Abstract
The maize (Zea mays) aleurone layer occupies the single outermost layer of the endosperm. The defective kernel1 (dek1) gene is a central regulator required for aleurone cell fate specification. dek1 mutants have pleiotropic phenotypes including lack of aleurone cells, aborted embryos, carotenoid deficiency, and a soft, floury endosperm deficient in zeins. Here we describe the thick aleurone1 (thk1) mutant that defines a novel negative function in the regulation of aleurone differentiation. Mutants possess multiple layers of aleurone cells as well as aborted embryos. Clonal sectors of thk1 mutant tissue in otherwise normal endosperm showed localized expression of the phenotype with sharp boundaries, indicating a localized cellular function for the gene. Sectors in leaves showed expanded epidermal cell morphology but the mutant epidermis generally remained in a single cell layer. Double mutant analysis indicated that the thk1 mutant is epistatic to dek1 for several aspects of the pleiotropic dek1 phenotype. dek1 mutant endosperm that was mosaic for thk1 mutant sectors showed localized patches of multilayered aleurone. Localized sectors were surrounded by halos of carotenoid pigments and double mutant kernels had restored zein profiles. In sum, loss of thk1 function restored the ability of dek1 mutant endosperm to accumulate carotenoids and zeins and to differentiate aleurone. Therefore the thk1 mutation defines a negative regulator that functions downstream of dek1 in the signaling system that controls aleurone specification and other aspects of endosperm development. The thk1 mutation was found to be caused by a deletion of approximately 2 megabases.
The importance of cereal grain is increasing due to demands for food, feed, energy, and other industrial applications. Different seed components contribute various biological functions as well as properties important for these diverse grain uses. The aleurone is important for mineral storage and the remobilization of storage compounds during germination. In Arabidopsis (Arabidopsis thaliana), the aleurone controls seed dormancy (Bethke et al., 2007). Aleurone is also the major source of amylase enzymes needed for the malting process and is thought to be responsible for many of the dietary benefits of cereal bran (Becraft and Yi, 2011).
Most cereal grains including maize (Zea mays) have a single cell layer of aleurone that forms at the periphery of the endosperm. Barley (Hordeum vulgare) and some varieties of rice (Oryza sativa) have multilayer aleurones (for review, see Becraft et al., 2001). In maize, the Coroico landrace was reported to have a multilayer aleurone (Wolf et al., 1972). In barley, aleurone layer number is under genetic control and is inherited as a quantitative trait (Jestin et al., 2008).
Aleurone cell fate specification and differentiation has been recently reviewed (Becraft and Yi, 2011). Plants such as cereals that undergo nuclear-type endosperm development show distinct cellular behaviors in the peripheral cell layer compared to internal cells from the onset of cellularization (Brown et al., 1994, 1996, 1999). The peripheral cells assume a typical plant cell division cycle, including the formation of a preprophase band of microtubules that predicts the plane of mitotic cell division. In contrast, the internal cells do not form preprophase bands. This demonstrates a differential response to cellular position at the earliest cellular stages of endosperm development. Nonetheless, aleurone cell fate remains plastic throughout development. The defective kernel1 (dek1) gene is required for aleurone cell identity, indicating it is required for the response of endosperm cells to peripheral position. In dek1 mutant kernels, the peripheral cell layer assumes starchy endosperm cell fate instead of aleurone (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Lid et al., 2002; Wisniewski and Rogowsky, 2004). Reversion of an unstable dek1 mutant late in development results in the formation of somatic sectors of aleurone cells in a dek1 mutant background lacking aleurone (Becraft and Asuncion-Crabb, 2000). These represent transdifferentiation of starchy endosperm cells to aleurone. Conversely, somatic loss of dek1 function late in development results in the transdifferentiation of aleurone cells to starchy endosperm. These results demonstrate that the positional cues that specify aleurone cell identity are present throughout endosperm development, that the cells retain the ability to respond to those cues throughout development, and that the cues are required to maintain as well as specify aleurone cell identity (Becraft and Asuncion-Crabb, 2000).
Mutant analyses suggest that aleurone cell differentiation is under a hierarchical genetic control (Becraft and Asuncion-Crabb, 2000; Wisniewski and Rogowsky, 2004). As described, the dek1 gene is one of the key components in this regulatory system (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Lid et al., 2002; Wisniewski and Rogowsky, 2004). The dek1 gene encodes a plasma membrane protein with 21 predicted transmembrane domains, an extracellular loop region, and a cytoplasmic domain containing a calpain protease (Lid et al., 2002; Wang et al., 2003). Mutants of crinkly4 (cr4) show similar phenotypes to dek1, suggesting they function in the same regulatory system (Becraft et al., 1996, 2002; Becraft and Asuncion-Crabb, 2000). CR4 is a receptor-like kinase (Becraft et al., 1996; Jin et al., 2000), and as such, both CR4 and DEK1 could potentially function as receptors for the positional cues that induce and maintain aleurone cell fate.
The supernumerary aleurone1 (sal1) mutant produces multiple aleurone layers, indicating it is a negative regulator of aleurone cell fate. The gene encodes a class E vacuolar sorting protein, suggesting it is involved in membrane vesicle trafficking (Shen et al., 2003). SAL1 is proposed to negatively regulate CR4 and/or DEK1 by directing their retrograde cycling from the plasma membrane and thereby dampening their levels of signaling. Thus, the sal1 mutant would have increased DEK1 or CR4 signaling, inducing extra layers of aleurone cells. SAL1, DEK1, and CR4 proteins all colocalize in endocytic vesicles, consistent with this hypothesis (Tian et al., 2007). As such, it appears that SAL1 functions upstream of DEK1 and CR4.
Here we report a novel multilayer aleurone mutant, thick aleurone1 (thk1). Genetic mosaic analysis indicates the gene functions locally to inhibit aleurone cell fate in subaleurone cells. Double mutant analysis demonstrates that the thk1 mutant is epistatic to dek1, suggesting that Thk1+ functions downstream of Dek1+ to regulate cell layer number. We propose a model where Dek1+ functions as a negative regulator of Thk1+.
RESULTS
The thk1 Mutant Causes Specification of Extra Aleurone Layers
In genetic backgrounds that confer anthocyanin pigmentation to the aleurone, recessive thk1 mutant kernels can be recognized on a segregating ear by their dark pigmentation (Fig. 1; Supplemental Fig. S1). The thk1 mutant is inherited as a recessive trait that shows complete penetrance and is fully transmissible through both male and female. Sectioning revealed that mutant kernels contain an abnormally thick aleurone layer, typically four to six cells thick, in contrast to the single cell layer of the wild type. The conclusion that these cells possess aleurone identity is based on their small cuboidal geometry, thick autofluorescent cell walls, accumulation of anthocyanin, lack of starch accumulation, and expression of a Vp1-GUS transgene, all markers of aleurone identity (Fig. 1; Supplemental Fig. S1).
