Abstract
The KNOXI transcription factor SHOOT MERISTEMLESS (STM) is required to establish and maintain the Arabidopsis (Arabidopsis thaliana) apical meristem, yet little is known about its direct targets. Using different approaches we demonstrate that the induction of STM causes a significant up-regulation of the organ boundary gene CUP SHAPED COTYLEDON1 (CUC1), which is specific and independent of other meristem regulators. We further show that the regulation of CUC1 by STM is direct and identify putative binding sites in its promoter. Continuous expression of STM in Arabidopsis leaf primordia also causes the activation of CUC2-3, as well as microRNA MIR164a, which provides a negative feedback loop by posttranscriptionally regulating CUC1 and CUC2. The results bring new insights into the mechanistic links between KNOXI and CUC transcription factors and contribute to the understanding of the regulatory network controlled by STM.
In contrast to animals, plants continue to produce new organs throughout their life cycle. All aboveground parts of plants are derived from a small number of stem cells located at the shoot apical meristem. Two homeodomain transcription factors play key roles in meristem formation and maintenance. WUSCHEL (WUS) is expressed at the core of the meristem and defines the stem cell niche in the overlaying cells (Tucker and Laux, 2007), while SHOOT MERISTEMLESS (STM) is expressed throughout the meristem and prevents the differentiation of the meristematic cells (for review, see Hake et al., 2004; Hamant and Pautot, 2010; Hay and Tsiantis, 2010).
STM belongs to class I of KNOX homeodomain transcription factors. In Arabidopsis (Arabidopsis thaliana), the KNOXI subclass comprises STM, KNAT1 (also called BREVIPEDICELLUS), KNAT2, and KNAT6, which can have partially overlapping functions in the shoot meristem (Scofield and Murray, 2006). KNOXI proteins interact with BELL1-like homedomain transcription factors, and these interactions determine their target affinity and subcellular localization (for review, see Hake et al., 2004; Hay and Tsiantis, 2010).
At least part of the functions of WUS and STM are performed through the control of hormone homeostasis and signaling. WUS directly represses a group of type-A ARABIDOPSIS RESPONSE REGULATORs involved in a negative feedback loop during cytokinin response (Leibfried et al., 2005; Busch et al., 2010). STM expression induces ISOPENTENYL TRANSFERASE7 (IPT7), which encodes a key enzyme involved in cytokinin biosynthesis (Jasinski et al., 2005; Yanai et al., 2005). Conversely, ectopic IPT expression or exogenous cytokinin can partially rescue weak stm mutants (Jasinski et al., 2005; Yanai et al., 2005). STM also represses gibberellin activity by reducing the levels of the biosynthetic enzyme GA 20-oxidase1 and increasing the levels of the catabolic enzyme GA 2-oxidase1, thus providing an environment of high cytokinin and low gibberellin (Sakamoto et al., 2001; Hay et al., 2002; Chen et al., 2004; Jasinski et al., 2005; Yanai et al., 2005).
The boundaries of the meristem are defined by members of the NAC family of transcription factors (Aida and Tasaka, 2006). In Arabidopsis, this function is redundantly performed by CUP SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3 (Aida et al., 1997, 1999; Vroemen et al., 2003; Hibara et al., 2006). CUC1 and CUC2 are also posttranscriptionally regulated by microRNA (miRNA) miR164, which is encoded by a small gene family comprising three members, MIR164a-c (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005; Nikovics et al., 2006; Sieber et al., 2007; Raman et al., 2008).
In Arabidopsis, CUC genes are required for the activation of STM during embryogenesis (Long et al., 1996; Aida et al., 1999), and it has been proposed that STM can in turn activate CUC expression (Aida et al., 1999, 2002; Takada et al., 2001; Kwon et al., 2006). Mutations in STM or two CUC genes compromise the population of self-renewing stem cells and cause fusions of the cotyledons. KNOX and CUC genes are recruited again at later stages of Arabidopsis development and are required for carpel and ovule development (Ishida et al., 2000; Pautot et al., 2001; Scofield et al., 2007). In many species, KNOXI and CUC genes are expressed in the leaf primordia and act in concert to sculpt the organ shape and generate compound leaves (Bharathan et al., 2002; Blein et al., 2008; Berger et al., 2009).
CUC1 and CUC2 share a common ancestor, but have diverged significantly within the Brassicaceae (Hasson et al., 2011). While both of them are required for organ separation, specialization has been acquired for certain functions, such as the control of the serrations of an Arabidopsis simple leaf, which is regulated by the balance between CUC2 and MIR164a genes (Nikovics et al., 2006; Hasson et al., 2010). STM expression also diverges within the Brassicaceae. While it is confined to the meristem in Arabidopsis, closely related species express STM in the leaf primordia and have more complex organs (Piazza et al., 2010).
Although the biological roles of the versatile developmental regulator STM are well characterized, little is known about its direct targets. In an attempt to bring insights into the network regulated by STM, we performed microarray experiments shortly after the induction of the transcription factor. We found that STM activates CUC1, and demonstrated that it directly binds to its promoter. Additionally, the long-lasting expression of STM also promotes the expression of CUC2-3, and MIR164a, which provides a negative feedback loop to adjust the final CUC level. These results provide new mechanistic insights into the regulatory network comprised by KNOXI and CUC transcription factors and miRNA miR164.
