Abstract
The assembly, sorting signals, and turnover of the tonoplast potassium channel AtTPK1 of Arabidopsis (Arabidopsis thaliana) were studied. We used transgenic Arabidopsis expressing a TPK1-green fluorescent protein (GFP) fusion or protoplasts transiently transformed with chimeric constructs based on domain exchange between TPK1 and TPK4, the only TPK family member not located at the tonoplast. The results show that TPK1-GFP is a dimer and that the newly synthesized polypeptides transiently interact with a thus-far unidentified 20-kD polypeptide. A subset of the TPK1-TPK4 chimeras were unable to assemble correctly and these remained located in the endoplasmic reticulum where they interacted with the binding protein chaperone. Therefore, TPK1 must assemble correctly to pass endoplasmic reticulum quality control. Substitution of the cytosolic C terminus of TPK4 with the corresponding domain of TPK1 was sufficient to allow tonoplast delivery, indicating that this domain contains tonoplast sorting information. Pulse-chase labeling indicated that TPK1-GFP has a half-life of at least 24 h. Turnover of the fusion protein involves internalization into the vacuole where the GFP domain is released. This indicates a possible mechanism for the turnover of tonoplast proteins.
Proteins located at the tonoplast are major regulators of the content, volume, and functions of plant vacuoles. To maintain optimal tonoplast functions and provide adapted response to environmental stimuli, these proteins must be correctly managed by the secretory pathway, sorted to their destination, and eventually turned over. Here, we have investigated the mechanisms underlying these controls using the Arabidopsis (Arabidopsis thaliana) tonoplast potassium channel AtTPK1 as a model protein.
AtTPK1 (formerly termed AtKCO1) belongs to the tandem-pore potassium channel (TPK) family, composed of five members in Arabidopsis (Czempinski et al., 1997). Like many mammalian TPK channels (Lotshaw, 2007), the Arabidopsis polypeptides have four putative transmembrane domains (TMDs) and two pore domains (Ps). The N-terminal portion of AtTPK1 interacts with the cytosolic 14-3-3 protein GRF6 (Latz et al., 2007a), indicating a topology similar to that of the animal counterparts, in which both the N and C termini are located in the cytosol (Fig. 1A). The observations that AtTPK1 undergoes homotypic interactions (Voelker et al., 2006) and that an active selective pore must be formed by four P domains (Lesage et al., 1996; Daram et al., 1997; Perozo et al., 1998) suggest that TPKs should be dimers (Latz et al., 2007a). However a second model has been proposed where only the first P domain would take part to the formation of the selective pore and thus functional channels would involve four subunits (Hamamoto et al., 2008; Marcel et al., 2010).
Figure 1.
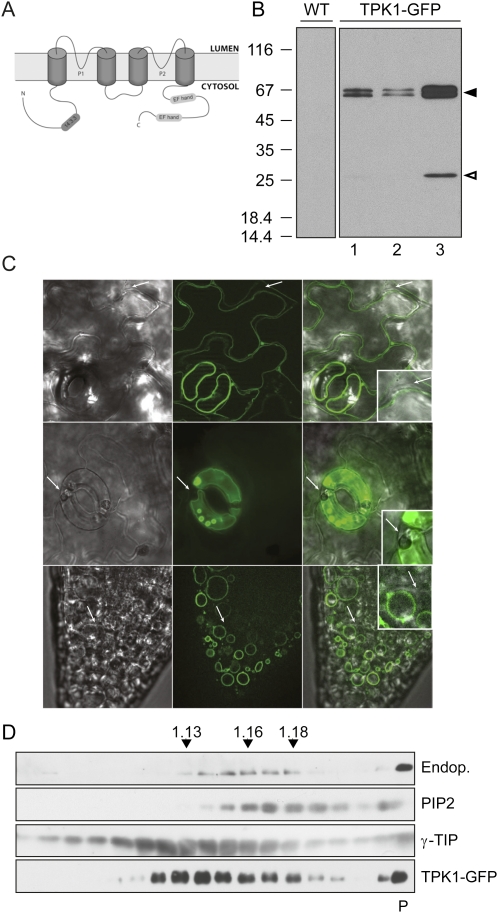
TPK1-GFP is located at the tonoplast in transgenic Arabidopsis. A, Predicted topology for AtTPK1, with four TMDs and two P domains. The N- and C-terminal domains contain a 14.3.3 protein binding site and two putative EF hands, respectively. B, Proteins extracted from leaves of wild type (WT) or three T2 plants expressing TPK1-GFP from transformation event number 9 (lanes 1–3) were analyzed by SDS-PAGE and protein blot using anti-GFP antiserum. The positions of TPK1-GFP (black arrowhead) and free GFP (white arrowhead) are indicated at right. Numbers at left indicate the positions of molecular mass markers, in kD. C, Leaves (top and center sections) or roots (bottom sections) were analyzed by epifluorescence microscopy. In each lane, bright-field (left) GFP fluorescence (center) and merge of the two (right) are shown. Arrows indicate plasma membranes. D, Subcellular compartments were fractionated by ultracentrifugation of leaf homogenate on isopycnic Suc gradient. Proteins in each fraction were analyzed by SDS-PAGE and protein blot with antibodies against endoplasmin (Endop., ER marker), PIP2 (plasma membrane marker), γ-TIP (tonoplast marker), or GFP (to detect TPK1-GFP). Top of the gradient is at left; numbers on top indicate fraction density (g/mL); P indicates the precipitate recovered from the bottom of the tube.
Four Arabidopsis TPKs (AtTPK1, 2, 3, and 5) are located at the tonoplast when expressed in transgenic tobacco (Nicotiana tabacum) BY2 cells or in Arabidopsis protoplasts transiently transformed with GFP fusions (TPK-GFP for short; Czempinski et al., 2002; Schönknecht et al., 2002; Voelker et al., 2006). Transient expression in onion (Allium cepa) epidermal cells also confirmed the tonoplast localization of a TPK1-GFP fusion and indicated that TPK4-GFP was instead located at the plasma membrane and other compartments of the secretory pathway but not at the tonoplast (Becker et al., 2004; Dunkel et al., 2008). Protein traffic and sorting along the secretory pathway require correct folding and assembly, which are under the control of the endoplasmic reticulum (ER) quality control mechanism (Vitale and Boston, 2008), and the presence of sorting signals (for review, see Rojo and Denecke, 2008; Richter et al., 2009). ER quality control, traffic routes, signals, and receptors have been characterized to relevant extent for vacuolar soluble proteins, but data on tonoplast proteins are much more limited. Similarly to integral membrane proteins of other compartments of the secretory pathway (Hanton et al., 2005; Zelazny et al., 2009), TPK1 requires a cytosol-exposed D/E-X-D/E diacidic motif, present in its C-terminal domain, to traffic from the ER (Dunkel et al., 2008). This, or other ER-export motifs in cytosolic domains, promotes interaction with the coat protein complex II (COPII) that mediates vesicle formation from the ER en route to the Golgi complex (Barlowe, 2003; Mikosch and Homann, 2009). Consistently, TPK1-mRFP1 remains located in the ER in the presence of brefeldin A, an inhibitor of Golgi-mediated vesicular traffic (Dunkel et al., 2008).
Proteins that pass ER quality control are sorted to their different destinations along the secretory pathway. A number of studies have addressed the question of tonoplast versus plasma membrane sorting. Expression of different deletion mutants of plasma membrane H+-ATPase of Nicotiana plumbaginifolia suggested that neither the plasma membrane nor the tonoplast is a default destination (Lefebvre et al., 2004). Exchange of domains between AtTPK1 and AtTPK4 showed that the N-terminal domain of the former does not contain information for tonoplast sorting. Furthermore, AtTPK1/AtTPK4 chimeras that were unable to reach the tonoplast remained in the ER or the Golgi complex but never reached the plasma membrane, not allowing a clear definition of sorting signals (Dunkel et al., 2008). The sixth (last) TMD of bean (Phaseolus vulgaris) α-TIP contains information for tonoplast sorting that can be transferred to reporter proteins (Höfte and Chrispeels, 1992). However, this bean TIP and Arabidopsis TIP3;1 seem to reach the tonoplast along a COPII-independent, brefeldin A-insensitive route (Gomez and Chrispeels, 1993; Park et al., 2004). The cytosolic domains of rice (Oryza sativa) TPKa and TPKb contain information that discriminates between brefeldin A-sensitive sorting to the lytic vacuole and brefeldin A-insensitive sorting to the storage vacuole (Isayenkov et al., 2011). Finally, exchange experiments between two Arabidopsis syntaxins (that are tail-anchored proteins; Pedrazzini, 2009) of the plasma membrane and the tonoplast, indicated that a cytosolic N-terminal domain of about 120 residues (called longin domain and present in some but not all syntaxins) of the latter is sufficient to redirect the former to the tonoplast, whereas the TMDs do not discriminate between the two membranes (Uemura et al., 2005). Thus, the general picture regarding the sorting signals that discriminate between plasma membrane and the tonoplast is not clear, especially for multispanning (type III) membrane proteins.