Figure 1.
Analysis of the thk1 mutant phenotype. A, Mutant kernel (right) shows increased anthocyanin pigmentation and lack of a well-developed embryo. WT, Wild type. B and C, Longitudinal sections of wild-type (B) and thk1 (C) embryos at 12 DAP. D to G, Microscopic sections showing wild-type (D and F) and thk1 (E and G) aleurone. D and E, Histologically stained sections. Starch grains stained pink with PAS and protein-dense aleurone cells (arrow) stained intensely with toluidine blue. F and G, Expression of the Vp1-GUS transgene as determined by X-gluc histochemical stain. Scale bars = 100 μm.
The thk1 Gene Is Required for Embryo Development
Mutant kernels lack well-developed embryos and fail to germinate when sewn. At the macroscopic level, normal kernels have a large embryo that occupies most of the adaxial face of the kernel. The embryo is greatly reduced or absent in thk1 mutant kernels (Fig. 1, B and C; Supplemental Fig. S1). Microscopic examination showed that mutant embryos are sometimes able to initiate basic embryonic structures, including shoot and root apical meristems and a scutellum. Other times variable morphological abnormalities manifest. Development always lags behind the wild type and arrests shortly after the transition stage. A novel aspect of the thk1 mutant phenotype is the lack of a well-developed embryo cavity in the endosperm. embryoless (emb) mutants typically display a cavity in the endosperm at the site vacated by the aborted embryo (Clark and Sheridan, 1991). In thk1 mutant kernels, the cavity is much less pronounced.
The thk1 Locus Maps to Chromosome 1S
Recessive mutants can be mapped to chromosome arm using B-A translocations, which undergo nondisjunction in the second pollen mitosis (Beckett, 1978). The thk1 locus was localized to chromosome 1S when the mutant phenotype was revealed in segmental monosomic endosperms generated by nondisjunction of the B-A translocation, TB-1Sb. Bulk segregant single nucleotide polymorphism assays using Sequenom Mass Array (Liu et al., 2010) confirmed the chromosomal location and suggested the locus mapped toward the distal end.
Further mapping was conducted in an F2 population generated by crossing thk1/+, in a W22 inbred background, to the B73 inbred. A summary of markers and recombination frequencies is presented in Supplemental Table S1. Tight linkage was detected with markers in the subdistal region of 1S, however nine markers to the distalmost region did not amplify from the mutant, even though they amplified from W22, the background in which thk1 arose, and B73 (Supplemental Tables S1 and S2). Among the markers tested, the distalmost to amplify from mutant DNA was 363D20-3,4 located at position 2.09 megabase pairs from the chromosome end, according to B73 genome assembly version 2.0. We interpret this to mean that the thk1 mutant is likely caused by a deletion encompassing approximately 2 megabases. It is therefore formally possible that the phenotype is caused by loss of multiple genes.
The thk1 Gene Functions Locally to Regulate Aleurone Fate
Several potential mechanisms for the action of the thk1 gene in aleurone cell fate specification give different predictions for the behavior of mutant sectors in genetic mosaics. For example, it is possible that the thk1 mutant alters the concentration of a hormone or other morphogen that forms a gradient at the endosperm periphery. In such a case, then the gene would be expected to act cell nonautonomously and mutant sectors would show diffuse boundaries. On the other hand, if the thk1 gene were involved in the interpretation or response to positional information, gene action would more likely be cell autonomous and sector boundaries would be sharp.
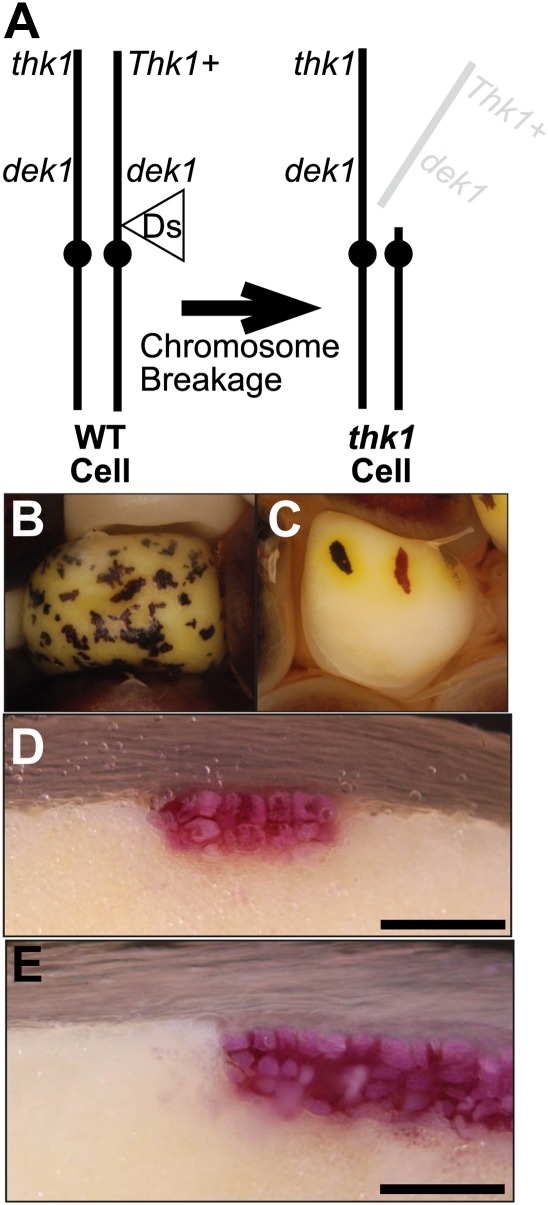
The Ds1S4 chromosome-breaking Ds transposable element was used to generate thk1 mutant sectors in a background of wild-type tissue (Neuffer, 1995; Becraft and Asuncion-Crabb, 2000). As shown in Figure 2, aberrant transposition of the Ds element leads to chromosome breakage followed by loss of the acentric chromosome fragment carrying the wild-type Thk1+ allele. Daughter cells from such an event do not inherit the wild-type allele and therefore form a clonal sector of hemizygous thk1 mutant cells. The Ds1S4 line carrying Ac at the P1 locus was crossed as a male to heterozygous thk1/+ mutant females. Among the F1 were kernels containing darkly pigmented sectors in an otherwise normally pigmented endosperm (Fig. 2; Table I). Sectioning of these dark sectors revealed that they displayed the multilayered aleurone phenotype typical of thk1 mutants.
