Abstract
Arabidopsis (Arabidopsis thaliana) SALT TOLERANCE/B-BOX ZINC FINGER PROTEIN24 (STO/BBX24) is a negative regulator of the light signal transduction that localizes to the nucleus of plant cells and interacts with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) in the yeast (Saccharomyces cerevisiae) two-hybrid system. The protein contains two B-box zinc-finger motives at the N terminus and a conserved motif at the C-terminal part required for the interaction with COP1. BBX24 accumulates during deetiolation of young seedlings in the first hours of exposure to light. However, this accumulation is transient and decreases after prolonged light irradiation. Here, we identified the amino acidic residues necessary for the nuclear import of the protein. In addition, we created mutated forms of the protein, and analyzed them by overexpression in the bbx24-1 mutant background. Our results indicate that the degradation of BBX24 occurs, or at least is initiated in the nucleus, and this nuclear localization is a prerequisite to fulfill its function in light signaling. Moreover, mutations in the region responsible for the interaction with COP1 revealed that a physical interaction of the proteins is also required for degradation of BBX24 in the light and for normal photomorphogenesis.
Plants have to adapt to a number of environmental factors during their life cycle. Among them, light is the most important cue that directs different developmental processes throughout the whole life (Franklin et al., 2005). The effect of light is especially prominent during seedling growth. Whereas dark-grown seedlings are pale in color, exhibit long hypocotyls, closed cotyledons, and an apical hook, light-grown seedlings undergo the photomorphogenesis process and are green, with shorter hypocotyls and expanded cotyledons (Yi and Deng, 2005). During skotomorphogenesis, light signaling components are suppressed by the activities of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), SUPPRESSOR OF PHYTOCHROME A proteins, PHYTOCHROME INTERACTING FACTORS, DEETIOLATED1, and the COP9 signalosome (Yi and Deng, 2005; Leivar et al., 2008, 2009). Mutations in COP1 cause constitutive photomorphogenesis phenotypes, with dark-grown mutant seedlings displaying features of light-grown seedlings (Kim et al., 2002).
COP1 functions as an E3 ubiquitin ligase. In darkness, COP1 is mainly localized to the nucleus, where it presumably targets photomorphogenesis-promoting transcription factors, such as ELONGATED HYPOCOTYL5 (HY5), LONG AFTER FAR-RED LIGHT1, and LONG HYPOCOTYL IN FAR-RED for ubiquitination and degradation, thereby repressing the expression of photomorphogenesis genes. In the light, active photoreceptors inhibit COP1 function and activators of the light response are no longer ubiquitylated and degraded. However, even in the light, there is residual COP1 function that prevents overstimulation by light (Dieterle et al., 2003; Yi and Deng, 2005).
SALT TOLERANCE/B-BOX ZINC FINGER PROTEIN24 (STO/BBX24) was initially characterized as a protein conferring salt tolerance to yeast (Saccharomyces cerevisiae; Lippuner et al., 1996). In Arabidopsis (Arabidopsis thaliana), salt treatment does not induce STO/BBX24 gene expression (Lippuner et al., 1996; Nagaoka and Takano, 2003) although, it was reported that overexpression of the protein confers enhanced salt tolerance to Arabidopsis transgenic plants (Nagaoka and Takano, 2003). Recent studies revealed a function of STO/BBX24 as a negative regulator during photomorphogenesis (Indorf et al., 2007).
STO/BBX24 belongs to an Arabidopsis family of proteins, BBX, characterized for presenting one or two B-box zinc (Zn)-finger motives at the N terminus (Kumagai et al., 2008; Khanna et al., 2009). In addition to STO/BBX24, there are other members of the double B-box Zn-finger subfamily that function as positive or negative regulators of light signaling (Datta et al., 2007, 2008; Chang et al., 2008; Kumagai et al., 2008). The BBX family also include CONSTANS (CO) and CO-LIKE (COL) proteins (Putterill et al., 1995; Lagercrantz and Axelsson, 2000; Griffiths et al., 2003), and another subfamily of single B-box proteins. About this last subfamily, almost no information on the function of its members is available.
BBX24 interacts with COP1 in the yeast two-hybrid system (Holm et al., 2002) and colocalizes with it in plant cells (Indorf et al., 2007). It contains two B-box Zn fingers situated in tandem in the N-terminal part of the protein, whereas at the C terminus, several amino acid residues necessary for the interaction with COP1 have been identified (Holm et al., 2001). Furthermore, two different point mutations, exchanging BBX24 amino acid residues V244A and P245A are sufficient to prevent the interaction between BBX24 and COP1 in the yeast two-hybrid system (Holm et al., 2001). BBX24 accumulates in the nucleus of cells during seedling deetiolation. This accumulation occurs only during the first hours of exposure to white light and decreases after prolonged light irradiation. In darkness, COP1 mediates BBX24 degradation (Indorf et al., 2007).
To further characterize BBX24 function, we identified the nuclear localization signal (NLS) of the protein and produced mutated versions that either prevent the nuclear import of the protein or the interaction with COP1. We further investigated the effect of these mutations by overexpressing them in the bbx24-1 mutant background and analyzed the light-dependent inhibition of the hypocotyl elongation in the resulting transgenic plants. In addition, we analyzed the overexpression of a truncated version of the protein containing only the two B-box Zn-finger domains.
