Abstract
A key floral activator, FT, integrates stimuli from long-day, vernalization, and autonomous pathways and triggers flowering by directly regulating floral meristem identity genes in Arabidopsis (Arabidopsis thaliana). Since a small amount of FT transcript is sufficient for flowering, the FT level is strictly regulated by diverse genes. In this study, we show that WEREWOLF (WER), a MYB transcription factor regulating root hair pattern, is another regulator of FT. The mutant wer flowers late in long days but normal in short days and shows a weak sensitivity to vernalization, which indicates that WER controls flowering time through the photoperiod pathway. The expression and double mutant analyses showed that WER modulates FT transcript level independent of CONSTANS and FLOWERING LOCUS C. The histological analysis of WER shows that it is expressed in the epidermis of leaves, where FT is not expressed. Consistently, WER regulates not the transcription but the stability of FT mRNA. Our results reveal a novel regulatory mechanism of FT that is non cell autonomous.
As for other plants, the correct timing of flowering is essential for reproductive success in Arabidopsis (Arabidopsis thaliana). Flowering time in Arabidopsis is regulated by complex genetic networks monitoring various environmental and endogenous signals. Four major genetic pathways for these signals have been revealed: the photoperiod and the vernalization pathways responding to environmental stimuli; and the autonomous and the GA-dependent pathways monitoring internal conditions (Mouradov et al., 2002; Simpson and Dean, 2002; Boss et al., 2004). These pathways are converged on common downstream target genes, FT, SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), and LEAFY (LFY), so-called flowering pathway integrators (Simpson and Dean, 2002). Flowering time in Arabidopsis is quantitatively controlled by the transcript level of these integrators (Kardailsky et al., 1999; Kobayashi et al., 1999; Blázquez and Weigel, 2000; Onouchi et al., 2000; Samach et al., 2000; Moon et al., 2003, 2005).
FT, encoding a small approximately 20-kD protein with homology to Raf kinase inhibitor protein, is one of the key floral activators integrating multiple floral inductive pathways (Kardailsky et al., 1999; Kobayashi et al., 1999). FT promotes the transition to flowering by activating other floral integrator, SOC1, and floral meristem identity genes such as APETALA1 (AP1), FRUITFULL, CAULIFLOWER, and SEPALLATA3 (Ruiz-García et al., 1997; Abe et al., 2005; Teper-Bamnolker and Samach, 2005; Wigge et al., 2005; Yoo et al., 2005). Recently, many laboratories have shown that FT protein produced in the leaf phloem moves to the shoot apex and executes its role through the interaction with the bZIP transcription factor FD, which is expressed in the shoot apex (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). Consequently, FT protein is considered a graft-transmissible florigen or at least a component of the floral stimuli.
Although the abundance of FT transcripts in the wild type is so low as not to be detected by in situ hybridization, FT overexpression plants or loss-of-function alleles show dramatic changes in flowering time (Koornneef et al., 1991; Kardailsky et al., 1999; Kobayashi et al., 1999). This suggests that a small amount of FT is sufficient for flowering in wild-type plants, and inappropriate FT expression causes disorder in flowering time. Hence, FT expression is regulated strictly by a diverse range of regulators. CONSTANS (CO) directly activates FT through the photoperiod pathway (Kobayashi et al., 1999; Samach et al., 2000; Suárez-López et al., 2001; Valverde et al., 2004). CO expression, of which transcription arises around 12 h after dawn and stays high until the following dawn, is activated in both long days and short days (Suárez-López et al., 2001). However, CO mRNA expressed at night does not cause the activation of FT, because CO protein is degraded in the dark (Valverde et al., 2004). Therefore, flowering is delayed in short days due to the absence of FT. In contrast, mutations in genes involved in the photoperiod pathway, such as gi, co, and ft, delay flowering only in long days but not in short days, since the signal from the photoperiod pathway is mainly mediated through FT (Koornneef et al., 1991; Suárez-López et al., 2001).
The MADS box transcription factor FLOWERING LOCUS C (FLC), a central floral repressor in autonomous and vernalization pathways, represses FT expression. FT expression repressed by FLC in winter annuals is important because this repression prevents flowering until the following spring (Searle et al., 2006). Another negative flowering repressor, SHORT VEGETATIVE PHASE (SVP), regulates FT expression through ambient temperature signaling in the thermosensory pathway (Lee et al., 2007b). Recently, it has been reported that SVP acts in a repressor complex together with FLC protein, and this complex binds directly to the CArG box DNA motifs in the first intron of the FT gene (Helliwell et al., 2006; Lee et al., 2007b; Li et al., 2008). Additionally, the chromatin-associated proteins, TERMINAL FLOWER2 and EARLY BOLTING IN SHORT DAYS, repress FT transcription by direct binding in FT chromatin (Piñeiro et al., 2003; Takada and Goto, 2003). In addition, CURLY LEAF and FERTILIZATION INDEPENDENT ENDOSPERM, the subunits of Arabidopsis Polycomb Repressive Complex2, strongly repress FT during vegetative development (Jiang et al., 2008). Many other genes, such as PHYTOCHROME AND FLOWERING TIME1 (PFT1), PHYTOCHROME-INTERACTING FACTOR3, and TEMPRANILLO genes (TEM1 and TEM2), which are involved in light signaling and circadian rhythm, are also known to be involved in FT regulation (Cerdán and Chory, 2003; Oda et al., 2004; Castillejo and Pelaz, 2008). Although various functions of upstream genes for FT regulation are revealed, little is known about the posttranscriptional regulation of FT mRNA.