Figure 2.
Genetically mosaic kernels with thk1 sectors. A, The chromosomal basis of thk1 sector generation. Endosperm cells were heterozygous for the recessive thk1 mutant and therefore phenotypically normal. The chromosome-breaking Ds1S4 element is located proximal to the wild-type (WT) Thk1+ allele. In the presence of an Ac element, aberrant Ds1S4 transposition results in chromosome breakage and loss of the distal chromosome 1S arm containing the Thk1+ allele, uncovering the thk1 mutant allele. Such events occur in individual cells occasionally during kernel development, and subsequent mitotic divisions generate clonal sectors of hemizygous thk1 mutant cells in a background of otherwise normal endosperm. B, Example of a kernel containing darkly pigmented thk1 mutant sectors (lower kernel, arrow) compared to a normal unsectored kernel (top). C and D, Kernel cross sections showing examples of thk1/− mutant sectors with adjacent normal endosperm. Sector boundaries appear sharp, consistent with a laterally cell-autonomous function. E, Several small sectors producing one or a few subperipheral aleurone cells (arrows). All are in direct contact with the peripheral layer of aleurone cells. Scale bars B to D = 200 μm; E = 50 μm.
Table I. Summary of genetic mosaic analysis.
WT, Wild type; K, kernels.
| Femalea | No. Ears | No. WT K | No. dek1 K | No. dek1 Sectored K | No. thk Sectored K | No. dek1 K with thk Sectors |
| thk1,dek1/+,+ | 10 | 1,684 | 947 | 386 | 930 | 115 |
| dek1/+ | 11 | 2,642 | 1,286 | 0 | 1,303 | 0 |
| thk1/+ | 8 | 2,656 | 0 | 0 | 772 | 0 |
| Wild type | 8 | 3,364 | 0 | 0 | 0 | 0 |
All males in these crosses were of the genotype Thk1+,dek1-1394/Thk1+,Dek1+,Ds1S4.
Sector borders were sharp as would be expected for cell-autonomous gene action. A stringent test of cell autonomy requires that sectors be marked with an independent marker. Sectors were generated that contained thk1 marked with the linked vp5 carotenoid-deficient mutant but unfortunately vp5 sectors were not discernable in the endosperm. Therefore, it can be concluded that the thk1 gene functions locally in a manner consistent with cell autonomy but because sector boundaries could not be assessed independent of the thk1 mutant phenotype, cell autonomy remains equivocal.
It was of interest to test whether the thk1 mutation could uncouple aleurone cell fate from surface position. Transpositions that occur late in development result in the formation of small sectors and the Ds1S4 line has been shown to generate single-celled sectors (Becraft and Asuncion-Crabb, 2000). Therefore, if thk1 uncoupled aleurone fate from surface position, it should be possible to detect internally isolated aleurone cells. To explore this, 60 kernels were sectioned and a total of 421 sectors examined. Late thk1 sectors were readily observed in most mosaic kernels but none were observed where there was not a continuity of aleurone cell contacts to the surface of the endosperm (Fig. 2; Supplemental Fig. S2). That is, no internal aleurone cells were observed where all their surrounding neighbors were starchy endosperm cells.
The thk1 Deletion Affects Leaf Epidermal Cell Size
These genetic stocks also afforded the opportunity to examine the phenotype of thk1 mutant cells in sporophyte tissues, which cannot otherwise be studied because thk1 mutants are embryo lethal. For these experiments, the thk1 mutant allele was linked in coupling to vp5, which confers cell-autonomous albino sectors in leaves due to carotenoid deficiency (Becraft et al., 2002). As a control, the vp5 mutant linked to the normal Thk1+ allele was used. These chromosomes were made heterozygous with the Ds1S4 chromosome breaker and leaves were examined for thk1 mutant sectors. In particular, we were interested in testing the hypothesis that the thk1 mutant might confer multiple leaf epidermal layers because many genes such as Extra Cell Layers (Xcl) similarly affect the aleurone and leaf epidermis (Kessler et al., 2002; Becraft and Yi, 2011). Ds1S4 leaves carrying the thk1 mutant allele showed prominent ridges that often caused the leaves to crease (Supplemental Fig. S3). These were not present in control vp5-sectored leaves, nor did they form in dek1 sectors (Becraft et al., 2002). Examination of these sectors revealed irregular epidermal cells that often appeared enlarged in section. In most instances, mutant epidermis was clearly a single cell layer, even when the sector covered an extensive area of the epidermis (e.g. Fig. 3B). Several instances were observed where internal cells displayed characteristics consistent with epidermal cell identities. Figure 3C shows an example where internal cells either showed features of sclerenchyma or bulliform-like cells. Sclerenchyma cells normally occur internally in localized foci associated with major and intermediate veins (Fig. 3A). In thk1 mutant sectors, internal sclerenchyma cells were observed interveinally. Other internal cells were observed that did not have characteristics of mesophyll or other normal internal cell types but resembled bulliform cells.
Figure 3.
Effect of the thk1 mutant on leaf cells. A, Control leaf with vp5 sector. The arrow designates a vascular bundle where the right half of the bundle sheath is devoid of well-developed chloroplasts due to the carotenoid deficiency conferred by the vp5 mutation. No morphological defects were observed. The brackets highlight an intermediate vascular bundle with foci of sclerenchyma cells (small, thick-walled cells, turquoise stained online) at the adaxial and abaxial poles. B, A thk1/− mutant sector covering an extensive area of upper leaf epidermis, including the entire area shown. The cells are enlarged but only occupy a single cell layer. C, Sector where internal cells show possible epidermal identities. The internal cells denoted with asterisks have attributes distinct from normal mesophyll or bundle sheath, but that resemble bulliform cells. Also, the lower (abaxial) epidermal cells resemble bulliform cells, which are normally restricted to the upper surface. Cells with clearly recognizable mesophyll or vascular bundle identities are outlined. Scale bars A and B = 100 μm; C = 50 μm. [See online article for color version of this figure.]
The thk1 Mutant Is Epistatic to dek1
Of the various mutants that disrupt aleurone cell fate specification, dek1 is the strongest and most consistent. The weak dek1-D allele causes a partial and somewhat variable loss of aleurone identity while the severe dek1-1394 allele causes a complete loss of aleurone (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002). To test the genetic relationship between the two genes, thk1/+ was crossed to dek1-D/+ and the resultant F1 plants were self pollinated to generate ears segregating F2 kernels. Six full ears were obtained that segregated both mutant (Table II, Experiment 1).