RESULTS
Characterization of STO/BBX24 NLS
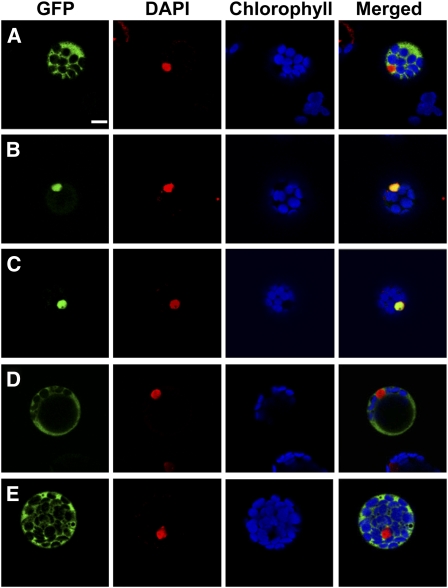
BBX24 localizes in the nucleus (Indorf et al., 2007). To determine its NLS, constructs carrying the full-length coding region of BBX24, or different deletions of the cDNA were fused in frame to the EGFP cDNA under the control of the cauliflower mosaic virus 35S promoter. To avoid nuclear accumulation of small Mr proteins by diffusion through the nuclear pores, the cDNA coding for GUS was placed between the EGFP and the BBX24 deletions. These deletion constructs were used to transfect Arabidopsis protoplasts and the subcellular localization of the transiently expressed proteins was analyzed by confocal laser-scanning microscopy (CLSM). As previously reported (Indorf et al., 2007), the EGFP-GUS-BBX24 protein fusion was detected exclusively in the nucleus (Fig. 1B), whereas the EGFP-GUS fusion used as control, was detected entirely in the cytosol (Fig. 1A). Analysis of the amino acid sequence of BBX24 revealed a putative NLS comprising the amino acids 226 to 229 (KKPR) in the C-terminal part of the protein. According to this analysis, a fragment containing the last 26 amino acids of the protein (residues 223–248; C26-BBX24), and another fragment containing only the last 16 amino acids (residues 233–248; C16-BBX24) and therefore excluding the KKPR motif, were fused to the EGFP-GUS chimeric protein. The 26-amino acid fragment was sufficient to direct the fusion protein into the nucleus (Fig. 1C) whereas in the absence of the KKPR motif, the fusion protein localized exclusively in the cytosol (Fig. 1D). Further detailed analysis of the NLS motif was performed by introducing a point mutation into the full-length BBX24 cDNA. The exchange of Lys-226 to Asn (bbx24NLS) was sufficient to prevent nuclear import of the fusion protein (Fig. 1E).
Figure 1.
Subcellular localization of BBX24 deletion fragments. Arabidopsis protoplasts were transfected with constructs carrying different EGFP fusions and analyzed after 24 h by CLSM. Sections show fluorescence images from the control EGFP-GUS, and translational fusions of BBX24 fragments and mutations (A): EGFP-GUS-BBX24 (B); EGFP-GUS-C26-BBX24 including the KKPR motif (C); EGFP-GUS-C16-BBX24 excluding the KKPR motif (D); and EGFP-bbx24NLS (E). Bar = 10 μm.
Nuclear Localization Is Required for the Degradation and the Function of BBX24
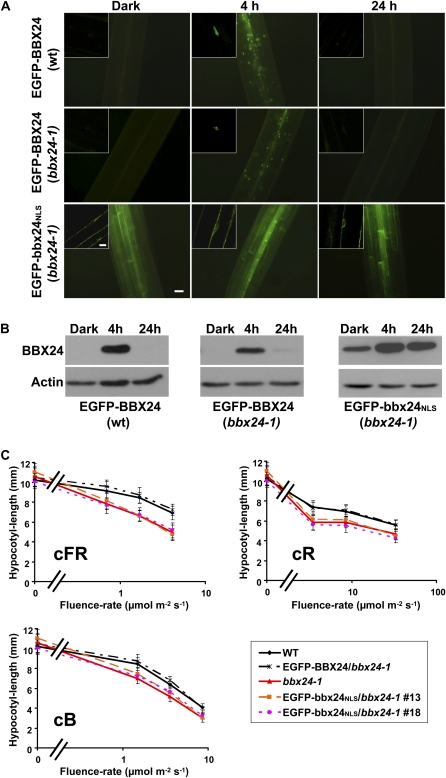
Accumulation of EGFP-BBX24 occurs during deetiolation in the first hours of exposure to white light but this accumulation stops after prolonged light exposure (Indorf et al., 2007). Transgenic lines carrying N-terminal or C-terminal EGFP fusions showed the same dynamics of protein accumulation during deetiolation treatments, varying only in the intensity of the GFP signal in different overexpressing lines (Supplemental Fig. S1A). In addition, both types of transgenic lines display similar photomorphogenic phenotypes under monochromatic light (Supplemental Fig. S1B). To investigate the biological relevance of the nuclear localization of BBX24, we generated transgenic homozygous bbx24-1 lines constitutively expressing EGFP-BBX24 or EGFP-bbx24NLS. The subcellular localization of EGFP-BBX24 and EGFP-bbx24NLS in 5-d-old etiolated seedlings was analyzed immediately after the period of growth in darkness, or after transfer to continuous white light (cWL; 120 μmol m−2 s−1). White light treatment of the overexpressor EGFP-BBX24, or the EGFP-BBX24/bbx24-1 lines led to the accumulation of the fusion protein in the nucleus (Fig. 2A). However, in the line overexpressing the mutated version of the protein (EGFP-bbx24NLS) cytosolic, but not nuclear GFP already appeared in the dark-grown seedlings. Moreover, after prolonged light exposure, even at time points where the EGFP-BBX24 signal diminished, the EGFP-bbx24NLS expressing plants still displayed a clear fluorescence signal (Fig. 2A).
Figure 2.
Analysis of transgenic bbx24-1 lines expressing EGFP-bbx24NLS. A, Five-day-old dark-grown wild-type or bbx24-1 seedlings overexpressing EGFP-BBX24, and transgenic bbx24-1 seedlings overexpressing EGFP-bbx24NLS were analyzed by fluorescence microscopy before (Dark), and after being transferred to cWL at the indicated time points. Images taken from hypocotyls are shown. Bar = 50 μm. Inserted sections are single-cell images analyzed by CLSM. Bar = 10 μm. B, Western-blot analysis of similarly treated plant material for the detection of EGFP-BBX24 or EGFP-bbx24NLS fusions. Antibodies against BBX24 or actin were used for the immune detection. C, Hypocotyl-length measurements of 5-d-old wild-type, bbx24-1, and bbx24-1 overexpressing EGFP-BBX24 or EGFP-bbx24NLS, grown under different fluence rates of cR, cFR, and cB light.
Western-blot analysis on protein extracts of the 5-d-old transgenic seedlings revealed an approximately 60-kD mass band that corresponds to the expressed fusion protein. The expression pattern resembled the microscopy observations, except for the weaker band present in the EGFP-bbx24NLS dark samples (Fig. 2B). This band correlates to the lower amount of EGFP-bbx24NLS transcripts in the dark (Supplemental Fig. S2). These results indicate that degradation of BBX24 in the dark and after prolonged light exposure mostly takes place, or at least initiates in the nucleus.
