Abstract
The Arabidopsis (Arabidopsis thaliana) genome encodes nine Salt Overly Sensitive3 (SOS3)-like calcium-binding proteins (SCaBPs; also named calcineurin B-like protein [CBL]) and 24 SOS2-like protein kinases (PKSs; also named as CBL-interacting protein kinases [CIPKs]). A general regulatory mechanism between these two families is that SCaBP calcium sensors activate PKS kinases by interacting with their FISL motif. In this study, we demonstrated that phosphorylation of SCaBPs by their functional interacting PKSs is another common regulatory mechanism. The phosphorylation site serine-216 at the C terminus of SCaBP1 by PKS24 was identified by liquid chromatography-quadrupole mass spectrometry analysis. This serine residue is conserved within the PFPF motif at the C terminus of SCaBP proteins. Phosphorylation of this site of SCaBP8 by SOS2 has been determined previously. We further showed that CIPK23/PKS17 phosphorylated CBL1/SCaBP5 and CBL9/SCaBP7 and PKS5 phosphorylated SCaBP1 at the same site in vitro and in vivo. Furthermore, the phosphorylation stabilized the interaction between SCaBP and PKS proteins. This tight interaction neutralized the inhibitory effect of PKS5 on plasma membrane H+-ATPase activity. These data indicate that SCaBP phosphorylation by their interacting PKS kinases is a critical component of the SCaBP-PKS regulatory pathway in Arabidopsis.
Calcium is a uniform second messenger involved in many plant responses to environmental stimuli. There are many different types of calcium-binding proteins identified in plants (Luan et al., 2002; Harper et al., 2004). One of them shares significant sequence similarity with the yeast (Saccharomyces cerevisiae) calcineurine B subunit (Luan et al., 2002; Gong et al., 2004). The first gene cloned from this family is SOS3 (for Salt Overly Sensitive3) by genetic screening of Arabidopsis (Arabidopsis thaliana) salt-sensitive mutants and map-based cloning (Liu and Zhu, 1998). SOS3 physically interacts with and activates SOS2, a protein kinase (Halfter et al., 2000), and this complex in turn activates SOS1, a plasma membrane Na+/H+ antiporter (Shi et al., 2000; Qiu et al., 2002). SOS3 interacts with the FISL motif of SOS2 at its C-terminal regulatory domain, a SOS2 kinase-inhibiting domain (Guo et al., 2001). Recently a SOS3 homolog, SCaBP8 (for SOS3-like calcium-binding protein8), was identified that interacts with and activates SOS2 to protect Arabidopsis shoots from salt stress, while SOS3 primarily protects roots (Quan et al., 2007). SOS3 has an N-terminal myristoylation signal peptide and it is required for SOS3 function in plant salt tolerance (Ishitani et al., 2000). SCaBP8 shares most of the biochemical features of SOS3 in regulating SOS2 kinase activity and recruiting SOS2 to plasma membrane, however it lacks such myristyolation signal sequence, suggesting that multiple regulatory processes exist in the SCaBP family (Quan et al., 2007). Although capable of performing similar functions in biochemical and cellular tests, SCaBP8 and SOS3 must fulfill distinct regulatory functions in the salt stress response, as they could not replace each other in genetic complementation experiments. SOS2 phosphorylates SCaBP8 at its C-terminal Ser-237 but does not phosphorylate SOS3. This phosphorylation is induced by salt stress, occurs at the plasma membrane, stabilizes the SCaBP8-SOS2 interaction, and enhances plasma membrane Na+/H+ exchange activity (Lin et al., 2009).
In Arabidopsis, there are nine SCaBPs/calcineurin B-like proteins (CBLs) and 24 SOS2-like protein kinases (PKSs)/CBL-interacting protein kinases (CIPKs; Luan et al., 2002; Gong et al., 2004). The regulation of PKS kinase activity by SCaBP calcium sensors is essential for their function in vivo (Gong et al., 2002; Guo et al., 2004). It is a general understanding that the PKS kinase domain interacts with the C-terminal regulatory domain and this intramolecular interaction blocks the substrates accessing into the kinase domain (Guo et al., 2001; Gong et al., 2004). Activation of PKS kinases requires both the phosphorylation of their kinase activation loop by an upstream kinase (Guo et al., 2001; Gong et al., 2002) and interaction with SCaBPs that releases the kinase domain for the target access (Gong et al., 2004).
Similar to the SOS regulatory process, two other pathways have been identified related to these two families. The CBL1- and CBL9-CIPK23 complexes phosphorylate and regulate AKT1 to protect plants from low potassium stress (Xu et al., 2006). SCaBP1 action is required for PKS5 to phosphorylate and negatively regulate AHA2 to mediate the cytoplasmic pH homeostasis (Fuglsang et al., 2007; Yang et al., 2010). In addition to regulating ion transporter activity, PKS kinases have been found to interact with protein phosphatases (Guo et al., 2002; Ohta et al., 2003; Lee et al., 2007), however the regulatory mechanism is not well understood.
In this study, we have demonstrated that phosphorylation of SCaBP calcium sensors by their functional interacting PKS kinases enhances the complex stability.