Eukaryotic protein turnover takes place mainly in the cytosol, via the ubiquitin/proteasome pathway, or in the inner hydrolytic compartments (vacuoles in plants and yeast [Saccharomyces cerevisiae], lysosomes in animals). In general, the former is more active on short-lived protein whereas long-lived ones are turned over by hydrolytic compartments through autophagy (Ohsumi, 2006). The latter mechanism is certainly involved in the turnover of a number of integral membrane proteins of the plasma membrane, a process that in yeast and mammalian cells is mainly signaled by monoubiquitination and Lys-63-linked polyubiquitination (Raiborg and Stenmark, 2009). The ubiquitinated cargo undergoes endocytosis and incorporation into early endosomes, similarly to plasma membrane proteins that after endocytosis undergo recycling instead of degradation. Ubiquitination however inhibits recycling to the plasma membrane and promotes further delivery and internalization into multivesicular bodies (MVBs) by membrane invagination, an event mediated by the endosomal sorting complex for transport (ESCRT) machinery. When MVBs fuse with the vacuole or lysosome, the internalized vesicles are finally degraded by lipases and proteases (Raiborg and Stenmark, 2009). Plant plasma membrane proteins are also recycled and in some cases eventually targeted for degradation to the vacuole in a MVB-mediated process that probably involves the ESCRT machinery (Otegui and Spitzer, 2008). Examples are the boron efflux transporter BOR1 (Takano et al., 2005) and the ferrous iron transporter IRT1 (Kerkeb et al., 2008). Mutation of two Lys residues in a cytoplasmic loop inhibits iron-induced turnover of IRT1, suggesting a role of ubiquitination (Kerkeb et al., 2008). To our knowledge, it has not been determined how the turnover of tonoplast proteins occurs.
Here, we have used transgenic Arabidopsis plants and transient expression in Arabidopsis protoplasts to study the assembly, tonoplast sorting, and turnover of AtTPK1. The results indicate that this protein forms dimers, is subjected to ER quality control, and contains tonoplast sorting information in its cytosolic C-terminal domain. We also show that its turnover mechanism involves internalization into the vacuole.
RESULTS
TPK1-GFP Localizes at the Tonoplast in Transgenic Arabidopsis Plants
TPK1 has been localized at the tonoplast when expressed transiently or in transgenic BY2 tobacco cells (Czempinski at el., 2002; Schönknecht et al., 2002; Dunkel et al., 2008). We first wanted to confirm this localization in transgenic Arabidopsis. Endogenous expression of TPK1 is rather low in general and very low in tissues other than leaves (Voelker et al., 2006). We reasoned that expression under endogenous promoter would have made detection of the recombinant protein very difficult. As the addition of a GFP tag at the C terminus does not modify the tonoplast localization of TPK1 in tobacco BY2 cells (Czempinski et al., 2002), we generated transgenic Arabidopsis plants constitutively expressing TPK1-GFP under the control of the cauliflower mosaic virus 35S promoter. These plants do not have any evident phenotype alteration when grown in normal conditions.
To verify whether TPK1-GFP was correctly synthesized, we first analyzed total leaf protein extracts by SDS-PAGE and protein blot using anti-GFP antiserum (Fig. 1B). Two closely migrating polypeptides with apparent molecular mass around 66 kD (black arrowhead) and one around 28 kD (white arrowhead) were specifically recognized in the transgenic plants. This pattern was consistently produced in independent transgenic lines (not shown). T2 plants from the same transformation event showed variable extents of TPK1-GFP accumulation (Fig. 1B), but in T3 plants the level was constant. The two larger polypeptides around 66 kD most probably correspond to full-length TPK1-GFP, as its predicted molecular mass is 67.6 kD. The close migration of the two bands suggests that TPK1-GFP can have two different conformations or undergo covalent modifications. TPK1 contains one potential N-glycosylation site (Asn131SerSer). However TPK1-GFP is not N-glycosylated (Supplemental Fig. S1). The difference between the two closely related polypeptides must therefore have another explanation.
The 28-kD polypeptide (Fig. 1B) is clearly detectable only in plants that have high accumulation of the 66-kD forms and may correspond to a degradation product containing the GFP epitopes recognized by the antibody. Its apparent molecular mass resembles that of GFP, indicating that this TPK1-GFP fragment contains most of the GFP sequence. For brevity, hereon this polypeptide will be referred to as free GFP.
The subcellular localization of TPK1-GFP was determined in leaf epidermal cells and root tissues by epifluorescence microscopy (Fig. 1C). In both cases, fluorescence was at the tonoplast. No signal was detected at the plasma membrane (arrows) or other endomembranes. To further verify this result, microsomes were prepared from leaf homogenates and fractionated by isopycnic ultracentrifugation on continuous Suc gradient (Fig. 1D). Intact TPK1-GFP cofractionated with the tonoplast marker γ-TIP but not with the plasma membrane intrinsic protein2 (PIP2; Santoni et al., 2003) or the ER chaperone endoplasmin (Klein et al., 2006). It can be concluded that TPK1-GFP is located at the tonoplast in transgenic Arabidopsis plants and that expression under the 35S promoter does not lead to saturation of its sorting machinery (which may lead to double localization, see for example Frigerio et al., 1998).
TPK1-GFP Forms Dimers
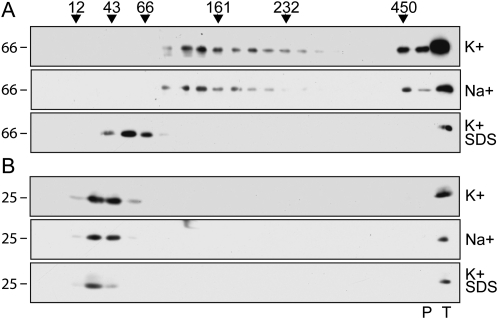
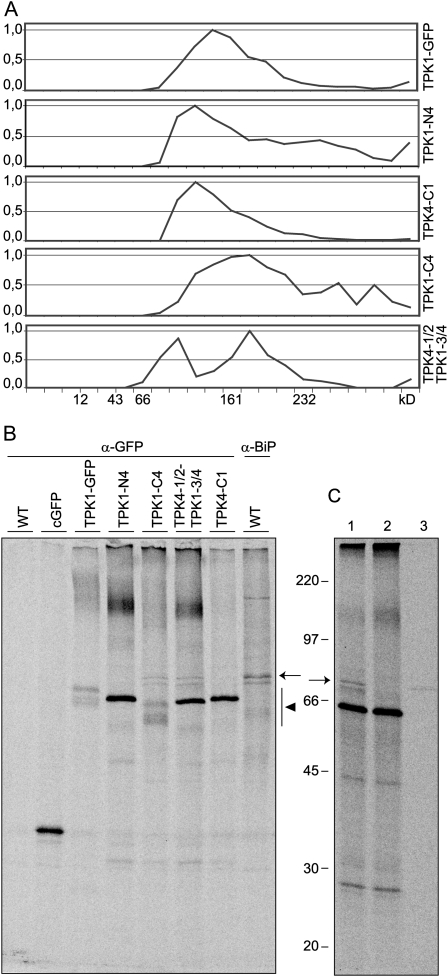
The assembly state of TPK1-GFP was analyzed by velocity Suc gradient centrifugations of leaf homogenates prepared in the presence of nonionic detergent. TPK1-GFP sedimented slightly slower than the 161-kD standard, indicating an apparent molecular mass consistent with a dimer of the 66-kD polypeptide (Fig. 2A, top section). Free GFP originating from TPK1-GFP migrated between the 12- and 43-kD markers, indicating that it is monomeric and thus strongly suggesting that it does not contribute to dimerization when it is part of TPK1-GFP (Fig. 2B, top section).
Figure 2.
TPK1-GFP is a dimer. Transgenic Arabidopsis leaves were homogenized in the following different conditions: nonionic detergent and low concentration of potassium (K+); nonionic detergent and high concentration of sodium (Na+); SDS and low concentration of potassium followed by denaturation at 100°C for 5 min (K+ SDS). The homogenates were then subjected to sedimentation velocity centrifugation on continuous 5% to 25% (w/v) Suc gradient. Aliquots of gradient fractions, material precipitated at the bottom of the tubes (P), and an aliquot of total, nonfractionated homogenate (T) were analyzed by SDS-PAGE and protein blot using anti-GFP antiserum. The regions of the blots containing proteins in the molecular mass range of TPK1-GFP (A) or free GFP (B) are shown. Top of gradients is at left. Numbers on top indicate the position along the gradient and the molecular mass of sedimentation markers, in kD. Numbers at left indicate the positions of SDS-PAGE molecular mass markers, in kD.
The intersubunit interactions of the viral K+ channel Kcv are disrupted in the presence of high concentration of sodium ions (Pagliuca et al., 2007). We analyzed the assembly of TPK1-GFP in the presence of 200 mm Na+ instead of 40 mm K+ or, as a control, after denaturation with 1% SDS (Fig. 2A, center and bottom sections). The sedimentation of TPK1-GFP was not affected by the high sodium conditions. On the opposite, SDS-denatured TPK1-GFP migrated as a monomer, between the 43- and 66-kD markers. Treatment with SDS did not cause any general protein migration artifact, since the SDS-denatured protein standards sedimented according to the molecular mass of their monomeric forms (not shown). Similarly, the migration of free GFP was not affected by high sodium or SDS (Fig. 2B, center and bottom sections).
We conclude that TPK1-GFP is a homodimer and that its assembly is not disrupted by high sodium concentration.
Newly Synthesized TPK1-GFP Transiently Interacts with a 20-kD Polypeptide
We investigated the dynamics of TPK1-GFP dimer formation by pulse-chase analysis. Attempts to use intact transgenic plantlets were unsuccessful, because incorporation of radioactive amino acids into TPK1-GFP continued during the chase, indicating slow uptake and depletion (not shown). Pulse chase was therefore performed on protoplasts isolated from transgenic leaves.