BBX24 is a negative regulator of red, far-red, and blue-light-mediated inhibition of hypocotyl elongation (Indorf et al., 2007). To investigate whether the nuclear localization of the protein is a prerequisite for its function, we carried out light fluence-rate response experiments with the above-described transgenic lines. Seeds were germinated on filter paper and grown for 5 d at 22°C under continuous red (cR; 4.7–44 μmol m−2 s−1), far-red (cFR; 0.43–2.5 μmol m−2 s−1), or blue light (cB; 1.3–7.3 μmol m−2 s−1). As previously reported (Indorf et al., 2007), bbx24-1 exhibits under monochromatic light fluences a pronounced inhibition of hypocotyl growth compared to the wild type. Independent EGFP-bbx24NLS/bbx24-1 transgenic lines displayed a similar reduction of hypocotyl elongation, as did bbx24-1, whereas dark-grown seedlings exhibited similar hypocotyl length as the wild type. This indicates that the mutant phenotype is not rescued by the overexpression of EGFP-bbx24NLS (Fig. 2C). As positive control, a line overexpressing the wild-type BBX24 fused to the EGFP in the mutant background (p35S:EGFP-BBX24/bbx24-1) was used throughout the experiment. The amount of BBX24-EGFP was reduced in the bbx24-1 background relative to the corresponding overexpressing line (Fig. 2B). Nevertheless, the expression level was sufficient to rescue the bbx24-1 mutation, indicating that ectopic expression of the wild-type protein complements BBX24 function.
These results provide evidence that overexpression of BBX24 in the cytosol does not rescue the bbx24-1 phenotype, and therefore argue that the localization of BBX24 in the nucleus is essential to fulfill its function in the light-dependent hypocotyl elongation.
As already mentioned, BBX24 confers tolerance to yeast cells growing on lithium chloride (LiCl) media (Lippuner et al., 1996). To verify into which extent the introduced point mutation might affect the function of the protein, we expressed BBX24, EGFP-bbx24NLS, or a new protein fusion containing EGFP-bbx24NLS with an additional SV40 NLS (EGFP-bbx24NLS-SV40NLS) in yeast. Yeast cells expressing either the negative control or EGFP-bbx24NLS did not grow on media containing 350 mm LiCl but expression of EGFP-bbx24NLS-SV40NLS conferred similar tolerance to high LiCl concentrations as the wild-type protein (Supplemental Fig. S3A). In addition, we also found that BBX24 hosts intrinsic transcription activity (autoactivation) when used in the yeast two-hybrid system. We also used this property to test the function of EGFP-bbx24NLS and found no difference in the growth capacity on selection media between cells expressing BBX24 and those expressing EGFP-bbx24NLS in the nucleus. In both cases, the fusion to the DNA binding domain provided in the bait vector of the yeast two hybrid allowed the growth of the cells on SD−Trp/−His (selective drop-out) medium containing 3-amino-1,2,4-triazole (Supplemental Fig. S3B), demonstrating that the introduced point mutation does not affect the autoactivation activity of the protein. These findings indicate that also in yeast the function of BBX24 is strongly linked to its nuclear localization.
BBX24 C-Terminal Part Is Necessary for the Protein Turnover
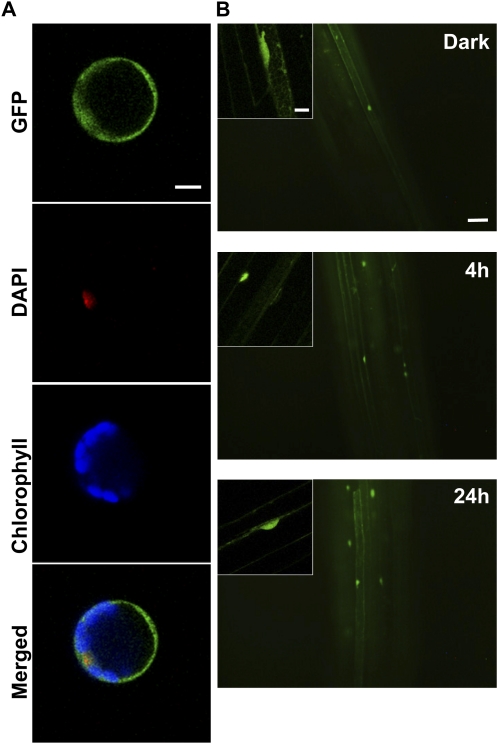
Cys-rich B-box Zn-finger domains mediate protein-protein, and protein-DNA interactions (Khanna et al., 2009). BBX24 contains two tandem repeat B-box Zn fingers at the N-terminal part of the protein. Transient expression assays in Arabidopsis protoplasts revealed that the EGFP fusion protein containing the N terminus of BBX24 (EGFP-N-BBX24) with the two B-box Zn fingers (amino acid 1–144) is present in the nucleus and the cytosol (Fig. 3A). To unravel possible roles for these domains, we produced homozygous transgenic bbx24-1 lines containing EGFP-N-BBX24 under the control of the 35S cauliflower mosaic virus promoter. Out of 10 independent monogenic lines, two showing the highest GFP signal were selected for further experiments. In 5-d-old etiolated seedlings, EGFP-N-BBX24 was present in the cytosol and in the nucleus of cells. Furthermore, the fluorescence signal remained in the cells after prolonged light treatment, indicating that the C-terminal part of BBX24 is required for the rapid turnover of the protein (Fig. 3B). We analyzed the level of expression of the chimeric protein by western blot using polyclonal antibodies raised against GFP. Seedlings of wild type, p35S:EGFP-BBX24/bbx24-1, and p35S:EGFP-N-BBX24/bbx24-1 were grown in darkness and transferred to cWL for 4 or 24 h. The results showed an approximately 60-kD mass band after 4 h induction in the p35S:EGFP-BBX24/bbx24-1 sample but no band could be detected in the p35S:EGFP-N-BBX24/bbx24-1 lines (data not shown). We performed quantitative reverse transcription PCR analysis to estimate the amount of transcripts of the transgene, and to compare it to the endogenous amount of transcripts in the wild type. The level of EGFP-N-BBX24 transcripts was evidently much lower than the amount of BBX24 (Supplemental Fig. S4). In addition, we carried out light fluence-rate response experiments to investigate whether the small fraction of the fusion protein that accumulated in the nucleus was sufficient to complement BBX24 function. However, the results showed that the bbx24-1 phenotype in those lines was not rescued (data not shown).
Figure 3.
Analysis of the expression of EGFP-N-BBX24. A, Arabidopsis protoplasts were transfected with the N-terminal region of BBX24 fused to EGFP and analyzed after 24 h. The representative picture shows the EGFP signal observed by confocal microscopy. Bar = 10 μm. B, Five-day-old dark-grown transgenic bbx24-1 seedlings overexpressing EGFP-N-BBX24 were analyzed by fluorescence microscopy before (Dark), and at the indicated time points after being transferred to cWL. Images taken from hypocotyls are shown. Bar = 50 μm. Inserted sections are single-cell images analyzed by CLSM. Bar = 10 μm.