RESULTS
Genome-Wide Response to STM Levels
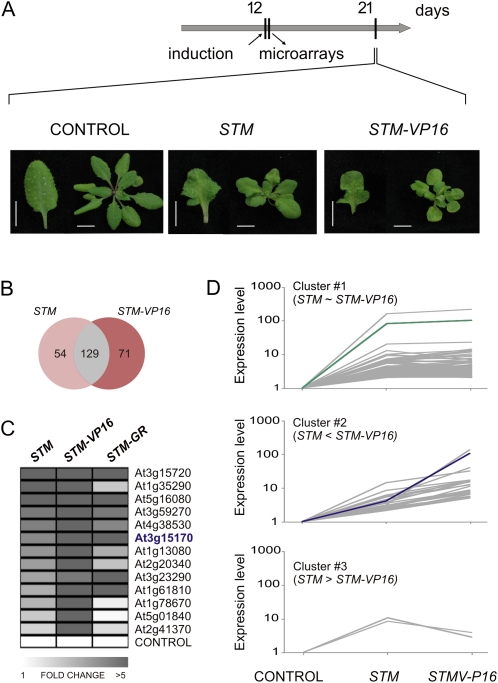
To start to explore the network controlled by STM, we analyzed the transcriptome of plants harboring an ethanol-inducible version of the transcription factor, an approach already used to identify targets of WUS (Leibfried et al., 2005). The selected transgenic plants did not show any obvious phenotypes when grown under normal conditions. However, one single treatment with ethanol was sufficient to cause leaf lobing, as expected for the ectopic expression of STM (Fig. 1A). These morphological changes were obvious 1 week after the induction (Fig. 1A).
Figure 1.
Genome-wide response to STM levels. A, Phenotype of STM and STM-VP16 inducible lines 9 d after treatment with 0.6% ethanol. Samples for microarray experiments were collected 12 h after ethanol induction. The transgenic line used as control expresses GUS under the ethanol inducible promoter. Bars, 1 cm. B, Venn diagram showing the overlap of STM and STM-VP16 up-regulated genes. C, Heat map representing relative expression levels in grayscale of 13 genes in three STM inducible systems: ethanol inducible STM and STM-VP16, and STM-GR. The genes were selected from those induced in STM and STM-VP16 (Fig. 1B). CUC1 (At3g15170) is depicted in blue. The data shown are mean of two biological replicates ± sem for microarray data (STM and STM-VP16) and three biological replicates ± sem for RT-qPCR experiments in the case of STM-GR (see Supplemental Fig. S1 for details). As a control, we used the constitutively expressed gene PROTEIN PHOSPHATASE2A. D, The 129 genes induced by both STM and STM-VP16 were classified in three clusters according to their relative expression levels in the Affimetrix microarrays. Cluster number 1 contains genes with similar levels in STM and STM-VP16 transgenic plants (101 genes); cluster number 2 has genes with higher expression in STM-VP16 than in STM (25 genes); cluster number 3 contains genes with higher expression in STM than in STM-VP16 (three genes) using the criteria described in the text to select differentially expressed genes (logit-T 0.05; 2-fold change with GCRMA). STM is highlighted in green and CUC1 in blue.
It is known that KNOXI transcription factors interact with other proteins that regulate their activity (for review, see Hake et al., 2004; Hay and Tsiantis, 2010). Therefore, we generated an activated version of STM by preparing transgenic plants where the transcription factor is fused to the transactivation domain from the herpes simplex virus VP16. This strategy has been previously used in plants to detect transcription factor activity independently of the presence of coactivators (e.g. Parcy et al., 1998). Treatment with ethanol of plants harboring an inducible STM-VP16 transgene caused a higher degree of leaf lobing than that observed for STM alone (Fig. 1A).
We performed a transcriptome analysis on ATH1 microarrays, 12 h after the induction of STM and STM-VP16. For the profiling experiments we used the shoot apex that includes the meristem and developing leaves, as has been described previously (Leibfried et al., 2005). Genes that showed per-gene variance P > 0.05 (logit-T; Lemon et al., 2003) and more than 2-fold change (GeneChip Robust Multiarray Averaging [GCRMA]; Irizarry et al., 2003) compared to control plants were considered as differentially expressed and were selected for further studies (Supplemental Tables S1 and S2).
Analysis of the STM-modified genes using Gene Ontology term enrichment revealed that there were no strong overrepresented functional categories among them (Supplemental Table S3). We observed, however, that At1g50960, which encodes a GA 2-oxidase7 involved in the catabolism of gibberellin, was significantly induced by both STM and STM-VP16 (Supplemental Tables S1 and S2). IPT7, which participates in cytokinin biosynthesis, was induced nearly three times by STM, although it did not pass the logit-T filter used for the analysis of the arrays (not shown). These observations are in agreement with previous results showing that STM increases cytokinin levels while reducing the gibberellins (Sakamoto et al., 2001; Hay et al., 2002; Jasinski et al., 2005; Yanai et al., 2005).
With the stringent selection criteria applied, 183 genes were induced by STM and 200 by STM-VP16 (Fig. 1B). Most of them (129 genes; Supplemental Table S4) were induced in both conditions. The higher activation capacity of STM-VP16, which is also correlated with the stronger leaf phenotypes observed, is likely responsible for the genes that are differentially expressed between STM and STM-VP16 transgenic plants.
To validate our transcriptome analysis we turned to another inducible system where STM is fused to the glucocorticoid receptor (GR; Gallois et al., 2002). The STM-GR fusion protein is retained in the cytoplasm of transgenic plants, but moves into the nucleus once the cells are treated with dexamethasone (DEX). We selected 13 genes induced by both STM and STM-VP16 (approximately 10% of the genes induced by both constructs) and tested their response to STM-GR (Prom35S:STM-GR construct) by real-time quantitative PCR (RT-qPCR). We found that 10 out of 13 genes were also induced by this system after 12 h of DEX application (Fig. 1C; Supplemental Fig. S1; Supplemental Table S4). These results highlight at least a reasonable reproducibility of the microarray data.
