Abstract
The cactus Opuntia ficus-indica is a constitutive Crassulacean acid metabolism (CAM) species. Current knowledge of CAM metabolism suggests that the enzyme phosphoenolpyruvate carboxylase kinase (PPCK) is circadian regulated at the transcriptional level, whereas phosphoenolpyruvate carboxylase (PEPC), malate dehydrogenase (MDH), NADP-malic enzyme (NADP-ME), and pyruvate phosphate dikinase (PPDK) are posttranslationally controlled. As little transcriptomic data are available from obligate CAM plants, we created an expressed sequence tag database derived from different organs and developmental stages. Sequences were assembled, compared with sequences in the National Center for Biotechnology Information nonredundant database for identification of putative orthologs, and mapped using Kyoto Encyclopedia of Genes and Genomes Orthology and Gene Ontology. We identified genes involved in circadian regulation and CAM metabolism for transcriptomic analysis in plants grown in long days. We identified stable reference genes for quantitative polymerase chain reaction and found that OfiSAND, like its counterpart in Arabidopsis (Arabidopsis thaliana), and OfiTUB are generally appropriate standards for use in the quantification of gene expression in O. ficus-indica. Three kinds of expression profiles were found: transcripts of OfiPPCK oscillated with a 24-h periodicity; transcripts of the light-active OfiNADP-ME and OfiPPDK genes adapted to 12-h cycles, while transcript accumulation patterns of OfiPEPC and OfiMDH were arrhythmic. Expression of the circadian clock gene OfiTOC1, similar to Arabidopsis, oscillated with a 24-h periodicity, peaking at night. Expression of OfiCCA1 and OfiPRR9, unlike in Arabidopsis, adapted best to a 12-h rhythm, suggesting that circadian clock gene interactions differ from those of Arabidopsis. Our results indicate that the evolution of CAM metabolism could be the result of modified circadian regulation at both the transcriptional and posttranscriptional levels.
Opuntia species are constitutive Crassulacean acid metabolism (CAM) plants, in which the capacity to induce CAM metabolism is developmentally regulated, with a progression from C3 to CAM metabolism occurring during cladode development. The CAM cycle is expressed under essentially all growing conditions (Winter et al., 2008). Opuntia can take up relatively large amounts of CO2 with respect to water loss by transpiration (4–10 mmol CO2 mol−1 water compared with 1–1.5 mmol in C3 plants), and the annual aboveground dry mass can be increased by 37% to 40% for Opuntia ficus-indica when the CO2 level is doubled (Cui et al., 1993; Nobel et al., 1994).
The CAM type of CO2 fixation is thought of as integrating into the day/night cycle in four phases. In phase I, nocturnal CO2 fixation and accumulation of malic enzyme (ME) occurs in the central vacuole. During phase II, a transition phase early in the light period, malic acid is remobilized from the vacuole and decarboxylated, generating high internal CO2 concentrations that lead to stomatal closure. Later, in phase III, the released CO2 is refixed by Rubisco and assimilated via the Calvin cycle of photosynthesis. Finally, in phase IV, the later light period, malic acid stores are exhausted, stomata may open, and CO2 may be taken up and assimilated directly via Rubisco (Rascher et al., 2001). Key enzymes of CAM metabolism include phosphoenolpyruvate carboxylase (PEPC), which catalyzes CO2 fixation into phosphoenolpyruvate (PEP) under the formation of oxaloacetate at night; phosphoenolpyruvate carboxylase kinase (PPCK), which controls the phosphorylation state of PEPC; malate dehydrogenase (MDH), required for the reduction of oxaloacetate to malate at night; NADP-ME, catalyzing the decarboxylation of malate during the day in the cytoplasm; and pyruvate phosphate dikinase (PPDK), which is only active in chloroplasts of the malic acid type of CAM species (Cushman and Bohnert, 1997; Scott and Mercer, 1997; Honda et al., 2000) and catalyzes the formation of PEP from pyruvate.
Many photosynthesis-related processes, including starch metabolism, are regulated by a circadian rhythm (Piechulla, 1988, Merida et al., 1999), and the circadian clock is related to gas exchange and CAM metabolism through the anticipation of cycles at dawn and dusk (Resco et al., 2009). CAM metabolism displays an endogenous rhythmicity, with circadian oscillations of CO2 uptake, stomatal conductance, and internal concentrations of CO2. There is empirical evidence that the circadian transcriptional regulation of PPCK, leading to a modulated sensitivity of PEPC to inhibition by malic acid (Nimmo et al., 1987; Hartwell et al., 1999; Taybi et al., 2000), plays a critical role in the control of CAM metabolism (Wyka et al., 2004). PPCK is the only CAM-related gene for which a circadian oscillation has been observed at the transcriptional level (Nimmo et al., 1987; Taybi et al., 2000). Phytochrome, acting as a photoreceptor, and the corresponding phytochrome-responsive transcription factors have been suggested to be involved in PPCK circadian regulation (Taybi et al., 2000). In general, circadian regulation of enzyme activities during the day/night cycle is mediated largely by posttranslational changes (Cushman et al., 2000) such as the reversible phosphorylation of PEPC by PPCK (Chollet et al., 1996; Vidal and Chollet, 1997; Nimmo, 2000), thus connecting PEPC activity to diurnal CAM regulation. PPDK activity is light/dark modulated by reversible phosphorylation in both C3 and C4 plants (Chastain et al., 2002), and chloroplastic NADP-MDH activity is light regulated via changing NADPH-to-NADP ratios and the thioredoxin redox state (Rebeille and Hatch, 1986; Li et al., 1994). The study of transcriptional patterns of key CAM metabolic enzymes for a plant of the Cactaceae family is novel and offers knowledge about enzyme regulation in CAM plants that can serve as a basis for comparison with other CAM types and C4 plants.
In Arabidopsis (Arabidopsis thaliana), a central circadian oscillator complex is formed by the genes CIRCADIAN CLOCK ASSOCIATED1/LATE ELONGATED HYPOCOTYL (CCA1/LHY) acting together with TIMING OF CAB EXPRESSION1 (TOC1) in a feedback-regulating system, operating throughout the life of the plant in a diversity of tissues (Locke et al., 2005, 2006). It controls stomatal conductance rhythms, hypocotyl elongation, and flowering time via the integration of photoperiodic timing and temperature (Somers et al., 1998; Strayer et al., 2000; Ding et al., 2007). Products of these genes interact with those from a set of other clock genes, like PSEUDO-RESPONSE REGULATOR9 (PRR9), a critical component of the circadian system and an integrator of temperature signals (Salomé and McClung, 2005). Although the circadian clock has been extensively studied in Arabidopsis (de Montaigu et al., 2010), the genetic contributors and their expression patterns have not been described in more than a few species (Beales et al., 2007; Iwamoto et al., 2009; Hayes et al., 2010). Studying these circadian oscillator genes in CAM plants should help us to understand the circadian regulation of key CAM metabolic enzymes.
As little transcriptomic data are available from obligate CAM plants, we developed a large collection of Opuntia ESTs. It is accessible through the OpuntiaESTdb, providing information about clusters, annotations, tandem repeats, and metabolic pathways. As a direct application, we used the OpuntiaESTdb to retrieve the sequences of CAM-related and circadian clock genes. There are multiple isoforms for photosynthetic genes with differing functions in plants with C3, C4, and CAM metabolism, and as expected, we obtained several contigs for each gene studied. We isolated a set of possible reference genes as a prerequisite for validating transcriptomic changes by quantitative (Q) real-time PCR. Reference genes, traditionally thought of as housekeeping genes, are needed in order to normalize mRNA levels between different samples, correcting for differences in the amount of starting material or RNA isolation efficiencies, thus allowing for exact interpretation of gene expression data (Vandesompele et al., 2002). Housekeeping genes, whose products are involved in basic functions needed for cell maintenance, were initially assumed to be constitutively expressed, but the expression levels of these genes can vary among tissues or cells (Mallona et al., 2010) and may be modified by environmental conditions. It can therefore be concluded that no gene can be presumed a priori to be “constitutively expressed” (Czechowski et al., 2005; Gutierrez et al., 2008). The selection of appropriate reference genes for the types of tissues and circumstances under investigation is required for accurate gene expression studies. We identified OfiSAND and OfiTUB as suitable genes for the normalization of transcription levels in a variety of tissues. After validating the adequacy of reference genes, we performed Q-PCR relative quantification and found several canonical gene expression profiles, including rhythmic expression of OfiPPCK and OfiTOC1 and arrhythmic expression of OfiPEPC and OfiMDH. Surprisingly OfiNADP-ME, OfiPPDK, OfiCCA1, and OfiPRR9 adopted a 12-h cycle with two peaks during the day, indicating significant differences in the structure of chronobiological regulation in CAM plants.