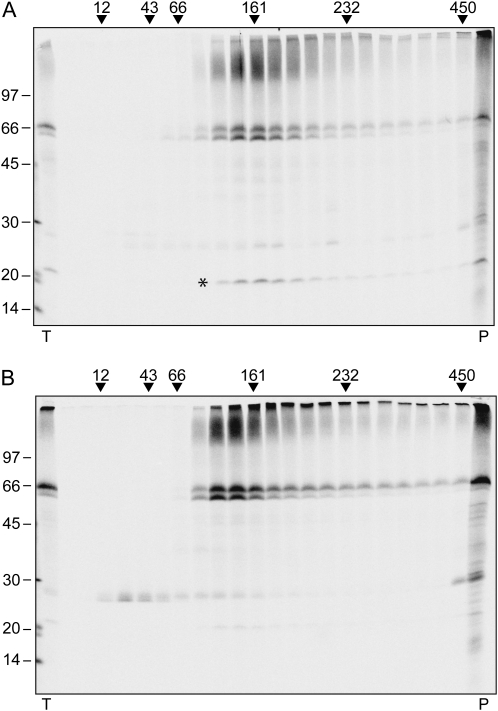
Protoplasts were subjected to 30-min pulse or to 1-h pulse followed by 4-h chase. Protoplast homogenates were then analyzed by velocity gradient centrifugation followed by immunoprecipitation of each fraction with anti-GFP antiserum, SDS-PAGE, and radiography scanning. After the 30-min pulse, no TPK1-GFP peak was detectable in the gradient region between the 43- and 66-kD markers, where monomers are expected to migrate (Fig. 3A). Newly synthesized TPK1-GFP was however in a complex of about 161 kD, slightly larger than the one detectable at steady state by protein blot. After 4-h chase, radioactive TPK1-GFP had shifted to lighter fractions, indicating that the expected dimers had been formed (Fig. 3B, compare with 3A and with 2A, top section). At 4-h chase free GFP becomes also clearly detectable, as radioactive monomers, indicating that GFP release is a posttranslational event occurring in vivo, and it is not due to in vitro proteolytic degradation. The time-dependent shift of intact TPK1-GFP is accounted for by the coimmunoselection of a radioactive polypeptide of about 20 kD after the short pulse but not after the chase (Fig. 3A, asterisk, and compare with 3B). This unidentified polypeptide comigrated along the gradient with full-length TPK1-GFP and was therefore transiently associated with it. Coselection of the 20-kD component with TPK1-GFP was also clearly detectable when immunoprecipitation was performed on unfractionated protoplast homogenates after pulse labeling, and gradually decreased during the chase (see Fig. 6A, asterisk). The component was not selected at late chase points or from labeled wild-type protoplasts (Fig. 6A). Therefore, it is not an unspecific contaminant of immunoprecipitation but rather a protein that transiently interacts with newly synthesized TPK1-GFP.
Figure 3.
Dynamics of TPK1-GFP assembly. Protoplasts prepared from transgenic Arabidopsis leaves were pulse labeled with a mixture of 35S-Met/Cys either for 30 min (A) or for 1 h followed by 4-h chase (B). Homogenates prepared in the presence of nonionic detergent and low concentration of potassium were subjected to sedimentation velocity centrifugation on a continuous 5% to 25% (w/v) Suc gradient. Aliquots of gradient fractions, proportional amounts of the material precipitated at the bottom of the tubes (P), and an aliquot of total, nonfractionated homogenate (T) were subjected to immunoprecipitation with anti-GFP antiserum and analyzed by SDS-PAGE and radiography scanning. Top of gradients is at left, numbers on the top indicate the position along the gradient and the molecular mass of sedimentation markers, in kD. The asterisk indicates the 20-kD polypeptide that cofractionates with newly synthesized TPK1-GFP. Numbers at left indicate the positions of SDS-PAGE molecular mass markers, in kD.
Figure 6.
Stability of TPK1-GFP and posttranslational release of free GFP. Protoplasts prepared from wild-type or transgenic (TPK1-GFP) Arabidopsis leaves were pulse labeled for 1 h with a mixture of 35S-Met/Cys and subjected to chase for the indicated hours. Homogenates were immunoselected with anti-GFP antiserum and analyzed by SDS-PAGE and fluorography. B is from a shorter gel exposure of the gel in A. The positions of TPK1-GFP (black arrowhead), free GFP (white arrowhead), and 20-kD polypeptide transiently associated to TPK1-GFP (asterisk) are indicated at right. Numbers at left indicate the positions of molecular mass markers, in kD.
A simple explanation of these results is that at 30-min pulse newly synthesized TPK1-GFP is already assembled into dimers that form a transient complex with the 20-kD polypeptide. As there is no information about the stoichiometry of the complex, it cannot be ruled out that this is instead composed of a single, not-yet-assembled TPK1-GFP subunit and several copies of the 20-kD polypeptide.
The TPK1 Cytosolic C-Terminal Domain Contains Tonoplast Sorting Information
To identify sorting signals present in AtTPK1, we exchanged various domains between TPK1-GFP and AtTPK4, the only member of the TPK family not located at the tonoplast (Becker et al., 2004; Latz et al., 2007b; Dunkel et al., 2008). We performed transient expression in protoplasts isolated from Arabidopsis cell culture and analyzed the samples by confocal microscopy 40 h after transfection.
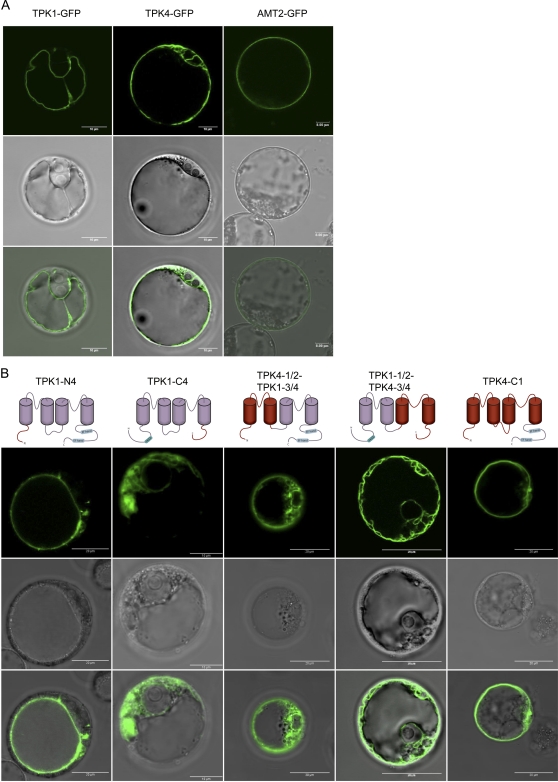
TPK1-GFP was located at the tonoplast and occasionally in internal bulbs, but never at the plasma membrane, identified by FM4-64 staining (Fig. 4A; Supplemental Fig. S2). TPK4-GFP was mostly in not-well-defined internal membranes but not at the tonoplast. Its frequent perinuclear distribution indicated at least partial retention in the ER, but the overall pattern was not typical of this compartment, perhaps suggesting a distortion of internal endomembranes (Fig. 4; Supplemental Fig. S2). FM4-64 staining indicated that TPK4-GFP delivery to the plasma membrane is very inefficient in Arabidopsis protoplasts. When apparent overlapping of the two signals was observed, this was in patches and never along an entire protoplast (Supplemental Fig. S2). The overall pattern is similar to those observed when TPK4-GFP constructs were transiently expressed in onion epidermis (Becker et al., 2004; Dunkel et al., 2008) or Nicotiana benthamiana mesophyll cells (Latz et al., 2007b). To further verify the efficiency of traffic in our expression system, we transformed protoplasts with a GFP fusion of the plasma membrane ammonium transporter AtAMT2 (Sohlenkamp et al., 2002). This chimeric construct was efficiently delivered to its correct destination (Fig. 4A), indicating that the peculiar distribution of TPK4-GFP is not due to a general inefficiency of traffic to the plasma membrane.
Figure 4.
Subcellular localization of TPK1-GFP, TPK4-GFP, and TPK1-TPK4 chimeras. TPK1-GFP, TPK4-GFP, the plasma membrane marker AMT2-GFP (A), or different TPK1-TPK4 chimeras (B) were transiently expressed in protoplasts prepared from Arabidopsis cultured cells. Protoplasts were observed 40 h after transformation by confocal fluorescence microscopy. For each construct, fluorescence (top section), bright field (middle), or merge (bottom) is shown.
The relative contribution of TPK1 and TPK4 domains to each of the TPK1/TPK4 chimeras is illustrated in the cartoons at top of Figure 4B. All the resulting chimeras contained GFP at their C terminus, but the GFP acronym is omitted, for brevity. The amino acid sequences are reported in Supplemental Figure S3. While this project was undergoing, results obtained by an almost identical approach were reported, using transient expression in onion epidermal cells and localization of fluorescent TPK1-TPK4 chimeras by confocal microscopy (Dunkel et al., 2008). For this reason, we will underline here only some aspects of our results. TPK1-N4 located at the tonoplast (with about 65% efficiency, similarly to TPK1-GFP), whereas TPK1-C4 and TPK4-1/2-TPK1-3/4 did not show any tonoplast labeling and were mainly located in typical reticular structures, indicating ER retention (Fig. 4B). These subcellular localizations are similar to those of the roughly corresponding constructs studied by Dunkel et al. (2008). TPK1-1/2-TPK4-3/4 was present in most protoplasts (between 70% and 90% in fully independent experiments) in extended membranes (Fig. 4B; Supplemental Fig. S2). This pattern was similar to that of TPK4-GFP, but TPK1-1/2-TPK4-3/4 could also be detected at the tonoplast. The reticular ER was labeled in only a minor percentage of protoplasts. The similar construct TPK1-P1-TPK4-P2 in Dunkel et al. (2008) gave an ER pattern. Finally, TPK4-C1, similar to the ER-located TPK4-TPK1CT in Dunkel et al. (2008), clearly exited the ER: Almost no protoplasts (5%) exhibited the GFP signal in this compartment. More interestingly, the signal was in most cases (55%) observed at the tonoplast (Fig. 4B), although the fusion is mainly based on TPK4 backbone. Minor proportions were located in bulbs (20%) or in dots possibly corresponding to the Golgi complex (15%). This indicates that the TPK1 C-terminal, cytosolic domain is sufficient to sort TPK4 to the tonoplast of Arabidopsis protoplasts.