In this study, we show that WEREWOLF (WER), known as a regulator of root hair patterning, is involved in posttranscriptional regulation of FT. WER, which is classified in the same subgroup with GLABROUS1 (GL1) and AtMYB23 (MYB23), encodes an R2R3 MYB transcription factor (Lee and Schiefelbein, 1999; Stracke et al., 2001). In Arabidopsis, root epidermal cells differentiate into either root hair cells or hairless cells in a position-dependent manner: epidermal cells between two cortical cells differentiate into root hair cells (called H-cells), whereas epidermal cells in contact with a single cortical cell usually become hairless cells (called N-cells; Dolan et al., 1994). WER is highly expressed in N-cells, whereas it is suppressed in H-cells (Lee and Schiefelbein, 1999; Kwak et al., 2005; Kwak and Schiefelbein, 2007). In N-cells, WER protein forms a transcriptional complex with a WD40 protein, TRANSPARENT TESTA GLABRA1 (TTG1), and a bHLH transcription factor, GLABRA3 (GL3), which functions redundantly with ENHANCER OF GLABRA3 (EGL3). This complex positively regulates GL2, which inhibits the generation of root hair and thus makes cells differentiate into N-cells (Lee and Schiefelbein, 1999; Walker et al., 1999; Payne et al., 2000; Bernhardt et al., 2003; Zhang et al., 2003; Koshino-Kimura et al., 2005).
Here, we report the late-flowering phenotype of the wer mutant, which has been previously reported to have hairy roots. Delayed flowering occurred in long days but not in short days; thus, wer can be classified as a photoperiod pathway mutant. The transcript level of FT was reduced in the wer mutant in long days, which was independent of CO and FLC. In addition, such a decrease of FT transcript level is due not to the altered transcription but to the reduced stability of mRNA. This study suggests that WER in epidermis modulates the FT transcript level in phloem through a novel mechanism.
RESULTS
WER Regulates Flowering Time through the Photoperiod Pathway
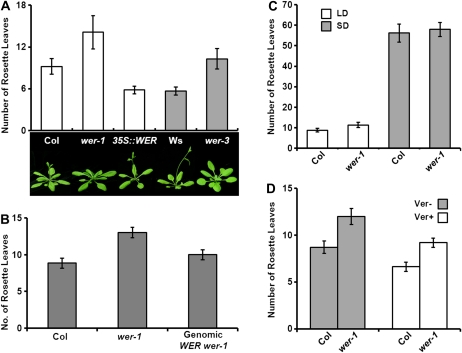
While exploring if WER acts in other developmental processes, we observed that the flowering time of wer loss-of-function mutant and WER overexpression transgenic plants is changed. The wer-1 allele in the Columbia (Col) background and the wer-3 allele in the Wassilewskija (Ws) background have nonsense mutations in the region of the second MYB domain; thus, both alleles apparently produce nonfunctional proteins (Lee and Schiefelbein, 1999). Under long-day (16 h of light/8 h of dark) conditions, both wer-1 and wer-3 plants produced more rosette leaves than the wild type at the time of bolting (Fig. 1A). In contrast, the transgenic plants containing WER genomic DNA under the control of the strong cauliflower mosaic virus 35S promoter (35S::WER) flowered earlier than the wild type (Fig. 1A). To verify that the late-flowering phenotype of wer-1 was caused by the loss of WER function, the WER genomic construct containing a 5-kb WER genomic fragment that includes a 2.5-kb upstream sequence was transformed into wer-1. Most of the resulting transformants showed comparable flowering time to the wild type, indicating that late flowering is caused by the loss of WER (Fig. 1B). In addition, the heterozygous lines obtained from the cross between Col and wer-1 showed similar flowering time with Col, confirming that wer is a recessive late-flowering mutant (Supplemental Fig. S1).
Figure 1.
Flowering time of wer loss-of-function mutants and WER overexpression transgenic plants. A, Comparison of flowering time in wild-type, wer mutant, and 35S::WER transgenic plants under long days. White bars show the flowering time of wer-1 and 35S::WER compared with that of Col, whereas gray bars show the flowering time of wer-3 compared with that of Ws. Twenty-five plants were used to measure the flowering time, and the error bars represent sd. Photographs display the phenotype of each plant when wer-1 or wer-3 initiated its flowering. B, Complementation analysis of wer-1-inserted WER genomic DNA. Thirty-six plants were used to measure the flowering time in long days, and the error bars represent sd. C, Flowering time of wer-1 mutants compared with Col under long days and short days. At least 30 plants were used to measure the flowering time, and the error bars represent sd. D, Flowering time of Col and wer-1 grown in long days after 0 weeks (Ver−) or 8 weeks (Ver+) of vernalization treatment. At least 25 plants were used to measure the flowering time, and the error bars represent sd. [See online article for color version of this figure.]
The flowering time in Arabidopsis is regulated by four major pathways: photoperiod, vernalization, autonomous, and GA pathways (Boss et al., 2004; Bäurle and Dean, 2006; Oh and Lee, 2007). To determine in which pathway WER regulates flowering, the flowering characteristics of wer were checked in response to photoperiod and vernalization. Unlike in long days, wer-1 flowered similarly to the wild type in short days (Fig. 1C). After 8 weeks of vernalization treatment, all genotypes showed acceleration of flowering compared with the nonvernalization treated control. Although wer-1 showed slight acceleration of flowering, the responsiveness was weaker than in the wild type (Fig. 1D). These observations demonstrated that loss of function in WER results in a delay of flowering only under long days and weak sensitivity to vernalization, which is similar in flowering characteristics to the photoperiod pathway mutants co and gi (Koornneef et al., 1991).