Colocalization Analysis of Mutated BBX24 Proteins with COP1
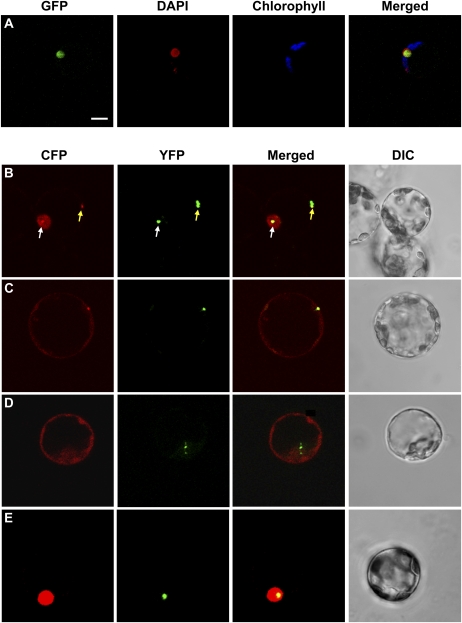
As mentioned before, a physical interaction occurs in the yeast two-hybrid system assay via the WD40 repeat of COP1 and identified conserved amino acid residues present in BBX24, HY5, and STH/BBX25 (Holm et al., 2001; Ma et al., 2002; Yi and Deng, 2005). To analyze whether the interaction with COP1 affects BBX24 function, we introduced two point mutations that exchange the residues V244A and P245A of BBX24 into the BBX24 cDNA (EGFP-bbx24COP1). The introduced amino acidic changes, already described by Holm et al. (2001), prevent the interaction of both proteins in the yeast two-hybrid system but do not change the subcellular localization pattern in protoplasts (Fig. 4A). To confirm that in fact those mutations hamper the interaction with COP1 in plant cells, we coexpressed ECFP-bbx24COP1 and EYFP-COP1 in Arabidopsis protoplasts. In addition, we also tested the localization of ECFP-bbx24NLS and ECFP-N-BBX24 together with EYFP-COP1. COP1 accumulates strongly in both the nucleus and the cytosol, forming aggregates of different sizes in both cellular compartments (Stacey et al., 1999). As previously reported (Indorf et al., 2007), single transfection of ECFP-BBX24 led to a diffuse distribution of the protein in the nucleus (Fig. 1B), whereas coexpression of BBX24-ECFP and EYFP-COP1 resulted in colocalization of both fusion proteins in the same nuclear, and cytosolic aggregates (Fig. 4B). The mutation introduced in the NLS led to the localization of ECFP-bbx24NLS exclusively in the cytosol in a diffuse manner (Fig. 1E). However, coexpression of ECFP-bbx24NLS and EYFP-COP1 resulted in the colocalization of both proteins to the same cytosolic aggregations (Fig. 4C). Coexpression of ECFP-N-BBX24 and EYFP-COP1 did not display any colocalization signal, and ECFP-N-BBX24 still distributed diffusely in both cell compartments (Fig. 4D). In the case of ECFP-bbx24COP1, single transfection or cotransfection with EYFP-COP1 invariably resulted in a diffuse distribution of the ECFP in the nucleus (Fig. 4, A and E), demonstrating that the described point mutations introduced into BBX24 were sufficient to prevent COP1 colocalization also in plant cells.
Figure 4.
Subcellular localization of bbx24COP1, and colocalization analysis of BBX24 variants and COP1 by transient expression. Arabidopsis protoplasts were transfected with EGFP-bbx24COP1 (A), or cotransfected with BBX24 wild-type or mutant proteins (B–E), and COP1 fused to ECFP and EYFP, respectively. Representative images of the fluorescence signals detected by CLSM of cells coexpressing ECFP-BBX24 and EYFP-COP1 (nuclear or cytosolic colocalization are indicated with a white or a yellow arrow, respectively; B); ECFP-bbx24NLS and EYFP-COP1 (C); ECFP-N-BBX24 and EYFP-COP1 (D); and ECFP-bbx24COP1 and EYFP-COP1 (E). Bar = 10 μm.
Interaction with COP1 Is Required for the Degradation of BBX24 in Light
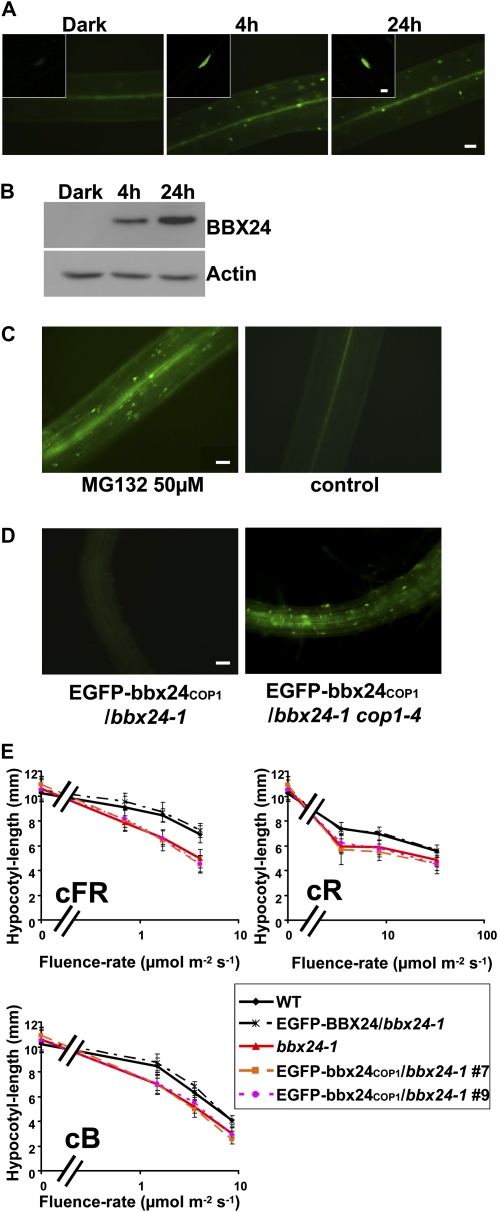
To investigate in planta the effect of the bbx24COP1 mutation, we generated transgenic lines overexpressing EGFP-bbx24COP1 in the bbx24-1 mutant, and analyzed the subcellular localization in 5-d-old seedlings. In etiolated seedlings, and in seedlings treated for 4 h with cWL, EGFP-bbx24COP1 displayed similar behavior as the wild-type protein fusion EGFP-BBX24. Thus, we did not observe any GFP fluorescence in the dark whereas after few hours of light treatment fluorescent nuclei appeared in the cells (Fig. 5A). However, after 24 h of continuous light exposure, only the EGFP-bbx24COP1/bbx24-1 lines showed a fluorescence signal (Fig. 5A). Western-blot analysis confirmed these results (Fig. 5B), indicating that mutations affecting the interaction with COP1 stabilize the protein in prolonged light exposure.
