Abstract
Plant root development is mediated by the concerted action of the auxin and cytokinin phytohormones, with cytokinin serving as an antagonist of auxin transport. Here, we identify the AUXIN UP-REGULATED F-BOX PROTEIN1 (AUF1) and its potential paralog AUF2 as important positive modifiers of root elongation that tether auxin movements to cytokinin signaling in Arabidopsis (Arabidopsis thaliana). The AUF1 mRNA level in roots is strongly up-regulated by auxin but not by other phytohormones. Whereas the auf1 single and auf1 auf2 double mutant roots grow normally without exogenous auxin and respond similarly to the wild type upon auxin application, their growth is hypersensitive to auxin transport inhibitors, with the mutant roots also having reduced basipetal and acropetal auxin transport. The effects of auf1 on auxin movements may be mediated in part by the misexpression of several PIN-FORMED (PIN) auxin efflux proteins, which for PIN2 reduces its abundance on the plasma membrane of root cells. auf1 roots are also hypersensitive to cytokinin and have increased expression of several components of cytokinin signaling. Kinematic analyses of root growth and localization of the cyclin B mitotic marker showed that AUF1 does not affect root cell division but promotes cytokinin-mediated cell expansion in the elongation/differentiation zone. Epistasis analyses implicate the cytokinin regulator ARR1 or its effector(s) as the target of the SKP1-Cullin1-F Box (SCF) ubiquitin ligases assembled with AUF1/2. Given the wide distribution of AUF1/2-type proteins among land plants, we propose that SCFAUF1/2 provides additional cross talk between auxin and cytokinin, which modifies auxin distribution and ultimately root elongation.
A collection of small-molecule phytohormones, alone or in concert, coordinates plant growth and development in response to internal and external cues and helps plants survive biotic and abiotic challenges. Auxin and cytokinin in particular have critical antagonistic roles in directing plant cell division, differentiation, and elongation, which eventually determine division planes, cell polarity, and the processes that define tissue and organ morphologies (Dello Ioio et al., 2008; Bishopp et al., 2011).
Auxin (or indole-3-acetic acid [IAA]) has a major influence on plant meristem maintenance, cell division, and cell elongation. It works in part by derepressing the expression of a battery of developmental regulators via the directed turnover through the ubiquitin/26S proteasome system (UPS) of a set of AUXIN (AUX)/IAA transcriptional repressors (Vierstra, 2009). Central to this signaling in Arabidopsis (Arabidopsis thaliana) is the family of TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN-BINDING F-BOX PROTEIN1-3 (ABF1-3) F-BOX (FBX) recognition factors (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). These FBX proteins assemble with the CULLIN1, RBX1, and SKP1 (ASK in Arabidopsis) subunits to form SKP1-Cullin1-F Box (SCF)-type ubiquitin-protein ligase (E3) complexes that bind to and ubiquitylate AUX/IAA proteins, thus triggering their degradation by the 26S proteasome (Tan et al., 2007). Recognition of AUX/IAA proteins requires prior docking of auxin with TIR1/ABF1-3; in this capacity, SCFTIR1/ABF1-3 E3s serve as the main auxin receptors.
Gradients of auxin are also essential to its functions with respect to stem cell differentiation, the patterning in developing tissues, tropic responses, and the initiation of lateral organs (Woodward and Bartel, 2005; Leyser, 2006; Petrasek and Friml, 2009). The establishment and maintenance of these gradients are coordinated by a network of membrane-localized proteins that facilitate directed auxin influx and efflux from individual cells. These facilitators include the AUX/LAX family of auxin influx proteins (Marchant et al., 1999), the PIN-FORMED (PIN) family of auxin efflux proteins (Chen et al., 1998; Galweiler et al., 1998; Friml et al., 2002; Blilou et al., 2005; Wisniewska et al., 2006), and the P-glycoprotein/multidrug resistance B family of ATP-binding cassette transporter B-type (ABCB) proteins (Noh et al., 2001; Lewis et al., 2007; Wu et al., 2007). Accordingly, interference with these transport facilitators induces dramatic defects in shoot and root architecture.