RESULTS
Phosphorylation of SCaBP Calcium Sensors by Their Interacting PKS Protein Kinases
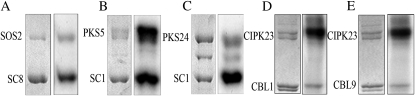
Previously we showed that SCaBP8 is specifically phosphorylated by SOS2 under salt stress in Arabidopsis (Lin et al., 2009). To determine if this phosphorylation regulation occurs in other functional interacting SCaBP-PKS pairs, in vitro kinase assays were performed between SCaBP1 and PKS5 (Fuglsang et al., 2007; Yang et al., 2010), CBL1/SCaBP5 or CBL9/SCaBP7 and CIPK23/PKS17 (Xu et al., 2006), and SCaBP1 and PKS24 (Akaboshi et al., 2008). PKS24, CIPK23, CBL1, and CBL9 cDNA was fused to a GST gene. The GST fusion proteins were purified from Escherichia coli and then subjected to a kinase assay. As a positive control, we first confirmed that SOS2 phosphorylated SCaBP8 (Fig. 1A). When we tested the other pairs, we found that both PKS5 and PKS24 phosphorylated SCaBP1 and CIPK23 phosphorylated both CBL1 and CBL9 (Fig. 1, B–E). Our results indicate that the phosphorylation of SCaBP proteins by their functional interacting PKS kinases may exist in plants.
Figure 1.
SCaBP proteins were phosphorylated by their functional interacting PKS kinases. A, Phosphorylation of SCaBP8 by SOS2. Left, Coomassie Brilliant Blue-stained polyacrylamide gel; right, SOS2 auto- and SCaBP8 transphosphorylation activity. B, Phosphorylation of SCaBP1 by PKS5. Left, Coomassie Brilliant Blue-stained polyacrylamide gel; right, PKS5 auto- and SCaBP1 transphosphorylation activity. C, Phosphorylation of SCaBP1 by PKS24. Left, Coomassie Brilliant Blue-stained polyacrylamide gel; right, PKS24 auto- and SCaBP1 transphosphorylation activity. D, Phosphorylation of CBL1 by CIPK23. Left, Coomassie Brilliant Blue-stained polyacrylamide gel; right, CIPK23 auto- and CBL1 transphosphorylation activity. E, Phosphorylation of CBL9 by CIPK23. Left, Coomassie Brilliant Blue-stained polyacrylamide gel; right, CIPK23 auto- and CBL9 transphosphorylation activity. SC, SCaBP.
Mapping of SCaBP1 Phosphorylation Site(s)
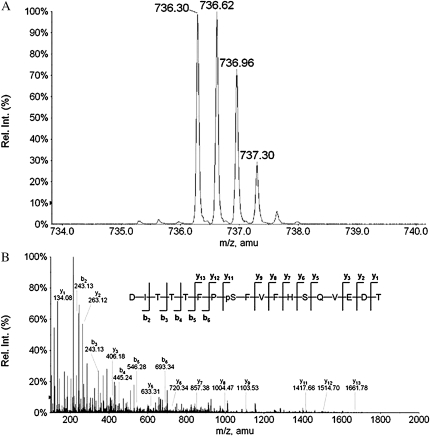
To map the phosphorylation site(s) in SCaBP1 by PKS24, we incubated GST-PKS24 that was on the GST beads and SCaBP1 in kinase buffer with cold ATP and digested the SCaBP1 protein with trypsin and enriched the phosphopeptides through immobilized metal affinity chromatography (IMAC) technology. The phosphorylation site(s) was identified by liquid chromatography-quadrupole mass spectrometry (LC/MS/MS) analysis subsequently. Figure 2 shows a MS spectrum of a triply charged phosphopeptide after IMAC enrichment. The measured mass of this peptide corresponds to the theoretical mass of peptide DITTTFPSFVFHSQVEDT plus one phosphate group. The collision-induced dissociation spectrum of this phosphopeptide was identified as DITTTFPpSFVFHSQVEDT. This phosphorylation site is Ser-216 at the C terminus.
Figure 2.
The phosphorylation site in SCaBP1 is mapped to the PFPF motif at Ser-216 by LC/MS/MS. A, Parent ion of the triply charged peptide DITTTFPSFVFHSQVEDT with one phosphate group. B, Collision-induced dissociation spectrum of DITTTFPpSFVFHSQVEDT. The major y- and b-type product ions are indicated.
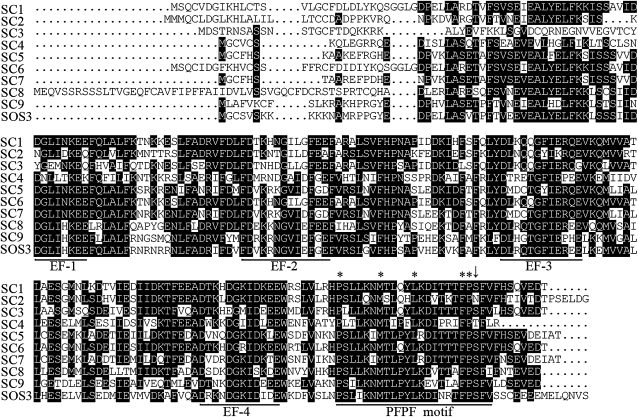
The Arabidopsis genome encodes 10 SCaBP calcium sensors. All of them contain four EF-hand calcium-binding motifs (Akaboshi et al., 2008) and a conserved 23-amino acid motif at the C terminus (Fig. 3). It seems that this motif is found only in the SCaBP subfamily of calcium sensors. Interestingly, both SCaBP8S237, phosphorylated by SOS2, and SCaBP1S216, phosphorlated by PKS24, are located and conserved in this motif. Sequence analyses of these putative motifs indicate that residues P, M, L, F, P, and F are conserved absolutely. Hence we name this motif as the PFPF motif. The putative phosphorylation Ser residue is conserved in the PFPF motif of eight out of 10 SCaBP proteins and surrounded by FP and F residues. SCaBP4 contains a Thr residue at this position. However, SCaBP2 is the only one without this putative phosphorylated residue in the PFPF motif.