ER-Retained Chimeric TPK1-TPK4 Chimeras Have Structural Defects Recognized by ER Quality Control
As also suggested by Dunkel et al. (2008), the ER localization of a number of TPK1-TPK4 exchange chimeras could be due to conformational defects rather than to the lack of signals for ER exit or tonoplast sorting. Assembly state and possible interactions with the ER machinery were therefore analyzed (Fig. 5A). Velocity gradient centrifugation of the tonoplast-located constructs TPK1-GFP, TPK1-N4, and TPK4-C1 indicated correct assembly into dimers (the latter two polypeptides have a smaller molecular mass than TPK1-GFP and therefore their dimers are also smaller). Conversely, the two ER-retained constructs TPK1-C4 and TPK4-1/2-TPK1-3/4 formed complexes that migrated either faster or markedly slower than the 161-kD marker. In fully independent experiments, the ER-retained fusion constructs never formed a single well-defined peak and showed variable distribution along the gradient, indicating disordered interactions and suggesting aggregation.
Figure 5.
ER-retained chimeras are assembly defective and interact with the chaperone BiP. Protoplasts isolated from Arabidopsis cultured cells were transformed with vectors encoding the indicated TPK1-TPK4 chimeras, cytosolic GFP (cGFP), or were not transformed (WT). A, Forty hours after transformation, protoplasts were homogenated in the presence of nonionic detergent and low concentration of potassium. After sedimentation velocity centrifugation on a continuous 5% to 25% (w/v) Suc gradient, aliquots of gradient fractions were analyzed by SDS-PAGE and protein blot using anti-GFP antiserum. For each gradient, the intensity of the bands representing the intact construct was quantified in each fraction and its value normalized relatively to the maximum. Top of gradients is at left. Numbers at bottom indicate the position along the gradient and the molecular mass, in kD, of sedimentation markers. B, Protoplasts were pulse labeled for 1 h with a mixture of 35S-Met/Cys and homogenized. Equal aliquots of homogenates were subjected to immunoprecipitation with anti-GFP (α-GFP) or anti-BiP (α-BiP) antiserum and analyzed by SDS-PAGE and radiography scanning. The region of migration of the TPK1-TPK4 chimeras (black arrowhead) and the position of BiP (arrow) are indicated. C, The homogenate of pulse labeled, TPK4-1/2-TPK1-3/4 expressing protoplasts was immunoprecipitated with anti-GFP antiserum (lane 1). An aliquot of this immunoprecipitate was treated with ATP (lane 2) and the released material was then immunoprecipitated with anti-BiP antiserum (lane 3). Samples were analyzed by SDS-PAGE and radiography scanning. The position of BiP (arrow) is indicated. Numbers at left indicate the positions of molecular mass markers, in kD.
Proteins with defective folding or assembly can be found in abnormally extensive association with the ER chaperone binding protein (BiP), a major component of the ER quality control mechanism (Pedrazzini et al., 1997; Hong et al., 2008). We therefore analyzed whether BiP associates with any of the TPK1-TPK4 chimeras. Transfected protoplasts or untransfected control were pulse labeled for 1 h. Proteins were immunoprecipitated from cell homogenates with anti-GFP antiserum and analyzed by SDS-PAGE and radiography scanning (Fig. 5B). No radioactive polypeptide was immunoprecipitated from control protoplasts (wild type). As expected, newly synthesized TPK1-GFP migrated as a doublet around 66 kD. The TPK1-TPK4 chimeras had variable banding patterns in the same molecular mass region (Fig. 5B, black arrowhead). The level of expression (judged from band intensities) was also variable, although the higher intensity of the bands representing TPK1-N4, TPK4-1/2-TPK1-3/4, and TPK4-C1 could in part be ascribed to the fact that each of them migrated as a single band, unlike TPK1-GFP and TPK1-C4. It therefore appears that the synthesis of two different isoforms requires the N-terminal domain of TPK1, possibly because the unknown modification occurs within this domain. Several additional faintly labeled polypeptides were coimmunoselected together with the different constructs, most of them being equally present in all samples. However, two closely migrating polypeptides of about 75 to 80 kD were exclusively coselected with the ER-retained constructs (Fig. 5B, position indicated by the arrow). They were not coselected with TPK1-GFP, with the similarly traffic-competent TPK1-N4 and TPK4-C1, or with cytosolic GFP that was also transiently expressed as a further control (cGFP). Immunoselection of control protoplasts using antiserum against BiP strongly suggested that at least one of the two bands represents the chaperone (Fig. 5B, α-BiP). Like all chaperones of the heat-shock 70 protein family, BiP is an ATPase and can be released from its ligands by treatment with ATP (Pedrazzini et al., 1997). In vitro treatment with ATP fully released the two closely migrating 75- to 80-kD polypeptides from the anti-GFP immunoprecipitate of protoplasts expressing TPK4-1/2-TPK1-3/4 (Fig. 5C, lanes 1–2). When the supernatant released by ATP treatment was immunoprecipitated with anti-BiP antiserum, the larger of the 75- to 80-kD polypeptides was selected, confirming that it is BiP and demonstrating that it specifically binds to the potassium channel chimeras due to its chaperone activity (Fig. 5C, lane 3).
The detectable interaction of BiP with the two ER-retained TPK1-TPK4 chimeras, but not with those sorted to the tonoplast, confirmed that the former have conformational defects and are retained in the ER by quality control.
GFP Is Cleaved from TPK1-GFP upon Vacuolar Internalization
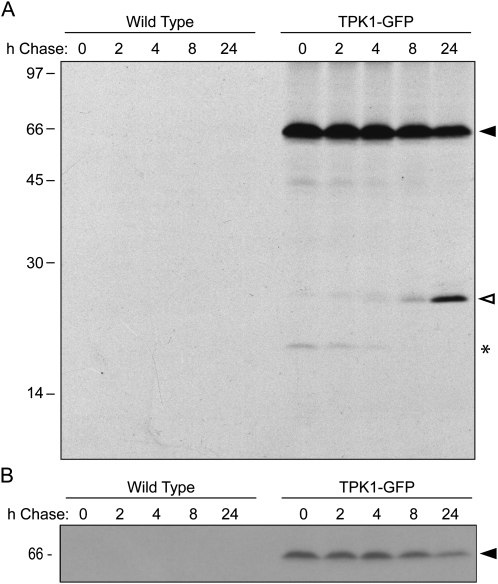
The turnover mechanism of tonoplast proteins has not yet been specifically investigated. The long-term destiny of TPK1-GFP was therefore studied by pulse chase of protoplasts isolated from leaves of transgenic Arabidopsis. No radioactive polypeptide was immunoprecipitated from wild-type protoplasts used as a control (Fig. 6A). Radioactive, intact TPK1-GFP synthesized during 1-h pulse slowly decreased in amount during the chase, to reach about 50% of the original signal after 24 h (Fig. 6A, black arrowhead). This time-dependent decline is better appreciated when the gel is exposed for a short time (Fig. 6B). Free GFP became detectable only during the chase (Fig. 6A, white arrowhead), as already shown in Figure 3. Its intensity increased with time in parallel with the decrease of radioactive intact TPK1-GFP. Since we have no tools to follow the destiny of TPK1 after GFP release, it can be concluded that TPK1 half-life is of at least 24 h. Although chase in intact tissue is inefficient, not allowing an accurate measure of turnover rates, when pulse chase was performed on young transgenic plantlets free GFP was first detectable at 8-h chase (not shown), suggesting that the turnover rate is not influenced by protoplast isolation.
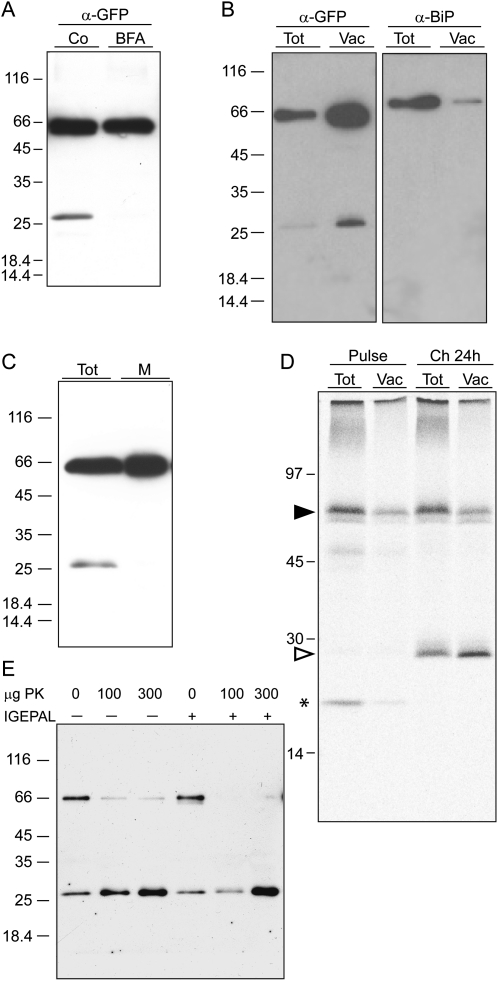
Establishing the subcellular compartment where free GFP is located could provide information on the turnover mechanism of TPK1-GFP. Traffic of TPK1-GFP fusions from the ER can be prevented by brefeldin A in transiently transfected onion epidermal cells (Dunkel et al., 2008) and in our transgenic plants (Supplemental Fig. S4). When transgenic leaves were incubated for 24 h in the presence of the drug, the release of free GFP from TPK1-GFP was fully inhibited (Fig. 7A). Thus, the proteolytic event requires traffic and possibly occurs in the vacuole. Both intact TPK1-GFP and free GFP were detected by protein blot in vacuoles isolated from mesophyll protoplasts (Fig. 7B). Analysis of the same preparations with anti-BiP antiserum showed that the ratio between intact TPK1-GFP and BiP was greatly reversed when the protoplast and vacuole preparations were compared, indicating only minor contamination of vacuoles by the ER, which is the most abundant endomembrane. Free GFP was absent from a leaf microsomal preparation (Fig. 7C), confirming that it is a soluble polypeptide and that its presence in the isolated vacuoles cannot be due to contamination by other compartments. Finally, when protoplasts were subjected to pulse chase followed by vacuole isolation, radioactive free GFP immunoprecipitated at 24-h chase was quantitatively recovered with vacuoles and, more importantly, the ratio between free GFP and TPK1-GFP was higher in the vacuole sample compared to the total protoplast sample (Fig. 7D). This is in agreement with a scenario in which free GFP would be totally vacuolar and newly synthesized intact TPK1-GFP would also be in part in other compartments, en route to its tonoplast destination.