Then, we decided to study in more detail the genes induced by both STM and STM-VP16 (Fig. 1D). We classified these genes in three groups depending on their relative expression in STM and STM-VP16 samples, using the criteria depicted above, variance P > 0.05 (logit-T), and more than 2-fold change (GCRMA). Using this criteria, most genes (101 genes) were similarly induced by STM and STM-VP16 (cluster no. 1), 25 genes were more expressed in STM-VP16 than in STM (cluster no. 2), and only three genes were higher in STM than in STM-VP16 (cluster no. 3).
As expected, STM was detected as significantly up-regulated in both samples. STM levels were, however, similar in STM and STM-VP16 arrays (Fig. 1D, cluster no. 1), indicating that both plants express their transgenes at similar levels and differences between their transcriptomes are likely caused by the presence of the VP16 domain. That the VP16 activated version caused stronger phenotypic defects than STM alone, suggested that the group of genes moderately induced by STM but strongly by STM-VP16 (Fig. 1D, cluster no. 2; Supplemental Table S4) might be particularly related to the KNOXI pathway. CUC1 (At3g15170) was included in this group and stood out as a particularly attractive candidate to study in more detail due to its known roles in the establishment of Arabidopsis meristem.
Specific Response of CUC1 to STM Levels
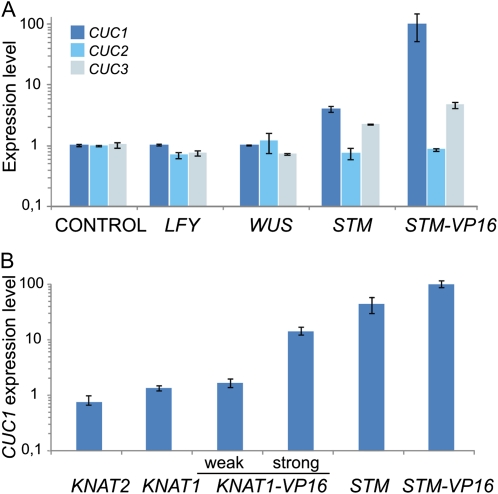
The induction of CUC1 by STM prompted us to study the effects on CUC expression of other transcription factors known to regulate the meristem function. First, we compared the induction of CUC1 by STM with the ones caused by other meristem regulators such as WUS and LFY that were prepared as similar inducible versions (Leibfried et al., 2005). STM was able to induce CUC1 levels 4-fold, while STM-VP16 further enhanced the response to more than 100-fold in the microarray experiments (Fig. 2A). In contrast to STM, LFY and WUS were not able to induce CUC1 (Fig. 2A). These transcription factors also failed to modify the levels of CUC2 and CUC3, while STM-VP16 caused a moderate up-regulation of CUC3 (Fig. 2A).
Figure 2.
Specific response of CUC1 to STM levels. A, Expression levels of CUC1-3 after the induction of LFY, WUS, STM, and STM-VP16 from Affimetrix microarrays (GCRMA normalization). Expression levels were normalized to plants carrying inducible GUS grown in the same conditions, which were used as a control. The data shown are mean of two biological replicates ± sem. B, Regulation of CUC1 expression by Arabidopsis KNOXI genes. The level of CUC1 was determined in transgenic lines harboring an inducible version of KNAT1, KNAT1-VP16, KNAT2, STM, and STM-VP16, 12 h after ethanol treatment. CUC1 levels were determined by RT-qPCR and expressed relative to plants expressing inducible GUS (Control). The data shown are mean of three biological replicates ± sem. [See online article for color version of this figure.]
We then tested the specificity of CUC1 induction inside the KNOXI family of transcription factors, which comprises STM, KNAT1, KNAT2, and KNAT6, being KNAT1 the more closely related to STM, as judged by phylogenetic analyses (Scofield and Murray, 2006). To test whether other KNOXI genes could activate CUC1 expression we prepared ethanol-inducible transgenic lines harboring KNAT1 and KNAT2. In contrast to STM, these other KNOXI genes failed to up-regulate CUC1 (Fig. 2B). These results indicate that there is at least certain degree of specificity for its induction in planta.
We then prepared an activated version of KNAT1, by fusing to it the VP16 domain. In this case, we observed that the induction of KNAT1-VP16 caused the activation of CUC1 (Fig. 2B). It is known that KNOXI proteins can interact with different partners (for review, see Hake et al., 2004; Hay and Tsiantis, 2010). The enhanced activity of the VP16 fusions might indicate that other factors could be required in vivo for a maximum activation of CUC1 by KNOXI proteins.
Direct Regulation of CUC1 by STM
To test whether STM was directly regulating the expression of CUC1, we turned again to the STM-GR system. Direct targets of STM-GR should be induced after DEX treatment, even in the presence of the translational inhibitor, cycloheximide (CYC).