Table II. F2 segregation of dek1 and thk1 mutant kernel phenotypes.
| Phenotype | Observeda | Expectedb | Expectedb |
| Experiment 1 | (9:3:3:1) | (9:3:4) | |
| Wild type | 1,701 | 1,635 | 1,635 |
| dek1-D | 484 | 545 | 545 |
| thk1 | 714 | 545 | 727 |
| dek1-D; thk1 | (7)c | 182 | 0 |
| Experiment 2 | (9:3:3:1) | (9:3:4) | |
| Wild type | 793 | 840 | 840 |
| dek1-1394 | 290 | 280 | 280 |
| thk1 | 364 | 280 | 373 |
| dek1-1394; thk1 | (14)c | 93 | 0 |
| Experiment 3 | (3:1) | ||
| Wild type | 737 | 719 | |
| dek1-1394 | 221 | 239 |
Observed number of kernels for each phenotype.
Expected number of kernels for the given phenotypic segregation ratio (parentheses).
Kernels displayed aborted kernel phenotypes indistinguishable as to their thk1 or dek1 phenotypes.
A summary of the totals of each phenotypic class are presented in Table II. Kernels with dek1-D phenotypes were examined for the presence of thick aleurone but none was found. Ears contained three major phenotypic classes, wild type, dek1-D, and thk, with a few malformed and aborted kernels of questionable phenotype with respect to dek1 and thk. Notably, there was an excess of thk mutant kernels relative to dek1-D. A χ2 test for goodness of fit of the data to a 9:3:3:1 ratio, expected for independent assortment, produced a value of 229.89, indicating that this hypothesis should be rejected. When tested for fit to a 9:4:3 ratio, a χ2 value of 9.68 indicated that the data fit this hypothesis. The thk1 mutant was then crossed to dek1-1394 to test whether the same segregation ratios would hold for a strong dek1 allele (Table II, Experiment 2). Three ears were obtained that segregated both mutants, and again thk1 mutants were in excess relative to dek1 and the data fit a 9:4:3 ratio (χ2 = 3.31) but not 9:3:3:1 (χ2 = 95.74). Thus, segregation ratios suggested that the excess thk1 mutants were in fact thk1,dek1 double mutants and that thk1 is epistatic to dek1.
Two alternative explanations other than epistasis were considered to explain the data. The first was linkage; both thk1 and dek1 loci are on chromosome 1S. However, dek1 maps proximally and thk1 maps distally, and in a cross of thk1,dek1-1394/+,+ to B73, the mutants were transmitted to progeny independently, indicating that the loci are genetically unlinked. The second possibility considered was inefficient transmission of the dek1 mutant alleles. The cross that generated the ears segregating both thk1 and dek1-1394 also generated ears segregating for each mutant individually. As summarized in Table II, Experiment 3, ears segregating only dek1-1394 contained mutant kernels in a ratio that did not differ significantly from 3:1 (χ2 = 1.91), indicating the mutant allele was transmitted with full efficiency.
Finally, kernels with a thk1 mutant phenotype were selected from an F2 ear segregating for both the thk1 and dek1 mutants. These kernels were genotyped using a DNA marker, IDP112, located approximately 2 cM from the dek1 locus (Fig. 4A). Among 18 kernels genotyped, five were homozygous Dek1+, eight were heterozygous Dek1+/dek1-1394, and five were homozygous mutant dek1-1394 (barring recombination). Therefore, double homozygous mutants have a thk1 phenotype and thk1 is epistatic to dek1-D and dek1-1394.
Figure 4.
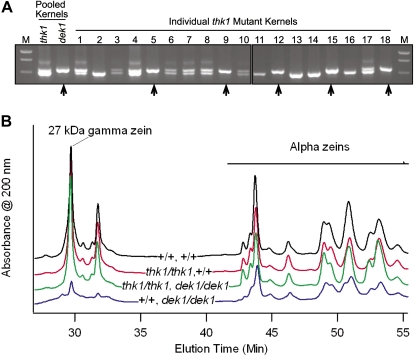
thk1 is epistatic for multiple pleiotropic dek1 phenotypes. A, Marker genotyping demonstrated the epistasis of thk1 over dek1. Kernels from an F2 ear segregating for the thk1 and dek1-1394 mutants were genotyped for the marker IDP112 (Fu et al., 2006). DNA from five pooled kernels with thk1 mutant phenotypes showed two polymorphic alleles. DNA from 12 pooled dek1 kernels only showed the upper band, indicating all were homozygous for the upper allele and that this marker allele is tightly linked to the dek1-1394 mutant allele. Among 18 individual kernels with thk1 mutant phenotypes, five were homozygous for the marker allele linked to dek1-1394 (arrows), indicating that these individuals were likely homozygous for the dek1 mutation. M, Molecular weight marker. B, Zein profiles were examined by HPLC analysis. The dek1 single mutant (bottom trace) is deficient in zein proteins compared to the wild type (top trace), particularly for the γ zein peaks. Both the thk1 single mutant and thk1,dek1 double traces (second and third from the top, respectively) are similar to the wild type. [See online article for color version of this figure.]
The Epistatic Action of thk1 Revealed in Genetic Mosaics
To further test the epistatic relationship of thk1 to dek1, we generated genetic mosaics that contained thk1 mutant sectors in a dek1-1394 background (Fig. 5; Supplemental Fig. S4). More precisely, thk1,dek1-1394/−,− hemizygous sectors were generated in a dek1-1394/dek1-1394 background. The epistatic action of thk1 predicted that such mosaics should contain sectors of thick aleurone on kernels otherwise devoid of aleurone. The strategy for generating these mosaics is shown in Figure 5A and the cross used to generate the appropriate genotypes is described in Supplemental Figure S4. Data are summarized in Table I.
Figure 5.
Genetic mosaics with thk1 sectors in dek1 mutant kernels. A, The chromosomal basis of these mosaics. Kernels were homozygous for the recessive dek1-1394 mutant allele and heterozygous thk1/Thk1+. Breakage and loss of the chromosome arm carrying the Thk1+ allele resulted in thk1,dek1/−,− double mutant sectors in a background of dek1 single mutant endosperm cells. WT, Wild type. B, An example of a dek1 mutant kernel with thk1 mutant sectors. The purple sectors are thk1,dek1/−,− double mutant cells. C, Yellow halos surrounding isolated thk1 sectors in a dek1 mutant kernel suggest carotenoids are produced by the thk1,dek1/−,− double mutant cells, while the surrounding dek1 single mutant tissue is carotenoid deficient. D and E, Sectioned kernels showing the multiple aleurone phenotype of thk1 sectors even though cells are also mutant for dek1. The red pigment is anthocyanin. Scale bars = 200 μm.