Cytokinin appears to antagonize auxin action in plant meristems by promoting cell differentiation (Miller et al., 1956; Riefler et al., 2006; Salome et al., 2006; Shani et al., 2006; Dello Ioio et al., 2007; Muller and Sheen, 2008; Moubayidin et al., 2010). In roots, increased cytokinin concentrations by either exogenous application or overexpression of the biosynthetic pathway inhibits root growth and reduces root apical meristem (RM) size (Medford et al., 1989; Kuderova et al., 2008), whereas decreased endogenous cytokinin levels have opposite effects (Werner et al., 2003). Cytokinin signal transduction is achieved by a multistep phosphorelay system similar to the bacterial two-component pathways (To et al., 2007). The hormone is perceived by a small family of membrane-localized Arabidopsis Histidine Kinase2 (AHK2), AHK3, and AHK4/WOL1/CRE1 receptors (Heyl and Schmuling, 2003; Higuchi et al., 2004) that direct a phosphorelay into the nucleus, which activates the type A and type B classes of primary Arabidopsis Response Regulators (ARRs; Argyros et al., 2008; To and Kieber, 2008; Zhao et al., 2010). The type B ARRs are transcription factors that serve as positive effectors of cytokinin signaling. They activate a battery of cytokinin-responsive genes, including type A ARRs (Sakai et al., 2001; To et al., 2004; Yokoyama et al., 2007). The type A ARRs, in contrast, negatively regulate cytokinin signaling by binding to and interfering with type B ARRs. The transient transcriptional induction of type A ARRs by type B ARRs serves to dampen cytokinin responses by negative feedback (To et al., 2007; To and Kieber, 2008).
To maintain the correct balance between meristem maintenance and cell differentiation, an auxin/cytokinin cross talk is used to adjust the influence of these two hormones, especially with respect to root and shoot identity. Auxin regulates the size of the RM by promoting cell division, whereas cytokinin acts in the transitional region overlapping between the distil RM zone and the proximal elongation/differentiation zone (EDZ) to promote root cell elongation/differentiation (Blilou et al., 2005; Dello Ioio et al., 2007). The connection between these two competing processes is primarily mediated by SHORT HYPOCOTYL2 (SHY2), an AUX/IAA transcriptional repressor whose expression is activated by cytokinin through the AHK3/ARR1 pathway (Dello Ioio et al., 2008). Increased SHY2 down-regulates the expression of multiple PIN proteins in the root, thus limiting the formation of auxin maxima and subsequent cell divisions. By contrast, auxin promotes SHY2 degradation through the SCFTIR1/AFB1-3 ubiquitylation machinery, thus relieving SHY2 repression on auxin redistribution. SHY2 also attenuates cytokinin synthesis to provide a second feedback loop (Dello Ioio et al., 2008). Through these flexible interconnected circuits, auxin and cytokinin responses are delicately balanced to antagonistically regulate root cell development and organogenesis.
Here, we describe a second FBX type encoded by the AUXIN UP-REGULATED F-BOX PROTEIN1 (AUF1) and possibly the AUF2 loci within the Arabidopsis UPS that connects auxin and cytokinin during root development. As the name implies, AUF1 was first noticed by the strong increase in its mRNA level upon treatment of seedlings with auxin. auf1 loss-of-function mutants have normal responsiveness to exogenous auxin but are hypersensitive to the auxin transport inhibitors 1-naphthylphthalamic acid (NPA) and 2,3,5-triiodobenzoic acid (TIBA) with respect to root elongation, and they have reduced rates of acropetal and basipetal auxin transport in roots. The transcript abundance for several PIN genes is altered in homozygous auf1 plants, which, at least for PIN2, decreases the accumulation of this efflux facilitator on root cell plasma membranes. auf1 root elongation is also hypersensitive to exogenous cytokinin, with auf1 roots expressing higher levels of the type A response regulators ARR5 and ARR15 in response to the hormone. Kinematic analyses pinpointed the root elongation defect to the zone of rapid expansion in the EDZ. Cytokinin-treated auf1 cells exited this zone earlier than the wild type. Given the widespread distribution of AUF1/2 proteins among land plants, the SCF complexes assembled with these FBX proteins likely target a conserved positive effector in the cross talk between auxin and cytokinin that regulates auxin movements and ultimately root elongation.