RESULTS
EST Annotation Overview
The sequencing of cDNA derived from RNA pools of various tissues (Fig. 1) generated a total of 604,176 ESTs with an average read length of 344 bp (Supplemental Table S2). Clustering of these ESTs produced a total of 43,066 contigs and 407,253 unassembled singletons with average lengths of 612 and 1,385 bp, respectively. Annotation against the National Center for Biotechnology Information (NCBI) nonredundant database showed that 29,835 contigs produced the lowest BLAST hit scores against sequences from a CAM species (69.3%) as listed by Sayed (2001), whereas 1,015 were closer to those of Arabidopsis (2.4%).
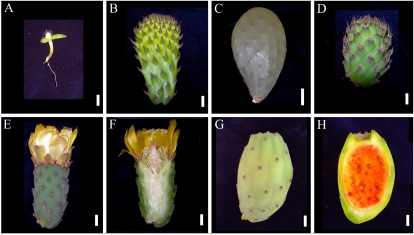
Figure 1.
Organs and developmental stages used for RNA extraction prior to EST sequencing and real-time Q-PCR. A, Three-month-old seedling. B, Young cladode. C, Fully grown cladode. D, Young flower bud. E and F, Flower buds at anthesis. G and H, Fruits. Bars = 1 cm, except for C, where bar = 5 cm.
The OpuntiaESTdb Web interface includes a searchable database through BLASTN, TBLASTN, BLASTX, TBLASTX, and BLASTP assembled contigs and singletons. Preexistent ESTs from CAM species were recovered from The Institute for Genomic Research assemblies and included in the database to allow for more comprehensive searches. Contig sequence recovery includes functional annotation fetching of the annotations obtained from its RefSeq/UniProtKB putative orthologs. Thus, Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologs and Pathways, PubMed publications, ExPaSy, InterPro, and Gene Ontology (GO) annotations, Tandem Repeats, and UniProtKB cellular locations and keywords are retrieved if present. KEGG Pathways including O. ficus-indica contigs are offered as highlighted maps. Additional information on data analysis and functional categorization can be found in the Supplemental Data, including a flow chart of database construction and its applications (Supplemental Fig. S1), the comparative distribution of a selection of functional categories between the classified genes from the Arabidopsis genome and the Opuntia EST clusters as detected by WEGO (Supplemental Fig. S2A), the distribution of functional categories among contigs showing the 10 most common GO terms in the EST database for each of the three ontology domains, molecular function, cellular component, and biological process (Supplemental Fig. S2B), and the KEGG pathway for carbon fixation in photosynthetic organisms with highlighted orthologs from the Opuntia ESTdb (Supplemental Fig. S3).
Validation of Reference Genes for Q-PCR in O. ficus-indica
The cDNA samples obtained from six different organ types of O. ficus-indica were used for real-time Q-PCR with the eight primer pairs designed for amplification of the candidate reference genes (Supplemental Table S1). For each run, the cycle threshold (CT) value (the number of cycles required for normalized fluorescence to reach a threshold), reaction efficiency, and product melting temperature were evaluated. Single melting peaks for all reference gene products indicated single-product amplification.
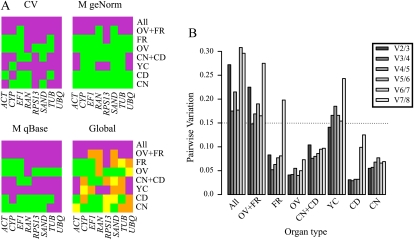
The CT values associated with each technical replicate were quality checked, and those differing from the average by more than one cycle were discarded. Values for 90% of the technical replicates differed by less than one cycle. Figure 2 as well as Supplemental Table S3 show the normalization results. Global results from stability and coefficient of variation values of qBasePlus and stability values from geNorm, as well as a pairwise variation cutoff of 0.15, indicate that for all single organs, a combination of two reference genes is sufficient and necessary for normalizing target gene expression (Fig. 2B). Combining two organ types requires two (for cladodes sampled at both day and night) and three (for ovules and fruits) reference genes, while for combining all tissue types, no pairwise variation value underneath the cutoff limit could be determined. Figure 2A indicates the reference gene ranking for single and mixed organs based on cutoff values of 0.5 for stability and 0.2 for coefficient of variation. Supplemental Figure S4 shows the relative quantities of reference gene expression for each biological replicate.
Figure 2.
Reference gene expression analysis. Organ types are as follows: YC, young cladodes; CD, cladodes sampled during the day; CD, cladodes sampled at night; OV, ovule tissue of fully developed flowers before anthesis; FR, ripe fruits. A, Reference gene expression stability heat map. geNorm stability (M) and qBasePlus stability and coefficient of variation (CV) expression values are shown for single and mixed organs. Each heat map spot represents a reference gene-organ or organ mix combination; spots with values below a stability threshold are represented in green (meaning reference gene suitability), whereas those above that threshold are plotted in magenta (not suitable). Specific thresholds are 0.2 for qBasePlus coefficient of variation values and 0.5 for geNorm and qBasePlus stability values. A superposed heat map, combining results from the three previous layers, is labeled as Global. Green reflects combinations that had three out of three suitable stability measures, yellow reflects two out of three, orange reflects one out of three, and magenta reflects none. B, Minimum number of genes necessary for reliable and accurate normalization. GeNorm pairwise variation values are computed by an algorithm that measures pairwise variation (Vn/n + 1) between two sequential normalization factors, NFn and NFn + 1, where n is the number of genes involved in the normalization factor. A pairwise variation threshold of 0.15 is plotted. [See online article for color version of this figure.]
Identification of CAM and Circadian Clock Homologs
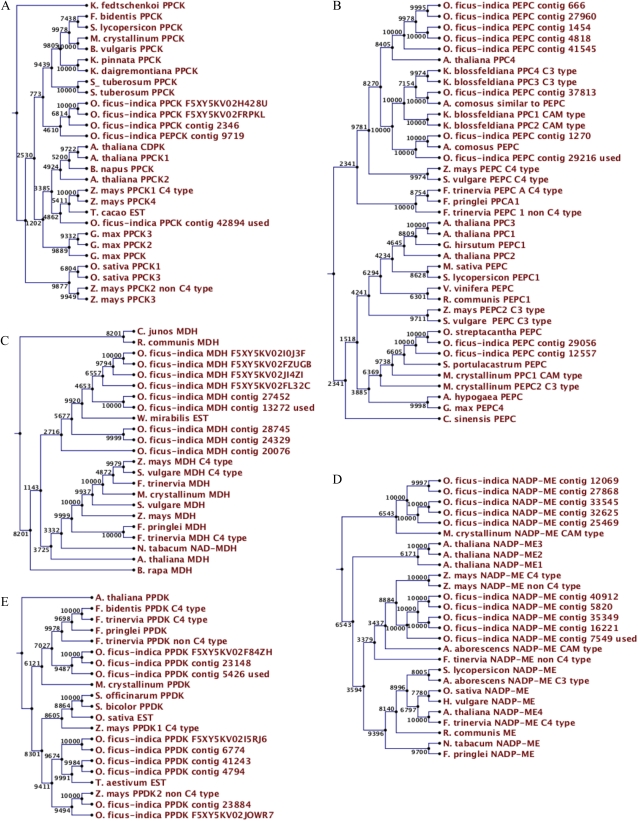
Several genes encoding CAM-related enzymes (e.g. PEPC, ME, and MDH) belong to small gene families (Cushman and Bohnert, 1989a, 1989b; Cushman et al., 1993; Honda et al., 2000). For example, up to four members of PEPC exist in Mesembryanthemum crystallinum and Kalanchoe blossfeldiana, with one or two CAM-specific isoforms (Cushman and Bohnert, 1997). As the Arabidopsis genome is thoroughly annotated, we used Arabidopsis ab initio to estimate the gene family structure of all genes analyzed. We found several contigs that fulfilled the criteria of homology and performed phylogenetic analysis of the contigs against available sequences from C3, C4, and CAM plants (Fig. 3).