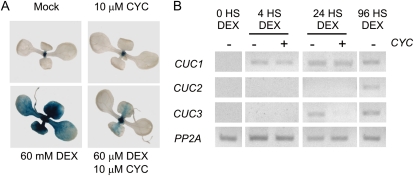
First, we crossed STM-GR transgenics to a CUC1 reporter line (PromCUC1:GUS). We used 1.4-kb CUC1 upstream sequences that have already been described to be sufficient to complement a cuc1 cuc2 mutant when fused to its coding sequence (Baker et al., 2005). The CUC1 promoter is normally expressed in the apical region, but DEX treatment during 24 h caused a strong induction in whole seedlings (Fig. 3A). As a control of the experimental approach, we observed that the supplemental addition of CYC largely prevented the burst of GUS protein activity, which is expected from the inhibition of the translational machinery (Fig. 3A). Note that these experiments were carried out under long induction and staining periods to ensure the saturation of the system.
Figure 3.
Direct regulation of CUC1 by STM. A, GUS expression in seedlings of transgenic plants expressing PromCUC1:GUS crossed to STM-GR after 24 h treatment with or without DEX and CYC. B, CUC1-3 expression levels determined by RT-PCR in STM-GR transgenics treated with or without DEX and CYC. The data shown is representative of at least three biological replicates. [See online article for color version of this figure.]
We then analyzed the expression of CUC genes at the RNA level. As CUC1 is posttranscriptionally regulated by miR164 in a quantitative way (Baker et al., 2005; Nikovics et al., 2006; Sieber et al., 2007), the potential induction caused by STM should overcome this repression to be detectable. We observed that 4 h of induction of STM-GR caused the up-regulation of CUC1 (Fig. 3B). CUC3 was also activated, but after 24 h. Supplemental addition of CYC prevented the induction of CUC3, while CUC1 remained unaffected, confirming CUC1 as a direct target of STM (Fig. 3B). We observed an effect on CUC2 only 96 h after DEX treatment (Fig. 3B). The longer activation time, which is prevented by incubation with CYC suggests that both CUC2 and CUC3 are indirectly regulated by STM. That CUC2 and CUC3 lack obvious STM binding sites in their promoters is in agreement with this possibility.
In Vitro Binding of STM to the CUC1 Promoter
Next, we searched for potential STM regulatory motifs by analyzing the promoters of genes up-regulated in the microarray data, as described previously (Schommer et al., 2008). We only found a potential candidate box when we analyzed genes induced at least 5-fold by STM-VP16, GTCACT (P = 0.06; Supplemental Table S5). Even though the enrichment of this site was not particularly high, it suggestively overlapped with the preferred binding site of STM, which has already been investigated in vitro and was found to be CTGTCA (Krusell et al., 1997; Smith et al., 2002; Viola and Gonzalez, 2006). These sequences share the minimal sequence recognized by KNOX homeodomains, a GTCA core (for review, see Hake et al., 2004).
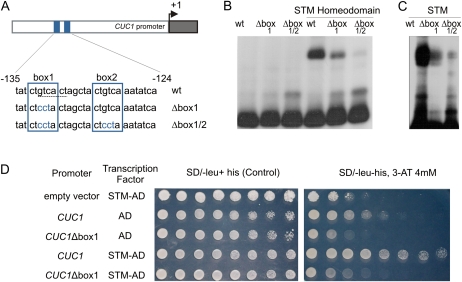
Interestingly, both 6-mer sequences are present in the CUC1 promoter in a narrow region at −135 (box1, CTGTCA and GTCACT) and −124 (box2, CTGTCA; Fig. 4A), which prompted us to perform a more detailed study. We tested whether a recombinant STM protein could recognize the CUC1 promoter in vitro. Electrophoretic mobility shift assays (EMSAs) showed a strong and specific interaction between a promoter fragment and the STM homeodomain alone or the complete recombinant transcription factor (Fig. 4, B and C). Binding was competed by a 50-fold molar excess of the same unlabeled fragment but not by a similar amount of a different fragment, thus showing the specificity of the interaction (Supplemental Fig. S2). Mutating box1 caused a significant decrease in the binding efficiency and a further mutation in box2 almost completely abolished the interaction between the CUC1 promoter and STM in vitro (Fig. 4, B and C).
Figure 4.
In vitro binding of STM to the CUC1 promoter. A, Scheme representing the CUC1 promoter. Putative STM-binding sites identified by SELEX are highlighted with blue squares and the sequence identified to be overrepresented in STM-VP16 induced genes (Supplemental Table S5) is indicated with a dashed line. B and C, EMSA with CUC1 promoter using recombinant STM homeodomain (B) or whole protein (C). D, One-hybrid experiment in yeast using wild-type and mutated CUC1 promoter. Growth in the absence of His due to activation of the HIS3 gene under the control of the CUC1 promoter was monitored using serial dilutions of the corresponding yeast strains. 3AT, 3-Amino-1,2,4-triazole. [See online article for color version of this figure.]
We also analyzed the interaction between STM and the CUC1 promoter in a yeast (Saccharomyces cerevisiae) one-hybrid assay. STM directed CUC1 expression also in this system (Fig. 4D), expression which was lost when the putative binding box1 was mutated. In summary, these results confirmed that STM directly regulates CUC1, likely through these two specific binding boxes, and provide a mechanistic scheme for the regulation of these transcription factors.