RESULTS
Genomic Analysis of an FBX Gene Pair Potentially Regulated by Auxin
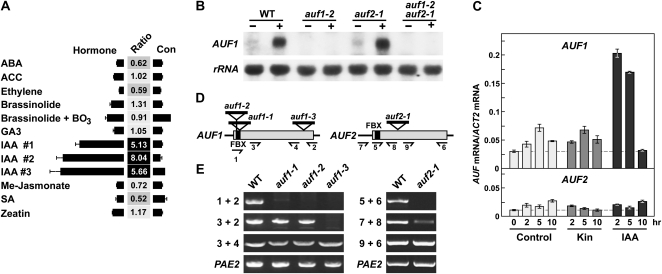
During our attempts to define the functions of the nearly 900 FBX loci in Arabidopsis by various “omic” approaches (Gagne et al., 2002; Hua et al., 2011), we noticed that the expression of one FBX gene designated AUF1 (At1g78100 in the C1 subclade) was shown in the Genevestigator (https://www.genevestigator.ethz.ch; Hruz et al., 2008) and eFP DNA microarray data browsers (www.Arabidopsis.org) to be strongly up-regulated by the natural auxin IAA but not by several other phytohormones or their precursors. The increase in AUF1 mRNA abundance ranged between 5- and 8-fold when whole seedlings were treated with 1 μm IAA (Fig. 1A). Further analyses of the microarray data sets revealed that AUF1 is expressed in most tissues, with the root-specific data sets in particular revealing high expression in the maturing cortical and epidermal cell files (Brady et al., 2007). To confirm the auxin up-regulation, we exposed Arabidopsis seedlings to 0.1 μm IAA for 5 h and then subjected root total RNA to RNA gel-blot analysis with an AUF1-gene specific probe. A strong increase in the expected 1-kb AUF1 transcript was evident, suggesting that the corresponding protein regulates auxin-dependent processes (Fig. 1B).
Figure 1.
AUF1 and AUF2 expression and description of auf1 and auf2 mutants. A, Microarray analysis of AUF1 transcript levels in response to various hormones as revealed by inspection of the Genevestigator Arabidopsis database (https://www.genevestigator.ethz.ch). The numbers reflect the ratio of expression between control and treated plants. ABA, Abscisic acid; ACC, 1-aminocyclopropane-2-carboxylic acid; Me, methyl; SA, salicylic acid. B, RNA gel-blot analysis of AUF1 mRNA after IAA treatment. Ten micrograms of total RNA was extracted from 7-d-old roots from auf1-2, auf2-1, and auf1-2 auf2-1 seedlings treated with (+) or without (−) 0.1 μm IAA for 5 h. 18S rRNA was used as a loading control. WT, Wild type. C, qRT-PCR of the AUF1 and AUF2 mRNAs in response to kinetin (Kin) and IAA. Seven-day-old seedlings were treated with 0.5 μm kinetin or 0.1 μm IAA for the indicated times. Root total RNA was then subjected to qRT-PCR using gene-specific primers. Transcript levels were normalized using the ACT2 transcript as a control. D, Organization of the AUF1 and AUF2 genes and insertion positions of the T-DNA mutations. Black lines and gray boxes indicate untranslated and coding regions, respectively. The black boxes mark the FBX domain. T-DNA insertion positions are marked by triangles. Primers used in RT-PCR analyses in E are indicated with half arrows; their nucleotide sequences can be found in Supplemental Table S1. E, RT-PCR analyses of auf mutants. RNA was subjected to first-strand cDNA synthesis with gene-specific primers 2 and 6 for AUF1 and AUF2, respectively, and then subjected to PCR using the indicated primers. RT-PCR of PAE2 mRNA was included as a control.
Sequence searches of the Arabidopsis ecotype Columbia (Col-0) genome revealed that AUF1 has an obvious paralog designated AUF2 (At1g22220). It is also on chromosome 1 but on the opposite side of the centromere relative to AUF1. Representative AUF2 transcripts are available in the Arabidopsis EST database (www.Arabidopsis.org), and we could generate a sequence-confirmed AUF2 cDNA by reverse transcription (RT)-PCR, indicating that the locus is expressed. Whereas 105 ESTs have been reported for AUF1, only nine have been reported for AUF2, suggesting that AUF2 is expressed at considerably lower levels than AUF1. Other details about AUF2 expression patterns are not yet known, mainly because of its omission from the Affymetrics ATH1 DNA microarrays commonly used to analyze the Arabidopsis transcriptome (www.Arabidopsis.org). However, quantitative real-time (q)RT-PCR showed that AUF2 expression is not affected by IAA (Fig. 1C). This is in contrast to the strong and transient up-regulation of AUF1 expression, which peaks between 2 and 5 h after IAA exposure and declines back to normal levels after 10 h.