Figure 3.
Phylogenetic analyses of PPCK (A), PEPC (B), MDH (C), NADP-ME (D), and PPDK (E). Bootstrapping values (percentage from 10,000 sampled trees) are shown at the branch points. O. ficus-indica clones labeled with “contig” or “F” plus letter/number code are contigs and singletons, respectively, identified from the EST database. Clones labeled as O. ficus-indica contig used were chosen for expression analysis. [See online article for color version of this figure.]
Several isoforms of PPCK have been associated with C4, CAM, or housekeeping C3 types. There are two PPCK genes in Arabidopsis; using PPCK1 as query, we identified three contigs and two singletons. Phylogenetic analysis of the O. ficus-indica sequences revealed two singletons and two contigs clustered together in a branch that includes genes from CAM and non-CAM plants, whereas a fifth clone clusters with Arabidopsis and other plants, including C4 plants (Fig. 3A). The data from this phylogenetic reconstruction did not show any obvious separation of CAM and non-CAM sequences (i.e. those from the C3 species Beta vulgaris, Solanum lycopersicum, and the CAM species Kalanchoe daigremontiana and M. crystallinum). A different O. ficus-indica sequence grouped together with Arabidopsis, Zea mays, Theobroma cacao, and Glycine max, reinforcing the scattered pattern of PPCK sequences.
We found 10 contigs of PEPC using AtPEPC2, a member of the small gene family comprising four loci in Arabidopsis, as query. The tree topology shows four clusters of OfiPEPC genes, including one close to AtPEPC4, and another group highly homologous to the CAM plant Ananas comosus (Fig. 3B). There is yet another group that clusters together with CAM- and C3-type clones from M. crystallinum, suggesting again a complex evolutionary history for the Opuntia gene family that cannot be resolved purely by sequence similarity criteria.
A total of 17 loci code for MDH in Arabidopsis, with apoplastic, chloroplastic, mitochondrial, and peroxisomal enzyme activities. The sequence used as query encodes an enzyme that is apoplastic as well as chloroplastic, with NAD+ or NADP+ (chloroplastic) as acceptor. It has been suggested that combined activities of cytosolic NAD+-MDH and chloroplastic NADP-MDH may be involved in nocturnal malic acid formation in CAM plants (Cushman, 1993). We identified five OfiMDH contigs and four OfiMDH singletons (Fig. 3C). We observed two major branches, with all O. ficus-indica clones clustering within one branch that also includes MDH from the CAM species Welwitschia mirabilis. The second main branch includes MDH isoforms of C4 plants like Z. mays and Sorghum vulgare as well as one from the CAM species M. crystallinum and C3 plants. The clustering of all O. ficus-indica clones indicates high similarity among possible isoforms in Opuntia. Nevertheless, CAM-specific isoforms fail to cluster together.
Malate decarboxylation in Cactaceae species, which belong to the ME group of CAM plants, mainly occurs via the activity of cytoplasmic NADP-ME (Scott and Mercer, 1997), although substantial activities of mitochondrial NAD-ME might also contribute to decarboxylation in CAM plants (Cushman and Bohnert, 1997). Four NADP-ME isoforms exist in Arabidopsis, cytosolic except for NADP-ME4, which is chloroplastic. We used as query the cytosolic Arabidopsis NADP-ME2 and identified 10 O. ficus-indica contigs satisfying the homology cutoff score (Fig. 3D). These contigs cluster on two branches. One branch includes five O. ficus-indica contigs as well as the CAM isoform of NADP-ME from M. crystallinum. The second branch including O. ficus-indica clones clusters with isoforms of C4 (Z. mays and Flaveria trinervia) and CAM (Aloe arborescens) plants. A third major branch contains isoforms from both C3 and C4 plants. Tree topology indicates that NADP-ME isoforms from CAM plants tend not to gather with those from C3 plants but are more similar to C4 isoforms.
The Arabidopsis genome contains one chloroplastic PPDK gene, while in C4 plants, both C4 and non-C4 isoforms have been identified (Chastain et al., 2002). Six O. ficus-indica contigs and three singletons are highly homologous to PPDK from Arabidopsis, clustering on two branches (Fig. 3E). One branch also contains PPDK of M. crystallinum as well as isoforms of C4 and C3 Flaveria species. The second major cluster carries O. ficus-indica sequences as well as PPDK isoforms from several C4 plants (Z. mays, Saccharum officinarum, and Sorghum bicolor) as well as Oryza sativa and Triticum aestivum. The AtPPDK was the most divergent sequence in the PPDK tree.
In summary, except for NADP-ME, which separates O. ficus-indica clones clearly from those of C3 species, the CAM-related genes do not clearly cluster separately from those encoding C3 or C4 isoforms or their host species.
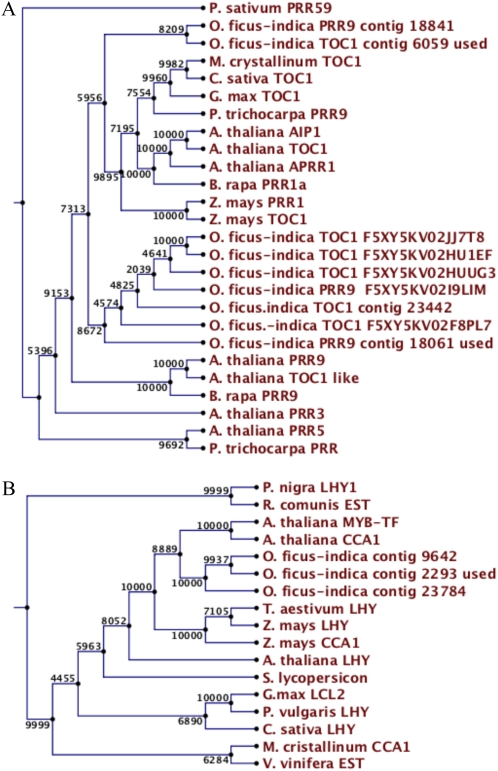
There are five PRR genes in the Arabidopsis genome that form three clades, corresponding to AtPRR1-TOC1, PRR3 and PRR7, and PRR5 and PRR9 (Takata et al., 2010). Our phylogenetic reconstruction (Fig. 4A) shows two branches, one containing the O. ficus-indica genes clearly isolated from the rest and a second containing Arabidopsis, G. max, Populus trichocarpa, and two O. ficus-indica genes, suggesting rapid evolution of a group of PRR genes in the genus Opuntia. An additional group of genes involved in circadian regulation are those encoding MYB transcription factors, out of which CCA1 and LHY are considered the morning oscillators (de Montaigu et al., 2010). We found only three genes with high homology to Arabidopsis LHY and CCA1 (Fig. 4B), all three clustering closer to CCA1 than to LHY. In contrast, the CCA1 gene from M. crystallinum was more similar to that of Vitis vinifera, indicating a lack of CAM specificity in the clustering of these genes.
Figure 4.
Phylogenetic analysis of TOC (A) and PRR (B) genes. Bootstrapping and clone labeling are as described for Figure 3. [See online article for color version of this figure.]
As our results showed no clustering of CAM, C3, or C4 genes, we used the homolog closest to the Arabidopsis query sequence for further analysis.