CUC1 Expression in Plants
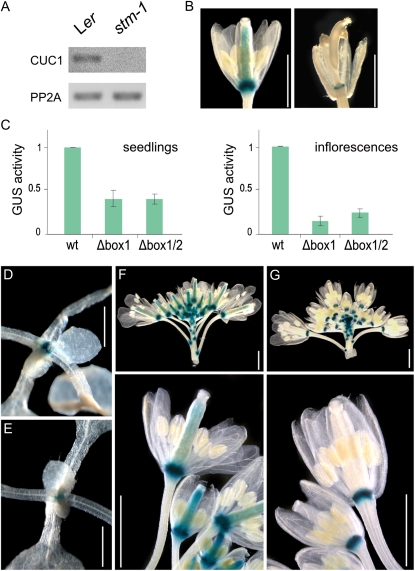
We then analyzed the expression of CUC1 in the strong stm-1 mutant. As expected, we found that the levels of CUC1 were reduced in this mutant (Fig. 5A). We also crossed the PromCUC1:GUS plants to the weak stm allele bum1-3 and found a reduction in the reporter expression in flowers (Fig. 5B). We tried to rescue the stm-1 mutant by overexpressing a miR164-resistant version of CUC1. However, the expression of CUC1 alone was not sufficient to complement the STM deficiency (not shown), as has been seen before when overexpressing a wild-type version of CUC1 in stm-1 mutants (Hibara et al., 2003).
Figure 5.
CUC1 expression in plants. A, CUC1 expression levels determined by RT-PCR in apices of wild-type (Ler) and stm-1 plants grown for 6 d in Murashige and Skoog media. The data shown is representative of at least three biological replicates. B, CUC1 reporter in a developing wild-type (left) or bum1-3 (right) flower. C, GUS activity of transgenic plants expressing wild-type and mutant CUC1 reporters in 12-d-old seedlings and inflorescences. The values correspond to the average of seven independent lines for each promoter version ± sem. D to G, GUS expression in seedlings (D and E) and inflorescences (F and G) of transgenic plants harboring the wild-type (D and F) and mutant (Δbox1/2; E and G) CUC1 reporter. Bars, 10 mm.
To study the role of the STM-binding sites on CUC1 transcription, we turned to reporters. Previously described transcriptional reporters for CUC1 and CUC2 are expressed in a broader domain inside the meristem while in situ hybridization assays have shown that CUC RNA accumulates in the boundaries (Nikovics et al., 2006; Sieber et al., 2007; Raman et al., 2008). This is at least partially achieved by the posttranscriptional repression carried out by miR164 (Sieber et al., 2007).
We analyzed the transcription of a wild-type reporter and two mutated versions where one or both STM-binding sites were removed. Mutations in the putative STM-binding sites quantitatively decreased its expression levels during vegetative development more than 2-fold by assaying seven independent transgenic lines for each construct (Fig. 5, C–E).
The expression of the mutated reporter was reduced 4-fold in inflorescences (Fig. 5C). Whole-mount stainings showed that the wild-type reporter was expressed at the base of the flowers and in the carpels (Fig. 5F), while the mutated version showed a significantly reduced staining in the carpels (Fig. 5G).
We then down-regulated CUC1 and other miR164 targets by expressing MIR164b from different promoters. Expression of MIR164b from the CUC1 promoter (PromCUC1:MIR164b) caused cotyledon fusions (20 out of 48 T1 plants; Fig. 6A) and most of them had severe stem-cauline leaf (Fig. 6B) and sepal fusions (Fig. 6C). In contrast, expression of MIR164b from the mutated CUC1 promoter did not cause any cotyledon fusions and the defects during reproductive development were weaker (Fig. 6, A–C). Additionally, expression of MIR164b from a STM promoter (PromSTM:MIR164b) also caused organ fusions (Fig. 6, D–F; see Supplemental Fig. S3 for PromSTM:GUS stainings).
Figure 6.
Misexpression of MIR164b using different promoters. A to C, Phenotypes of plants expressing MIR164b from a CUC1 and CUC1 Δbox1/2 promoter. D to F, Phenotypes of PromSTM:MIR164b transgenic plants. F, Wild-type (left) and PromSTM:MIR164b flowers. Bars, 10 mm (A, C, F) and 1 cm (B, D, E). Organ fusions are indicated with arrowheads.
These developmental defects are similar to some of the phenotypes observed in Prom35S:MIR164 plants (Laufs et al., 2004; Mallory et al., 2004), but still highlight the importance of the STM-binding sites on the quantitative regulation of CUC1 and the importance of CUC activity inside the STM domain. We also tried to complement the cuc1/cuc2 double mutant with PromCUC1:CUC1 and PromCUC1Δbox:CUC1 constructs. Unfortunately, the transgenes were silenced in the mutant background.
A Feedback Regulatory Loop Mediated by MIR164a
In many plant species with compound leaves, KNOXI transcription factors are expressed in the leaf primordia (Hareven et al., 1996; Bharathan et al., 2002; Hay and Tsiantis, 2006) where they interact with CUC genes (Blein et al., 2008). Therefore, we prepared Arabidopsis plants expressing STM from the LEAFY promoter (PromLFY:STM), which is active in the primordia of leaves and flowers (Blázquez and Weigel, 2000), and studied the effects on CUC activity.
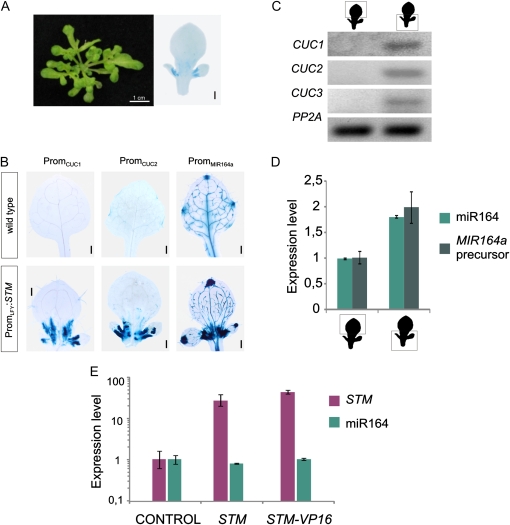
PromLFY:STM transgenics had lobed leaves as expected from ectopic expression of a KNOXI gene in leaf primordia (Fig. 7A). Analysis of CUC1 and CUC2 reporters in PromLFY:STM plants revealed that they were both ectopically expressed in young leaves, especially at the base of leaf lobes (Fig. 7B; Supplemental Fig. S4) resembling the ectopic pattern of expression of STM itself (Fig. 7A, right). These plants have constitutively altered levels of STM, so the induction of CUC2 is likely an indirect effect of STM as we have observed after 96 h of DEX treatment of STM-GR plants (Fig. 3B).