Figure 5 shows examples of clonal sectors of thk1,dek1-1394/−,− double mutant cells in otherwise dek1-1394 single mutant kernels. Whereas dek1-1394 kernels are devoid of aleurone cells, the loss of Thk1+ function restored the ability of dek1 mutant endosperm cells to form aleurone, and conferred the multilayered aleurone phenotype of thk1 mutants.
In addition to the sector types described above, kernels were frequently observed that contained separate dek1 and thk1 sectors (Supplemental Fig. S4). Such kernels most likely result from interstitial chromosome deletions or rearrangements as has been described for chromosome-breaking double Ds elements (English et al., 1995). Because the exact chromosomal position of the Ds1S4 is not known, the experiment was repeated with two well-characterized fAc derivatives of P1-vv. P1-wwB54 has been shown to cause various chromosome rearrangements, including interstitial deletions and rearrangements, while P1-vv9D9A generates primarily distal deletions (Zhang and Peterson, 2004; Yu et al., 2011). Both are located at the P1 locus, 1 cM proximal to dek1. Crosses with P1-wwB54 generated the same array of kernel types as Ds1S4 (data not shown). In contrast, p1-vv9D9A generated all the expected sector types but very few kernels with mixed thk1 and dek1 sectors. Among five ears from crosses to dek1-1394/+ females, 320 dek1 mutant kernels were observed and none contained any aleurone cells. From 18 crosses to thk1,dek1-1394/+,+ females, 14 kernels were recovered that had thk1 phenotypic sectors in dek1 mutant kernels. Thus, for all three chromosome breakers, crosses to thk1,dek1-1394/+,+ females produced dek1 mutant kernels with thk1 sectors, while when crossed to dek1-1394/+ alone, no instance of a pigmented sector or aleurone cell was observed among 1,586 dek1 mutant kernels examined, validating the interpretation of sector types.
thk1 Epistasis Is Pleiotropic
Mutant dek1 kernels show a variety of phenotypic defects in addition to the lack of aleurone. These include a deficiency of carotenoid pigments as well as opaque endosperm with a soft floury texture. Mosaic kernels with isolated thk1 sectors in a dek1 mutant background showed yellow halos of carotenoid pigments surrounding the thk1 sectors (Fig. 5C), suggesting that thk1 is epistatic to dek1 for this trait as well.
Floury endosperm can result from a deficiency in zein storage protein accumulation. The double mutant kernels genotyped above did not show a floury texture. To test the hypotheses that the floury texture of dek1 endosperm reflected a zein deficiency and that the thk1 mutant suppressed this deficiency in double mutants, zein profiles were analyzed by HPLC (Fig. 4). Compared to the wild type, dek1 mutants showed dramatic reductions in γ zeins including the 27-kD γ zeins. In contrast, the thk1 mutant endosperms showed zein profiles very similar to the wild type. The dek1,thk1 double mutant endosperms also showed zein profiles remarkably similar to the wild type. Thus, the thk1 mutant is epistatic to dek1 for the regulation of aleurone formation, carotenoid accumulation, and γ zein accumulation.
DISCUSSION
We described a new recessive loss-of-function mutant that causes an increased number of aleurone layers in maize endosperm. In addition the mutant results in embryo lethality. Other than the effect on the aleurone layer, the overall morphology of the endosperm is generally normal. One aspect of the thk1 mutant kernels that is novel compared to most emb mutants is that they do not form a prominent cavity in the region of the endosperm vacated by the aborted embryo (Clark and Sheridan, 1991).
The thk1 mutant is likely caused by a deletion encompassing approximately 2 megabases. It is therefore formally possible that the phenotype is caused by loss of multiple genes. Regardless of whether the thk1 mutant phenotype results from loss of one or multiple genes, the analysis presented here clearly demonstrates a function in the aleurone signaling system encoded by the distal region of chromosome 1S.
The thk1 Gene Functions Locally in the Cellular Response to Position
The thk1 gene functions locally within the endosperm as evident by the distinct sector boundaries. Similarly clear boundary delineations were observed both for thk1 mutant sectors in fields of normal endosperm cells and for double mutant sectors in fields of dek1 mutant cells. Because mutant sectors were not marked with an independent cell-autonomous marker, it cannot be definitely concluded that thk1 function is cell autonomous but this is likely the case.
The dek1 gene produces similarly sharp sector boundaries and has been proposed to function in the cellular response to positional cues (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Lid et al., 2002; Tian et al., 2007). The results reported here strengthen that model. The sharp boundaries of thk1,dek1 double mutant sectors in dek1 mutant endosperm indicate that the epistatic relationship of thk1 to dek1 is likely to function at the cell-autonomous level. As such, the signaling system defined by these two genes would appear to function in the response of endosperm cells to position rather than in the generation of those positional cues.
Aleurone forms on the surface of endosperm that is isolated from contacts with surrounding kernel tissues either by in vitro culture or as a consequence of a mutant phenotype (Olsen, 2004; Gruis et al., 2006; Reyes et al., 2010). These observations led to the proposal of the surface rule that posits that the source of the positional information that specifies aleurone identity is intrinsic to the endosperm. The Ds1S4 line generates late-occurring transposition events that produce sectors as small as a single cell (Becraft and Asuncion-Crabb, 2000). Late thk1 sectors were readily observed but interestingly, none were observed where there was not a continuity of aleurone cell contacts to the surface layer of the endosperm (Fig. 2; Supplemental Fig. S2). That is, no internal aleurone cells were observed that were completely surrounded on all sides by starchy endosperm cell neighbors. In contrast, isolated single cells at the surface can be completely surrounded by starchy endosperm and still assume aleurone identity (Supplemental Fig. S2; Becraft and Asuncion-Crabb, 2000). So while the thk1 mutant produces aleurone cells in subperipheral cell layers, there still appears to be adherence to the surface rule. It may be that determinants of aleurone fate are transmitted from the surface only via aleurone cells. This transmission could potentially be through plasmodesmatal connections, which are distinct between aleurone cells compared to other endosperm cell types (Tian et al., 2007), via lineage, or by an aleurone-specific signaling mechanism. These results imply that Thk1+ function is normally required for the transdifferentiation event that occurs in the internal daughters of periclinal aleurone cell divisions. Such internal cells normally redifferentiate to starchy endosperm cells (Becraft and Asuncion-Crabb, 2000) and this might be the primary site of action for Thk1+. The formation of small isolated thk1 sectors in dek1 kernels indicates there is no requirement for aleurone continuity in the lateral dimension (Supplemental Fig. S2).