WER Expression in Root Does Not Affect the Regulation of Flowering Time
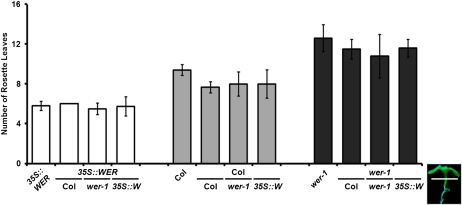
Floral evocation occurs in the shoot apex by inducing floral initiation genes such as AP1 and LFY (Weigel and Nilsson, 1995; Hempel et al., 1997). However, it was previously reported that WER is expressed mainly in roots (Lee and Schiefelbein, 1999). To determine whether a root-derived signal induced by WER affects flowering time, graft chimeras among wer-1, 35S::WER, and the wild type were produced by a transverse-cut grafting method described before (Turnbull et al., 2002). All grafts were denoted as scion/rootstock genotypes. Self-grafted plants, in which the scion and rootstock were from the same genotype, were also produced as controls. These plants appeared to flower slightly earlier than ungrafted plants, probably due to mechanical stress (Fig. 2). The results showed that grafting failed to rescue the late flowering of wer-1 scions regardless of whether Col or 35S::WER was used as rootstock. All grafted plants showed similar flowering time as the plants used as scion (Fig. 2). These results suggest that WER expressed in root does not affect flowering time.
Figure 2.
Flowering time of grafts among Col, wer-1, and 35S::WER. The graft type is a 90° transverse-cut graft using 5-d-old seedlings grown in long days, shown in the photograph on the right side. Plants on the left in each panel are ungrafted controls. Genotype notation for the stock is shown below the lines at bottom, and that of the scion is shown above. [See online article for color version of this figure.]
Expression of WER
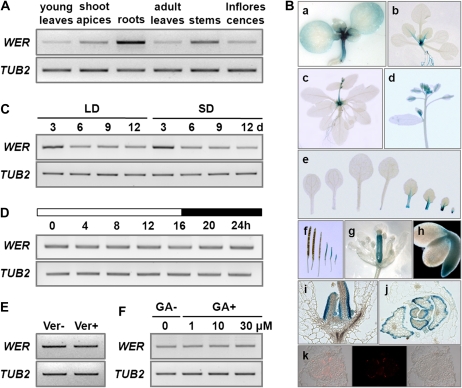
To examine the tissue expression pattern of WER, reverse transcription (RT)-PCR analysis was performed with total RNA extracted from various tissues. Although WER was highly expressed in roots, as reported previously (Lee and Schiefelbein, 1999), transcripts were also detectable in young leaves, shoot apices, adult rosette leaves, stems, and inflorescences, including floral buds and mature flowers (Fig. 3A).
Figure 3.
Expression of WER. A, RT-PCR analysis of WER in diverse tissue. RNAs of young leaves, shoot apices, and roots were isolated from 11-d-old Col seedlings grown in long days, while RNAs of rosette leaves, stems, and inflorescences were isolated from 28-d-old Col plants. TUB2 was used as a quantitative control. B, Spatial expression patterns of WER. a to i, GUS staining in WERp::GUS transgenic plants: a, 6-d-old seedling; b, 12-d-old plant; c, 22-d-old plant; d, an inflorescence with flowers; e, leaves obtained from a 12-d-old plant (the leaves are shown in order of production from cotyledon at left); f, siliques of different stages; g, a mature flower; h, a mature embryo; i, longitudinal section. j, Transverse section through the shoot apex of a 7-d-old seedling. k, MYC-WER protein expression in WERp::MYC-WER transgenic plants, with immunohistochemical data obtained from a 6-d-old seedling. C, Temporal expression of WER detected by RT-PCR analysis. RNA was isolated from the shoot of Col plants grown for 3, 6, 9, and 12 d in both long days and short days. TUB2 was used as a quantitative control. D, RT-PCR analysis of WER expression in Col plants during a 24-h cycle in long days. Shoot of 11-d-old seedlings was harvested every 4 h for RNA isolation. The zero time corresponds to right after dawn, and white or black boxes indicate light on or light off, respectively. TUB2 was used as a quantitative control. E, Comparison of WER expression in Col:FRISF2 grown for 11 d in long days with (Ver+) and without (Ver−) vernalization at 4°C for 8 weeks. TUB2 was used as a quantitative control. F, RT-PCR analysis of WER expression with (GA+) or without (GA−) exogenous GA3 treatment. TUB2 was used as a quantitative control. The whole seedling was used for RNA extraction.
To inspect the spatial pattern of WER expression, WERp::GUS transgenic plants, in which the GUS reporter is driven by the WER promoter with a 4-kb DNA fragment upstream of the WER coding sequence, were used for histochemical GUS staining. This transgenic line was used before for the spatial expression analysis because GUS staining faithfully followed the endogenous expression in roots (Lee and Schiefelbein, 1999). In 6-d-old seedlings, GUS expression was most notable in the hypocotyl and the shoot apex as well as in the root tip, whereas weak GUS expression was detected in the margin of the cotyledons (Fig. 3Ba). GUS was also detected in petiole, stem, stigma, and siliques (Fig. 3B, b–g). In shoot and root apices, GUS was detected from the embryo stage (Fig. 3Bh). Interestingly, GUS expression gradually disappeared while leaves matured (Fig. 3B, b and e). A detailed examination in the shoot apex by longitudinal sectioning of WERp::GUS revealed that WER is concentrated in the epidermis of leaf primordia (Fig. 3B, i and j). Immunohistochemistry using WERp::MYC-WER transgenic plants, in which WER protein fused with MYC epitope is driven by the WER promoter, showed that WER proteins are also expressed along the epidermis (Fig. 3Bk).