Temporal Gene Expression Analysis by Q-PCR
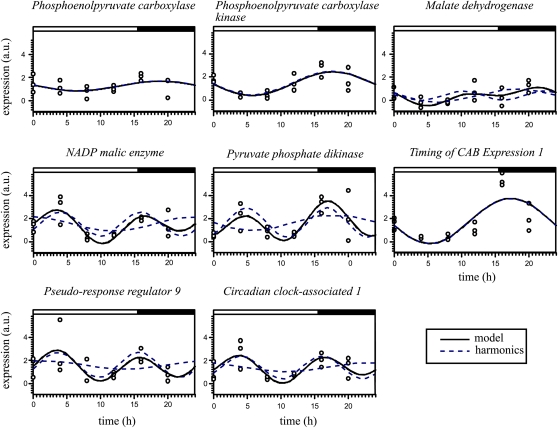
We followed the expression pattern of three genes coding for enzymes active in the cytoplasm during the night, OfiPEPC, OfiPPCK, and OfiMDH , and two-light active enzymes, OfiNADP-ME and OfiPPDK (Fig. 5). OfiPEPC and OfiMDH expression patterns did not fit to a circadian rhythm (Supplemental Table S3). In contrast, OfiNADP-ME and OfiPPDK showed significant adjustment to a 12-h rhythm, while OfiPPCK oscillated with a 24-h periodicity. OfiPPCK expression was highest about 3 h after the beginning of the dark period; OfiNADP-ME and OfiPPDK showed increases in expression during the morning and again in the afternoon approaching the dark period. For core clock genes, OfiTOC1 adapted to a circadian rhythm with lowest expression during the morning and the acrophase at dusk. In contrast, homologs to the Arabidopsis clock genes OfiCCA1 and OfiPRR9 both followed statistically significant 12-h oscillations. Peak expression of these two genes coincided in the morning and again at dusk. This 12-h period has not been observed in Arabidopsis, where CCA1 and PRR9 follow a circadian rhythm. Similar to TOC expression in Arabidopsis, OfiTOC1 showed an evening peak slightly later than the evening peaks of CCA1 and PRR9.
Figure 5.
Gene expression chronograms of CAM and circadian rhythm pathways during the day/night cycle in cladodes of O. ficus-indica. Three normalized, relative gene expression quantities corresponding to three biological replicates were collected every 4 h (circles). Oscillation (represented as a continuous curve) was modeled, via Fourier analysis, decomposing it in two harmonic or sinusoidal components: one circadian (24-h harmonic) and one ultradian (12 h). Dotted curves indicate harmonic adjustment. The solid line indicates the proposed model including all significant harmonics. a.u., Arbitrary units. [See online article for color version of this figure.]
DISCUSSION
EST Cluster Generation and Sequence Annotation
ESTs provide a shortcut to the transcribed portions of the genome, allowing a rapid screen for novel genes based on BLAST searches. The sequencing of O. ficus-indica cDNA performed here generated 604,176 sequences. Sixty-seven percent of these sequences were unassembled singletons, not clustering with any other sequences. These stand-alone ESTs may be from regions of low coverage; nevertheless, similar numbers were reported for other EST collections (Cervigni et al., 2008). The OpuntiaESTdb presented here is, to our knowledge, the first online database of a plant species performing CAM metabolism whose sequences can be freely accessed and downloaded, with valuable information about whole-plant gene expression.
From the contigs whose sequences showed homology to those present in the public databases, 29,835 (69%) had significant matches with categorized proteins, and putative functions were assigned on this basis. Nonmatching contigs might indicate specific roles for the products of these sequences in constitutive CAM plants. The Arabidopsis genome is predicted to contain more than 25,000 protein-coding genes and the Z. mays genome at least 32,540 (Schnable et al., 2009). Assuming a number in O. ficus-indica similar to that in Z. mays, the identified 29,835 protein-coding genes represents close to 90% of the O. ficus-indica gene complement (Segura et al., 2007).
Validation of Reference Genes for Q-PCR in O. ficus-indica
The identification of O. ficus-indica gene homologs to well-categorized protein-coding genes from other organisms opens up the possibility for broad-scale transcriptional analysis. Quantitative analysis relies upon reference genes with essentially invariant transcript levels in different tissues under different conditions; the OpuntiaESTdb allowed us to identify O. ficus-indica gene homologs to known reference genes. Because CO2 absorption changes from C3 to CAM metabolism during cladode development (Winter et al., 2008), basing our analysis on both young and fully grown cladodes allowed us to analyze CO2 uptake-related gene expression during this developmental step. We also used adult cladodes sampled during day and night as prerequisite for analyzing the transcript levels of genes showing circadian oscillation. Finally, the determination of optimal reference genes in floral and fruit tissue is crucial for investigating the genes involved in flower and fruit development, for example those related to parthenocarpic fruit development (Weiss et al., 1993) and fruit ripening (Collazo-Siqués et al., 2003). Our results show that the use of two reference genes is sufficient and necessary for single and mixed organs except for the combination “ovules plus fruits,” which requires three reference genes. Interestingly, the SAND protein (SAND), proposed as a reference gene for Arabidopsis (Czechowski et al., 2005), proved the most reliable reference gene for the set of organs examined here, followed by b-Tubulin6 (TUB).
Gene Identification and Phylogenetic Analysis
Phylogenetic analysis of CAM-related and circadian clock-related homologs from O. ficus-indica showed no specific clustering with genes and/or CAM-specific isoforms of other CAM plants. With the exception of O. ficus-indica clones for NADP-ME, which did not seem to cluster with C3 isoforms of this enzyme, all the genes under investigation had contigs and singletons in mixed branches that included C3 and C4 isoforms.
Irrespective of their photosynthetic metabolism, C4 and CAM plants have C3 housekeeping as well as C4- and CAM-specific isoforms of photosynthetic enzymes. It has been postulated that C4 photosynthesis arose in a multistep process starting with the duplication of genes for ubiquitous metabolic enzymes, which then acquired specific functions in a light-, development-, cell-, and tissue-specific manner (Ku et al., 1996; Besnard et al., 2003; Monson, 2003). Our phylogenetic analysis for PEPC showed that, in the case of the CAM plants M. crystallinum and K. blossfeldiana, the C3 and CAM isoforms were more similar within the species than they were to C3 and CAM isoforms of the other CAM species. We conclude that O. ficus-indica CAM-specific isoforms cannot be identified based on phylogenetic analysis. Lack of clear grouping among CAM-type isoforms, as observed in our analysis, has also been reported by Taybi et al. (2000) for PPCK from M. crystallinum, whose most closely related PPCKs were those from S. lycopersicum followed by Kalanchoe fedtschenkoi and Arabidopsis. Concerning PPDK, Chastain et al. (2011) found only minor differences between C3 and C4 isoforms from Arabidopsis and Z. mays in key catalytic and regulatory properties. Those authors propose a transition from a functionally C3 isoform into a C4 pathway enzyme involving only minor changes in enzyme properties. The suggested mechanism could account for the close relationships between CAM and C3 and C4 isoforms observed in our phylogenetic analysis. CAM metabolism is widespread, occurring within 33 plant families, and water stress is proposed as the ultimate selective factor for terrestrial CAM plants (Keeley and Rundel, 2003). The widespread systematic occurrence hints at multiple independent evolutionary events (Smith and Winter, 1996) and might explain the phylogenetic tree structures observed here.
Rhythmic Expression of CAM and Clock Genes
We studied the temporal expression pattern of O. ficus-indica genes homologous to circadian clock- and CAM-related genes using the identified reference genes. The objectives were (1) to compare the expression patterns of several CAM-related genes in the cactus O. ficus-indica with those known from other CAM species, with special focus on the circadian oscillation pattern, and (2) to determine whether central oscillator components in the circadian cycle known from Arabidopsis show the typical circadian rhythm of transcription in O. ficus-indica.
While the enzymatic machinery required to perform CAM is present in all higher plants, activities of the key enzymes are much higher in CAM and C4 plants, and it is proposed that they have acquired diurnal patterns of expression and regulation to meet nighttime CO2 fixation and daytime decarboxylation of C4 acids (Cushman and Bohnert, 1997). It has also been suggested that posttranslational control mediates circadian regulation of enzyme activities during the day/night cycle, while transcriptional, posttranscriptional, and translational control is primarily responsible for the buildup of CAM-related enzymes (Cushman et al., 2000).