Figure 3.
Amino acid sequence alignment of the Arabidopsis SCaBP proteins. The phospho-Ser residue is conserved in the PFPF motif at the C terminus of SCaBP proteins. The alignment is generated with DNAMAN software. Conserved and identical amino acid sequences are shown on a black background. The EF hand and the PFPF motif were underlined in black line. Arrow indicates the conserved Ser/Thr residue and asterisks indicate the conserved amino acids in the PFPF motif. SC, SCaBP.
PKS Kinases Phosphorylate SCaBP Calcium Sensors at the Conserved Ser Residue of the PFPF Motif
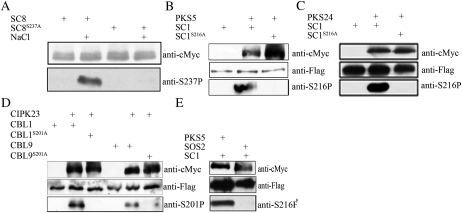
To determine if SCaBP calcium sensors are phosphorylated by PKS kinases in planta, phosphorylation site-specific antibodies were generated by immunizing rabbits with the chemically synthesized phosphorylated peptide Cys-TFPpSFVFH-NH2 (phosphorylated form) for SCaBP1, CBL1, and CBL9. To select the phospho-specific antibodies, the serum was first incubated with the phosphopeptides that had been coupled to SulfoLink resin. After washing the column, the phospho-specific antibodies were eluted at pH 2.7 and immediately neutralized. The antibodies were then run over a column containing the unphosphorylated polypeptide to remove antibodies not specific for the phospho-Ser. The flow through was collected and characterized (Lin et al., 2009). To evaluate the specificity of the antibodies for the Ser phosphorylation site in the PFPF motif, we mutated the Ser residue in the PFPF to Ala. SCaBP1, CBL1, CBL9, and their mutant SCaBP1S216A, CBL1S201A, and CBL9S201A proteins were incubated with their functional interacting PKS proteins in kinase buffer in the presence or absence of ATP. The proteins were then separated using SDS-PAGE and detected by immunoblotting. A strong signal was detected only when SCaBP proteins were incubated with both PKS kinases and ATP (Supplemental Fig. S1, A–D). In the absence of ATP or when the Ser was changed to Ala, only a very weak signal appeared (Supplemental Fig. S1, A–D). These results suggest that the antibodies specifically recognize the phosphorylated Ser in the PFPF motif of SCaBP calcium sensors and the mutant SCaBP proteins were no longer phosphorylated by the PKS kinases. Consistent with our previous finding, SOS2 phosphorylated SCaBP8 at Ser-237 (Supplemental Fig. S1E; Lin et al., 2009).
SOS2 phosphorylated SCaBP8 Ser-237 under salt stress (Fig. 4A), which is consistent with our previous finding (Lin et al., 2009). To examine SCaBP proteins phosphorylated by their interacting PKS kinases in vivo, we generated 35S:3flag-SCaBP1, 35S:3flag-SCaBP1S216A, 35S:3flag-CBL1, 35S:3flag-CBL1S201A, 35S:3flag-CBL9, 35S:3flag-CBL9S201A, 35S:6myc-PKS5, 35S:6myc-PKS24, and 35S:6myc-CIPK23 plasmids. Each pair of SCaBP-PKS plasmids was cotransformed into Arabidopsis protoplasts. Total proteins were extracted from the transformed protoplasts and subjected to immunoblots. Anti-cMyc antibody readily detected myc-labeled PKS kinases and anti-Flag antibody detected Flag-labeled SCaBP proteins in the transfected protoplasts. The SCaBP proteins were immunoprecipitated by anti-Flag and the phosphorylation was detected by the anti-phospho-Ser antibodies. The phospho-Ser-216 signal of SCaBP1 was detected only in protein extracts from plants coexpressing Flag-SCaBP1 and myc-PKS5 or Flag-SCaBP1 and myc-PKS24 (Fig. 4, B and C). The phospho-Ser-201 signal of CBL1 and CBL9 was detected in protein extracts from plants coexpressing myc-CIPK23 and Flag-CBL1 or myc-CIPK23 and Flag-CBL9 (Fig. 4D). No phospho-Ser signal was seen in plants expressing Flag-SCaBPS/A mutants and their interacting myc-PKS (Fig. 4, B–D). As a control, SOS2 did not phosphorylate SCaBP1S216 when Flag-SCaBP1 and myc-SOS2 were coexpressed in Arabidopsis protoplasts (Fig. 4E). Nearly equal amounts of Flag-SCaBP or myc-PKS proteins were present in the assays as demonstrated by immunoblots with anti-cMyc or anti-Flag antibodies (Fig. 4). Our data demonstrate that PKS kinases specifically phosphorylate their functional interacting SCaBP calcium sensors in planta at the Ser residue in the PFPF motif.