Figure 7.
Free GFP is released upon internalization of TPK1-GFP in the vacuole. A, Transgenic leaves were incubated for 24 h in the presence (BFA) or absence (Co) of brefeldin A. Proteins were extracted and equal volumes of homogenates were analyzed by SDS-PAGE and protein blot using anti-GFP antiserum. B, Vacuoles were isolated from protoplasts prepared from transgenic leaves. Protoplast (Tot) and vacuole (Vac) aliquots corresponding to equal α-mannosidase activity were analyzed by SDS-PAGE and protein blot using anti-GFP (α-GFP) or anti-BiP (α-BiP) antiserum. C, Transgenic leaf tissue was homogenized in the absence of detergent and microsomes were isolated. Equal aliquots of total homogenate (Tot) and microsomes (M) were analyzed by SDS-PAGE and protein blot using anti-GFP antiserum. D, Protoplasts prepared from transgenic leaves were pulse labeled for 1 h with 35S-Met/Cys and subjected to chase for 0 (Pulse) or 24 h (Ch 24 h). Equal aliquots of protoplast or vacuole homogenates were immunoprecipitated with anti-GFP antiserum and the selected proteins analyzed by SDS-PAGE and radiography scanning. The positions of TPK1-GFP (black arrowhead), free GFP (white arrowhead), and 20-kD polypeptide transiently associated to TPK1-GFP (asterisk) are indicated. E, Vacuoles were isolated as in B and the preparation was divided into six equal aliquots, which were treated with the indicated amount of proteinase K (PK) in the absence (−) or presence (+) of nonionic detergent (IGEPAL). Analysis was by SDS-PAGE and protein blot using anti-GFP antiserum. In each section, numbers at left indicate the positions of molecular mass markers, in kD.
Free GFP released posttranslationally from TPK1-GFP is therefore located in vacuoles. However, GFP was fused at the C terminus of AtTPK1, which is predicted to be cytosolic (Fig. 1A). Isolated vacuoles were thus subjected to in vitro digestion with proteinase K to test the predicted orientation of the GFP tag (Fig. 7E). Intact TPK1-GFP was fully digested by proteinase K in the presence of nonionic detergent, leading to a marked increase in the amount of free GFP (that is rather resistant to proteolytic attack; see Chiang et al., 2001). In the absence of detergent, proteinase K released GFP from most TPK1-GFP molecules, indicating that the C-terminal domain has the expected cytosolic topology. However, a small proportion of TPK1-GFP polypeptides remained intact, indicating that they are located inside the vacuolar lumen, where the cleavage of GFP from TPK1-GFP eventually occurs in vivo. We often detected internal vacuolar structures labeled by TPK1-GFP (Fig. 1C, central section). These are very similar to the vacuolar bulbs described by Saito et al. (2002), and could represent the first step of TPK1-GFP internalization from the tonoplast, en route to its physiological turnover.
As described, delivery of plasma membrane proteins to the vacuole for degradation is signaled by ubiquitination (Raiborg and Stenmark, 2009). We tested whether ubiquitinated TPK1-GFP could be detected by antiubiquitin antibodies or by mass spectrometry analysis of TPK1-GFP tryptic peptides. The results indicate that monoubiquitination is not the basis of the difference in molecular mass between the two TPK1-GFP isoforms (Supplemental Table S1) but suggest that a minor proportion of TPK1-GFP molecules, below the detection limit of our mass spectrometry assay, may be ubiquitinated (Supplemental Fig. S5), probably en route to degradation.
DISCUSSION
Assembly and Intracellular Traffic
We have shown here that TPK1-GFP synthesized in transgenic Arabidopsis is detected as a dimer. Although we cannot rule out a higher assembly grade, involving weak interactions disrupted in the conditions used for analysis, dimerization is in agreement with the model of an active selective filter formed by four P domains. Dimerization of animal two P K+ channels has been attributed to the approximately 45-amino acid long luminal loop between TMD1 and P1, which is N-glycosylated (Lotshaw, 2007). Our topology prediction indicates that the corresponding AtTPK1 segment is formed by eight residues only and that other luminal loops are not longer than 11 residues. These observations suggest that the interchain interactions of AtTPK1 may be different from those observed for the animal channels. Pulse chase indicates the transient interaction with an unidentified 20-kD polypeptide that could have a helper function in AtTPK1 folding, assembly, or activation.
TPK1/TPK4 chimeras that fail to traffic to the tonoplast have assembly defects and undergo detectable, ATP-sensitive interactions with BiP, a major component of the ER quality control mechanism. We conclude that correct assembly is a prerequisite for AtTPK1 traffic. The relationship between assembly and traffic has been established for many animal and viral multimeric proteins (Hurtley and Helenius, 1989; Christis et al., 2008) and a few plant proteins. The soluble, homotrimeric vacuolar protein phaseolin requires correct trimerization to pass ER quality control (Vitale et al., 1995; Pedrazzini et al., 1997). Heteromeric assembly is necessary for the ER exit of the plasma membrane proteins AtKC1 (Duby et al., 2008) and ZmPIP1 (Zelazny et al., 2007). The results presented here indicate that a requirement of correct assembly also functions in the traffic of a homomeric tonoplast protein.
Interactions of traffic-competent chimeras or TPK1-GFP with BiP were not detected. This does not rule out an involvement of the chaperone in the folding and assembly of AtTPK1. Interactions between BiP and newly synthesized phaseolin were detectable in developing bean cotyledons, the natural tissue of phaseolin synthesis (Vitale et al., 1995) but not in transformed tobacco protoplasts, unless mutations that inhibit phaseolin trimerization were introduced (Pedrazzini et al., 1994) or BiP was overexpressed (Foresti et al., 2003). This can be explained considering that phaseolin assembly is a fast process that conceals BiP binding sites (Foresti et al., 2003). Newly synthesized monomers can be detected in cotyledons due to the massive synthesis of this storage protein but are below our detection limit in transformed tobacco protoplasts. It is therefore possible that TPK1-GFP also interacts with BiP before dimer formation in a process that is too transient to be detected. We have therefore analyzed the luminal domains of AtTPK1 for the presence of potential BiP binding sites (Supplemental Fig. S6). The only luminal peptide of AtTPK1 with high probability of recognition by BiP, LYFCIVT, is most probably implicated in the formation of P1: its exposure to the ER lumen, necessary for BiP binding, is therefore uncertain during AtTPK1 synthesis. It is thus possible that BiP is not involved in wild-type AtTPK1 folding and assembly. Conversely, BiP was found in association with newly synthesized, partially assembled wild-type tonoplast V-ATPase of oat (Avena sativa) seedlings (Li et al., 1998).
The association of BiP with the traffic- and assembly defective TPK1-C4 and TPK4-1/2-TPK1-3/4 chimeras could be the result of altered interactions of their TMDs with the membrane and consequent abnormal exposure of hydrophobic sequences recognized by BiP in the ER lumen. Similar, although not identical, TPK1/TPK4 chimeras produced by Dunkel et al. (2008) were also unable to traffic from the ER. It is very tempting to speculate that these are similarly assembly defective and recognized by ER quality control, as hypothesized by the authors. Domain exchanges have also been performed on rice TPKa and TPKb, two homologs of AtTPKs located at the tonoplast of lytic and storage vacuoles, respectively (Isayenkov et al., 2011). A number of these hybrid constructs were blocked at the ER. Therefore, the failure in producing TPK exchange chimeras delivered to the plasma membrane can be at least in part explained by the apparently high probability of generating fusions with structural defects recognized by ER quality control.
Tonoplast Sorting
The available data on the delivery of integral membrane proteins to the tonoplast imply that, after approval by ER quality control, at least two further requirements must be met: (1) promotion of traffic from the ER by ER-export motifs, (2) sorting of tonoplast proteins from those destined to the plasma membrane. The length of TMDs also influences the traffic of single-spanning membrane proteins, because bilayer thickness increases along the secretory pathway (Brandizzi et al., 2002; Saint-Jore-Dupas et al., 2006), however it is not clear how this holds for multispanning membrane proteins. Incorrect folding or assembly could also affect proper exposure of ER-export motifs, suggesting that, besides chaperone interactions, poor affinity to COPII may contribute the ER retention of defective proteins (Barlowe, 2003). We do not know whether the functional diacidic motif present in the C-terminal tail of AtTPK1 is properly exposed in the assembly defective TPK1-C4 and TPK4-1/2-TPK1-3/4 fusions.