Mutant sectors in leaves produced enlarged epidermal cells that resembled bulliform cells in morphology and histological staining characteristics. In most sectors observed, these cells occupied a single epidermal layer. In cr4 mutants, cells with unmistakable epidermal identities, such as epidermal hair cells, were observed internally (Jin et al., 2000). A few examples were observed in thk1 sectors where internal cells had characteristics consistent with epidermal identity, but not unequivocal. Thus, it seems possible that thk1 may function to regulate leaf epidermal cell layer number, but this remains tenuous. It is clear that a thk1 deficiency in epidermal cells does not directly cause them to form a multilayered epidermis. Furthermore, because multiple genes are deficient due to the deletion, it is not possible to formally conclude that the gene(s) regulating leaf epidermal cells is the same as that regulating aleurone development, although this seems likely given the similar functions of other genes in aleurone and leaf epidermis (Kessler et al., 2002; Becraft and Yi, 2011).
The Regulation of Aleurone Layer Number
The genetic regulation of aleurone cell layer number does not appear to be simple. Typical barley cultivars contain three to four aleurone layers, with layer number inherited as a quantitative trait (Jestin et al., 2008). The recessive barley des5 mutant possesses a single aleurone layer, indicating this gene normally functions to increase layer number, opposite Thk1+ (Olsen et al., 2008). The multiple aleurone layer trait present in Coroico maize landraces is inherited as a partially dominant, but complex, trait (Wolf et al., 1972; P. Becraft, unpublished data). Xcl is another dominant maize mutant that produces double aleurone cell layers (Kessler et al., 2002). Transgenic knockdown of two rice transcription factors also affected aleurone layer number. RISBZ1 and RPBF are bZIP and DOF zinc finger transcription factors, respectively, that function together to regulate endosperm storage protein gene expression (Yamamoto et al., 2006). Knockdown of either factor singly had mild or no effects on the aleurone but the double knockdown of both genes produced a multilayered, disordered aleurone (Kawakatsu et al., 2009).
Maize, sal1 mutants also produce extra layers of aleurone cells. The sal1 gene maps to chromosome 9 and encodes a class E vacuolar sorting protein. It appears to act upstream as a negative regulator of dek1 (Shen et al., 2003; Tian et al., 2007). In contrast, the epistatic relationship of thk1 over dek1 suggests that thk1 functions downstream of dek1. dek1 encodes a plasma membrane localized protein with a predicted extracellular domain and a cytoplasmic calpain protease domain (Lid et al., 2002; Wang et al., 2003). It functions in the signaling system that specifies aleurone fate (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Lid et al., 2002; Tian et al., 2007). As such, the thk1 gene product represents a likely target of regulation by DEK1, potentially a direct target.
The calpain protease function of DEK1 suggests that targets of regulation would likely be proteolytic substrates, which could provide a clue to the identity of the thk1 gene. However, substrate recognition by calpains is complex and involves three-dimensional structures that are not easily recognized in primary sequences (for review, see Goll et al., 2003; Croall and Ersfeld, 2007). Known calpain substrates in mammalian systems include many membrane proteins including receptors, but also diverse other proteins such as cytoskeletal proteins and transcription factors. Therefore, it is not feasible to predict a candidate thk1 gene from within the deleted region based on potential calpain substrates.
A Model for the Regulation of Aleurone Cell Fate
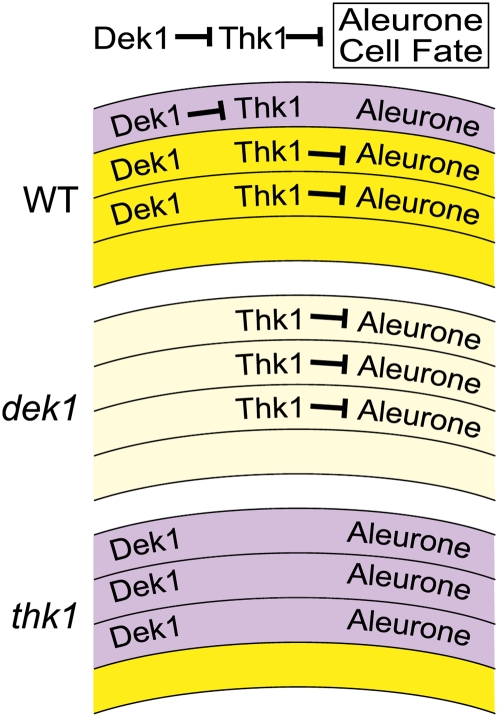
The loss-of-function thk1 mutant results in the specification of extra layers of aleurone cells, indicating that the normal gene function is to negatively regulate aleurone fate in the subperipheral layers. The function of the normal dek1 gene is to promote aleurone identity in the peripheral cell layer. The epistatic relationship of thk1 over dek1 suggests that the thk1 gene functions downstream of dek1 in the specification of aleurone fate (and in the control of other endosperm traits). As such, a model where Dek1+ promotes aleurone identity by negatively regulating Thk1+ accounts for all the available data. As shown in Figure 6, Dek1+ could normally function in the peripheral layer to inhibit Thk1+ function and thereby permit aleurone formation. In dek1 mutants, Thk1+ function would be derepressed in the peripheral cell layer and block aleurone differentiation. In thk1 mutants, there would be no repressive function regardless of whether dek1 were functional. As discussed below, additional factors must be involved that limit aleurone fate to the peripheral regions of the endosperm.
Figure 6.
Model for thk1 function in dek1 regulation of aleurone cell fate. Thk1+ functions as a negative regulator of aleurone identity. In the outer layer, Dek1+ inhibits Thk+ function to derepress aleurone fate. In internal layers, Thk+ is not inhibited and therefore represses aleurone fate. In dek1 mutants, Thk+ function would not be inhibited and aleurone fate would be repressed in all cell layers. In thk1 mutants, there is no repression of aleurone identity in any of the cell layers, permitting the formation of aleurone, independent of Dek1+ function. WT, Wild type.
Pleiotropic Epistasis
DEK1 function is not restricted to the regulation of aleurone cell fate as is evident from the pleiotropy of the dek1 mutant phenotype. Mutants are carotenoid deficient, have floury, opaque endosperms, and are embryo lethal. The opaque endosperm phenotype likely relates to a zein storage protein deficiency (Fig. 4). Carotenoid and zein content are important grain quality traits. Interestingly, thk1 mutants appear to rescue these aspects of the dek1 mutant phenotype, indicating that Thk1+ has functions in addition to aleurone regulation.
That dek1 and thk1 appear to function in a regulatory pathway that controls zein and carotenoid accumulation in internal starchy endosperm cells indicates that the function of this pathway to regulate aleurone specification is context dependent and only occurs in the peripheral few cell layers of the endosperm. It is therefore evident that additional factors are ultimately involved in patterning the endosperm. One possible source of positional information could be auxin, which was recently reported to be at highest concentrations in the periphery of maize endosperm and to possibly influence aleurone fate specification (Forestan et al., 2010).