Figure 4.
Phosphorylation of the conserved Ser in the PFPF motif by PKS kinases. A, Phosphorylation of SCaBP8S237 by SOS2 is induced by salt stress. SCaBP8 proteins were immunoprecipitated from the transgenic plants expressing 6myc-SCaBP8 and 6myc-SCaBP8S237A and detected with anti-cMyc antibody (top) or anti-phospho-Ser-237 antibodies (bottom). Seedlings were treated with water or 100 mm NaCl for 12 h. B, Phosphorylation of SCaBP1S216 by PKS5 in vivo. Myc-PKS5 was cotransformed with flag-SCaBP1 or flag-SCaBP1S216A into Arabidopsis protoplasts. PKS5 was detected by anti-cMyc antibody (top) and SCaBP1 was detected by anti-Flag antibody (middle). Flag-SCaBP1 and Flag-SCaBP1S216A were immunoprecipitated and phosphorylation of Ser-216 was detected by anti-phospho-Ser-216 antibodies (bottom). C, Phosphorylation of SCaBP1S216 by PKS24 in vivo. Myc-PKS24 was transformed or cotransformed with flag-SCaBP1 or flag-SCaBP1S216A into Arabidopsis protoplasts. SCaBP1 was detected by anti-Flag antibody (middle) and PKS24 was detected by anti-cMyc antibody (top). Flag-SCaBP1 and Flag-SCaBP1S216A were immunoprecipitated and phosphorylation of Ser-216 was detected by anti-phospho-Ser-216 antibodies (bottom). D, Phosphorylation of CBL1S201 and CBL9S201 by CIPK23 in vivo. Myc-CIPK23 was cotransformed with flag-CBL1, flag-CBL1S201A, flag-CBL9, or flag-CBL9S201A into Arabidopsis protoplasts. CIPK23 was detected by anti-cMyc antibody (top) and CBL1/9 was detected by anti-Flag antibody (middle). Flag-CBL1, Flag-CBL1S201A, Flag-CBL9, and Flag-CBL9S201A were immunoprecipitated and phosphorylations of Ser-201 were detected by anti-phospho-Ser-201 antibodies (bottom). E, SCaBP1 was not phosphorylated by SOS2 in vivo. Flag-SCaBP1 was cotransformed with myc-SOS2 or myc-PSK5 into Arabidopsis protoplast. PKS5 and SOS2 were detected by anti-cMyc antibody (top) and SCaBP1 was detected by anti-Flag antibody (middle). Flag-SCaBP1 was immunoprecipitated and phosphorylation of Ser-216 was detected by anti-phospho-Ser-216 antibodies (bottom). SC, SCaBP.
Phosphorylation of SCaBP Proteins Enhances Their Interaction with PKS Kinases
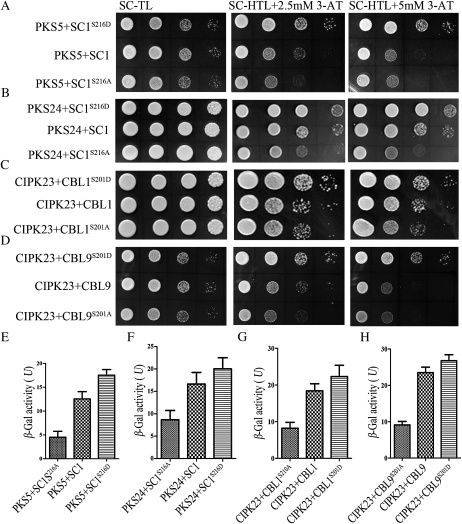
SCaBP8 phosphorylation enhances its interaction with SOS2 (Lin et al., 2009). To determine if PKS phosphorylation of SCaBP has a similar biological function as SOS2 and SCaBP8, we performed yeast two-hybrid assays. As shown in Figure 5, mutation of Ser-216 to Ala in SCaBP1 significantly decreased the interaction between SCaBP1 and PKS5 (Fig. 5A) or PKS24 (Fig. 5B), and mutation of Ser-201 to Ala in CBL1 or CBL9 significantly decreased the interaction between CBL1 or CBL9 and CIPK23 (Fig. 5, C and D). In contrast, when the conserved Ser was substituted to an Asp to mimic a phosphorylated Ser residue, the SCaBPS/D mutation enhanced the interaction with the PKS kinases (Fig. 5, A–D). To further examine the quantitative binding affinities between phosphorylated and nonphosphorylated SCaBP forms with PKS kinases, a β-galactosidase activity assay in yeast for LacZ reporter expression was performed (Fig. 5, E–H). These interaction differences were not due to differences in levels of SCaBP, SCaBPS/A, or SCaBPS/D expression (Supplemental Fig. S2). Our results indicate that the interaction between PKS and phosphorylated SCaBP is stronger than that between PKS and nonphosphorylated SCaBP.