The temporal changes in WER transcript level were further determined by RT-PCR to investigate how WER expression is regulated during the flowering process. WER expression in the aerial part of seedlings peaked in 3 d; afterward, it was reduced and became steady under both long days and short days (Fig. 3C). Such temporal expression was not affected by photoperiod. Moreover, WER expression did not show any daily rhythm under long days (Fig. 3D).
When the vernalization effect on WER expression was checked in Col:FRISF2 plants, which have a strong flowering response to vernalization, there was no change (Fig. 3E). In addition, GA treatment did not change the expression level (Fig. 3F). These results indicate that the vernalization or GA-dependent pathway does not affect WER expression.
WER Regulates FT Transcript Level Independent of CO and FLC
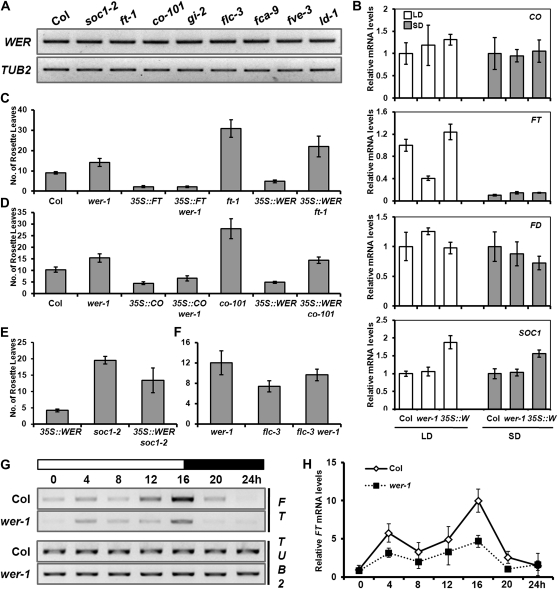
WER expression was further studied in various flowering-time mutants to elucidate the possible involvement of WER in the previously known flowering pathways. Although wer showed flowering characteristics similar to the mutants of the long-day pathway, WER expression was unchanged in these mutants, such as ft-1, co-101, and gi-2 (Fig. 4A). In addition, the expression was not affected by mutations in the autonomous pathway or flc, indicating that WER does not act downstream of the previously known flowering pathways.
Figure 4.
Regulation of flowering-time genes by WER. A, RT-PCR analysis of WER expression in various flowering-time mutants. For RNA isolation, shoot of seedlings grown for 11 d under long days was harvested. TUB2 was used as a quantitative control. B, The expression of various flowering-time genes in Col, wer-1, and 35S::WER was detected by qRT-PCR. White bars show gene expression in plants grown for 9 d under long days (LD), while gray bars show gene expression in plants grown for 21 d under short days (SD). The values and error bars represent means and sd, respectively, from three technical replicates. C, Flowering time of Col, wer-1, 35S::FT, 35S::FT wer-1, 35S::WER, ft-1, and 35S::WER ft-1 grown in long days. D, Flowering time of Col, wer-1, 35S::CO, 35S::CO wer-1, co-101, 35S::WER, and 35S::WER co-101 grown in long days. E, Flowering time of 35S::WER, soc1-2, and 35S::WER soc1-2 grown in long days. F, Flowering time of wer-1, flc-3, and flc-3 wer-1 grown in long days. G and H, Daily rhythm of FT in Col and wer-1 plants grown under long days was detected by RT-PCR analysis (G) or calculated by ImageJ from three independent RT-PCR results (H). Nine-day-old seedlings grown under long-day conditions were harvested every 4 h for RNA isolation. The zero time corresponds to right after dawn, and white or black boxes indicate light on or light off, respectively.
To examine the molecular basis of the late flowering of wer, we checked the expression of genes in the long-day pathway or genes acting on the shoot meristem, such as CO, FT, FD, and SOC1 (Fig. 4B). Real-time quantitative (q) RT-PCR showed that only FT expression was reduced in wer mutants under long days, whereas the levels of CO, FD, and SOC1 were similar to the wild type. In short days, there was no difference between the wild type and wer in the transcript level of all four genes. Such a result is consistent with the flowering characteristics of wer, which is late flowering only under long days. It also strongly suggests that WER regulates FT independent of CO. The double mutant analyses also support this hypothesis, such that 35S::FT completely suppresses but 35S::CO partially suppresses the late-flowering phenotype of wer-1 (Fig. 4, C and D). Double mutant analysis also shows that flc and wer are additive (Fig. 4F), suggesting that WER acts independent of FLC. It is noteworthy that the molecular basis of the early flowering of 35S::WER was a little complicated, because ectopic overexpression of WER caused increases of both FT and SOC1 (Fig. 4B). Thus, neither ft nor soc1 mutation was epistatic to 35S::WER (Fig. 4, C and E).