Figure 7.
CUC and MIR164 genes expression pattern in PromLFY:STM plants. A, Altered leaf shape of plants expressing STM under the control of the LFY promoter, PromLFY:STM. Right section: PromLFY:STM crossed to PromLFY:GUS, which indicates the domain of expression of STM in these transgenics. B, Expression of CUC1, CUC2, and MIR164a reporters in wild-type and PromLFY:STM plants. C, CUC1-3 expression levels determined by RT-PCR in the proximal and distal parts of leaves 3 and 4 of PromLFY:STM plants. The data shown is representative of at least three biological replicates. D, miR164 and MIR164a precursor expression levels determined by RT-qPCR in PromLFY:STM leaves dissected as in C. The data shown are mean of three biological replicates ± sem. E, Expression levels of miR164 determined by RT-qPCR after 12 h of induction of STM and STM-VP16 with ethanol. Expression levels were normalized as described in Figure 1. The data shown are mean of three biological replicates ± sem. Bars, 2 mm unless otherwise noted.
MIR164a has previously been implicated in the regulation of CUC activity during Arabidopsis leaf development (Nikovics et al., 2006; Hasson et al., 2010), so we crossed PromLFY:STM plants to a MIR164a reporter. We found that MIR164a was also activated by STM, in a similar way to the CUC reporters (Fig. 7B; Supplemental Fig. S4).
To validate these results we performed sections of wild-type and PromLFY:STM developing leaves. We then determined the levels of CUC genes and miR164 by RT-PCR in the proximal and distal region of the organ. The levels of CUC1-3 as well as miR164 were increased in the proximal part of the organ, as expected from the whole-mount staining (Fig. 7, C and D). We also determined the levels of the precursor of MIR164a and found that it was also activated, demonstrating its increased transcription is at least partially responsible for the elevated miR164 levels (Fig. 7D).
We then tested the short-term response of miR164 to STM levels. We measured miR164 12 h after the induction of STM (Fig. 7E). However, in this case we did not observe an obvious change in the levels of the miRNA.
These results suggest that MIR164a operates in a negative feedback loop to adjust the final CUC levels. The lack of change in miR164 levels when STM is transiently induced suggests that the activation of MIR164a is an indirect modification caused by STM and the consequence of the long-lasting expression of the KNOXI transcription factor in the leaf primordia.
We have also performed crosses between PromLFY:STM and cuc1-1 mutants and observed that the plants still have lobes (Supplemental Fig. S5), which is in good agreement with the ability of PromLFY:STM to activate CUC1, CUC2, and CUC3 in leaf primordia (Fig. 7C). These results also indicate that the activation of CUC2 and CUC3 by STM is independent of CUC1.
DISCUSSION
Targets of the KNOXI Transcription Factors
The class I of KNOX family of transcription factors comprises a small family of TALE homeobox genes that are widely distributed in plants. They regulate diverse developmental processes throughout the Arabidopsis life cycle. KNOXI transcription factors maintain the activity of the meristem, the boundaries between the stem and the meristem, and diverse aspects of flower development and leaf morphology (for review, see Hake et al., 2004; Hamant and Pautot, 2010; Hay and Tsiantis, 2010).
Despite their central role as developmental regulators, few downstream effectors of KNOXI activities are currently known. They regulate the levels of cytokinins and gibberelins (Sakamoto et al., 2001; Hay et al., 2002; Chen et al., 2004; Jasinski et al., 2005; Yanai et al., 2005) and the deposition of lignin (Mele et al., 2003). Still, precise mechanistic insights into KNOXI action are lacking in most cases. Here, we identified a direct target of STM in Arabidopsis. We show that STM directly activates the organ-boundary gene CUC1 and characterize the process at the biochemical level.
Genetic analyses revealed that KNOX genes conforms a complex network exhibiting both overlapping and antagonistic activities (Hamant and Pautot, 2010). To add further complexity to the network, a recently discovered KNOX protein lacking the DNA-binding domain competes for interacting factors (Magnani and Hake, 2008). Our results indicate that the induction of CUC1 is relatively specific to STM levels, although the closely related KNAT1 could also rapidly induce CUC1 when fused to the VP16 domain.
KNOXI transcription factors form complexes with BELL1-like proteins (for review, see Hake et al., 2004; Hay and Tsiantis, 2010). Our results cannot rule out that the activation of CUC1 by STM requires additional factors in vivo, or that the STM-binding sites are also recognized by other TALE transcription factors in certain tissues. Actually, the higher activation capacity of STM and KNAT1 on CUC1 levels conferred by the addition of VP16 might suggest that additional factors could be involved in plants.
It has been suggested that KNOXI transcription factors might exert their functions through the regulation of a few targets at least in certain situations (Hay and Tsiantis, 2010), as it has been shown for animal homeodomain transcription factors (Lovegrove et al., 2006). Genome-wide experiments will likely be required to understand the complex KNOX networks. The transcriptome analysis that we have performed here might also aid in the identification of other STM regulated genes.