Figure 5.
Phosphorylation of SCaBP proteins by PKS kinases enhances their interaction in yeast two-hybrid assays. PKS coding sequences in the pGBKT7 vector were cotransformed with their interacting SCaBP, SCaBPS/D, or SCaBP8S/A in the pGADT7 vector into yeast strain AH109. A, PKS5-SCaBP1. B, PKS24-SCaBP1. C, CIPK23-CBL1. D, CIPK23-CBL9. Growth of the transformed yeast was assayed on media minus Trp and Leu (SC-TL; left); minus Trp, Leu, and His (SC-HTL) with 2.5 mm 3-amino-1,2,4-triazole (3-AT; middle); or minus Trp, Leu, and His with 5 mm 3-amino-1,2,4-triazole (right). E to H, Quantitative β-galactosidase assays were performed. Yeast containing various PKS and SCaBP proteins was tested for LacZ activity using O-nitrophenyl-β-d-galactopyranoside as substrate. E, PKS5-SCaBP1. F, PKS24-SCaBP1. G, CIPK23-CBL1. H, CIPK23-CBL9. All data represent means ± sem (n = 3). SC, SCaBP.
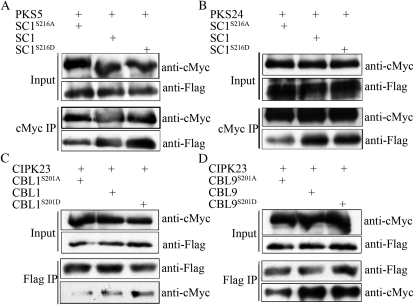
To examine this interaction in Arabidopsis cell, we generated 35S:3flag-SCaBP1S216D, 35S:3flag-CBL1S201D, and 35S:3flag-CBL9S201D plasmids and transformed the combinations of 35S:6myc-PKS5 with 35S:3flag-SCaBP1, 35S:3flag-SCaBP1S216A, or 35S:3flag-SCaBP1S216D, the combinations of 35S:6myc-PKS24 with 35S:3flag-SCaBP1, 35S:3flag-SCaBP1S216A, or 35S:3flag-SCaBP1S216D, the combinations of 35S:6myc-CIPK23 with 35S:3flag-CBL1, 35S:3flag-CBL1S201A, or 35S:3flag-CBL1S201D, and the combinations of 35S:6myc-CIPK23 with 35S:3flag-CBL9, 35S:3flag-CBL9S201A, or 35S:3flag-CBL9S201D into Arabidopsis protoplasts. Myc-PKS (Fig. 6, A and B) or Flag-SCaBP (Fig. 6, C and D) was immunoprecipitated and Flag-SCaBP or myc-PKS was detected via immunoblot analysis. In comparison to SCaBP and SCaBPS/D mutant proteins, SCaBPS/A mutation significantly reduced the interaction with PKSs (Fig. 6). Our results suggest that the phosphorylation of SCaBP calcium sensors by their interacting PKS kinases shares the similar biological function that enhances the interaction between PKS and SCaBP proteins.
Figure 6.
Phosphorylation of SCaBP proteins by PKS kinases enhances their interaction in Arabidopsis. Myc-PKS plasmids were cotransformed with flag-SCaBP, flag-SCaBPS/D, or flag-SCaBP8S/A plasmids into Arabidopsis protoplasts, respectively. A, Transformation of PKS5 and SCaBP1. B, Transformation of PKS24 and SCaBP1. C, Transformation of CIPK23 and CBL1. D, Transformation of CIPK23 and CBL9. Total protein was analyzed using anti-cMyc and anti-Flag antibodies (Input). After immunoprecipitation (IP) with anti-Flag or anti-cMyc conjugated agarose as labeled, the PKS or SCaBP proteins in the pellet were detected via immunoblot analysis using anti-cMyc or anti-Flag antibody (Output). SC, SCaBP.
Phosphorylation of SCaBP1 by PKS5 Is Required for AHA2 Regulation in Yeast
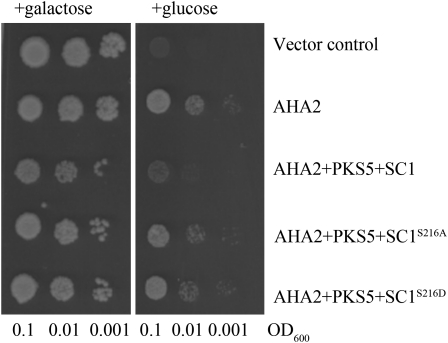
Previously we demonstrated that PKS5 and SCaBP1 can inactivate plasma membrane H+-ATPase AHA2 in yeast and plant (Fuglsang et al., 2007; Yang et al., 2010). To test the effect of the phosphorylation SCaBP1S216 by PKS5 on the regulation of AHA2 activity, we reconstituted the SCaBP1-PKS5-AHA2 regulatory pathway in the yeast strain RS-72. The detailed information for the reconstitution was described by Fuglsang et al. (2007). When we expressed AHA2 in RS-72, the function of the endogenous H+ pump was complemented and the yeast was able to grow on Glc medium (Fig. 7). Consistent with our previous finding, when PKS5 and SCaBP1 were coexpressed with AHA2, the growth of the yeast was significantly inhibited compared to expression AHA2 alone. However, when PKS5 and SCaBP1S216D were coexpressed with AHA2, the inhibition of AHA2 activity was abolished compared with coexpression of PKS5 and SCaBP1 (Fig. 7). PKS5 and SCaBP1S216A had a similar effect on AHA2 as PKS5 and SCaBP1S216D (Fig. 7). These results demonstrate that residue Ser-216 in SCaBP1 is essential for PKS5-mediated inactivation of AHA2 in the yeast system.