Because FT is known to be regulated by the circadian clock, we wondered whether the daily rhythm of FT is affected by wer. To address this, FT expression was analyzed every 4 h over a 24-h period under long days by RT-PCR. The result showed that the daily rhythm of FT expression is not affected by wer, although the amplitude is lower than in the wild type (Fig. 4, G and H).
wer Mutation Affects the FT mRNA Stability
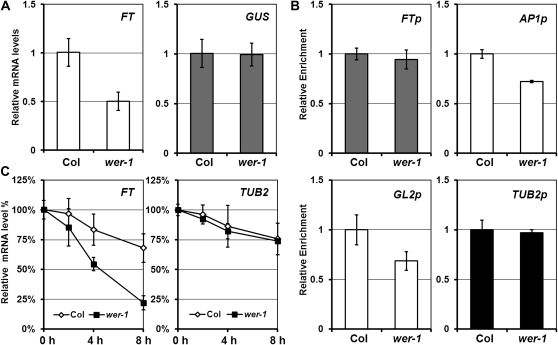
Although WER is expressed in the epidermis (Fig. 3B, i and j), FT is expressed in the vasculature (Takada and Goto, 2003). Thus, it is not likely that WER directly regulates FT. However, it is still possible that WER acts non cell autonomously through mRNA or protein transport. To check if wer mutation affects the transcription of FT, the expression of the GUS reporter gene driven by the FT promoter in wer-1 was analyzed (Fig. 5A). The result showed that GUS expression level in wer-1 was similar to that in Col, whereas endogenous FT level in wer-1 was less than that in Col. Since initiation of transcription is achieved by binding of RNA polymerase II to the promoter DNA in eukaryotes, we checked if wer mutation affects the binding capacity of RNA polymerase II to the proximal region of the FT promoter by chromatin immunoprecipitation (ChIP) assay. In wer-1 mutants, the enrichment of RNA polymerase II at the FT promoter was similar to that in Col, demonstrating that the wer mutation does not affect FT transcription (Fig. 5B). Unlike at the FT promoter, RNA polymerase II enrichment was reduced by wer-1 approximately 40% at promoters of GL2 and AP1, which are a direct target of WER and a downstream gene of FT, respectively. This suggests that the transcription rate of these two genes is reduced in wer-1. Taken together, these results strongly suggest that FT transcript level is reduced in wer-1 by posttranscriptional regulation such as mRNA decay.
Figure 5.
Posttranscriptional regulation of FT mRNA. A, Expression levels of the GUS gene driven by the FT promoter and endogenous FT mRNA in FTp::GUS Col (Col) and FTp::GUS wer-1 (wer-1) plants detected by qRT-PCR. The values and error bars represent means and sd, respectively, from three technical replicates. B, ChIP assay with RNA polymerase II antibody. Enrichment in each promoter was confirmed by ChIP-qPCR analysis. Values are normalized against Col, and means of triplicate experiments are presented with error bars representing sd. C, RNA was isolated from 7-d-old Col and wer-1 grown in continuous light (24 h of light) after 200 μm actinomycin D treatment for 0, 2, 4, or 8 h. Expression of FT and TUB2 in each sample was detected by RT-qPCR, and values are normalized against the expression level of untreated sample (0 h). Mean values from three technical replicates are shown with error bars representing sd.
To directly assess whether the reduced FT mRNA level in wer-1 was a result of altered mRNA stability, the half-life of FT mRNA was compared between the wild type and wer-1 after actinomycin D, a transcription inhibitor, was given. Total RNA of 7-d-old seedlings grown under continuous light was isolated after incubation with actinomycin D for 0, 2, 4, or 8 h. After transcriptional block with actinomycin D treatment, FT mRNA abundance in wer-1 was more rapidly reduced than that in the wild type (Fig. 5C). However, the half-life of control mRNA, TUB2, was not different in the wild type and wer-1, suggesting that the wer mutation does not cause a general RNA instability. Therefore, our results suggest that WER regulates the stability of FT mRNA.
Mutation Affects the FT mRNA Stability
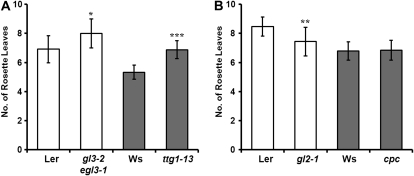
For the root hair pattern formation, WER forms a transcriptional protein complex with TTG1 and GL3/EGL3 and positively regulates the transcription of GL2 and CPC (Lee and Schiefelbein, 2002; Schellmann et al., 2002; Wada et al., 2002; Zhang et al., 2003; Koshino-Kimura et al., 2005; Ryu et al., 2005). We wondered if the genetic tool kit regulating root-hair pattern is also involved in the determination of flowering time. Interestingly, ttg1-13 and gl3-2 egl3-1, mutants of components in the WER protein complex, flowered as late as wer mutants (Fig. 6A), suggesting that the WER complex regulates flowering time also. In contrast, gl2 and cpc showed similar flowering time to the wild type (Fig. 6B). Therefore, our results indicate that WER downstream signaling does not regulate flowering although the WER protein complex does, through the regulation of FT mRNA stability.
Figure 6.
Flowering time of root hair patterning mutants. A, Mutation in components of the same complex with WER delayed flowering time. The double mutant gl3-2 egl3-1 is in the Ler background, while the ttg1-13 mutant is in the Ws background. At least 20 plants were used to measure the flowering time in long days, and the error bars represent sd. The asterisks denote statistical significance: * P < 0.05, *** P < 0.0001 (t test). B, Mutants of WER downstream target genes showed similar flowering time with their wild-type plants. The gl2-1 mutant is in the Ler background, while the cpc mutant is in the Ws background. At least 20 plants were used to measure the flowering time in long days, and the error bars represent sd. The asterisks denote statistical significance: ** P < 0.01 (t test).