AtTPK4 is not a tonoplast protein, but the TPK4-C1 fusion was rather efficiently delivered to the tonoplast. Thus, the AtTPK1 C-terminal domain contains tonoplast sorting information that can be transferred to TPK4. TPK4-C1 is very similar to TPK4-TPK1CT expressed by Dunkel et al. (2008) but the latter is retained in the ER, in spite of the diacidic ER export motif that is also present in the AtTPK1 C-terminal domain. This strikingly different behavior can be due to the structural differences of the two constructs that could lead to ER quality control retention of the latter but not the former. Alternatively, differences in the quality control sensitivity of tobacco protoplasts and onion epidermal cells may exist. Tissue specificity of ER quality control has been found in mammals (Sekijima et al., 2005) and could occur in plants: Perturbation of secretory protein folding enhances the synthesis of the ER chaperone endoplasmin in tobacco seedlings but not in protoplasts (Klein et al., 2006).
Turnover
Intact TPK1-GFP has a half-life of about 24 h. Bean α-TIP expressed in tobacco protoplasts did not undergo marked degradation after 24-h chase (Gomez and Chrispeels, 1993). To our knowledge, these are the available data on tonoplast protein stability. Whether these half-lives are representative of the average lifespan of tonoplast proteins will probably be established by large-scale analysis of protein turnover rates, now possible using quantitative mass spectrometry (Oeljeklaus et al., 2009). We cannot rule out that strong expression driven by the 35S promoter stimulates enhanced degradation of these recombinant tonoplast proteins. In general, studies on secretory proteins destined to different locations of the endomembrane system have however shown that turnover rates are mainly determined by the compartment of destination rather than by expression levels (Benchabane et al., 2008).
The time-dependent decrease of intact TPK1-GFP is accompanied by the appearance of a fragment that includes most or all the GFP sequence. Posttranslational proteolysis that generates such a GFP core is typical of GFP-tagged soluble secretory constructs delivered to the vacuole (Frigerio et al., 2001; daSilva et al., 2005; Foresti et al., 2008). Pulse-chase, vacuole isolation, and proteolytic protection experiments indeed indicate that the C-terminal tail of TPK1-GFP starts its life in the cytosol, consistent with the expected AtTPK1 topology, but the GFP core ends up in the vacuolar lumen. Therefore, TPK1-GFP fragmentation involves an internalization event.
In plants as well as other eukaryotes the turnover of many plasma membrane proteins can occur via MVB-mediated delivery of vesicles to the lumen of vacuoles or lysosomes (Takano et al., 2005; Kerkeb et al., 2008; Otegui and Spitzer, 2008; Raiborg and Stenmark, 2009). Our data suggest that a similar mechanism could operate for tonoplast proteins. Vesicles could bud from the tonoplast into the cytoplasm and fuse to MVBs. An alternative model would involve direct internalization of the tonoplast into the vacuolar lumen. Whatever the mechanism, we have shown that a minority of TPK1-GFP molecules could be ubiquitinated and therefore possibly recognized by the ESCRT machinery as a step toward turnover. Invagination-like structures or bulbs at the tonoplast similar to the ones labeled by TPK1-GFP have been observed in protoplasts transiently expressing fluorescent-tagged Arabidopsis TPK or KCO3 (Voelker et al., 2006) and in several different tissues of transgenic Arabidopsis expressing fluorescent-tagged AtTIP1 under constitutive or native promoter (Saito et al., 2002; Beebo et al., 2009; Gattolin et al., 2009). It is however still unclear whether these structures are the consequence of membrane invagination, engulfment of another vacuole, or fusion between different vacuoles (Saito et al., 2002). Interestingly, a GFP fusion of the endosome binding domain of human early endosome antigen1 that was transiently expressed in Arabidopsis protoplasts also moved with time from a prevacuolar compartment to the tonoplast and then in internal vacuolar vesicles before disappearing, indicating its turnover by internalization into the vacuole (Kim et al., 2001).
The internalization event shown in this study could be indicative of a general turnover pathway of tonoplast proteins. Whether this occurs directly by invagination of the tonoplast or prior sorting into MVB remains to be determined, but we now have a tool to investigate this process.
MATERIALS AND METHODS
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Plant Material
Wild-type or transgenic plants of Arabidopsis (Arabidopsis thaliana), ecotype Columbia, were grown in sterile conditions on half-concentrated Murashige and Skoog media (Duchefa Biochemie) supplemented with 10 g/L Suc and 0.8% (w/v) phyto agar (Duchefa Biochemie) at 23°C under a 16/8-h light/dark cycle. Transgenic Arabidopsis plants expressing TPK1-GFP were generated by vacuum infiltration with Agrobacterium tumefaciens strain GV3101 and the construct p35S-TPK1-GFP (Czempinski et al., 2002). Kanamycin-resistant plants (T0) were identified. Experiments were conducted using T2 or T3 plants. Arabidopsis (ecotype Columbia) suspension-cultured T87 cells (Axelos et al., 1992) were grown in JPL medium (Jouanneau and Péaud-Lenoël, 1967) at 25°C under 16-h light/8-h dark cycle with 120 rpm agitation. Cell culture was maintained by weekly 1/10 dilution.
Plasmid and Constructs
The transient expression vector pA7-GFP (Voelker et al., 2006) was used for expression of cytosolic GFP. Wild-type TPK1 fused to GFP was expressed transiently using the pA7-TPK1-GFP construct (Voelker et al., 2006). Standard PCR reactions and subcloning procedures were used to engineer chimeric TPK1-TPK4 sequences. cDNA clones were isolated from cDNA preparations from seedlings or flowers of Arabidopsis (Columbia-0 ecotype). Complementary DNAs were cloned into plasmid pCRScript (Stratagene) or pCR2.1 (Invitrogen). C-terminal GFP fusion constructs were produced using pA7-GFP. The amino acid sequence of each chimeric protein is illustrated in Supplemental Figure S2.
Protoplast Isolation, Transformation, and Vacuole Isolation
Protoplasts for transient expression were prepared from 7-d-old cell culture. Cells were washed twice with 0.5 m sorbitol, 1 mm CaCl2, 10 mm MES pH 5.6 and digested in the same medium containing 1% (w/v) cellulose, 0.25% (w/v) macerozyme, and 0.1% (w/v) bovine serum albumin for 4 to 5 h at 26°C in darkness under shaking (60 rpm). The digestion solution was then brought to 15% (v/v) Percoll with Percoll solution (500 mm sorbitol, 1 mm CaCl2, 20 mm MES, pH 6.0 in Percoll [GE Healthcare]). Protoplasts were then isolated using a step gradient made as follows, from bottom to top: 5 mL 100% Percoll solution, digestion solution brought to 15% Percoll, 2 mL betaine solution (500 mm betaine, 1 mm CaCl2, 10 mm MES, pH 6.0). After centrifugation at 60g, 4°C for 8 min (brake off), protoplasts were recovered at the interface between 15% (v/v) Percoll and betaine solutions and washed with 20 mL W5 buffer (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm Glc, 0.03% MES, pH 5.8), centrifuged at 60g as above, and finally resuspended in 20 mL W5 buffer. Protoplasts were then transformed with 50 μg plasmid DNA using a polyethylene glycol-mediated procedure (Pedrazzini et al., 1994) and analyzed 20 to 40 h after transformation.
Leaf protoplasts were prepared from 4- to 6-week-old, axenic Arabidopsis plants. Leaves were cut on the lower epidermis side and laid down, first on 0.5 m mannitol for 1 h, then overnight on digestion solution (0.33% cellulase, 0.17% macerozyme, 0.4 m mannitol, 7 mm CaCl2, 3 mm MES pH 5.8). Protoplasts were then filtrated through a 0.63 μm inox filter and washed twice with W5 buffer. Protoplasts were finally resuspended at a concentration of 106 cell/mL in K3 medium (Gamborg’s basal media with minimal organics [Duchefa Biochemie] supplemented with 5 mm CaCl2, 3 mm NH4NO3, 0.4 m Suc, 1.5 mm Xyl, 1 mg/L 6-benzylaminopurine, and 1 mg/L 1-naphtalenacetic acid, pH 5.5).
Vacuoles were isolated from leaf protoplasts. About 7 million protoplasts were resuspended in 7-mL prewarmed (42°C) lysis buffer (10% [w/v] Ficoll 400, 0.2 m sorbitol, 10 mm HEPES pH 7.5). After 10-min incubation at room temperature, released vacuoles were isolated using a step gradient containing the vacuole-containing lysis buffer, 3 mL Ficoll solution (5% [w/v] Ficoll 400, 0.3 m sorbitol, 0.3 m betaine, 10 mm HEPES, pH 7.5), and 0.4-mL betaine solution (0.6 m betaine, 10 mm HEPES, pH 7.5). After centrifugation at 5,000g, 10°C for 30 min, vacuoles were collected at the interface between the Ficoll and betaine solutions. The activity of α-mannosidase was quantified by incubating samples with 0.6 mm p-nitrophenyl-α-d-mannopiranoside (Sigma-Aldrich) in 50 mm CH3COONa pH 5.0 at 30°C for 10, 30, or 60 min in a total volume of 500 μL. Reactions were stopped by adding 800 μL 1 m Na2CO3 and reaction product was quantified by reading A410. Protoplast or vacuole homogenates corresponding to equal activity were then analyzed by SDS-PAGE and protein blot.
Protease protection assay was performed by treating isolated vacuoles for 60 min on ice with 0, 100, or 300 μg proteinase K (Sigma-Aldrich) in 400 μL 0.6 m betaine, 10 mm CaCl2, 50 mm Tris-Cl pH 7.5 with or without 0.75% Igepal (Sigma-Aldrich). The reaction was stopped by adding phenylmethanesulfonylfluoride to final concentration of 5 mm.