In the case of PEPC, short-term responses are posttranslationally controlled, as shown for M. crystallinum (Li and Chollet, 1994) and K. fedtschenkoi (Carter et al., 1996) as well as in Crassula argentea, where two PEPC configurations exist and modification of the configuration involves phosphorylation and dephosphorylation (Lambers, 2008). This observation is in accordance with our data showing that the OfiPEPC homolog did not show major transcriptional changes over a 24-h period (Fig. 5; Supplemental Tables S4 and S5).
The only CAM-related enzyme for which a circadian oscillation rhythm in transcription has been reported in different CAM species is PPCK. A diurnal oscillation in transcript abundance was reported for the constitutive CAM plant K. fedtschenkoi (Hartwell et al., 1999) and the facultative CAM plant M. crystallinum (Taybi et al., 2000). The circadian transcription of the gene for PPCK, which leads to a modulated sensitivity of PEPC to inhibition by malic acid, is proposed to be the key clock system responsible for circadian regulation of CAM metabolism (Nimmo et al., 1987; Hartwell et al., 1999; Wyka et al., 2004). Our studies confirm the diurnal expression pattern of PPCK in O. ficus-indica, although the acrophase was observed at dusk, while highest transcript levels in M. crystallinum and K. fedtschenkoi occur toward dawn. Thus, activation of PEPC by PPCK seems to be regulated at the transcriptional level and might represent a crucial control point in CAM metabolism in O. ficus-indica, but with significant differences from the system reported for M. crystallinum and K. fedtschenkoi.
The enzyme MDH catalyzes the conversion of oxaloacetate to malate. In M. crystallinum, a gene encoding NADP+-dependent MDH was isolated that shows a salt stress-induced expression pattern, indicating its participation in the CO2 fixation pathway during CAM (Cushman, 1993). Circadian regulation of MDH activity was reported to be posttranslational, as chloroplastic NADP-MDH activity is light regulated in response to the NADPH/NADP+ ratio and the thioredoxin redox state (Rebeille and Hatch, 1986; Li et al., 1994). The selected OfiMDH gene, similar to OfiPEPC, did not follow a circadian expression rhythm but rather a constitutive transcription pattern during a 24-h course, suggesting a reliance on posttranscriptional regulation.
Species of the Cactaceae family belong to the ME type of CAM plant, and malate decarboxylation mainly occurs via the activity of cytoplasmic NADP-ME (Scott and Mercer, 1997). NADP-ME plants require PPDK in the chloroplast for gluconeogenic recovery of PEP. Its activity is under dark/light regulation by reversible phosphorylation in Z. mays (Budde et al., 1985; Chastain et al., 2002), involving posttranslational regulation through the PPDK regulatory protein (Chastain et al., 2002, 2011). It was reported for M. crystallinum that transcript levels of NADP-ME increase 8- to 10-fold in response to salt stress in the leaves as CAM metabolism is induced (Cushman, 1992). In our studies, transcription of the two enzymes adjusted to a 12-h rhythm, and acrophase almost coincided for these enzymes. A rise in transcript levels starts at the beginning of CAM phase III, when CO2 is released from malate, refixed by Rubisco, and assimilated via the Calvin cycle of photosynthesis, and the resulting pyruvate is converted into PEP. A second rise in expression during phase IV might be related to an increased demand of PEP for CO2 fixation. Our results show that in O. ficus-indica, transcription of genes for PPDK and NADP-ME changes during the day and these changes, together with posttranslational activations, might be relevant for the adaptation of enzyme activity during the CAM cycle.
Control of gene expression by the circadian clock involves some central oscillator genes. CCA1/LHY together with TOC1 form a core oscillator complex in Arabidopsis. CCA1 and LHY bind to the TOC1 promoter and negatively regulate its expression, while TOC1 positively regulates CCA1/LHY together with other genes, including GIGANTEA, EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRITHMO (McClung, 2006). PRR9 acts as an oscillator necessary for the clock to respond to temperature signals, anticipating diurnal cold stress and initiating a stress response by mediating cyclic expression of stress response genes, including DREB1/CBF (Salomé and McClung, 2005; Nakamichi et al., 2009). PRR9 is positively regulated by CCA1/LHY, but it negatively regulates CCA1/LHY (McClung, 2006).
Our results show that of the O. ficus-indica homologs to Arabidopsis central oscillator genes CCA1, TOC1, and PRR9, only OfiTOC1 shows circadian rhythmicity. OfiTOC1, similar to its expression profile in Arabidopsis, has its acrophase at the onset of the dark period. In Arabidopsis, the morning-activated oscillator CCA1 shows an opposite rhythm, as CCA1 inhibits TOC1 while TOC1 induces CCA1, creating an autoregulatory feedback loop (Locke et al., 2006). In O. ficus-indica, OfiCCA1 and OfiPRR9 seem to adapt to an ultradian 12-h rhythm and not to the 24-h cycle seen in Arabidopsis, with peak expression during the morning and a second expression peak at dusk before the rise in OfiTOC1 expression. Our results indicate that homologs to key oscillator genes from Arabidopsis do not build the same interconnected autoregulatory loops in O. ficus-indica, pointing to the involvement of other key oscillator genes in this CAM species. It remains to be determined if the evolution of CAM occurred by changes in the setup of the circadian clock rather than by a modification of downstream genes in their cis-regulatory regions.
MATERIALS AND METHODS
cDNA Library Construction and Sequencing
A normalized cDNA library was constructed from RNA isolated from six individuals of the species Opuntia ficus-indica, collected from different sites in the Mediterranean southeast of Murcia, Campo de Cartagena, Spain, where these plants are typically planted by commercial growers as small populations alongside other crops such as citrus or Vitis vinifera. Total RNA was extracted from seedlings, young cladodes, fully grown cladodes sampled during the day and night, young flower buds, fully developed flowers, and ripe fruits (Fig. 1). Total RNA was purified using the Nucleospin RNA Plant Kit (Macherey-Nagel), which includes a recombinant DNase I treatment in order to eliminate genomic DNA contamination. Sequencing was performed by Eurofins MWG Operon applying 454 sequencing technology.
EST Sequence Analysis
Raw sequence data were clustered using CAP3 with a stringency level of 95% similarity per 100 bp; clipping of poor regions regarded base quality values and similarity information (Huang and Madan, 1999). Assembled EST data sets were compared using BLASTX (e value < 1e-10) with the NCBI nonredundant database in order to identify putative homologs. In the case of significant BLASTX, the hit with minimal e value was ascribed to the contig description, and thus a RefSeq and NCBI GI identifier was assigned to each contig. These accessions were mapped to KEGG Orthology and GO. KEGG Orthology terms were assigned to KEGG Genes based on cross-database links to GI identifiers and compared with Arabidopsis (Arabidopsis thaliana) as background using KOBAS (for KEGG Orthology-Based Annotation System; Wu et al., 2006). Relevant metabolic reactions were plotted with the KEGG Pathway exploring tool. RefSeq accessions were mapped to UniProtKB identifiers and thereafter to GO using DAVID (for Database for Annotation, Visualization, and Integrated Discovery; Dennis et al., 2003; Da Wei Huang and Lempicki, 2008). GO abundance was compared with the Arabidopsis background and represented with WEGO (Ye et al., 2006). Orthologs for the selected pathways were obtained from the collection of plant annotated sequences available at the GO consortium, and they were accessed through the European Bioinformatics Institute QuickGO interface (Binns et al., 2009). Protein sequences involved in those processes were retrieved from the QuickGO GOA files and compared via TBLASTN against the OpuntiaESTdb (e value cutoff of 1e-10). For each significant Opuntia sequence, only the BLAST record hit with the lowest e value was taken, thus avoiding multiple assignments.
The database architecture runs on Ubuntu Linux with an apache2 HTTP server and MySQL as the database management system. Pages can be requested through several Common Gateway Interface python scripts. The source code is available at the server’s page.