Figure 5.
Analysis of bbx24-1 lines expressing EGFP-bbx24COP1. A, Five-day-old dark-grown bbx24-1 seedlings overexpressing EGFP-bbx24COP1 were analyzed with fluorescence microscopy in darkness (Dark), and at the indicated time points of cWL irradiation. The pictures represent images taken from hypocotyls. Bar = 50 μm. Small sections are confocal images of single cells. Bar = 10 μm. B, Western-blot analysis of the same material treated as above for the detection of EGFP-bbx24COP1. C, Five-day-old dark-grown EGFP-bbx24COP1/bbx24-1 seedlings were treated with a solution containing the proteasome inhibitor MG132, or without it as control. After 24 h of treatment, fluorescence microscopy was used to investigate the presence of the fusion protein. The pictures represent images taken from hypocotyls. Bar = 50 μm. D, Analysis of EGFP-bbx24COP1 expression in 5-d-old dark-grown bbx24-1 or cop1-4 seedlings with fluorescence microscopy. The pictures represent images taken from roots. Bar = 50 μm. E, Hypocotyl-length measurements of 5-d-old wild-type, bbx24-1, and bbx24-1 overexpressing EGFP-BBX24 or EGFP-bbx24COP1 grown under different fluence rates of cR, cFR, and cB light.
To investigate the nature of the degradation process that takes place in the dark, etiolated seedlings overexpressing EGFP-bbx24COP1 were treated with the proteasomal inhibitor MG132 and analyzed after 24 h by fluorescence microscopy. Whereas the control seedlings did not display any fluorescence signal, seedlings treated with the inhibitor exhibited several fluorescent nuclei in hypocotyls (Fig. 5C), demonstrating that the degradation of the protein in the dark depends on the 26S proteasome. BBX24-EGFP accumulates in the cop1-4 mutant in the dark and this result suggested a COP1-dependent degradation of the protein in darkness (Indorf et al., 2007). However, our results indicated that degradation of BBX24 occurs even if the direct interaction with COP1 is hampered (Fig. 5, A and B). We therefore investigated the stability of the mutant protein fusion EGFP-bbx24COP1 in the cop1-4 background. Crosses between cop1-4 and one of the EGFP-bbx24COP1/bbx24-1 transgenic lines were produced and the F2 generation was analyzed after growing the seedlings for 5 d in the dark. Approximately 25% of the siblings exhibited a cop1-4 phenotype in the dark. The analysis of the etiolated cop1-4 progeny revealed that around 75% of this population exhibited GFP signal in the nuclei of root cells (Fig. 5D). However, due to the strong autofluorescence displayed by these seedlings we were unable to detect clear GFP signal in other tissues. In the rest of the progeny (i.e. seedlings displaying long hypocotyls) the GFP signal could not be detected, suggesting that the degradation of EGFP-bbx24COP1 is COP1 dependent.
Interaction with COP1 Is Required for BBX24 Function in Photomorphogenesis
To test whether the interaction with COP1 is relevant for the function of BBX24, we carried out light fluence-rate response experiments using the above-mentioned transgenic lines overexpressing EGFP-bbx24COP1 in the bbx24-1 mutant. We could not observe differences between the hypocotyls length of the EGFP-bbx24COP1/bbx24-1 transgenic lines and the bbx24-1 mutant, whereas the positive control could rescue the bbx24-1 phenotype (Fig. 5E). This indicates that the overexpression of EGFP-bbx24COP1 does not complement BBX24 function. These results support the idea that the interaction with COP1 is necessary for the correct function of BBX24. To verify that the observed result is not due to an effect of the introduced mutations, causing for example misfolding of the protein, we performed yeast assays as described above. We found that the introduced point mutations did not affect either the salt tolerance capacity nor the autoactivation activity of the protein in yeast (Supplemental Fig. S5).
DISCUSSION
In this study, we identified the NLS of STO/BBX24 and investigated the biological functions of different domains of the protein by expressing mutant molecules in bbx24-1.
A detailed deletion analysis of BBX24 revealed a four-amino acid sequence (KKPR) that serves as functional NLS. Furthermore, a point mutation introduced in this motif led to the localization of the EGFP-bbx24NLS fusion protein exclusively to the cytosol, demonstrating that this NLS is necessary for the nuclear uptake of the protein. The KKPR motif, already characterized as NLS in mammalian systems (Krauer et al., 2004), was only suggested for plant cells (Hosoda et al., 2002). This analysis represents a detailed characterization of this motif as a functional NLS in plants. In eukaryotes, the functions of proteins correlate to their locations, and many proteins lose their functions upon disruption of targeting signals such as NLSs (Moes et al., 2008; Zhang et al., 2010). At the same time, there are also proteins that share functions in different compartments (Edgington and Futcher, 2001). In the case of BBX24, we could show that its function is placed exclusively in the nucleus, since the overexpression of the cytosolic version of the protein (EGFP-bbx24NLS) in bbx24-1 did not rescue the mutant phenotype. Taking advantage of the capacity of BBX24 to confer resistance to high salt concentrations in yeast, we could exclude that the introduced point mutation completely abolishes the functionality of the protein.
BBX24 belongs to a subfamily of B-box Zn-finger proteins of Arabidopsis whose members contain two B-box Zn-finger domains in tandem at the N terminus. This subfamily includes eight putative transcription factors recently renamed according to Khanna et al. (2009) as BBX18 to BBX25. Interestingly, most of these proteins are factors involved in photomorphogenesis (Datta et al., 2007, 2008; Indorf et al., 2007; Chang et al., 2008; Kumagai et al., 2008). Apart from STO/BBX24, at least other three members of this subfamily (STH2/BBX21, LZF1/STH3/BBX22, and STH/BBX25) localize to the nucleus of plant cells (Indorf et al., 2007; Datta et al., 2007, 2008; K. Marquardt, unpublished data). For two of them (STH2/BBX21 and LZF1/STH3/BBX22) transcriptional activity through G-box regulatory elements has been proposed (Datta et al., 2008). Our results showed that for a proper function of STO/BBX24, the protein requires nuclear localization. This, together with the described autoactivation activity in the yeast two-hybrid assay, would suggest also a regulation of transcriptional activity by STO/BBX24 through direct or indirect recognition of promoter regions of downstream target genes. In addition, the increased stability of bbx24NLS suggests that the protein requires nuclear localization to be targeted for degradation.