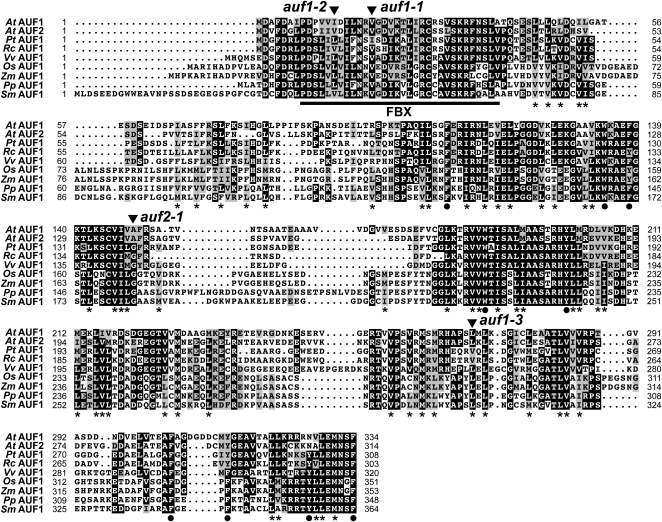
Transcripts from both AUF1 and AUF2 were predicted to be intronless, which was subsequently confirmed by DNA sequence analysis of cDNAs generated from seedling mRNA by RT-PCR. The single contiguous reading frames encode polypeptides of 334 and 311 residues, respectively, which share 71%/63% amino acid sequence similarity/identity (Fig. 2). Like other members of the FBX superfamily (Gagne et al., 2002; Hua et al., 2011), a signature FBX domain was predicted with high probability (1.2e-04 [AUF1] and 1.2e-04 [AUF2] by HMMER analysis) near the N terminus of both proteins. Typically, the region C terminal to the FBX domain contains one or more motifs that help recognize ubiquitylation targets. No known protein-protein interaction domains were obvious in AUF1 or AUF2 by searching with PFAM, but several stretches enriched in Leu, Ile, Met, and Val and interspersed with large bulky hydrophobic residues were evident, suggesting that Leu-rich-type repeats (LRRs) are present, but in a noncanonical arrangement (Fig. 2).
Figure 2.
Amino acid sequence comparisons of AUF proteins in land plants. Identical and similar residues are shown in black and gray boxes, respectively. Dots denote gaps. The numbers refer to the amino acid positions in each sequence. The residue length of each protein is shown at the end of the sequence. The FBX domain is underlined. The asterisks identify positions enriched in Leu, Ile, Met, and Val residues. The circles identify conserved Phe, Trp, and Tyr residues. The sites of Arabidopsis T-DNA insertions are indicated by the arrowheads. At, Arabidopsis thaliana; Pt, Populus trichocarpa; Rc, Ricinus communis; Vv, Vitis vinifera; Os, Oryza sativa; Zm, Zea mays; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
Subsequent searches detected AUF1/2-related genes in all available land plant genomes examined, including the moss Physcomitrella patens and the lycopod Selaginella moellendorffii, but not in any animal, fungal, or algal species, implying that the corresponding proteins have land plant-specific functions (Supplemental Fig. S1). In fact, AUF1/2 are members of a widely distributed collection of land plant FBX genes undergoing strong purifying selection (Hua et al., 2011), suggesting that the corresponding AUF1/2 proteins direct a conserved and likely essential ubiquitylation event important to terrestrial plant life. Amino acid alignment of AUF1/2-related proteins revealed strong conservation in the FBX domain as well as in the C-terminal region bearing the potential LRRs (Fig. 2). Multiple AUF1/2 genes are present in all species with well annotated genomes, including two each in P. patens, S. moellendorffii, and Pinus strobus, three in rice (Oryza sativa), and four in maize (Zea mays; Supplemental Fig. S1), raising the possibility that the AUF family expanded early in land plant evolution. While the clustering of monocot paralogs with close relatives from other species rather than with each other supports this scenario, the eudicot paralogs often clustered together with their paralogs, suggesting that the eudicot AUF genes expanded by more lineage-specific events (Supplemental Fig. S1).
Reverse Genetic Analysis of AUF1 and AUF2
To help define the functions of AUF1/2, we acquired several T-DNA insertion mutants affecting the coding regions that should strongly impair the synthesis of the corresponding proteins (Fig. 1C). The three auf1 alleles prevent the synthesis of the full-length AUF1 transcript, as detected by RT-PCR and/or RNA gel-blot analysis (Fig. 1, B and E). However, RT-PCR detected partial transcripts emanating downstream from the insertion sites in auf1-1 and auf2-1 plants and upstream of the insertion site in auf1-3 plants. The auf1-1 and auf1-2 transcripts likely reflect cryptic promoter activity within the T-DNA insertion. Nonetheless, these two mutants should represent strong alleles based on their disruption of the coding region for the FBX domain, which is required for docking AUF1 with the rest of the SCF complex. The auf1-3 mutant protein should be missing much of its predicted LRR-like target recognition module, likely rendering this truncation inactive even if expressed and assembled into an SCFauf1-3 complex via its intact FBX domain.
Only a single T-DNA insertion line was available for AUF2 (Fig. 1D). RT-PCR analyses showed that the auf2-1 allele also disrupts the synthesis of a full-length AUF2 mRNA but that partial transcripts before and after the T-DNA accumulate (Fig. 1E). Even if translated, the resulting truncated polypeptides should be missing either the FBX domain or much of the target recognition module, thus likely compromising both fragments if synthesized separately.