Protein Analysis
For total protein extraction from plant tissues, Arabidopsis leaves (3–6 weeks old) were homogenized in ice-cold homogenization buffer (200 mm NaCl, 1 mm EDTA, 0.2% Triton X-100, 2% 2-mercaptoethanol, 100 mm Tri-Cl pH 7.8, supplemented with Complete protease inhibitor cocktail [Roche]). After centrifugation at 5,000g, 4°C for 10 min, the resulting supernatant was considered as the total protein extract. For microsome preparation, leaf tissues were homogenized in ice-cold buffer containing 250 mm sorbitol, 2 mm EGTA, 2 mm EDTA, 2 mm dithiothreitol, 500 mm Tris-acetate pH 7.5 supplemented with Complete. After centrifugation at 5,000g, 4°C for 10 min, the supernatant was further centrifuged at 100,000g, 4°C for 30 min, and the resulting pellet resuspended in the original buffer before analysis by SDS-PAGE.
Brefeldin A (Invitrogen) treatment was performed on whole leaves cut on the lower epidermis, as for protoplast preparation. Leaves were laid on liquid half-concentrated Murashige and Skoog media (Duchefa Biochemie) supplemented with 10 g/L Suc and either 50 μg/mL Brefeldin A (from a 2 mg/mL stock in ethanol) or equal volume of ethanol and incubated for 24 h at 23°C under a 16/8 h light/dark cycle. Leaf tissues were then gently dried with paper and homogenized as described above.
Isopycnic gradient was performed as described in Ishikawa et al. (2005).
For velocity centrifugation on Suc gradients, Arabidopsis leaves (3–6 weeks old) or protoplasts were homogenized in ice-cold buffer containing 40 mm KCl, 0.2% Triton X-100, 50 mm Tris-Cl pH 7.8 (2 mL per g of leaf tissue), supplemented with Complete. The homogenate was laid on a linear 5% to 25% (w/v) Suc gradient (20 mm KCl, 0.1% Triton X-100, 25 mm Tris-Cl pH 7.5). An additional gradient was loaded with a mixture of markers containing 200 μg each of cytochrome C (12.4 kD), ovalbumin (43 kD), bovine serum albumin (67 kD), aldolase (161 kD), and catalase (232 kD). After centrifugation at 39,000 rpm, 4°C for 25 h in a Beckman SW40 rotor (200,000g average; Beckman Instruments), gradients were divided into 22 fractions of about 550 μL each and the precipitate at the bottom of the tube solubilized with 550 μL of SDS-PAGE loading buffer. Equal volumes of each fraction and solubilized precipitate were analyzed by SDS-PAGE.
For protein blot, proteins were separated by SDS-PAGE and then electrotransferred into nitrocellulose membrane (PerkinElmer). Membranes were then blotted as described in Pedrazzini et al. (1997) using the following antisera: anti-GFP (1:10,000 dilution; Invitrogen), anti-BiP (1:7,500; Pedrazzini et al., 1997), antiendoplasmin (1:500; Klein et al., 2006), anti-PiP2 (1:10,000; Santoni et al., 2003), or antiγ-TIP (1:1,000; prepared in chicken against a synthetic peptide corresponding to the C-terminal nine amino acids of Arabidopsis γ-TIP, a gift from N.V. Raikhel). Super West Pico chemiluminescence substrate (Pierce) was used according to the manufacturers’ protocol. Protein Mr Markers (Fermentas) were used as molecular mass markers.
Pulse-Chase Labeling and Immunoprecipitation
Pulse-chase labeling of protoplasts was performed as described by Pedrazzini et al. (1997) using Easytag mixture of 35S-labeled Met and Cys (PerkinElmer). Protoplasts were homogenized with 2 volumes of ice-cold 1.5× protoplast homogenization buffer (150 mm NaCl, 1.5 mm EDTA, 1.5% Triton X-100, 150 mm Tris-Cl pH 7.5, supplemented with Complete) and subjected to immunoprecipitation as described by D’Amico et al. (1992) using anti-GFP (Invitrogen) or anti-BiP antiserum. The immunoprecipitated proteins were analyzed by SDS-PAGE, using 14C-methylated proteins (Sigma) as molecular mass markers. Radioactive proteins were revealed either by scanning using Starion FLA-9000 phosphoimager system (Fujifilm) or by fluorography as described by Pedrazzini et al. (1997). Quantization of bands representing radioactive proteins was performed using TotalLab software (TotalLab). For the ATP-release assay, protein extract was subjected to immunoprecipitation with anti-GFP antiserum as described above. The protein A Sepharose beads were then incubated with 1-mL BiP release buffer (150 mm NaCl, 0.1% Triton X-100, 6 mm MgCl2, 3 mm ATP, 20 mm Tris-Cl pH 7.5) for 90 min at 4°C with gentle shaking. The supernatant was recovered, brought to 0.25% (w/v) gelatin, 50 mm Tris-Cl pH 7.5, and subjected to immunoprecipitation with anti-BiP antibody.
Microscopy
Confocal microscopy of protoplasts was performed as described by Voelker et al. (2006). In some experiments, protoplasts were incubated with 50 μm FM4-64 (Sigma) in K3 medium 30 min before microscopy (Ueda et al., 2001). Subcellular localization of transiently expressed proteins was determined in blind experiments. For each construct, at least 20 protoplasts per construct were randomly chosen and observed in at least two fully independent transfections. Epifluorescence microscopy of plant tissues was performed using a Zeiss Axiovert 200 microscope (Zeiss) equipped for epifluorescence, followed by the collection of optical sections using Zeiss Apotome and Axiovision 4.1 software.
Liquid Chromatography-Electrospray Ionization-Tandem Mass Spectrometry
Homogenate containing total proteins was prepared as described above from transgenic plants expressing TPK1-GFP. Proteins were immunoselected with anti-GFP antiserum and separated by SDS-PAGE. Upon Coomassie Blue staining, a major band corresponding to the larger AtTPK1-GFP isoform was identified and excised. After trypsin digestion (Prinsi et al., 2009), the sample was analyzed on an Agilent 6520 Q-TOF mass spectrometer with an HPLC Chip Cube source (Agilent Technologies). Analysis of tandem mass spectrometry spectra for peptides identification was performed by database searching with Spectrum Mill software (Agilent Technologies). See the Supplemental Materials and Methods S1 for further details.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. TPK1-GFP is not N-glycosylated.
Supplemental Figure S2. FM4-64 staining of transformed protoplasts.
Supplemental Figure S3. Amino acid sequences of the TPK1-TPK4 chimeric constructs.
Supplemental Figure S4. Brefeldin A treatment inhibits tonoplast delivery of TPK1-GFP in Arabidopsis root tissues.
Supplemental Figure S5. Detection of ubiquitinated proteins.
Supplemental Figure S6. Predicted BiP binding sites in TPK1-GFP.
Supplemental Table S1. Graphical visualization of AtTPK1-GFP sequencing and list of liquid chromatography-electrospray ionization-tandem mass spectrometry identified peptides.
Supplemental Materials and Methods S1. Materials and Methods for Supplemental Data.
Acknowledgments
We thank Karina Schulz, Eike Kamann, and Michele Bologna for technical assistance and Eugenia Maximova, Max-Planck-Institut für Molekulare Pflanzenphysiologie Golm for support in confocal microscopy. We also thank Michael Udvardi for providing the construct encoding AtAMT2-GFP, and Christophe Maurel and Natasha Raikhel for the gift of antisera against PIP2 and γ-TIP, respectively. We are thankful to the other colleagues of the Vacuolar Transport Equipment for Growth Regulation of Plants European Union network as well as to Ingo Dreyer, Aldo Ceriotti, Luca Espen, and Maurizio Cocucci for the stimulating discussions and suggestions.