DISCUSSION
Myriad genes are reported to regulate the transcript level of FT, since FT turns out to be a “florigen” as well as a key integrator of flowering signals. However, in most cases it is not clear if it is regulated at the transcriptional level or the posttranscriptional level. In this study, we clearly show that wer, producing ectopic root hairs, is a typical photoperiod pathway mutant and that WER regulates FT mRNA stability in a non-cell-autonomous way. Therefore, our study provides a novel mechanism regulating FT transcript level.
WER Regulates FT Non Cell Autonomously
Although WER expression was mostly detected in root, grafting analysis showed that flowering time was not affected by WER in root (Fig. 2). This result indicates that WER expressed in aerial parts are involved in the regulation of flowering time. Consistent with this, WER expression was observed in diverse aerial parts such as young leaves, stems, flowers, and siliques (Fig. 3, A and B). In addition, it is expressed in young, developing leaves where FT is expressed (Fig. 3B; Takada and Goto, 2003). The leaf consists of three distinct tissues: mesophyll, vascular bundle, and epidermis. Histological analysis showed that WER is expressed in leaf epidermis, whereas FT is expressed in vascular bundles (Fig. 3B; Takada and Goto, 2003). Therefore, WER is most likely to regulate FT non cell autonomously. Alternatively, WER may be transported to vascular bundles for the regulation of FT. However, this is not likely, because it produces a protein complex, as discussed below, and our preliminary result showed that WER protein is located in the leaf epidermis.
It is noteworthy that root hair pattern formation shares the same genetic tool kit with trichome formation in the leaves (Schiefelbein, 2003). The many glabrous mutants with no trichome have defects in root hair formation as well (Masucci et al., 1996; Payne et al., 2000; Ohashi et al., 2002; Bernhardt et al., 2003; Zhang et al., 2003). In addition, such genes are expressed in both root and leaf epidermis. Therefore, it provides good evidence indicating that trichome and root hair are evolutionarily homologous organs, as suggested before (Kellogg, 2001). However, our results also show how functional divergence occurs in WER activity. Although the same WER protein complex regulates root hair pattern formation and flowering in roots and leaves, respectively, the downstream factors involved in each process are different (Fig. 6). Thus, the divergence occurs at the downstream target genes. Future analysis to search for the factors mediating signals between the epidermis and vascular bundles for FT regulation would be interesting.
Non-cell-autonomous regulation of FT is not unprecedented. It has been reported that phytochrome B located in mesophyll suppresses FT expression through a downstream gene, PFT1 (Cerdán and Chory, 2003; Endo et al., 2005). Interestingly, the PFT1 mediating phyB signaling also regulates FT independent of CO, similar to WER. This may indicate that non-cell-autonomous regulation of FT in the leaf is a common process. Therefore, it is possible to identify the intertissue signals regulating FT, which is critical to understanding florigen entity.
WER Regulates FT Posttranscriptionally
CO is known to directly regulate FT transcription by binding to the promoter (Samach et al., 2000; Tiwari et al., 2010). Although CO protein does not have a conspicuous domain for the transcription factor, much compelling evidence supports that it plays a transcriptional coactivator (Samach et al., 2000; Hepworth et al., 2002; Wenkel et al., 2006). In addition, FT is transcriptionally regulated by FLC, a central flowering repressor. FLC protein binds directly to the first intron of FT to prevent the induction of FT transcription (Helliwell et al., 2006). Therefore, transcriptional regulation of FT is relatively well studied, but posttranscriptional regulation is poorly studied. Here, we revealed that WER positively regulates FT by controlling mRNA stability at the posttranscriptional level (Fig. 5). This indicates that a FT mRNA decay pathway is involved in the regulation of flowering time. Transcript abundance is determined by the equilibrium between the rate of mRNA synthesis and the rate of degradation; yet, in the majority of gene expression analyses, the mRNA decay process has not been considered seriously. However, recent advances provide some knowledge about mRNA decay pathways, such as that the mRNAs involved in regulatory processes have shorter half-lives than those involved in the metabolic pathway in Arabidopsis (Gutierrez et al., 2002; Belostotsky and Sieburth, 2009). As was found in yeast and human studies, this implies that a rapid mRNA decay process is required for strict regulation of developmental processes. This is consistent with the result that FT transcript was less stable than TUB2 in this study (Fig. 5C). Because FT protein is considered a florigen, the FT protein level is directly linked to flowering. Therefore, it is probable that FT transcripts must be carefully monitored to produce FT protein in appropriate amounts. The regulation of FT mRNA stability proposed in this study may provide a new mechanism to control FT transcripts. Since WER encodes a transcription factor, WER would not be directly involved in the regulation of FT mRNA stability. Thus, it is likely that WER activates stabilizing factors or inhibits destabilizing factors for FT mRNA. The genetic components regulating FT mRNA stability will be pursued.