Identification of Genes with Stable Patterns of Gene Expression
We selected a list of eight genes considered to exhibit stable patterns of expression and therefore suitable for use as internal controls for Q-PCR (Andersen et al., 2004; Czechowski et al., 2005; Nicot et al., 2005; Mallona et al., 2010). The list consisted of Actin-11 (ACT), Cyclophilin-2, Elongation factor 1a (EF1), Ubiquitin GTP-binding protein RAN1, SAND, Ribosomal protein S13, and TUB (Supplemental Table S1). Highly homologous contigs or singletons to the corresponding Arabidopsis identifiers were retrieved from the OpuntiaESTdb using TBLASTN. Primers were designed using the application PRIMEGENS version 2 (which discards cross-hybridizations within the EST database; Srivastava et al., 2008), resulting in an amplicon size between 103 and 154 bp (Supplemental Table S1).
Total RNA was extracted as described above using young cladodes (6–8 cm length), fully grown cladodes sampled during the day and at night, ovule tissue of fully developed flowers before anthesis, and ripe fruits. Tissue samples were taken from one of the populations used for RNA sequencing. Three independent samples, representing pools of three samples per plant, were taken for each tissue and time course. RNA concentration and purity were estimated from the ratio of absorbance readings at 260 and 280 nm, and RNA integrity was tested by gel electrophoresis. cDNA synthesis was performed from equal amounts of total RNA using Moloney murine leukemia virus reverse transcriptase (SuperScript III First-Strand Synthesis SuperMix; Invitrogen). Real-time PCR was carried out in an Mx3000P Q-PCR system (Stratagene) using a SYBR Green-based PCR assay (with 5-carboxy-X-rhodamine as the optional reference dye; Brilliant II SYBR Green Q-PCR Master Mix; Stratagene), applying a PCR protocol of 95°C for 5 min, 40 cycles of 95°C for 30 s, 60°C for 30 min, and 72°C for 30 s, followed by a standard dissociation protocol to assess product uniqueness. All assays were performed in three technical replicates for each of the three biological replicates and included nontemplate controls.
For data mining and statistical analysis, PCR amplification kinetics were analyzed to calculate crossing points and reaction efficiencies with the qpcR R package version 1.3-3 (Spiess et al., 2008). The efficiency value (E) was defined as E = Fn/(Fn −1), in which n is determined as the 20% value of the normalized fluorescence at the maximum of the second derivative curve. Data generated for potential internal controls were quality tested and checked for expression stability using qBasePlus version 1.5 (Hellemans et al., 2007), which includes stability and coefficient of variation determination as well as the classical geNorm parameters such as stepwise stability and pairwise variation (Vandesompele et al., 2002).
Identification of O. ficus-indica Homologs and Phylogenetic Analysis
For each candidate circadian-regulated gene, we searched the OpuntiaESTdb via TBLASTN (e value < 1e-10) using an Arabidopsis accession as the query sequence. In order to analyze sequence dissimilarities between the putative O. ficus-indica homologous sequences and those corresponding to other C4, C3, and CAM species, we employed the same Arabidopsis sequence with TBLASTN against the NCBI nonredundant database. Significant hits were recovered, and further accessions based on literature searches were included in the phylogenetic work flow.
All sequences were translated and aligned using a progressive alignment algorithm with default parameters (Feng and Doolittle, 1987) and clustered via neighbor joining. Internal node support was assessed using 10,000 bootstrap replicates. Sequence alignment and clustering were performed with the CLC bioinformatic suite version 6. The accessions employed were as follows.
(1) Query: Arabidopsis PPCK1 AT1G08650; C3 plants: Arabidopsis CDPK AT1G08650, Glycine max PPCK3 AY144184.1, G. max PPCK2 AY144182.1, G. max PPCK AY143660.2, Theobroma cacao EST EST01262, Solanum lycopersicum PPCK AF203481.1, Beta vulgaris PPCK AJ309171.1, Solanum tuberosum PPCK AF453448, Oryza sativa PPCK1 AK101080.1, O. sativa PPCK2 XM46683.1, and O. sativa PPCK3 AK066885; CAM plants: Kalanchoe fedtschenkoi PPCK AF162661, Mesembryanthemum crystallinum PPCK AF158091, Kalanchoe pinnata PPCK EF157816.1, and Kalanchoe daigremontiana PPCK EF157817.1; C4 plants: Zea mays PPCK4 AY911416, Z. mays PPCK1 (C4 type) AY911413, Z. mays PPCK3 AY911413, Z. mays PPCK2 (non-C4 type) AY911415, and Flaveria bidentis PPCK AB065100.1.
(2) Query: Arabidopsis PEPC2 AT2G42600; C3 plants: Arachnea hypogea PEPC EV391629.1, G. max PEPC4 AY563044.1, Sesuvium portulacastrum PEPC DQ655705.1, S. lycopersicum PEPC1 AJ243416.1, Medicago sativa PEPC M83086.1, Citrus sinensis PEPC EF058158.2, Arabidopsis PPC4 AT1G68750, Arabidopsis PPC1 AT1G53310, Gossypium hirsutum PEPC1 AF008939.1, Ricinus communis PEPC1 EF634319.1, Arabidopsis PPC3 AT3G14940, and Flaveria pringlei PEPCA1 Q01647; CAM plants: Opuntia streptacantha PEPC EX720539, M. crystallinum PPC1 (CAM type) X13660.1, M. crystallinum PPC2 P16097.1, Kalanchoe blossfeldiana PPC1 (CAM type) CAA61083.1, K. blossfeldiana PPC2 (CAM type) CAA61084.1, K. blossfeldiana PPC3 (C3 type) CAA61085.1, and K. blossfeldiana PPC4 (C3 type) CAA61086.1; C4 plants: Flaveria trinervia PPC-A (C4 type) P30694, F. trinervia PEPC1 (non-C4 type) Q9FV66, Ananas comosus PEPC DV190685, A. comosus similar to PEPC DT338073, Z. mays PEPC1 NP 001105418.1, Z. mays PEPC2 (C3 type) X61489.1, Z. mays PEPC2 (C4 type) X03613.1, Sorghum vulgare PEPC (C3 type) X59925.1, and S. vulgare PEPC (C4 type) X17379.1.
(3) Query: Arabidopsis MDH AT3G47520.1; C3 plants: R. communis MDH XM002514704.1, Brassica rapa MDH FJ208590, Citrus junos MDH DQ901430.1, F. pringlei MDH P36444, and Nicotiana tabacum MDH AJ006574.1; CAM plants: Welwitschia mirabilis EST DT584016 and M. crystallinum MDH chloroplast Q05145; C4 plants: Z. mays MDH cytoplast Q08062, Z. mays MDH (C4 type) P15719, S. vulgare MDH (C4 type) P17606, S. vulgare MDH Q43830, F. trinervia MDH (C4 type) P22178, and F. trinervia MDH Q42737.
(4) Query: Arabidopsis cytosolic NADP-ME2 NM121205.3; C3 plants: N. tabacum cytosolic NADP-ME DQ923118.2, F. pringlei NADP-ME AF288921.1, Arabidopsis cytosolic NADP-ME3 AT5G25880, O. sativa NADP-ME AY435404.1, Hordeum vulgare NADP-ME EU977175.1, Arabidopsis cytosolic NADP-ME1 AT2G19900, R. communis ME XM002514184, S. lycopersicum chloroplast NADP-ME AF001269.1, and Arabidopsis NADP-ME4 chloroplast AAF68118.1; CAM plants: M. crystallinum NADP-ME (C4 type) CAA45772.1, Aloe arborescens NADP-ME (C3 type) AB005808.1, and A. arborescens NADP-ME (C4 type) BAA24950.1; C4 plants: Z. mays NADP-ME (non-C4 type) U39958.1, Z. mays NADP-ME (C4 type) J05130.1, F. trinervia NADP-ME (non-C4 type) EU118291.1, and F. trinervia NADP-ME (C4 type) X61304.1.