As shown earlier, a physical interaction between BBX24 and COP1 occurs in the yeast two-hybrid system assay (Holm et al., 2001). Also, transient expression of BBX24-ECFP and EYFP-COP1 in plant cells resulted in colocalization of both fusion proteins in the same nuclear and cytosolic aggregates, whereas single transfection of ECFP-BBX24 led to a diffused accumulation of the protein in the nucleus (Indorf et al., 2007). The coexpression of ECFP-bbx24NLS and EYFP-COP1 also resulted in the colocalization of both proteins. However, in this case, since BBX24 was not anymore imported into the nucleus, the colocalization occurred only in the cytosolic aggregates. Nevertheless, this cytosolic colocalization, possibly indicative of a direct or indirect interaction of the proteins, does not seem to have any physiological relevance, since the plants overexpressing the cytosolic version of BBX24 did not display any overt phenotype.
In the next step, we analyzed the two B-box Zn-finger domains of BBX24. Cys-rich B-box Zn fingers mediate protein-protein and/or protein-DNA interactions (Borden et al., 1995; Rushton et al., 1995; Yang et al., 2006; Fujioka et al., 2007; Khanna et al., 2009). As expected, this protein fragment did not colocalize with EYFP-COP1 neither in the cytosol nor in the nucleus. Transformation of bbx24-1 with p35S:EGFP-N-BBX24 resulted in transgenic lines with relatively low level of transcripts of the transgene. This could be due to a positional effect of the integrations, or to the instability of the transcript. Nevertheless, two transgenic lines that expressed higher protein levels, as judged by fluorescence microscopy, were analyzed in light fluence-rate response experiments. Although the lines displayed fluorescence signal in the nucleus and the cytosol, the chimeric protein failed to rescue the mutant phenotype, leaving open the question whether the N terminus might have a function on its own. However, CLSM analyses of the transgenic lines revealed that the protein was present in etiolated seedlings, and after 4 and 24 h of light treatment, demonstrating that the C-terminal part is essential for the degradation of the protein.
Finally, we focused on the biological relevance of the COP1 interaction with BBX24. COP1 is an E3 ubiquitin ligase. In darkness, it localizes in the nucleus where it targets photomorphogenesis-promoting transcription factors for degradation. Interestingly, COP1 also interacts and/or promotes degradation of other B-box Zn-finger proteins as BBX1/CO (Jang et al., 2008; Liu et al., 2008), BBX4/COL3 (Datta et al., 2006), and BBX22/LZF1 (Chang et al., 2011). Moreover, the dynamics of accumulation and degradation of BBX22/LZF1 (Chang et al., 2011) are very similar to those of BBX24, although both proteins have opposite functions during seedling photomorphogenesis. This indicates clearly that the tight regulation of the balance between positive and negative regulating factors during seedling deetiolation is crucial for proper seedling development. However, in contrast to BBX24, BBX22 does not directly interact with COP1 (Datta et al., 2008).
Previous results by Indorf et al. (2007), suggested that the functional significance of the interaction between COP1 and BBX24 was the degradation of BBX24 in darkness. However, the overexpression of BBX24 in the cop1-4 mutant did not result in an additive phenotype. Therefore, to distinguish whether this interaction is necessary for BBX24 function or if it is needed only for targeting BBX24 for degradation, we introduced two point mutations into the BBX24 cDNA (EGFP-bbx24COP1) that prevent interaction of both proteins in the yeast two-hybrid system (Holm et al., 2001). By transient expression in protoplasts, we could verify that the introduced mutations affected the colocalization of the proteins. However, we could not detect accumulation of this fusion protein in transgenic etiolated seedlings, which suggests that direct interaction with COP1 is not the reason for the destabilization of the protein in the dark. Nevertheless, the degradation of BBX24 in darkness is still occurring via the 26S proteasome, since the proteasome inhibitor MG132 blocks this process. As mentioned above, other BBX proteins interact with COP1 and it has been proposed that COP1 is directly responsible for their degradation in darkness (Jang et al., 2008; Liu et al., 2008; Chang et al., 2011). However, those results were obtained using in vitro assays, or by monitoring the accumulation of the wild-type proteins in planta. Our results overexpressing the mutated EGFP-bbx24COP1 in the cop1-4 mutant demonstrate that the degradation in the dark is not a result of the direct interaction with COP1 but is rather due to a downstream factor regulated by COP1.
In light, Indorf et al. (2007) did not observe a dramatic change of the GFP accumulation in the cop1-4 lines overexpressing BBX24-EGFP, but they monitored the GFP accumulation only during the first hours of light treatment. In our work, the analysis of EGFP-bbx24COP1/bbx24-1 transgenic lines revealed that COP1 is required for the degradation of BBX24 in light, since mutations in the COP1 binding region resulted in the accumulation of the protein during prolonged light exposure. This regulation of BBX24 expression by COP1 provides more evidence for COP1 function in the light, a phenomenon supported by numerous lines of evidence. For example, weak alleles of cop1 exhibit reverse photomorphogenesis responses and COP1 overexpressors present enhanced photomorphogenesis under red light (Boccalandro et al., 2004). In addition, COP1 was recently found to regulate UV-B-induced photomorphogenesis and stress acclimation together with UVR8 in Arabidopsis (Favory et al., 2009). However, the COP1-dependent degradation of BBX24 in the light does not explain the phenotypes observed in the EGFP-bbx24COP1/bbx24-1 transgenic lines. Light fluence-rate response experiments showed that the overexpression of bbx24COP1 does not rescue the bbx24-1 phenotype, implying that even if the protein is present in the nucleus and stable, the lack of interaction with COP1 renders it nonfunctional. Thus, the interaction with COP1 seems to be necessary not only for the rapid turnover of the protein but also for the proper function of BBX24. COP1 is a ubiquitin ligase and multiubiquitin chains are required for efficient recognition and degradation of ubiquitylated proteins by the proteasome (Yi and Deng, 2005). However, ubiquitin is a posttranslational modification that also regulates processes unrelated to degradation; these range from membrane transport to transcriptional regulation. Instead of targeting proteins for degradation through the proteasome, single ubiquitin or short ubiquitin chains mark proteins for distinct fates and functions (Hicke, 2001). It might thus be possible that COP1 confers not only a signal for the degradation of BBX24 in the light, but also regulates the function of the protein by such posttranslational modification.
In summary, the investigations here have revealed that the nuclear localization and the interaction with COP1 are crucial for BBX24 degradation and function. These findings now open further questions to understand in more detail the molecular mechanism by which COP1 regulates light signaling.