References
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension culture. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Barlowe C. (2003) Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol 13: 295–300 [DOI] [PubMed] [Google Scholar]
- Becker D, Geiger D, Dunkel M, Roller A, Bertl A, Latz A, Carpaneto A, Dietrich P, Roelfsema MRG, Voelker C, et al. (2004) AtTPK4, an Arabidopsis tandem-pore K+ channel, poised to control the pollen membrane voltage in a pH- and Ca2+-dependent manner. Proc Natl Acad Sci USA 101: 15621–15626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebo A, Thomas D, Der C, Sanchez L, Leborgne-Castel N, Marty F, Schoefs B, Bouhidel K. (2009) Life with and without AtTIP1;1, an Arabidopsis aquaporin preferentially localized in the apposing tonoplasts of adjacent vacuoles. Plant Mol Biol 70: 193–209 [DOI] [PubMed] [Google Scholar]
- Benchabane M, Goulet C, Rivard D, Faye L, Gomord V, Michaud D. (2008) Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol J 6: 633–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N. (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CF, Okou DT, Griffin TB, Verret CR, Williams MN. (2001) Green fluorescent protein rendered susceptible to proteolysis: positions for protease-sensitive insertions. Arch Biochem Biophys 394: 229–235 [DOI] [PubMed] [Google Scholar]
- Christis C, Lubsen NH, Braakman I. (2008) Protein folding includes oligomerization—examples from the endoplasmic reticulum and cytosol. FEBS J 275: 4700–4727 [DOI] [PubMed] [Google Scholar]
- Czempinski K, Frachisse JM, Maurel C, Barbier-Brygoo H, Mueller-Roeber B. (2002) Vacuolar membrane localization of the Arabidopsis ‘two-pore’ K+ channel KCO1. Plant J 29: 809–820 [DOI] [PubMed] [Google Scholar]
- Czempinski K, Zimmermann S, Ehrhardt T, Müller-Röber B. (1997) New structure and function in plant K+ channels: KCO1, an outward rectifier with a steep Ca2+ dependency. EMBO J 16: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico L, Valsasina B, Daminati MG, Fabbrini MS, Nitti G, Bollini R, Ceriotti A, Vitale A. (1992) Bean homologs of the mammalian glucose-regulated proteins: induction by tunicamycin and interaction with newly synthesized seed storage proteins in the endoplasmic reticulum. Plant J 2: 443–455 [DOI] [PubMed] [Google Scholar]
- Daram P, Urbach S, Gaymard F, Sentenac H, Chérel I. (1997) Tetramerization of the AKT1 plant potassium channel involves its C-terminal cytoplasmic domain. EMBO J 16: 3455–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva LL, Taylor JP, Hadlington JL, Hanton SL, Snowden CJ, Fox SJ, Foresti O, Brandizzi F, Denecke J. (2005) Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell 17: 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G, Hosy E, Fizames C, Alcon C, Costa A, Sentenac H, Thibaud JB. (2008) AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J 53: 115–123 [DOI] [PubMed] [Google Scholar]
- Dunkel M, Latz A, Schumacher K, Müller T, Becker D, Hedrich R. (2008) Targeting of vacuolar membrane localized members of the TPK channel family. Mol Plant 1: 938–949 [DOI] [PubMed] [Google Scholar]
- Foresti O, De Marchis F, de Virgilio M, Klein EM, Arcioni S, Bellucci M, Vitale A. (2008) Protein domains involved in assembly in the endoplasmic reticulum promote vacuolar delivery when fused to secretory GFP, indicating a protein quality control pathway for degradation in the plant vacuole. Mol Plant 1: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Foresti O, Frigerio L, Holkeri H, de Virgilio M, Vavassori S, Vitale A. (2003) A phaseolin domain directly involved in trimer assembly is a BiP binding determinant. Plant Cell 15: 2464–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L, de Virgilio M, Prada A, Faoro F, Vitale A. (1998) Sorting of phaseolin to the vacuole is saturable and requires a short C-terminal peptide. Plant Cell 10: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L, Foresti O, Hernández Felipe D, Neuhaus J-M, Vitale A. (2001) The C-terminal tetrapeptide of phaseolin is sufficient to target green fluorescent protein to the vacuole. Plant Physiol 158: 499–503 [Google Scholar]
- Gattolin S, Sorieul M, Hunter PR, Khonsari RH, Frigerio L. (2009) In vivo imaging of the tonoplast intrinsic protein family in Arabidopsis roots. BMC Plant Biol 9: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez L, Chrispeels MJ. (1993) Tonoplast and soluble vacuolar proteins are targeted by different mechanisms. Plant Cell 5: 1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto S, Marui J, Matsuoka K, Higashi K, Igarashi K, Nakagawa T, Kuroda T, Mori Y, Murata Y, Nakanishi Y, et al. (2008) Characterization of a tobacco TPK-type K+ channel as a novel tonoplast K+ channel using yeast tonoplasts. J Biol Chem 283: 1911–1920 [DOI] [PubMed] [Google Scholar]
- Hanton SL, Renna L, Bortolotti LE, Chatre L, Stefano G, Brandizzi F. (2005) Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell 17: 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfte H, Chrispeels MJ. (1992) Protein sorting to the vacuolar membrane. Plant Cell 4: 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtley SM, Helenius A. (1989) Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol 5: 277–307 [DOI] [PubMed] [Google Scholar]
- Isayenkov S, Isner JC, Maathuis FJ. (2011) Rice two-pore K+ channels are expressed in different types of vacuoles. Plant Cell 23: 756–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Suga S, Uemura T, Sato MH, Maeshima M. (2005) Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett 579: 5814–5820 [DOI] [PubMed] [Google Scholar]
- Jouanneau JP, Péaud-Lenoël C. (1967) Croissance et synthese des proteines de suspensions cellulaires de Tabac sensibles à la kinétine. Physiol Plant 20: 834–850 [Google Scholar]
- Kerkeb L, Mukherjee I, Chatterjee I, Lahner B, Salt DE, Connolly EL. (2008) Iron-induced turnover of the Arabidopsis IRON-REGULATED TRANSPORTER1 metal transporter requires lysine residues. Plant Physiol 146: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I. (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EM, Mascheroni L, Pompa A, Ragni L, Weimar T, Lilley KS, Dupree P, Vitale A. (2006) Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J 48: 657–673 [DOI] [PubMed] [Google Scholar]
- Latz A, Becker D, Hekman M, Müller T, Beyhl D, Marten I, Eing C, Fischer A, Dunkel M, Bertl A, et al. (2007a) TPK1, a Ca(2+)-regulated Arabidopsis vacuole two-pore K(+) channel is activated by 14-3-3 proteins. Plant J 52: 449–459 [DOI] [PubMed] [Google Scholar]
- Latz A, Ivashikina N, Fischer S, Ache P, Sano T, Becker D, Deeken R, Hedrich R. (2007b) In planta AKT2 subunits constitute a pH- and Ca2+-sensitive inward rectifying K+ channel. Planta 225: 1179–1191 [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Batoko H, Duby G, Boutry M. (2004) Targeting of a Nicotiana plumbaginifolia H+-ATPase to the plasma membrane is not by default and requires cytosolic structural determinants. Plant Cell 16: 1772–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage F, Reyes R, Fink M, Duprat F, Guillemare E, Lazdunski M. (1996) Dimerization of TWIK-1 K+ channel subunits via a disulfide bridge. EMBO J 15: 6400–6407 [PMC free article] [PubMed] [Google Scholar]
- Li XH, Su RTC, Hsu HT, Sze H. (1998) The molecular chaperone calnexin associates with the vacuolar H(+)-ATPase from oat seedlings. Plant Cell 10: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotshaw DP. (2007) Biophysical, pharmacological, and functional characteristics of cloned and native mammalian two-pore domain K+ channels. Cell Biochem Biophys 47: 209–256 [DOI] [PubMed] [Google Scholar]
- Marcel D, Müller T, Hedrich R, Geiger D. (2010) K+ transport characteristics of the plasma membrane tandem-pore channel TPK4 and pore chimeras with its vacuolar homologs. FEBS Lett 584: 2433–2439 [DOI] [PubMed] [Google Scholar]
- Mikosch M, Homann U. (2009) How do ER export motifs work on ion channel trafficking? Curr Opin Plant Biol 12: 685–689 [DOI] [PubMed] [Google Scholar]
- Oeljeklaus S, Meyer HE, Warscheid B. (2009) Advancements in plant proteomics using quantitative mass spectrometry. J Proteomics 72: 545–554 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2006) Protein turnover. IUBMB Life 58: 363–369 [DOI] [PubMed] [Google Scholar]
- Otegui MS, Spitzer C. (2008) Endosomal functions in plants. Traffic 9: 1589–1598 [DOI] [PubMed] [Google Scholar]
- Pagliuca C, Goetze TA, Wagner R, Thiel G, Moroni A, Parcej D. (2007) Molecular properties of Kcv, a virus encoded K+ channel. Biochemistry 46: 1079–1090 [DOI] [PubMed] [Google Scholar]
- Park M, Kim SJ, Vitale A, Hwang I. (2004) Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol 134: 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini E. (2009) Tail-anchored proteins in plants. J Plant Biol 52: 88–101 [Google Scholar]
- Pedrazzini E, Giovinazzo G, Bielli A, de Virgilio M, Frigerio L, Pesca M, Faoro F, Bollini R, Ceriotti A, Vitale A. (1997) Protein quality control along the route to the plant vacuole. Plant Cell 9: 1869–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini E, Giovinazzo G, Bollini R, Ceriotti A, Vitale A. (1994) Binding of BiP to an assembly-defective protein in plant cells. Plant J 5: 103–110 [Google Scholar]
- Perozo E, Cortes DM, Cuello LG. (1998) Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol 5: 459–469 [DOI] [PubMed] [Google Scholar]
- Prinsi B, Negri AS, Pesaresi P, Cocucci M, Espen L. (2009) Evaluation of protein pattern changes in roots and leaves of Zea mays plants in response to nitrate availability by two-dimensional gel electrophoresis analysis. BMC Plant Biol 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452 [DOI] [PubMed] [Google Scholar]
- Richter S, Voss U, Jürgens G. (2009) Post-Golgi traffic in plants. Traffic 10: 819–828 [DOI] [PubMed] [Google Scholar]
- Rojo E, Denecke J. (2008) What is moving in the secretory pathway of plants? Plant Physiol 147: 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye ML, Plasson C, Hawes C, Driouich A, Faye L, Gomord V. (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito C, Ueda T, Abe H, Wada Y, Kuroiwa T, Hisada A, Furuya M, Nakano A. (2002) A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J 29: 245–255 [DOI] [PubMed] [Google Scholar]
- Santoni V, Vinh J, Pflieger D, Sommerer N, Maurel C. (2003) A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem J 373: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht G, Spoormaker P, Steinmeyer R, Brüggeman L, Ache P, Dutta R, Reintanz B, Godde M, Hedrich R, Palme K. (2002) KCO1 is a component of the slow-vacuolar (SV) ion channel. FEBS Lett 511: 28–32 [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarström P, Miller SR, Sawkar AR, Balch WE, Kelly JW. (2005) The biological and chemical basis for tissue-selective amyloid disease. Cell 121: 73–85 [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK. (2002) Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol 130: 1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Miwa K, Yuan LX, von Wirén N, Fujiwara T. (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A. (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Sato MH, Takeyasu K. (2005) The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett 579: 2842–2846 [DOI] [PubMed] [Google Scholar]
- Vitale A, Bielli A, Ceriotti A. (1995) The binding protein associates with monomeric phaseolin. Plant Physiol 107: 1411–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Boston RS. (2008) Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Voelker C, Schmidt D, Mueller-Roeber B, Czempinski K. (2006) Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J 48: 296–306 [DOI] [PubMed] [Google Scholar]
- Zelazny E, Borst JW, Muylaert M, Batoko H, Hemminga MA, Chaumont F. (2007) FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc Natl Acad Sci USA 104: 12359–12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E, Miecielica U, Borst JW, Hemminga MA, Chaumont F. (2009) An N-terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2;4 and ZmPIP2;5 to the plasma membrane. Plant J 57: 346–355 [DOI] [PubMed] [Google Scholar]