(5) Query: Arabidopsis PPDK AT4G15530; C3 plants: O. sativa PPDK NM 001062042, Triticum aestivum EST CK208491.1, and F. pringlei PPDK Q42736; CAM plant: M. crystallinum PPDK CAA55143; C4 plants: Saccharum officinarum PPDK AF194026.1, Sorghum bicolor PPDK AY268138.1, Z. mays PPDK1-1 (C4 type) P11155-1, Z. mays PPDK1-2 (non-C4 type) P11155-2, F. bidentis PPDK (C4 type) Q39735, F. trinervia PPDK (C4 type) P22221-1, and F. trinervia PPDK (non-C4 type) P22221-4.
The clock genes used for phylogenetic reconstruction were as follows.
(1) Query: Arabidopsis TOC1 AT5G61380 and Arabidopsis APRR9 AT2G46790; C3 plants: Pisum sativum PRR59 FJ609179.1, Castanea sativa TOC1 AY611028.1, G. max TOC1 EU076435.1, Populus trichocarpa PRR9 XM002330094.1, Arabidopsis AIP1 AJ251086.1, Arabidopsis TOC1 AT5G61380, Arabidopsis APRR1 AB041530.1, B. rapa PRR1a GU219472.1, Arabidopsis TOC1-like AF272040.1, B. rapa PRR9 GU219476.1, Arabidopsis APRR3 NM125403.2, Arabidopsis APRR5 NM122355.3, and P. trichocarpa PRR XM002318466.1; CAM plant: M. crystallinum TOC1 AY371288.1; C4 plants: Z. mays TOC1 HQ003892.1 and Z. mays PRR1 HM452303.1.
(2) Query: Arabidopsis CCA1 AT2G4630; C3 plants: Populus nigra LHY1 AB429410.1, R. communis EST XM002515047.1, Arabidopsis MYP-TF AT2G46830, T. aestivum LHY HQ222606.1, G. max LCL2 EU076434.1, C. sativa LHY AY611029.1, Arabidopsis LHY NM_179237.1, S. lycopersicum EST BT012912.1, and Vitis vinifera EST XM002267684.1; CAM plant: M. crystallinum CCA1 AY371287.1; C4 plants: Z. mays LHY NM001138057 and Z. mays CCA1 HM452304.1.
Temporal Gene Expression Analysis by Q-PCR
Contigs highly homologous to Arabidopsis genes involved in CAM metabolism and the circadian cycle were retrieved from the OpuntiaESTdb using TBLASTN (Supplemental Table S1). Primers for an amplicon length between 266 and 349 bp were designed using the application PRIMEGENS version 2 in order to ensure unique hybridization within the EST database.
As plant material, 1-year-old O. ficus-indica cladodes, clonally derived from one mother plant, were grown in the greenhouse in a substrate of perlite:coconut fiber:floral substrate (ratio of 1:1:1) and watered as required. Cladodes were transferred to Sanyo MRL350 growth chambers for adaptation for 1 week and programmed for a light regime of 16 h of light and 8 h of darkness at day/night temperatures of 22°C/15°C and a photosynthetically active photon flux density of 250 mE s−1 m−2. Transversal, circular samples of 1 cm diameter were taken every 4 h from three plants. Total RNA extraction and cDNA synthesis as well as real-time PCR were performed as described above using the same PCR cycling conditions.
CT and efficiency values for each single PCR run were used as input for the qBasePlus software, which computed relative quantities against a normalization factor based on EF1 and ACT, which satisfied the geNorm stability measure. In order to detect rhythmic patterns, relative quantities were processed with the El Temps tool (http://www.el-temps.com/), which included a Fourier analysis comprising one or two harmonics. By this means, periodicity, Euler’s coefficients (a, b), acrophase, amplitude, phase, significance, Snedecor’s F, power, and cumulated power were obtained. Details about Fourier and Cosinor analyses are listed in Supplemental Tables S4 and S5.
The OpuntiaESTdb is freely available at http://srvgen.upct.es/opuntia/index.html (user: opuntia; password: Opuntia ficus-indica). All questions, comments, and requests may be sent by e-mail to julia.weiss@upct.es.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Work flow scheme of OpuntiaESTdb.
Supplemental Figure S2. EST functional categories.
Supplemental Figure S3. KEGG pathways for “carbon fixation in photosynthetic organisms.”
Supplemental Figure S4. Reference gene expression profiling.
Supplemental Table S1. Primer information and amplicon characteristics.
Supplemental Table S2. Characteristics of the OpuntiaESTdb sequences.
Supplemental Table S3. Optimal genes for expression quantification in individual and mixed organs.
Supplemental Table S4. Fourier analysis.
Supplemental Table S5. Cosinor analysis.
Acknowledgments
We thank Luis Pedro García and SEDIC for aid in conducting the computation-intensive part of this work as well as the Web server setup. Marta Pawluczyk is acknowledged for comments on the manuscript. Special thanks to Judith Strommer for help with editing.
References
- Andersen CL, Jensen JL, Ørntoft TF. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250 [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115: 721–733 [DOI] [PubMed] [Google Scholar]
- Besnard G, Pinçon G, D’Hont A, Hoarau JY, Cadet F, Offmann B. (2003) Characterisation of the phosphoenolpyruvate carboxylase gene family in sugarcane (Saccharum spp.). Theor Appl Genet 107: 470–478 [DOI] [PubMed] [Google Scholar]
- Binns D, Dimmer E, Huntley R, Barrell D, O’Donovan C, Apweiler R. (2009) QuickGO: a Web-based tool for Gene Ontology searching. Bioinformatics 25: 3045–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde RJ, Holbrook GP, Chollet R. (1985) Studies on the dark/light regulation of maize leaf pyruvate, orthophosphate dikinase by reversible phosphorylation. Arch Biochem Biophys 242: 283–290 [DOI] [PubMed] [Google Scholar]
- Carter PJ, Fewson CA, Nimmo GA, Nimmo HG, Wilkins MB. (1996) Role of circadian rhythms, light and temperature in the regulation of phosphoenolpyruvate carboxylase in Crassulacean acid metabolism. Winter K, Smith JAC, , Crassulacean Acid Metabolism: Biochemistry, Ecophysiology and Evolution. Springer, Berlin, pp 46–52 [Google Scholar]
- Cervigni GD, Paniego N, Díaz M, Selva JP, Zappacosta D, Zanazzi D, Landerreche I, Martelotto L, Felitti S, Pessino S, et al. (2008) Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostis curvula. Plant Mol Biol 67: 1–10 [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Failing CJ, Manandhar L, Zimmerman MA, Lakner MM, Nguyen TH. (2011) Functional evolution of C4 pyruvate,orthophosphate dikinase. J Exp Bot 62: 3083–3091 [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Fries JP, Vogel JA, Randklev CL, Vossen AP, Dittmer SK, Watkins EE, Fiedler LJ, Wacker SA, Meinhover KC, et al. (2002) Pyruvate,orthophosphate dikinase in leaves and chloroplasts of C3 plants undergoes light-/dark-induced reversible phosphorylation. Plant Physiol 128: 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O’Leary MH. (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 273–298 [DOI] [PubMed] [Google Scholar]
- Collazo-Siqués PV, Valverde ME, Paredes-López O, Guevara-Lara F. (2003) Expression of ripening-related genes in prickly pear (Opuntia sp.) fruits. Plant Foods Hum Nutr 58: 317–326 [DOI] [PubMed] [Google Scholar]
- Cui M, Miller PM, Nobel PS. (1993) CO2 exchange and growth of the Crassulacean acid metabolism plant Opuntia ficus-indica under elevated CO2 in open-top chambers. Plant Physiol 103: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC. (1992) Characterization and expression of a NADP-malic enzyme cDNA induced by salt stress from the facultative Crassulacean acid metabolism plant, Mesembryanthemum crystallinum. Eur J Biochem 208: 259–266 [DOI] [PubMed] [Google Scholar]
- Cushman JC. (1993) Molecular cloning and expression of chloroplast NADP-malate dehydrogenase during Crassulacean acid metabolism induction by salt stress. Photosynth Res 35: 15–27 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ. (1989a) Nucleotide sequence of the gene encoding a CAM specific isoform of phosphoenolpyruvate carboxylase from Mesembryanthemum crystallinum. Nucleic Acids Res 17: 6745–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ. (1989b) Nucleotide sequence of the Ppc2 gene encoding a housekeeping isoform of phosphoenolpyruvate carboxylase from Mesembryanthemum crystallinum. Nucleic Acids Res 17: 6743–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ. (1997) Molecular genetics of Crassulacean acid metabolism. Plant Physiol 113: 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Meiners MS, Bohnert HJ. (1993) Expression of a phosphoenolpyruvate carboxylase promoter from Mesembryanthemum crystallinum is not salt-inducible in mature transgenic tobacco. Plant Mol Biol 21: 561–566 [DOI] [PubMed] [Google Scholar]