The relationship between protein content and aleurone cell layer number is not currently understood but several previous reports have hinted at it. The Coroico landrace with the multiple aleurone layer trait is also high in protein content (Wolf et al., 1972). While it was recently shown that zeins are expressed in aleurone cells (Reyes et al., 2011), such a minor fraction of the endosperm is unlikely to have a major impact on the overall endosperm protein content, and it was concluded that the aleurone layer and protein content traits of Coroico were likely controlled separately (Nelson and Chang, 1974). As mentioned, the rice RISBZ1 and RPBF transcription factors function to promote prolamine storage protein gene expression in the endosperm as well as restrict aleurone layer number (Yamamoto et al., 2006; Kawakatsu et al., 2009). This relationship is different from thk1 that appears to negatively regulate both aleurone layers and zein accumulation.
Signaling has previously been implicated in the regulation of zein accumulation. Protein levels and activity, as well as transcript levels of maize OPAQUE2, a bZIP transcription factor that promotes α-zein gene expression, are regulated diurnally (Ciceri et al., 1997, 1999). The protein undergoes cyclic phosphorylation and dephosphorylation, with unphosphoryated forms having higher DNA-binding activity. Highest levels are present during the daytime phase of the diurnal cycles, but phosphorylation ratios do not appear to change during development. Genetic and cell biological evidence suggest that DEK1 functions in the same regulatory system as the CR4 receptor kinase to regulate aleurone cell fate as well as other aspects of development (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Tian et al., 2007). Mutants of cr4 show an opaque kernel phenotype, similar to dek1, consistent with the hypothesis that both genes may also function together to regulate storage proteins (Jin et al., 2000). This study now places thk1 in that system. It is not yet evident why the same signaling system responsible for aleurone cell fate specification would also be used to regulate zein accumulation. This will become clear when the regulatory networks of endosperm development are more completely understood.
MATERIALS AND METHODS
Genetic Stocks
The thk1 mutant arose in a Mutator transposon line in a maize (Zea mays) W22 inbred background and was a kind gift from Donald McCarty, University of Florida. The dek1-1394 and vp5 mutants and the Ds1S4 line were obtained from the Maize Genetics Cooperation Stock Center. The dek1-D mutant was a gift from Hugo Dooner, Waksman Institute. The P1-WWB54 and p1-vv9D9A lines were gifts from Thomas Peterson, Iowa State University. The thk1 and dek1 mutants were backcrossed five generations into a B73 inbred background.
Microscopy and Histology
For Figure 1, B and C, developing kernels were dissected from young ears, fixed in formaldehyde, alcohol, and acetic acid, and embedded in Paraplast Plus according to standard procedures (Berlyn and Miksche, 1976). Ten-micrometer sections were affixed to glass slides, deparaffinized, stained with fast green, and mounted. For Figure 1, D and E, 28 days after pollination (DAP) kernels were fixed in 2% paraformaldehyde and 3% gluteraldehyde buffered in 0.1 m cacodylate (pH 7.2) and embedded in LR White resin. Samples were sectioned to 1 μm on a Leica EM UC6 ultramicrotome and stained with periodic acid Schiff’s (PAS) and toluidine blue. Leaf sections (Fig. 3) were prepared the same way except no PAS stain was applied. For Figure 1, F and G, 24 DAP kernels were hand sectioned and incubated in GUS staining solution (50 mm sodium phosphate [pH 7.0], 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 10 mm EDTA, 0.05% [v/v] Triton X-100, 0.35 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) for 24 h at 37°C for several hours until blue precipitate was apparent (Jefferson et al., 1987). For Figure 2E, hand-sectioned mature kernels were examined under epifluorescence using a narrow violet filter (excitation 400–410 nm, dichroic mirror and barrier filter, 455 nm). Mosaic kernel sectors were examined using epi-illumination of hand sections. All micrography was performed with an Olympus BX-60 microscope equipped with a Jenoptik C-5 camera.
Genetic Mapping and Genotyping
Genomic DNA was extracted from mature dried kernels with modifications to a described protocol (Edwards et al., 1991). Briefly, kernels were cut with a razor blade and a half kernel was extracted for mutants and a quarter kernel for the wild type (approximately 100 mg). Kernels were soaked in 200 μL extraction buffer for 3 to 12 h at room temperature. Samples were then ground with a plastic pestle and a hand-held drill. Additional buffer was added to a total volume of 500 μL, followed by the addition of 400 μL chloroform. Samples were vortexed and then centrifuged 10 min at 13,000 rpm with a tabletop centrifuge. After transferring 250 μL of supernatant to a new tube, DNA was precipitated with an equal volume of isopropyl alcohol. Samples were centrifuged and the pellet was washed with 70% ethanol and air dried. The pellet was then dissolved in 500 μL distilled water. One microliter (20–50 ng genomic DNA) was used for PCR. All PCRs were performed with GoTag green mastermix (Promega) in 10 μL reactions containing 5 pmol of each primer. Cycling conditions were 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. After an additional 10 min at 72°C, reactions were kept at 4°C until visualized by agarose gel electrophoresis.
HPLC Analysis of Zein Proteins
Alcohol-soluble proteins were extracted from 25 mg of flour using 1,000 μL extraction buffer consisting of 70% ethanol, 61 mm NaOAc, and 5% β-mercaptoethanol. The mixture was vortexed briefly, shaken for 1 h at room temperature, and then centrifuged for 10 min at 13,000 rpm. An aliquot of 25 μL of each extract was injected into a C18 protein and peptide column heated to 55°C in a Waters 2695 separation module, and A200 was measured with a Waters 2487 dual absorbance detector. Separation of distinct proteins based on hydrophobicity was achieved with a gradient of ultrapure water and acetonitrile, both containing 0.01% trifluoroacetic acid. The complex gradient ranged from 20% to 60% acetonitrile for a total of 75 min excluding equilibration steps before and after elution. Gradient slopes were optimized for separation of peaks. A flow rate of 1 mL/min was used.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Early-stage embryo development in wild type versus thk1.
Supplemental Figure S2. Single-celled thk1 mutant sectors in kernels.
Supplemental Figure S3. Mutant thk1 sectors in leaves.
Supplemental Figure S4. Crossing scheme for generating thk1 genetic mosaic kernels.
Supplemental Table S1. Genetic mapping of the thk1 mutation
Supplemental Table S2. Genetic markers absent in the thk1 deletion.