Figure 7.
The phosphorylation of SCaBP1S216 is required for regulating AHA2 activity. SCaBP1, SCaBP1S216A, or SCaBP1S216D was expressed in the yeast RS-72 strain harboring two plasmids, AHA2 and PKS5. A suspension of each transformed strain was diluted in sterile water at three concentrations (optical density at 600 nm [OD600] = 0.1, 0.01, and 0.001) and spotted on solid media containing either Gal (left) or Glc (right), pH 6.5. The growth of the cells was monitored 4 d after transfer. All data represent means of three replicate experiments.
DISCUSSION
Outside stimuli increase cytoplasmic calcium concentration and this in turn triggers plant responses. SCaBP family proteins are a group of calcium sensors that physically interact with and activate PKS kinases. It is likely that ion transporters are the preferential targets of the SCaBP-PKS complex (Gong et al., 2002; Qiu et al., 2002; Quintero et al., 2002; Xu et al., 2006; Batelli et al., 2007; Fuglsang et al., 2007; Quan et al., 2007). In this study, we determined that phosphorylation of SCaBP proteins by PKS kinases is a common regulatory mechanism in Arabidopsis. The phospho-Ser is located in the conserved PFPF motif at the C terminus of SCaBP proteins and this phosphorylation enhances the interaction between PKS kinases and SCaBP calcium sensors.
Previous genetic studies suggested that SCaBP proteins function upstream of PKS kinases (Halfter et al., 2000; Xu et al., 2006; Fuglsang et al., 2007; Quan et al., 2007). The SOS pathway is described, as SOS3 and SCaBP8 perceive calcium signals evoked by salt stress, interact with and activate SOS2, recruit SOS2 to the plasma membrane, and further activate SOS1. It is believed that phosphorylation of SCaBP proteins by PKS kinases is the general regulatory mechanism for other SCaBP-PKS pathways. Upon the perception of a calcium signal, SCaBP calcium sensors interact with and activate PKS kinases and that in turn triggers downstream responses (Halfter et al., 2000; Xu et al., 2006; Fuglsang et al., 2007).
Phosphorylation modification is critical for protein precise function in vivo. Our finding of phosphorylation of SCaBP8 by SOS2 led to the discovery that the phosphorylation is important for SCaBP8 function in salt tolerance by stabilizing the plasma membrane localization of the SCaBP8-SOS2 complex (Lin et al., 2009). In this study, we further determined that the phosphorylation of SCaBP proteins by PKS kinases exists in other functional interacting SCaBP-PKS pairs. These results provide important insights into the regulation of activities of the PKS protein kinases by SCaBP calcium sensors.
SCaBP calcium sensors interact with PKS protein kinases through the FISL motif that is conserved in all PKS proteins (Guo et al., 2001; Gong et al., 2004). In this study, we found that the phosphorylation of SCaBP proteins by PKS kinases locates in the PFPF motif, a 23-amino acid fragment, which is found in all SCaBP proteins and located at their C terminus. It is known that various parts of SOS3 participate in its binding to SOS2 and no single motif was found to be sufficient for SOS2 binding (Ishitani et al., 2000). Analysis of SOS3-SOS2 (Sánchez-Barrena et al., 2007) and CBL2/SCaBP1-CIPK14/PKS24 (Akaboshi et al., 2008) complex structures suggest that the recognition process of PKS kinases by their interacting calcium sensor SCaBPs is similar: The FISL motif inserts into a hydrophobic cleft generated by SCaBP/CBL, however the specificity may be conferred by calcium-binding and protein-interaction activity.
Crystal structure analysis of the SOS3-SOS2 C-terminal domain revealed that calcium stabilizes the SOS3-SOS2 complex and that the C-terminal EF4 calcium-binding site of SOS3 may be responsible for activating SOS2 (Sánchez-Barrena et al., 2007). These findings suggest that calcium is involved in both the interaction of SOS3 with SOS2 and the activation of SOS2 by SOS3. The manner of SCaBPs/CBLs binding to calcium may provide the structural basis and specificity for the interaction between SCaBPs and PKSs (Nagae et al., 2003; Sánchez-Barrena et al., 2005, 2007; Akaboshi et al., 2008). Phosphorylation of SCaBP8 by SOS2 enhances their interaction. However, this is independent of calcium (Lin et al., 2009). These results suggest that calcium signal and phosphorylation modification may function in separate steps in regulating the PKS-SCaBP interaction. This is consistent with the fact that calcium has a lesser effect on the interactions between SCaBP1/CBL2 and the regulatory domain of PKS24/CIPK14 (Akaboshi et al., 2008). The structural differences between SCaBP1 and SOS3 are mainly in their N- and C-terminal domains. The crystal structure of the C terminus of SCaBP1 has been examined in detail (Nagae et al., 2003), whereas that of SOS3 was found to be disordered (Sánchez-Barrena et al., 2005). This may imply a reason that SCaBP1 is phosphorylated by both PKS5 and PKS24 and SOS3 is not phosphorylated by SOS2. These results suggest the PFPF motif of SCaBP proteins may confer specificity and strength to the regulation of their target protein kinases.
