater T, Fischer A, Saedler H. (1995) Structural characterization, chromosomal localization and phylogenetic evaluation of two pairs of AGAMOUS-like MADS-box genes from maize. Gene 156: 155–166 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, Hake S. (2009) bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraud G, Bill L, Piollat MT. (1997) Phyllodied panicles caused by head smut of maize and vegetative multiplication of maize. Plant Cell Tissue Org Cult 50: 19–26 [Google Scholar]
- Tsukuda T, Carleton S, Fotheringham S, Holloman WK. (1988) Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol Cell Biol 8: 3703–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit B, Greene B, Lowe B, Mathern J, Sinha N, Vollbrecht E, Walko R, Hake S. (1991) Genetic approaches to inflorescence and leaf development in maize. Development 113: 105–111 [Google Scholar]
- Veit B, Schmidt RJ, Hake S, Yanofsky MF. (1993) Maize floral development: new genes and old mutants. Plant Cell 5: 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES, IV, Martienssen R. (2005) Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126 [DOI] [PubMed] [Google Scholar]
- Walbot V, Skibbe DS. (2010) Maize host requirements for Ustilago maydis tumor induction. Sex Plant Reprod 23: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. (1994) The ABCs of floral homeotic genes. Cell 78: 203–209 [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. (2007) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 35: D5–D12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, McSteen P. (2007) The role of auxin transport during inflorescence development in maize (Zea mays, Poaceae). Am J Bot 94: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S. (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]