- Cushman CJ, Taybi T, Bohnert HJ. (2000) Advances in Photosynthesis, Vol 9. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Wei Huang B, Lempicki R. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- de Montaigu A, Tóth R, Coupland G. (2010) Plant development goes like clockwork. Trends Genet 26: 296–306 [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- Ding Z, Doyle MR, Amasino RM, Davis SJ. (2007) A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 176: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng DF, Doolittle RF. (1987) Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol 25: 351–360 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, et al. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG. (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20: 333–342 [DOI] [PubMed] [Google Scholar]
- Hayes KR, Beatty M, Meng X, Simmons CR, Habben JE, Danilevskaya ON. (2010) Maize global transcriptomics reveals pervasive leaf diurnal rhythms but rhythms in developing ears are largely limited to the core oscillator. PLoS ONE 5: e12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda H, Akagi H, Shimada H. (2000) An isozyme of the NADP-malic enzyme of a CAM plant, Aloe arborescens, with variation on conservative amino acid residues. Gene 243: 85–92 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Higo K, Takano M. (2009) Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ 32: 592–603 [DOI] [PubMed] [Google Scholar]
- Keeley JE, Rundel PW. (2003) Evolution of CAM and C4 carbon-concentrating mechanism. Int J Plant Sci 164: S55–S77 [Google Scholar]
- Ku MS, Kano-Murakami Y, Matsuoka M. (1996) Evolution and expression of C4 photosynthesis genes. Plant Physiol 111: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H. (2008) Plant Physiological Ecology. Springer, New York [Google Scholar]
- Li B, Chollet R. (1994) Salt induction and the partial purification/characterization of phosphoenolpyruvate carboxylase protein-serine kinase from an inducible Crassulacean-acid-metabolism (CAM) plant, Mesembryanthemum crystallinum L. Arch Biochem Biophys 314: 247–254 [DOI] [PubMed] [Google Scholar]
- Li D, Stevens FJ, Schiffer M, Anderson LE. (1994) Mechanism of light modulation: identification of potential redox-sensitive cysteines distal to catalytic site in light-activated chloroplast enzymes. Biophys J 67: 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Kozma-Bognár L, Gould PD, Fehér B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ. (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 2005.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I, Lischewsky S, Weiss J, Hause B, Egea-Cortines M. (2010) Validation of endogenous genes as controls for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merida A, Rodriguez-Galan JM, Vincent C, Romero JM. (1999) Expression of the granule-bound starch synthase I (Waxy) gene from snapdragon is developmentally and circadian clock regulated. Plant Physiol 120: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson RK. (2003) Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. Int J Plant Sci 164: 43–54 [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman J-F, Hoffmann L, Evers D. (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914 [DOI] [PubMed] [Google Scholar]
- Nimmo GA, Wilkins MB, Fewson CA, Nimmo HG. (1987) Persistent circadian rhythms in the phosphorylation state of phosphoenolpyruvate carboxylase from Bryophyllum fedtschenkoi leaves and its sensitivity to inhibition by malate. Planta 170: 408–415 [DOI] [PubMed] [Google Scholar]
- Nimmo HG. (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5: 75–80 [DOI] [PubMed] [Google Scholar]
- Nobel PS, Andrade JL, Wang N, North GB. (1994) Water potentials for developing cladodes and fruits of a succulent plant, including xylem-versus-phloem implications for water movement. J Exp Bot 45: 1801–1807 [Google Scholar]
- Piechulla B. (1988) Plastid and nuclear messenger-RNA fluctuations in tomato leaves: diurnal and circadian rhythms during extended dark and light periods. Plant Mol Biol 11: 345–353 [DOI] [PubMed] [Google Scholar]
- Rascher U, Hütt MT, Siebke K, Osmond B, Beck F, Lüttge U. (2001) Spatiotemporal variation of metabolism in a plant circadian rhythm: the biological clock as an assembly of coupled individual oscillators. Proc Natl Acad Sci USA 98: 11801–11805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeille F, Hatch MD. (1986) Regulation of NADP-malate dehydrogenase in C4 plants: relationship among enzyme activity, NADPH to NADP ratios, and thioredoxin redox states in intact maize mesophyll chloroplasts. Arch Biochem Biophys 249: 171–179 [DOI] [PubMed] [Google Scholar]
- Resco V, Hartwell J, Hall A. (2009) Ecological implications of plants’ ability to tell the time. Ecol Lett 12: 583–592 [DOI] [PubMed] [Google Scholar]
- Salomé PA, McClung CR. (2005) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed O. (2001) Crassulacean acid metabolism 1975–2000, a check list. Photosynthetica 39: 339–352 [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Scott T, Mercer EI. (1997) Concise Encyclopedia of Biochemistry and Molecular Biology, Ed 3. de Gruyter, New York [Google Scholar]
- Segura S, Scheinvar L, Olalde G, Leblanc O, Filardo S, Muratalla A, Gallegos C, Flores C. (2007) Genome sizes and ploidy levels in Mexican cactus pear species Opuntia (Tourn.) Mill. series Streptacanthae Britton et Rose, Leucotrichae DC., Heliabravoanae Scheinvar and Robustae Britton et Rose. Genet Resour Crop Evol 54: 1033–1041 [Google Scholar]
- Smith JAC, Winter K. (1996) Taxonomic distribution of Crassulacean acid metabolism. Winter K, Smith JAC, , Crassulacean Acid Metabolism: Biochemistry, Ecophysiology and Evolution. Springer, New York, pp 427–436 [Google Scholar]
- Somers DE, Devlin PF, Kay SA. (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Spiess AN, Feig C, Ritz C. (2008) Highly accurate sigmoidal fitting of real-time PCR data by introducing a parameter for asymmetry. BMC Bioinformatics 9: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava GP, Guo J, Shi H, Xu D. (2008) PRIMEGENS-v2: genome-wide primer design for analyzing DNA methylation patterns of CpG islands. Bioinformatics 24: 1837–1842 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Takata N, Saito S, Saito CT, Uemura M. (2010) Phylogenetic footprint of the plant clock system in angiosperms: evolutionary processes of Pseudo-Response Regulators. BMC Evol Biol 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC. (2000) A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in Crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol 123: 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal J, Chollet R. (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2: 230–237 [Google Scholar]
- Weiss J, Nerd A, Mizrahi Y. (1993) Vegetative parthenocarpy in the cactus pear Opuntia ficus-indica (L.) Mill. Ann Bot 72: 521–526 [Google Scholar]
- Winter K, Garcia M, Holtum JAM. (2008) On the nature of facultative and constitutive CAM: environmental and developmental control of CAM expression during early growth of Clusia, Kalanchöe, and Opuntia. J Exp Bot 59: 1829–1840 [DOI] [PubMed] [Google Scholar]
- Wu J, Mao X, Cai T, Luo J, Wei L. (2006) KOBAS server: a Web-based platform for automated annotation and pathway identification. Nucleic Acids Res (Web Server issue) 34: W720– W724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyka TP, Bohn A, Duarte HM, Kaiser F, Lüttge UE. (2004) Perturbations of malate accumulation and the endogenous rhythms of gas exchange in the Crassulacean acid metabolism plant Kalanchoë daigremontiana: testing the tonoplast-as-oscillator model. Planta 219: 705–713 [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, et al. (2006) WEGO: a Web tool for plotting GO annotations. Nucleic Acids Res (Web Server issue) 34: W293– W297 [DOI] [PMC free article] [PubMed] [Google Scholar]