The KNOXI-CUC Regulatory Network
It has been shown that both KNOXI and CUC systems reinforce each other, and that the ectopic expression of CUC1 induces STM (Aida et al., 2002; Hibara et al., 2003; Furutani et al., 2004; Kwon et al., 2006; Blein et al., 2008). The results obtained showing that STM directly induces CUC1, while indirectly activates CUC2-3 and MIR164a provide further insights into this regulatory network.
That STM can directly activate CUC1, while indirectly affecting CUC2-3 indicates that the CUC genes respond to different signals. Previous evidence has already shown that CUC genes can be regulated independently of each other by different factors. For instance, CUC1-2 are differentially affected by PIN1 (Aida et al., 2002), CUC1-2 are targets of miR164 but not CUC3 (Laufs et al., 2004; Mallory et al., 2004), and only CUC2 is regulated by SPLAYED (Kwon et al., 2006). The regulation of redundant factors by different pathways might confer robustness to a biological process, such as the formation of the meristem.
The activation of MIR164a by STM that we observed, which is likely indirect, might also contribute to fine tune the levels of CUC expression as a part of a negative feedback loop. A general homeostatic function has been already proposed for miR159 as its targets, the GA-MYB transcription factors, might activate the expression of the miRNA (Achard et al., 2004).
KNOXI and CUC genes are also versatile developmental regulators whose functions go beyond the establishment and maintenance of the meristem. They are recruited during carpel and ovule development in Arabidopsis (Ishida et al., 2000; Pautot et al., 2001; Scofield et al., 2007) and the formation of complex leaves in many species (Bharathan et al., 2002; Blein et al., 2008; Berger et al., 2009). Specific relationships between KNOXI, CUC, and MIR164 family members can be established during particular biological processes. The specific function of MIR164c in the regulation of petal number (Baker et al., 2005) and the role of CUC2 and MIR164a in the formation of leaf serrations (Nikovics et al., 2006) are in good agreement with this possibility.
CUC1 and CUC2 have diverged significantly within the Brassicaceae (Hasson et al., 2011). That CUC1, but not CUC2, responds directly to STM and has STM-binding sites in its promoter is also consistent with this data. The expression of STM also varies considerably in different species closely related to Arabidopsis. Many relatives express STM in the leaf primordia and have organs with more complex morphology (Piazza et al., 2010). Interestingly, the STM-binding sites are conserved in the CUC1 promoter in several Brassicaceae species (S. Spinelli and J. Palatnik, unpublished data). It might be interesting to determine whether the direct regulation of CUC1 by STM has a role in the formation of complex leaf morphologies within the Brassicaceae.
MATERIALS AND METHODS
Plant Material
Plants were grown in long days (16-h light/8-h dark) at 23°C. See Supplemental Table S6 for a list of transgenic lines and mutants. For Dex treatments, 2 week-old seedlings were transplanted to Murashige and Skoog plates containing 60 μm Dex or 60 μm Dex and 10 μm CYC. Control plates were treated in the same way without the addition of Dex or the translational inhibitor. Control plates treated only with CYC showed no significant differences with respect to untreated controls. CYC was also added before the DEX treatment as an additional control without any modification of the results. Seedlings were collected at different times after treatment for analysis.
Analytical Procedures
GUS stainings, microscopic observations, RNA extraction, and analysis by RT-qPCR was performed as described previously (Rodriguez et al., 2010). PROTEIN PHOSPHATASE2 was used as a control to normalize the data (Czechowski et al., 2005). In cases where no expression was detected in the reference sample after 35 cycles, results were shown as semiquantitative data (CUC1 was usually detected around cycles 26–29 with primers flanking the miRNA target site). GUS activity was assayed in protein extracts by a fluorescence method with 4-methylumbelliferyl glucuronide as a substrate (Jefferson et al., 1987). Mature miR164 levels were determined by stem-loop RT-qPCR (Chen et al., 2005). Primers are shown in Supplemental Table S7 and Supplemental Table S8 has a description of binary plasmids prepared for this study.
Microarray Analyses
Two-week-old seedlings were treated with 0.6% ethanol during 12 h. The shoot apex and the surrounding tissue was analyzed with Affymetrix ATH1 arrays (n = 2; E-MEXP-2550). Differentially expressed genes (Supplemental Tables S1 and S2) and overrepresented motifs (Supplemental Table S5) were identified as described before (Schommer et al., 2008).
DNA-Binding Assays
Proteins were obtained as fusions with glutathione S-transferase as described previously (Viola and Gonzalez, 2006, 2009). For EMSAs, purified recombinant proteins were incubated with 0.5 ng of labeled CUC1 promoter fragments (−177/−84 respective to the transcription start site). Binding reactions (20 μL) contained 20 mm HEPES (pH 7.5), 50 mm KCl, 2 mm MgCl2, 0.5 mm EDTA, 1.0 mm dithiothreitol, 0.5% Triton X-100, 22 ng/μL bovine serum albumin, 0.5 μg poly(dI-dC), and 10% glycerol. EMSAs were performed as described (Viola and Gonzalez, 2006). For the analysis of STM binding to the CUC1 promoter in yeast (Saccharomyces cerevisiae), the STM coding sequence was cloned in pGADT7 (Clontech) and introduced into yeast strains constructed using the pHIS3NX/pINT1 vector and carrying the CUC1 promoter inserted in the PDC6 locus in front of the HIS3 reporter gene preceded by its own minimal promoter.