MATERIALS AND METHODS
Plant Material, Growth, and Light Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study were in Columbia-0 background. The bbx24-1 T-DNA line (SALK_067473) was obtained from the Nottingham Arabidopsis Stock Centre, and it was previously described by Indorf et al. (2007) as sto. Seeds were sown on filter paper and treated as described by Indorf et al. (2007). For light fluence-rate response analyses, plates were then transferred to dark, or to specific light conditions described by Kircher et al. (1999) and Kaiser et al. (1995), and incubated for 4 d at 22°C. For deetiolation experiments, plants were grown for 5 d in the dark at 22°C and then transferred to cWL (120 μmol m−2 s−1).
Hypocotyl-Length Measurements
Hypocotyls length was measured using ZEISS AXIOVISION 4.0 software. All values are presented as the mean ± sd for minimum three experiments of around 30 seedlings per line. Comparisons were performed by two-tailed paired Student’s t test. P values < 0.01 were considered as statistically significant.
Plasmid Construction
BBX24 full-length cDNA and p35S:EYFP-COP1 used in this work were described by Indorf et al. (2007). Generation of N-BBX24 was achieved by PCR amplification of BBX24 using primers 5′-TTACCCGGGATGAAGATACAATGTGATGTGTGTG-3′ and 5′-CGGTGGATTCAATATCAGAGAAGTGA-3′. This PCR fragment was digested with SmaI and HincII and cloned in frame with the EGFP cDNA in the plasmid pBS 35S:EGFP+term. Generation of bbx24NLS was achieved in two steps. In the first step, a DNA fragment encoding the N terminus of BBX24 was amplified using the primers 5′-CGGAATTCATCCCACCTACTTGTTCCCCACA-3′ and 5′-AACCTAGGCTTGTTGTGTGAAACATTTGAC-3′, that introduced a mutation in nt 928 (provoking an exchange K226N in the NLS) and added an XmaJI restriction site. A second PCR to obtain the C terminus was performed using the primers 5′-GGGAAGCTTGAACAAAACTCAAACACAGACATTTGT-3′ and 5′-AAGCCTAGGTTTGAGACCAGATATGATG-3′ that introduces also an XmaJI restriction site. In the next step, the two fragments were cut with BglII and XmaJI and subcloned to create a vector that carried the whole BBX24 sequence containing the mutated NLS from KKPR to NKPR. The mutated coding sequence was then amplified by PCR using the primers 5′-TATCCCGGGATGAAGATACAGTGTGATGTGTG-3′ and 5′-GGGAAGCTTGAA-CAAAACTCAAACACAGACATTTGT-3′, digested with HindIII and XmaI, and cloned in frame with the EGFP in the plasmid pBS 35S:EGFP+term. Mutation of N-BBX24 was achieved by PCR using primers TTACCCGGGATGAAGATACAATGTGATGTGTGTG and CGGTGGATTCAATATCAGAGAAGTGA, digested with SmaI and HincII, and cloned in frame with the EGFP in the plasmid pBS 35S:EGFP+term. Mutation of the COP1 interaction residues was achieved by PCR amplification of the BBX24 cDNA using the forward primer 5′-TATCCCGGGATGAAGATACAGTGTGATGTGTG-3′ and the reverse primer 5′-ATGGATCCTTAGCCAAGATCAGCGGCAATGAAGTG-3′ that introduced a mutation at nt 926 and 928 and resulted into V244A and P245A exchanges of the BBX24 amino acid sequence. The PCR product was digested with BamHI and XmaI and cloned in frame with the EGFP cDNA in the plasmid pBS 35S EGFP+term. EGFP fusion constructs were subcloned into the plant binary vector pCambia-2300 (Cambia). For colocalization experiments, the EGFP cDNA was substituted by the ECFP cDNA. For expression in yeast (Saccharomyces cerevisiae), BBX24 or mutated bbx24 cDNAs were introduced in the yeast expression vector pFL61 (Minet et al., 1992) or cloned in frame to the C terminus of the GAL4 binding domain of the bait vector pGBT9 (Clontech).
Transient Expression in Protoplasts
Polyethylene glycol transformation of Arabidopsis protoplasts was performed as described by Yoo et al. (2007).
Plant Transformation and Selection of Transgenic Lines
Arabidopsis plants were transformed with Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) via the floral-dip method (Clough and Bent, 1998). Transgenic plants were selected on plates containing 50 μg/mL kanamycin. Selected plants were transferred to soil, grown to maturation, and T2 seeds were harvested. Only T2 populations showing a segregation ratio of 3:1 on selection media were chosen to obtain the homozygous T3 generation.
Production of Specific BBX24 Antibody
The C-terminal part (amino acids 125–248) of BBX24 was used to raise polyclonal antibodies against BBX24. Recombinant GST-C-BBX24 was generated in Escherichia coli BL21-Codon Plus (DE3)-Ril cells (Stratagene) and then purified with glutathione sepharose 4B (Amersham Pharmacia Biotech) according to manufacturer’s instructions. After thrombin digestion, C-BBX24 protein was used for rabbit immunization (Eurogentec). Affinity purification of antiserum was achieved by Affi Gel 10 activated immunoaffinity support (BioRAD).
Protein Extraction and Western Blotting
Plant tissue was frozen in liquid nitrogen and ground with a pestle. Total protein was isolated using PeqGOLD TriFast reagent (Peqlab) according to manufacturer’s instructions. Protein pellets were resuspended in 1% (w/v) SDS and protein concentration was quantified by the Qubit Fluorometer protein assay (Invitrogen). Equal amounts of denatured total protein were separated in SDS-PAGE using a 12% resolving gel and proteins were transferred by electro blotting onto a polyvinylidene difluoride membrane (Millipore). Immune detection was performed using anti-BBX24, anti-GFP antibodies for detecting the N-terminal fusion, or anti-actin (AtACT8) antibodies (Sigma) as primary antibodies. As secondary antibodies peroxidase-coupled anti-rabbit or anti-mouse antibodies (Sigma) were used. Detection of the proteins was performed using the ECL western blotting detection reagents kit (Amersham).
Microscopic Analysis of Plant Tissue
Epifluorescence and light microscopy of plant seedlings was performed with an Axiovision microscope with the appropriate filter settings (Zeiss). Pictures were taken with a digital camera system using the AXIOVISION software 4.6 (Zeiss). For confocal microscopy the LSM510 META microscopy system and software was used (Zeiss). Representative cells were chosen for digital images. Photographs were mounted with ADOBE PHOTOSHOP.