MATERIALS AND METHODS
Plant Materials and Genotyping
Arabidopsis (Arabidopsis thaliana) wer-1 in the Col background and wer-3 in the Ws background were used (Lee and Schiefelbein, 1999). 35S::WER is a transgenic line with WER genomic DNA (inserting from the start to the stop codon) driven by the cauliflower mosaic virus 35S promoter in wer-1 mutants (Lee and Schiefelbein, 1999). To confirm that genomic WER rescues the flowering phenotype of wer-1, a 5-kb genomic fragment including 2.5 kb of the upstream sequence of WER was cloned into the binary vector pPZP221 and transformed into wer-1. The WERp::GUS transcriptional reporter construct was reported previously (Lee and Schiefelbein, 1999). The gl2-1 mutant is in the Landsberg erecta (Ler) background, and cpc and ttg1-13 are in the Ws background (Masucci and Schiefelbein, 1996; Wada et al., 1997; Walker et al., 1999). The homozygous double mutant gl3-2 egl3-1 is generated by crossing two single mutants in the Ler background (Payne et al., 2000; Zhang et al., 2003; Bernhardt et al., 2005). The 35S::CO and 35S::FT transgenic lines and ft-1, co-101, gi-2, fca-9, fve-3, soc1-2, and ld-1 are in the Col background, as described before (Lee et al., 1994, 2006; Fowler et al., 1999; Kardailsky et al., 1999; Kobayashi et al., 1999; Page et al., 1999; Takada and Goto, 2003; Ausín et al., 2004). The flc-3 mutant is the line originally obtained from fast-neutron mutagenesis of Col:FRISF2, a Col line with FRIGIDA (FRI) from San Feliu-2 (SF2), by backcrossing eight times (Michaels and Amasino, 1999; Lee et al., 2000). However, the FRISF2 allele has been eliminated from flc-3 by backcrossing several times into Col. The FTp::GUS Col line containing an 8.9-kb upstream sequence from the start codon fused to GUS protein was used (Takada and Goto, 2003). FTp::GUS wer-1 was obtained from the cross between FTp::GUS Col and wer-1, and kanamycin-resistant F2 seedlings were genotyped with cleaved-amplified polymorphic sequence (CAPS) markers. To check the genotype of double mutants, the F2 plants were checked using PCR-based markers, simple sequence length polymorphism markers, and CAPS and degenerate CAPS markers, which are described in Supplemental Table S1.
Growth Conditions
Seeds were sterilized by 75% ethanol with 0.05% Triton X-100, then rinsed twice using absolute ethanol and dried. They were seeded on 0.85% plant agar (Duchefa) containing 1% Suc and half-strength Murashige and Skoog (MS; Duchefa) plates and incubated for 3 d at 4°C to break seed dormancy. Afterward, plants were transferred and grown at 22°C with 60% ± 10% relative humidity in long days (16 h of light/8 h of dark) or short days (8 h of light/16 h of dark) under cool-white fluorescent lights (100 μmol m–2 s–1). For vernalization treatment, seeds on the MS plates were incubated for 8 weeks at 4°C under short-day conditions. For exogenous application of GA, we transferred 7-d-old plants into MS medium with GA3 and incubated for 2 d under short-day conditions. At least 20 plants were used to measure the flowering time of each genotype. Flowering time was measured as the number of rosette leaves produced when flowering occurs.
Grafting
Transverse-cut grafting was performed as described previously (Turnbull et al., 2002). The grafting experiment was accomplished with a microscope using 5-d-old seedlings grown on Suc-free medium. Horizontal cuts were made in the upper region of the hypocotyl with small blades (Dorco T-300). For 5 d after grafting, grafts were monitored for whether they formed good unions without bending or any other growth problems. All successful grafts were transplanted to soil. The final proportion of successful grafts, which grow normally until flowering, was over 70%. At least 10 plants were used to measure the flowering time of each graft.
Analysis of Gene Expression
Total RNA extraction and RT-PCR were performed as described before (Lee et al., 2008). The RT-PCR analysis was repeated at least three times using separately harvested samples. The information of each primer for PCR is described in Supplemental Table S2. For semiquantitative RT-PCR analysis, RT-PCR products were analyzed using the ImageJ 1.42 (http://rsbweb.nih.gov/ij/) program to quantify the expression level of each gene. qRT-PCR analysis was performed in 96-well format using the 7300 Fast Real-Time PCR System (Applied Biosystems) and iQ SYBR Green supermix (Bio-Rad). Four microliters of cDNA was used in a 20-μL reaction. Primers were designed to amplify DNA fragments shorter than 150 bp. The details of each primer for PCR are described in Supplemental Table S3. Reaction conditions were as follows: 5 min at 95°C, 40 cycles of PCR (30 s at 95°C, 30 s at 60°C, and 30 s at 72°C), and a dissociation from 60°C to 95°C. Data were collected at 72°C in each cycle, and TUB2 was used as the reference gene. The half-life of mRNA was referenced by 18S rRNA levels. The qRT-PCR analysis was biologically repeated three times, and each consisted of three technical replicates.
Plasmid Construction
To make the chimeric genes of MYC-tagged WER, the DNA fragment for the MYC epitope was inserted in-frame into the 5′ end of the PCR-amplified coding region of WER genomic DNA. The insertion of this chimeric MYC-WER between a 2.4-kb 5′ flanking region DNA fragment and a 1.1-kb 3′ flanking region DNA fragment from the WER gene resulted in the WERp::MYC-WER construct.
GUS Staining and Histological Analysis
GUS staining and histological analysis were performed following standard methods described before (Choi et al., 2007). Embedded samples in paraffin were sectioned at a thickness of 8 μm with a microtome (Leica RM2135). Photographs were taken with a digital microscope (Dimis M) or a digital camera (Photometrics) connected to a microscope (Zeiss Axioskop 2 Plus).