Sequence data from this article can be found in the Array Express data libraries under accession number E-MEXP-2550.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression levels of genes analyzed using the STM-GR system by RT-qPCR.
Supplemental Figure S2. Binding of STM to the CUC1 promoter fragment in vitro.
Supplemental Figure S3. Expression pattern of the STM reporter.
Supplemental Figure S4. Expression pattern of CUC1, CUC2, and MIR164a reporters in different backgrounds.
Supplemental Figure S5. Phenotypes of PromLFY:STM crossed to cuc1-1 plants.
Supplemental Table S1. Genes modified by STM after 12 h of induction.
Supplemental Table S2. Genes modified by STM-VP16 after 12 h of induction.
Supplemental Table S3. Gene Ontology term enrichment for STM and STM-VP16 regulated genes.
Supplemental Table S4. Genes induced by both STM and STM-VP16.
Supplemental Table S5. Presence of putative STM-binding motifs in genes induced at least five times by STM-VP16.
Supplemental Table S6. List of previously described transgenic lines used in this article.
Supplemental Table S7. Relevant locus IDs and oligonucleotide primers used in RT-qPCR assays.
Supplemental Table S8. List of binary vectors prepared in this article.
Acknowledgments
We thank Detlef Weigel, in whose lab at the Max Planck Institute for Developmental Biology (Tübingen) some of these experiments were started; Jan Lohmann for access to comparative microarray data; Carla Schommer, Edgardo Bresso, Juan Debernardi, Jean-Luc Gallois, and Detlef Weigel for discussions; Patrick Laufs for CUC2 and MIR164a reporters; and Robert Sablowski for the Prom35S:STM-GR construct. S.S. and A.M. are Argentinian Research Council (CONICET) fellows, and J.P., I.L.V., and D.H.G. are members of CONICET.
References
- Achard P, Herr A, Baulcombe DC, Harberd NP. (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M. (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 [DOI] [PubMed] [Google Scholar]
- Aida M, Tasaka M. (2006) Genetic control of shoot organ boundaries. Curr Opin Plant Biol 9: 72–77 [DOI] [PubMed] [Google Scholar]
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M. (2002) Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129: 3965–3974 [DOI] [PubMed] [Google Scholar]
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM. (2005) The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15: 303–315 [DOI] [PubMed] [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. (2009) The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. (2002) Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Weigel D. (2000) Integration of floral inductive signals in Arabidopsis. Nature 404: 889–892 [DOI] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. (2008) A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 849–861 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ. (2004) The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J 38: 276–284 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M. (2004) PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131: 5021–5030 [DOI] [PubMed] [Google Scholar]
- Gallois JL, Woodward C, Reddy GV, Sablowski R. (2002) Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129: 3207–3217 [DOI] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hamant O, Pautot V. (2010) Plant development: a TALE story. C R Biol 333: 371–381 [DOI] [PubMed] [Google Scholar]
- Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. (1996) The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84: 735–744 [DOI] [PubMed] [Google Scholar]
- Hasson A, Blein T, Laufs P. (2010) Leaving the meristem behind: the genetic and molecular control of leaf patterning and morphogenesis. C R Biol 333: 350–360 [DOI] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. (2011) Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. (2006) The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M. (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K, Takada S, Tasaka M. (2003) CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J 36: 687–696 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Aida M, Takada S, Tasaka M. (2000) Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol 41: 60–67 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M. (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Rasmussen I, Gausing K. (1997) DNA binding sites recognised in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare). FEBS Lett 408: 25–29 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D. (2006) A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230 [DOI] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J. (2004) MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131: 4311–4322 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lemon WJ, Liyanarachchi S, You M. (2003) A high performance test of differential gene expression for oligonucleotide arrays. Genome Biol 4: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lovegrove B, Simões S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombría JC. (2006) Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol 16: 2206–2216 [DOI] [PubMed] [Google Scholar]
- Magnani E, Hake S. (2008) KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S. (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. (1998) A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Pautot V, Dockx J, Hamant O, Kronenberger J, Grandjean O, Jublot D, Traas J. (2001) KNAT2: evidence for a link between knotted-like genes and carpel development. Plant Cell 13: 1719–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza P, Bailey CD, Cartolano M, Krieger J, Cao J, Ossowski S, Schneeberger K, He F, de Meaux J, Hall N, et al. (2010) Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr Biol 20: 2223–2228 [DOI] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K. (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S, Dewitte W, Murray JA. (2007) The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J 50: 767–781 [DOI] [PubMed] [Google Scholar]
- Scofield S, Murray JA. (2006) KNOX gene function in plant stem cell niches. Plant Mol Biol 60: 929–946 [DOI] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Smith HM, Boschke I, Hake S. (2002) Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA 99: 9579–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M. (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Laux T. (2007) Connecting the paths in plant stem cell regulation. Trends Cell Biol 17: 403–410 [DOI] [PubMed] [Google Scholar]
- Viola IL, Gonzalez DH. (2006) Interaction of the BELL-like protein ATH1 with DNA: role of homeodomain residue 54 in specifying the different binding properties of BELL and KNOX proteins. Biol Chem 387: 31–40 [DOI] [PubMed] [Google Scholar]
- Viola IL, Gonzalez DH. (2009) Binding properties of the complex formed by the Arabidopsis TALE homeodomain proteins STM and BLH3 to DNA containing single and double target sites. Biochimie 91: 974–981 [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N. (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15: 1566–1571 [DOI] [PubMed] [Google Scholar]