AUF1 Mutants Display Defects in Auxin Transport
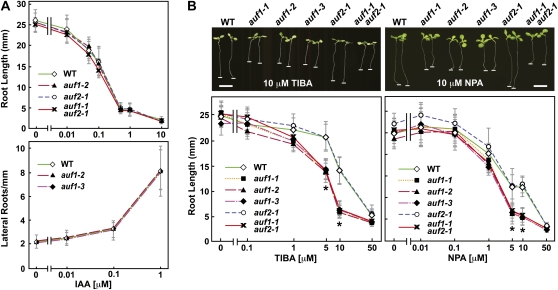
Under normal laboratory growth conditions, homozygous seedlings for each of the three auf1 alleles, auf2-1, and the auf1-2 auf2-1 double mutant were phenotypically indistinguishable from the wild-type Col-0 parent and showed normal fertility and genetic segregation. Thus, we conclude that the AUF1 and AUF2 proteins separately or together are not essential for most, if not all, aspects of Arabidopsis development and reproduction. Given the strong increase in AUF1 mRNA by IAA, we predicted that auf1-2 and auf1-2 auf2-1 plants would show defects in auxin perception and signaling. In contrast, the mutant plants responded normally to exogenous IAA and the synthetic auxin 1-naphthaleneacetic acid, as measured by auxin-induced inhibition of root elongation and promotion of lateral root emergence and by the rate of root curvature induced by gravity, a well-described response that depends upon local changes in the distribution of auxin (Fig. 3A; Supplemental Fig. S2, D and E). auf1-2 root elongation was similarly unaffected by several other growth regulators, including jasmonic acid, abscisic acid, and the ethylene precursor 1 aminocyclopropane-1-carboxylic acid (data not shown).
Figure 3.
Response of auf mutant roots to auxin and auxin transport inhibitors. For primary root length measurements, seedlings were germinated and exposed to the indicated concentrations of IAA and the inhibitors TIBA and NPA for 7 d in constant light before measurement. For lateral root densities, the plants were first grown without IAA for 4 d, transferred to medium containing various concentrations of IAA, and grown for an additional 6 d. Each point represents the average ± sd of more than 20 seedlings. A, Effect of IAA on root elongation and lateral root emergence. B, Effect of TIBA and NPA on root elongation. Asterisks identify points with significant differences between the wild type and the mutants containing the auf1 alleles (Student’s t test; P < 0.005). Images of representative plants exposed to 10 μm TIBA or NPA are shown above each graph. The white lines highlight root tips. WT, Wild type. Bars = 5 mm.
However, subtle differences in auxin signaling were detected for auf1 plants using several molecular markers of auxin signaling. For example, the expression of the AUX/IAA gene IAA1, which is up-regulated by 1 μm IAA (Abel et al., 1994), was poorly responsive in the auf1-2 background (Supplemental Fig. S2C). The well-characterized auxin-responsive reporter DR5pro::GUS (Ulmasov et al., 1997) also showed a dampened response to exogenous IAA in auf1-2 plants. In the absence of IAA, both wild-type and auf1-2 RMs harboring DR5pro::GUS expressed GUS, as observed by histochemical staining, in a small collection of cells comprising the quiescent center (QC) and the root stem cell niche (Supplemental Fig. S2A). IAA treatment (1 μm) greatly expanded the zone of expression into the EDZ and the mature zone (with root hairs), with the expression less robust in auf1-2 and auf1-2 auf2-1 roots as compared with wild-type and auf2-1 roots (Supplemental Fig. S2A). This dampened auxin response for auf1 plants could also be seen by quantitative 4-methyllumbelliferyl-β-d-glucuronide (MUG) assays measuring GUS activity in whole root tips treated with 1 μm IAA (Supplemental Fig. S2B).
To examine the ability of auf1/2 plants to maintain appropriate auxin maxima when auxin transport is compromised, we tested the response of the mutants to NPA and TIBA, two drugs that inhibit polar auxin transport (Lomax et al., 1995). NPA works by blocking the action of the ABCB and potentially PIN families (Noh et al., 2001; Bouchard et al., 2006; Petrasek et al., 2006), whereas TIBA appears to impair cycling of the PIN family between the plasma membrane and endosomes (Geldner et al., 2001). Strikingly, root elongation of seedlings homozygous for each of the three auf1 alleles and the auf1-2 auf2-1 combination, but not the auf2-1 allele, was hypersensitive to both inhibitors (Fig. 3B). The most significant effects were seen for 5 to 10 μm TIBA or NPA, where a greater than 2-fold difference in root length was observed after a 7-d exposure. The response appeared specific for roots, as no obvious differences in shoot growth were evident between NPA-treated wild-type and auf1 plants.