Treatment of Plants with the Proteasome Inhibitor MG132
The MG132 (carbobenzoxy-l-leucyle-l-leucyle-l-leucinale) powder was dissolved in dimethyl sulfoxide to a concentration of 20 mm. For the treatment, MG132 was diluted in water to a 50 μm final concentration. Dark-grown 5-d-old seedlings were vacuum infiltrated with the MG132 solution three times for 10 s. Fluorescence microscopy analysis was performed with samples taken after 24 h of incubation in the dark. Control plants were treated in parallel with dimethyl sulfoxide in distilled, deionized water (1:400).
Yeast Assays
For salt tolerance experiments, the pFL61 vector was used to transfect DBY689 (Beeler et al., 1994) using the method described by Gietz and Schiestl (1996). Transformed colonies were selected on yeast drop-out selection medium lacking uracil. Overnight cultures of three different colonies per construct together with a negative control were spotted on media supplemented with 250 μm or 350 μm LiCl. Growth of cells at 30°C was evaluated after 2 to 4 d. Autoactivation of wild-type BBX24 or BBX24 mutations was investigated by expressing the protein fused to the DNA binding domain of the GAL4 transcription factor in the yeast strain Y190. After transformation positive colonies were selected on plates lacking Trp (SD−Trp). Overnight cultures of the transformants were grown in selective liquid media and then spotted on SD−Trp/−His plates containing 0, 45, or 100 mm of 3-amino-1,2,4-triazole. Growth of the different clones was evaluated after 2 to 4 d.
Quantitative Real-Time PCR Analyses
Total RNA was isolated using QIAGEN RNeasy mini kit, according to manufacturer’s protocol. First-strand synthesis was performed using GeneAmp reverse transcription reagents (Applied Biosystems). SYBR-Green (Thermo Scientific) real-time reverse transcription PCR assays were carried out with an ABI Prism 7000 (Applied Biosystems) using actin rRNA levels for normalization. The relative quantity of the transcripts was calculated by means of the comparative threshold cycle method (Heid et al., 1996). The following primers were employed in this assay: BBX24 fw: 5′-CCACTTCCCTGGGCTGTT-3′, BBX24 rev: 5′-CCCAAGATCAAGC-TGTCCTTT-3′, N-BBX24 fw: 5′-CACGTGGCTAATTCTCGATCTG-3′, N-BBX24 rev: 5′-GGAAGGCTCAGGTTGATTCTTCTC-3′, Actin fw: 5′-CGCTATGTATGTCGCCA-3′, and Actin rev: 5′-CTTGCCCATCGGGTAA-3′.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G06040/NM_100484.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Functional analysis of N-terminal and C-terminal fusions of EGFP and BBX24.
Supplemental Figure S2. Expression analysis of BBX24NLS.
Supplemental Figure S3. Effect of EGFP-BBX24NLS expression in yeast.
Supplemental Figure S4. Expression analysis of N-BBX24 in the bbx24-1 transgenic line.
Supplemental Figure S5. Effect of EGFP-BBX24COP1 expression in yeast.
Acknowledgments
The cop1-4 mutant was obtained from Roman Ulm, and the anti-GFP antibodies were kindly provided by Tim Kunkel. We would like to thank Xin Yu for helping with the transient expression experiments, Eberhard Schäfer for critical reading of the work, and Eija Schulze and Rosula Hinnenberg for excellent technical assistance.
References
- Beeler T, Gable K, Zhao C, Dunn T. (1994) A novel protein, CSG2p, is required for Ca2+ regulation in Saccharomyces cerevisiae. J Biol Chem 269: 7279–7284 [PubMed] [Google Scholar]
- Boccalandro HE, Rossi MC, Saijo Y, Deng X-W, Casal JJ. (2004) Promotion of photomorphogenesis by COP1. Plant Mol Biol 56: 905–915 [DOI] [PubMed] [Google Scholar]
- Borden KL, Lally JM, Martin SR, O’Reilly NJ, Etkin LD, Freemont PS. (1995) Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development. EMBO J 14: 5947–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Li Y-H, Chen L-T, Chen W-C, Hsieh W-P, Shin J, Jane W-N, Chou S-J, Choi G, Hu J-M, et al. (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chang CS, Maloof JN, Wu SH. (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M. (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GH, Deng XW, Holm M. (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Buche C, Schafer E, Kretsch T. (2003) Characterization of a novel non-constitutive photomorphogenic cop1 allele. Plant Physiol 133: 1557–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington NP, Futcher B. (2001) Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci 114: 4599–4611 [DOI] [PubMed] [Google Scholar]
- Favory J-J, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Larner VS, Whitelam GC. (2005) The signal transducing photoreceptors of plants. Int J Dev Biol 49: 653–664 [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. (2007) Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol 48: 1243–1253 [DOI] [PubMed] [Google Scholar]
- Gietz R, Schiestl R. (1996) Transforming yeast with DNA. Methods Mol Cell Biol 5: 255–269 [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131: 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. (1996) Real time quantitative PCR. Genome Res 6: 986–994 [DOI] [PubMed] [Google Scholar]
- Hicke L. (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195–201 [DOI] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW. (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma L-G, Qu L-J, Deng X-W. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T. (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M. (2007) Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J 51: 563–574 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T, Emmler K, Kretsch T, Weisshaar B, Schäfer E, Batschauer A. (1995) Promoter elements of the mustard CHS1 gene are sufficient for light regulation in transgenic plants. Plant Mol Biol 28: 219–229 [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu S-H. (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-H, Kim B-H, von Arnim AG. (2002) Repressors of photomorphogenesis. Int Rev Cytol 220: 185–223 [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F. (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics MGG 204: 383–396 [Google Scholar]
- Krauer K, Buck M, Flanagan J, Belzer D, Sculley T. (2004) Identification of the nuclear localization signals within the Epstein-Barr virus EBNA-6 protein. J Gen Virol 85: 165–172 [DOI] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T. (2008) The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Lagercrantz U, Axelsson T. (2000) Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol Biol Evol 17: 1499–1507 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Cyert MS, Gasser CS. (1996) Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem 271: 12859–12866 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gao Y, Qu L, Chen Z, Li J, Zhao H, Deng XW. (2002) Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14: 2383–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F. (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. (2008) Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J 54: 806–819 [DOI] [PubMed] [Google Scholar]
- Nagaoka S, Takano T. (2003) Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot 54: 2231–2237 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R. (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Stacey MG, Hicks SN, von Arnim AG. (1999) Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11: 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW. (2005) COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen S, Mou Z. (2010) Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J Plant Physiol 167: 144–148 [DOI] [PubMed] [Google Scholar]