Immunohistochemistry
Immunohistochemistry was performed as described (Paciorek et al., 2006) with some modifications. Tissue samples were fixed in methanol:acetic acid (3:1) fixative solution. The fixed tissue was dehydrated in ethanol, cleared in Neo-clear (Merck), and embedded in ParaplastPlus. Tissue sections were made with 10 μm thickness and mounted on SuperfrostPlus slides. The sections were deparaffinized and rehydrated with 1× phosphate-buffered saline. Antigen unmasking was performed by heating the slides in 10 mm sodium citrate buffer boiled in a microwave oven and incubating the slides in 3% hydrogen peroxide solution. After blocking with MTSB solution (50 mm PIPES [pH 7.0], 5 mm EGTA, and 5 mm MgSO4) containing 3% bovine serum albumin and 5% goat serum, the sections were incubated with anti-MYC monoclonal antibodies (Calbiochem) at a 1:3,000 dilution in the blocking solution overnight. The slides were washed and incubated with Alexa Fluor 546 goat anti-mouse IgG secondary antibodies (Invitrogen) at a 1:5,000 dilution in MTSB. After washing with MTSB, the fluorescence signal was observed using an AxioImager fluorescence microscope (Zeiss) with a DsRed filter.
Analysis of mRNA Stability
Whole seedlings of the wild type and wer-1 grown for 7 d in continuous light were harvested and incubated in liquid MS medium with 200 μm actinomycin D (Sigma-Aldrich) for 0, 2, 4, or 8 h. Prior to this treatment, plants were soaked in actinomycin D for 30 min to allow proper distribution of the antibiotic solution.
ChIP Assay
A total of 1 g of Col and wer-1 seedlings grown under long days for 11 d was used for ChIP. Procedures for ChIP followed the method described before (Lee et al., 2007a, 2008), and antibody for the C-terminal domain of the RNA polymerase II (Abcam AB817) was used. Four microliters of ChIP products resuspended in 100 μL of Tris-Cl (pH 8.0, 10 mm) and EDTA (1 mm) was used for qPCR. In qPCR analysis, expression levels were normalized against expression in Col. The information of the primer pairs for ChIP-qPCR is presented in Supplemental Table S4.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: WER (AT5G14750), FT (AT1G65480), CO (AT5G15840), SOC1 (AT2G45660), FD (AT4G35900), FLC (AT5G10140), AP1 (AT1G69120), GL1 (AT3G27920), MYB23 (AT5G40330), TTG1 (AT5G24520), GL3 (AT1G11130), EGL3 (AT1G63650), GL2 (AT1G79840), CPC (AT2G46410), and TUB2 (AT5G62690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Flowering time of heterozygotes.
Supplemental Table S1. Primers for genotyping.
Supplemental Table S2. Primers for RT-PCR.
Supplemental Table S3. Primers for qRT-PCR.
Supplemental Table S4. Primers for ChIP-qPCR.
Acknowledgments
We thank Dr. J.H. Ahn for providing 35S::CO seeds.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM. (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Bäurle I, Dean C. (2006) The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Belostotsky DA, Sieburth LE. (2009) Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol 12: 96–102 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Zhao MZ, Gonzalez A, Lloyd A, Schiefelbein J. (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Weigel D. (2000) Integration of floral inductive signals in Arabidopsis. Nature 404: 889–892 [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell (Suppl) 16: S18–S31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I. (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A. (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ. (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, Peacock WJ, Dennis ES. (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. (1997) Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124: 3845–3853 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y. (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kellogg EA. (2001) Root hairs, trichomes and the evolution of duplicate genes. Trends Plant Sci 6: 550–552 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Koshino-Kimura Y, Wada T, Tachibana T, Tsugeki R, Ishiguro S, Okada K. (2005) Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol 46: 817–826 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Schiefelbein J. (2007) The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev Biol 302: 118–131 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J. (2005) Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM. (1994) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao HY, Lee I, Deng XW. (2007a) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I. (2008) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J 55: 832–843 [DOI] [PubMed] [Google Scholar]
- Lee JH, Hong SM, Yoo SJ, Park OK, Lee JS, Ahn JH. (2006) Integration of floral inductive signals by FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1. Physiol Plant 126: 475–483 [Google Scholar]
- Lee JH, Park SH, Lee JS, Ahn JH. (2007b) A conserved role of SHORT VEGETATIVE PHASE (SVP) in controlling flowering time of Brassica plants. Biochim Biophys Acta 1769: 455–461 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (2002) Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW. (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I. (2005) Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol 46: 292–299 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell (Suppl) 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda A, Fujiwara S, Kamada H, Coupland G, Mizoguchi T. (2004) Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett 557: 259–264 [DOI] [PubMed] [Google Scholar]
- Oh M, Lee I. (2007) Historical perspective on breakthroughs in flowering field. J Plant Biol 50: 249–256 [Google Scholar]
- Ohashi Y, Oka A, Ruberti I, Morelli G, Aoyama T. (2002) Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J 29: 359–369 [DOI] [PubMed] [Google Scholar]
- Onouchi H, Igeño MI, Périlleux C, Graves K, Coupland G. (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Sauer M, Balla J, Wiśniewska J, Friml J. (2006) Immunocytochemical technique for protein localization in sections of plant tissues. Nat Protoc 1: 104–107 [DOI] [PubMed] [Google Scholar]
- Page T, Macknight R, Yang CH, Dean C. (1999) Genetic interactions of the Arabidopsis flowering time gene FCA, with genes regulating floral initiation. Plant J 17: 231–239 [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro M, Gómez-Mena C, Schaffer R, Martínez-Zapater JM, Coupland G. (2003) EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15: 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-García L, Madueño F, Wilkinson M, Haughn G, Salinas J, Martínez-Zapater JM. (1997) Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9: 1921–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KH, Kang YH, Park YH, Hwang I, Schiefelbein J, Lee MM. (2005) The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132: 4765–4775 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. (2003) Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr Opin Plant Biol 6: 74–78 [DOI] [PubMed] [Google Scholar]
- Searle I, He YH, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C. (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K. (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou YL, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO. (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. (2002) Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419 [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377: 495–500 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]