We further confirmed the NPA-hypersensitive phenotype by rescuing auf1-2 plants with transgenes expressing full-length AUF1 under its native promoter, either as an N-terminal fusion to the Flag epitope tag or to GFP. Root growth in the presence of NPA for multiple independent AUF1pro::AUF1-Flag auf1-2 and AUF1pro::AUF1-GFP auf1-2 lines was restored to near the wild-type rate and significantly better than the auf1-2 parent (Supplemental Fig. S3). Unfortunately, while the tagged AUF1 transgenes appeared to be active phenotypically, we could not detect accumulation of the corresponding proteins. No signals at the appropriate apparent masses were detected in untreated seedlings or in seedlings treated with 1 μm IAA or 10 μm of the proteasome inhibitor MG132 by immunoblot analysis with either anti-Flag or anti-GFP antibodies or by confocal fluorescence microscopic examination of GFP-expressing roots.
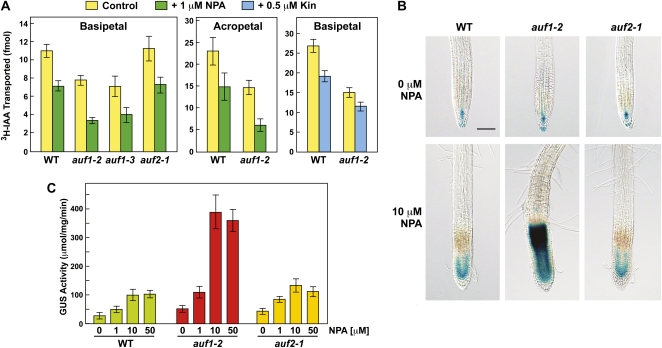
The hypersensitivity of auf1 plants to NPA and TIBA strongly suggested that AUF1 promotes auxin transport. To test this hypothesis, we measured the movement of [3H]IAA after localized application (Lewis and Muday, 2009). As can be seen in Figure 4A, IAA transport was significantly depressed in the basipetal (shootward) direction in auf1-2 and auf1-3 roots but not in auf2-1 roots, and it was significantly depressed in the acropetal (rootward) direction in auf1-2 roots. Transport in both directions was further dampened by 1 μm NPA, with the NPA-treated auf1-2 roots having approximately 30% of the auxin transport rate found in nontreated wild-type roots.
Figure 4.
Effect of auf1 mutants on auxin transport and the accumulation of auxin in roots. A, Effect of auf1-2 and auf1-3 mutants on basipetal (shootward) and acropetal (rootward) transport of [3H]IAA in roots incubated with or without 1 μm NPA or 5 μm kinetin (Kin). Each bar represents the mean ± se of three independent experiments each involving 15 to 20 individual root measurements. Differences with or without NPA and between the wild type/auf2-1 and the auf1 mutants were statistically significant by Student’s t test (P < 0.0013). B, Effect of the auf1-2 mutant on DR5pro::GUS expression in roots incubated with or without 10 μm NPA. Seedlings were grown on agar medium supplemented without or with NPA for 4 d and then stained for GUS activity. Bar = 0.5 mm. C, Quantitative measure of DR5pro::GUS expression in roots treated with increasing concentrations of NPA. Seedlings were grown on the indicated concentrations of NPA for 4 d before harvest. Each bar represents GUS activity from 50 excised root tips assayed in triplicate ± sd using the MUG assay. WT, Wild type.
To test the likelihood that NPA-treated auf1-2 plants have altered auxin distributions, we indirectly measured auxin maxima using the DR5pro::GUS reporter (Ulmasov et al., 1997). As with auxin (Supplemental Fig. S2A), NPA treatment of wild-type roots expanded the zone of GUS expression basally from the QC and stem cell niche into the RM (Fig. 4B). Notably, the NPA-induced expansion of auxin maxima was substantially more robust in auf1-2 roots and extended further basally into the region of root hair emergence that likely includes the EDZ (Fig. 4B). The edge of high DR5pro::GUS expression was remarkably sharp, with little or no staining evident in mature cells, suggesting that NPA traps basipetal auxin flow at a defined boundary in auf1-2 roots. Quantitative measure of DR5pro::GUS activity showed that auf1-2 root tips overall had 3- to 4-fold more GUS upon NPA treatment as compared with wild-type or auf2-1 roots (Fig. 4C). Taken together, these data suggest that AUF1 regulates auxin distribution primarily in the small region surrounding the RM/EDZ junction, a location consistent with its reported expression patterns (Brady et al., 2007).