SCaBP1 is phosphorylated by PKS5 and PKS24 at its C-terminal Ser-216 and only phosphorylation at Ser-216 was detected by LC/MS/MS. The SCaBP1S216D mutation enhances SCaBP1 interaction with PKS5, while the SCaBP1S216A mutation reduces this interaction. However, both of the mutations abolish the inhibition of AHA2 activity in the yeast system, suggesting that formation of a tight complex might change the effect of SCaBP1 and PKS5 on downstream targets. The SerS/A mutation in the PFPF motif reduces the interaction between SCaBP and PKS proteins but does not block the interaction and affect the PKS kinase activity (Lin et al., 2009), suggesting the phosphorylation mechanism may be involved in fine tuning the regulatory activity of SCaBP calcium sensors.
MATERIALS AND METHODS
Construction of Plasmids
PKS and SCaBP cDNAs were obtained by reverse transcription-PCR from wild-type Arabidopsis (Arabidopsis thaliana; Columbia ecotype). PKS5, PKS24, CIPK23, SCaBP1, SCaBP1S216A, SCaBP1S216D, CBL1, CBL1S201A, CBL1S201D, CBL9, CBL9S201A, and CBL9S201D coding sequences were cloned into the BamHI-SalI or SpeI-KpnI sites in pCAMBIA1307-6myc or pCAMBIA1307-3Flag binary vector, respectively. To produce PKS5, PKS24, SCaBP1, CBL1, CBL9, and the mutant GST fusion protein, the DNA fragments were excised from corresponding pCAMBIA1307-6myc or pCAMBIA1307-3Flag plasmid and subcloned into the BamHI-SalI sites in the pGEX-6p-1 vector. CIPK23 was amplified using 6myc-CIPK23 as template and cloned into BamHI-NotI sites in the pGEX-6p-1 or pET-28a-c vector. All primers and plasmid constructs used in these studies are listed in Supplemental Table S1.
Protein Purification and Kinase Assays
All GST or His fusion constructs were transformed into Escherichia coli BL21 (DE3). The transformed cells were grown at 37°C in Luria-Bertani medium with ampicillin (100 μg/mL) or kanamycin (100 μg/mL) until the optical density at 600 nm reached 1.0. Recombinant protein expression was induced by 1.0 mm isopropyl-β-d-thiogalactopyranoside for 12 h at 20°C. The recombinant proteins were affinity purified according to the manufacturer’s protocol (GE Healthcare Life Science) and analyzed by SDS-PAGE.
The kinase assays were performed as described previously (Lin et al., 2009). The kinase buffer included 20 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 1 mm CaCl2, 10 μm ATP, and 1 mm dithiothreitol. The kinase reaction was in a total volume of 20 μL and was started by adding 0.5 μL of γ-32P-ATP (5 μCi), and the mixtures were immediately incubated at 30°C for 0.5 h. Reactions were terminated by adding 6× SDS loading buffer and incubating at 95°C for 5 min. The reaction mixtures were separated by 12% (w/v) SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Then the gels were exposed to a phosphor screen (Amersham Biosciences). After 12 h of exposure, signals were captured with a Typhoon 9410 phosphor imager (Amersham Biosciences).
Phosphopeptide Enrichment and MS Analysis
The recombinant SCaBP1 protein was digested with sequencing grade modified trypsin (Promega) in 50 mm NH4HCO3 (pH 7.8) at 37°C overnight. The resulting peptide mixture was desalted through a C18 reversed-phase cartridge and then vacuum dried in a CentriVap concentrator (Labconco). The dry peptides were esterified in dry methanol:thionyl chloride (25:1, v/v) for 1 h at room temperature. The mixture was vacuum dried again and the resulting peptide methyl esters were reconstituted in acetonitril:methanol:0.01% acetic acid (1:1:1, v/v/v). The phosphorylated peptide methyl esters were enriched through a homemade IMAC column and submitted to a QSTAR XL mass spectrometer (MDS SCIEX) for LC/MS/MS analysis.
Preparation of Anti-Phospho-Ser SCaBP Polyclonal Antibodies
Anti-phospho-Ser-216 (SCaBP1) polyclonal antibodies were made by AbMart (www.ab-mart.com.cn). Two 8-amino acid peptides (corresponding to amino acids 213–220 of SCaBP1 and 198–205 of CBL1/9) with N-terminal Cys residues, Cys-TFPpSFVFH-NH2 (phosphorylated form) and Cys-TFPSFVFH-NH2 (nonphosphorylated form), were also synthesized by AbMart. The Ser phosphospecific peptide (Cys-TFPpSFVFH-NH2) was used to produce polyclonal phosphospecific antibodies and the nonphosphorylated peptide (Cys-TFPSFVFH-NH2) was used for screening and purification.