Acknowledgments
The authors thank Donald R. McCarty for the gift of the thk1 mutant and Thomas A. Peterson for the fAc lines. Amanda Ripley provided excellent technical assistance. The Iowa State University Microscopy and NanoImaging Facility assisted with some of the microscopic preparations. The Iowa State University Genomic Technologies Facility provided the mass array single nucleotide polymorphism genotyping service.
References
- Beckett JB. (1978) B-A translocations in maize. I. Use in locating genes by chromosome arms. J Hered 69: 27–36 [Google Scholar]
- Becraft PW, Asuncion-Crabb YT. (2000) Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127: 4039–4048 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Brown RC, Lemmon BE, Opsahl-Ferstad HG, Olsen O-A. (2001) Endosperm development. Bhojwani SS, , Current Trends in the Embryology of Angiosperms. Kluwer, Dordrecht, The Netherlands, pp 353–374 [Google Scholar]
- Becraft PW, Li K, Dey N, Asuncion-Crabb YT. (2002) The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR. (1996) CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273: 1406–1409 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Yi G. (2011) Regulation of aleurone development in cereal grains. J Exp Bot 62: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Berlyn GP, Miksche JP. (1976) Botanical Microtechnique and Cytochemistry. Iowa State University Press, Ames, IA [Google Scholar]
- Bethke PC, Libourel IGL, Aoyama N, Chung Y-Y, Still DW, Jones RL. (2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143: 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen O-A. (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12: 32–42 [Google Scholar]
- Brown RC, Lemmon BE, Olsen O-A. (1994) Endosperm development in barley: microtubule involvement in the morphogenetic pathway. Plant Cell 6: 1241–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Olsen OA. (1996) Development of the endosperm in rice (Oryza sativa L.): cellularization. J Plant Res 109: 301–313 [Google Scholar]
- Ciceri P, Gianazza E, Lazzari B, Lippoli G, Genga A, Hoscheck G, Schmidt RJ, Viotti A. (1997) Phosphorylation of Opaque2 changes diurnally and impacts its DNA binding activity. Plant Cell 9: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciceri P, Locatelli F, Genga A, Viotti A, Schmidt RJ. (1999) The activity of the maize Opaque2 transcriptional activator is regulated diurnally. Plant Physiol 121: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JK, Sheridan WF. (1991) Isolation and characterization of 51 embryo-specific mutations of maize. Plant Cell 3: 935–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croall DE, Ersfeld K. (2007) The calpains: modular designs and functional diversity. Genome Biol 8: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JJ, Harrison K, Jones J. (1995) Aberrant transpositions of maize double Ds-like elements usually involve Ds ends on sister chromatids. Plant Cell 7: 1235–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S. (2010) ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol 152: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wen T-J, Ronin YI, Chen HD, Guo L, Mester DI, Yang Y, Lee M, Korol AB, Ashlock DA, et al. (2006) Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics 174: 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll DE, Thompson VF, Li H, Wei W, Cong J. (2003) The calpain system. Physiol Rev 83: 731–801 [DOI] [PubMed] [Google Scholar]
- Gruis DF, Guo H, Selinger D, Tian Q, Olsen O-A. (2006) Surface position, not signaling from surrounding maternal tissues, specifies aleurone epidermal cell fate in maize. Plant Physiol 141: 898–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jestin L, Ravel C, Auroy S, Laubin B, Perretant M-R, Pont C, Charmet G. (2008) Inheritance of the number and thickness of cell layers in barley aleurone tissue (Hordeum vulgare L.): an approach using F2-F3 progeny. Theor Appl Genet 116: 991–1002 [DOI] [PubMed] [Google Scholar]
- Jin P, Guo T, Becraft PW. (2000) The maize CR4 receptor-like kinase mediates a growth factor-like differentiation response. Genesis 27: 104–116 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Touno SM, Yasuda H, Takaiwa F. (2009) Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J 59: 908–920 [DOI] [PubMed] [Google Scholar]
- Kessler S, Seiki S, Sinha N. (2002) Xcl1 causes delayed oblique periclinal cell divisions in developing maize leaves, leading to cellular differentiation by lineage instead of position. Development 129: 1859–1869 [DOI] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen O-A. (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci USA 99: 5460–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen HD, Makarevitch I, Shirmer R, Emrich SJ, Dietrich CR, Barbazuk WB, Springer NM, Schnable PS. (2010) High-throughput genetic mapping of mutants via quantitative single nucleotide polymorphism typing. Genetics 184: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson OE, Chang MT. (1974) Effect of multiple aleurone layers on the protein and amino acid content of maize endosperm. Crop Sci 14: 374–376 [Google Scholar]
- Neuffer MG. (1995) Chromosome breaking sites for genetic analysis in maize. Maydica 40: 99–116 [Google Scholar]
- Olsen LT, Divon HH, Al R, Fosnes K, Lid SE, Opsahl-Sorteberg HG. (2008) The defective seed5 (des5) mutant: effects on barley seed development and HvDek1, HvCr4, and HvSal1 gene regulation. J Exp Bot 59: 3753–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O-A. (2004) Dynamics of maize aleurone cell formation: the “surface” rule. Maydica 49: 37–40 [Google Scholar]
- Reyes FC, Chung T, Holding D, Jung R, Vierstra R, Otegui MS. (2011) Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell 23: 769–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FC, Sun B, Guo H, Gruis DF, Otegui MS. (2010) Agrobacterium tumefaciens-mediated transformation of maize endosperm as a tool to study endosperm cell biology. Plant Physiol 153: 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Li C, Min Z, Meeley RB, Tarczynski MC, Olsen OA. (2003) sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc Natl Acad Sci USA 100: 6552–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Olsen L, Sun B, Lid SE, Brown RC, Lemmon BE, Fosnes K, Gruis DF, Opsahl-Sorteberg HG, Otegui MS, et al. (2007) Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. Plant Cell 19: 3127–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Barry JK, Min Z, Tordsen G, Rao AG, Olsen O-A. (2003) The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J Biol Chem 278: 34467–34474 [DOI] [PubMed] [Google Scholar]
- Wisniewski JP, Rogowsky PM. (2004) Vacuolar H+-translocating inorganic pyrophosphatase (Vpp1) marks partial aleurone cell fate in cereal endosperm development. Plant Mol Biol 56: 325–337 [DOI] [PubMed] [Google Scholar]
- Wolf MJ, Cutler HC, Zuber MS, Khoo U. (1972) Maize with multilayer aleurone of high protein content. Crop Sci 12: 440–442 [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F. (2006) Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol 141: 1694–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zhang J, Peterson T. (2011) Genome rearrangements in maize induced by alternative transposition of reversed ac/ds termini. Genetics 188: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peterson T. (2004) Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167: 1929–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]