AUF1 Modifies PIN Gene Expression
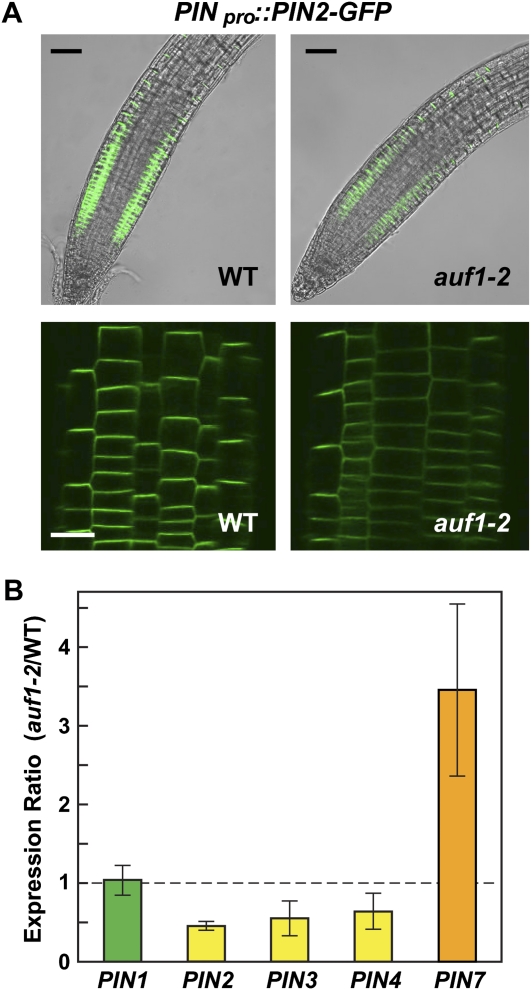
Unlike many other mutations that affect root auxin transport in either the acropetal or basipetal direction (Okada et al., 1991; Lewis et al., 2009), the auf1 mutants significantly reduced auxin transport in both directions, suggesting that AUF1 participates in a global auxin transport-regulating network genetically upstream of tissue-specific auxin transport facilitators. Obvious candidates for the downstream effectors are the PIN efflux facilitators, which play prominent and isoform-specific roles in directing auxin redistribution within the root (Blilou et al., 2005). Using as a reporter the PIN2-GFP translational fusion protein expressed under the control of the native PIN2 promoter (PIN2pro::PIN2-GFP; Pan et al., 2009), we found that AUF1 inactivation markedly reduced the accumulation of PIN2 (and possibly other PIN proteins) in roots. Upon comparison of wild-type and auf1-2 lines introgressed with the same PIN2pro::PIN2-GFP insertion event, considerably less GFP fluorescence was detected in the root tip in the absence of AUF1 (Fig. 5A). Lower PIN2-GFP signal in auf1-2 roots could have been generated by defects related to the asymmetric plasma membrane partitioning of PIN2 (Blilou et al., 2005) or its continuous auxin-dependent cycling between the plasma membrane and endosomes (Geldner et al., 2001; Pan et al., 2009). However, the intracellular distribution of PIN2-GFP in root epidermal cells appeared indistinguishable between the wild type and auf1-2, with a majority of the fluorescence in both lines localized to what appears to be the basal plasma membrane (Fig. 5A). Moreover, the cycling of PIN2-GFP between the plasma membrane and endosomes was unaffected. Upon treating roots with the drug brefeldin A (BFA), which inhibits endosome cycling of PIN proteins (Geldner et al., 2001), PIN2-GFP accumulated in the endosome-like “BFA bodies” of auf1-2 root epidermal cells at a rate comparable to that of the wild type (Supplemental Fig. S4).
Figure 5.
Distribution and expression of PIN2 in auf1 mutants. A, PIN2 localization in wild-type and auf1-2 roots using the PIN2-GFP fusion reporter. The top panels show merged views of representative 7-d-old roots imaged by light microscopy and by fluorescence microscopy of GFP. Identical microscopic settings were used to illustrate the reduced accumulation of PIN2-GFP in the auf1-2 background. The bottom panels show higher magnifications of PIN2-GFP localization in root epidermal cells. WT, Wild type. Bars = 0.5 mm (top) and 50 μm (bottom). B, Accumulation of PIN mRNAs in the auf1-2 mutant. Transcript levels were measured by qRT-PCR in wild-type and auf1-2 roots and expressed as a ratio. Each bar represents the average ± se of three independent experiments that each analyzed a pool of 120 roots. Student’s t test values are as follows: PIN1, P = 0.83; PIN2, P < 0.0001; PIN3, P = 0.093; PIN4, P = 0.13; and PIN7, P = 0.043.