Coimmunoprecipitation Assays and SCaBP Protein Phosphorylation in Planta
Transgenic plant seedlings expressing 35S:6myc-SCaBP8 and 35S:6myc-SCaBP8S237A were treated with 100 mm NaCl for 12 h. Plant protein was extracted using 2× cold extraction buffer containing 10 mm Tris-Cl, 150 mm NaCl, 2 mm EDTA, 0.5% (v/v) Nonidet P-40, 2× protease inhibitor (Roche), and 2× phosphatase inhibitor (Roche). The resulting samples were then analyzed by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Millipore). The blots were probed with primary anti-SCaBP8-phospho-Ser-237 or anti-cMyc (Sigma-Aldrich) antibodies. Plasmids of 35S:6myc-PKS5 or 35S:6myc-PKS24 were cotransformed with 35S:3flag-SCaBP1 or 35S:3flag-SCaBP1S216A, respectively, and 35S:6myc-CIPK23 was cotransformed with 35S:3flag-CBL1, 35S:3flag-CBL1S201A, 35S:3flag-CBL9, or 35S:3flag-CBL9S201A into Arabidopsis protoplasts. The protoplasts were lysed with the extraction buffer and immunoprecipitated with anti-Flag antibody-conjugated agarose (Sigma-Aldrich). The immunoprecipitates were analyzed by western blot with the anti-phospho-Ser antibodies.
For coimmunoprecipitation assays, the protoplasts were incubated for 12 h after transformation, and lysed, sonicated, and centrifuged. Ten microliters of anti-cMyc or anti-Flag conjugated agarose (Sigma) was incubated with the extract supernatant overnight at 4°C. The coimmunoprepcipitation products were detected via immunoblot analysis using anti-cMyc or anti-Flag antibodies.
Yeast Two-Hybrid Assays
PKS5, PKS24, and CIPK23 were amplified using the myc-tag plasmids as templates and cloned into the pGBKT7 vector. SCaBP1, SCaBP1S216A, SCaBP1S216D, CBL1, CBL1S201A, CBL1S201D, CBL9, CBL9S201A, and CBL9S201D were amplified using the Flag-tag plasmids as templates and cloned into the pGADT7 vector. All primers used for these constructs are listed in Supplemental Table S1.
Yeast (Saccharomyces cerevisiae) strain AH109 was used in the yeast two-hybrid assay. Yeast transformation, growth assays, and β-galactosidase activity assays were performed as described in the Yeast Protocols Handbook (Clontech).
Yeast Complementation Assays
SCaBP1S216A and SCaBP1S216D were amplified and cloned into NotI sites in the pMP1645 vector using the primer pairs SCaBP1 NotIF and SCaBP1 NotIR (see Supplemental Table S1). PKS5 and AHA2 were cotransformed with SCaBP1, SCaBP1S216A, or SCaBP1S216D, respectively, into yeast strain RS-72 (Mat a; ade1-100 his4-519 leu2-3,312 pPMA1::pGAL1) for complementation tests. Growth of the transformed yeast on solid and liquid medium was performed as described previously (Fuglsang et al., 2007).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT5G35410 (SOS2), AT2G30360 (PKS5), AT5G01820 (PKS24), AT1G30270 (CIPK23), AT5G55990 (SCaBP1), AT4G33000 (SCaBP8), AT4G17615 (CBL1), and AT5G47100 (CBL9).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phosphorylation of the conserved Ser in the PFPF motif by PKS kinases in vitro.
Supplemental Figure S2. Expression of PKS and SCaBP proteins in yeast.
Supplemental Table S1. Primers used for plasmid construction.
Acknowledgments
We thank Ms. Anette Lund for excellent technical assistance on the yeast complementation assay.
References
- Akaboshi M, Hashimoto H, Ishida H, Saijo S, Koizumi N, Sato M, Shimizu T. (2008) The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J Mol Biol 377: 246–257 [DOI] [PubMed] [Google Scholar]
- Batelli G, Verslues PE, Agius F, Qiu Q, Fujii H, Pan S, Schumaker KS, Grillo S, Zhu JK. (2007) SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol 27: 7781–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song CP, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Gong Z, Guo Y, Chen X, Zhu JK. (2002) Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J Biol Chem 277: 28340–28350 [DOI] [PubMed] [Google Scholar]
- Gong D, Guo Y, Schumaker KS, Zhu JK. (2004) The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol 134: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK. (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu JK. (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16: 435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK. (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK. (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. (2004) Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK. (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S. (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker K, Pardo JM, Guo Y. (2009) Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell (Suppl) 14: S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Nozawa A, Koizumi N, Sano H, Hashimoto H, Sato M, Shimizu T. (2003) The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J Biol Chem 278: 42240–42246 [DOI] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu JK. (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100: 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM. (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Barrena MJ, Fujii H, Angulo I, Martínez-Ripoll M, Zhu JK, Albert A. (2007) The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol Cell 26: 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Barrena MJ, Martínez-Ripoll M, Zhu JK, Albert A. (2005) The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. J Mol Biol 345: 1253–1264 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yang Y, Qin Y, Xie C, Zhao F, Zhao J, Liu D, Chen S, Fuglsang AT, Palmgren MG, Schumaker KS, et al. (2010) The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]