Another possibility was that auf1 mutants dampen PIN gene transcription, consistent with prior studies showing that PIN expression is dynamically regulated in RMs (Dello Ioio et al., 2008; Ruzicka et al., 2009). This effect was confirmed for several PIN members by qRT-PCR; here, the transcript levels of PIN2, PIN3, and possibly PIN4 were significantly decreased in the auf1-2 background (Fig. 6B). Similar to the report of Ruzicka et al. (2009) studying the effects of cytokinin on PIN expression, we found that not all PIN transcripts tested were affected similarly by auf1 mutations. The level of PIN7 mRNA was markedly elevated in auf1-2 plants, whereas the level of PIN1 mRNA was essentially unchanged, suggesting that AUF1 has an isoform-specific control on PIN protein accumulation (Fig. 6B). It was also possible that AUF1 affects the expression of other key auxin transporters, including AUX1, ABCB4, and ABCB19 (Yang et al., 2006; Lewis et al., 2007; Wu et al., 2007). However, qRT-PCR analyses of auf1-2 roots detected no significant change in their transcript levels as compared with the wild type (Supplemental Fig. S5).
Figure 6.
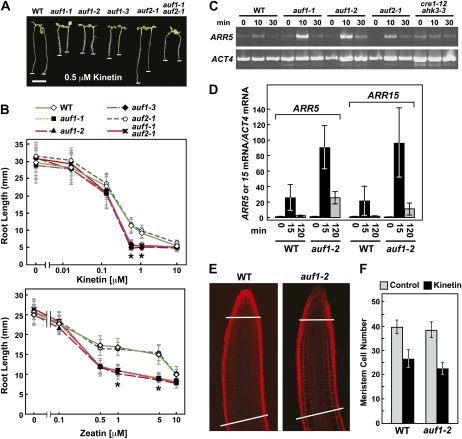
Effects of auf1 mutations on the response of roots to cytokinin. A and B, Effect of the cytokinins kinetin and zeatin on root elongation. A, Representative roots grown for 7 d on medium containing 0.5 μm kinetin. Bar = 5 mm. The white lines highlight root tips. B, Effect of various concentrations of kinetin and zeatin on root growth. Seedlings were grown for 7 d before measurement. Asterisks identify points with significant differences between the wild type (WT) and the mutants containing the auf1 alleles (Student’s t test; P < 1.5e-13). C, Accumulation of the cytokinin-regulated ARR5 transcript in 10-d-old seedlings treated for various times with 0.5 μm kinetin as determined by semiquantitative RT-PCR. RT-PCR of the ACT4 transcript was used to confirm the analysis of equal amounts of RNA. The cytokinin-insensitive cre1-12 ahk3-3 double mutant was used as a control. D, Accumulation of the cytokinin-regulated ARR5 and ARR15 transcripts in 10-d-old seedlings treated for various times with 0.5 μm kinetin, as determined by qRT-PCR using the ACT4 transcript as a control. Each bar represents the mean ± se of three independent biological replicates in which each RNA sample was analyzed in triplicate. E and F, Root meristem size is unaffected by the auf1-2 mutation. E, Representative root tips from 7-d-old wild-type and auf1-2 plants treated with 0.5 μm kinetin. The cell walls were stained with propidium iodide and visualized by confocal fluorescence microscopy. The lines highlight the approximate positions of the RM boundaries as defined by the zone between the stem cell niche and the first noticeably more elongated cortical cell in the proximal EDZ (Dello Ioio et al., 2007). F, Size of the RM from wild-type and auf1-2 plants exposed to 0.5 μm kinetin. Each bar represents the average cell number in the RM cortical file from 20 roots ± sd as defined in E.
auf1 Mutants Are Hypersensitive to Cytokinin
The aforementioned antagonistic connection between cytokinin and auxin transport through SHY2 (Dello Ioio et al., 2008), and our observations that AUF1 inactivation alters the expression of some PIN genes similar to that observed upon cytokinin treatment (Fig. 5; Ruzicka et al., 2009), raised the possibility that auf1 plants have altered cytokinin sensitivity. To test this scenario, we performed a dose-response analysis on auf1 root growth using two natural cytokinins, kinetin and zeatin. Like the response to auxin transport inhibitors, all three mutant alleles of auf1 and the auf1-2 auf2-1 double mutant, but not auf2-1, were significantly more sensitive to these cytokinins (Fig. 6, A and B). Moreover, this hypersensitivity could be reversed by introducing the AUF1pro::AUF1-Flag and AUF1pro:AUF1-GFP transgenes into the auf1-2 background (Supplemental Fig. S3B). Like NPA, exogenous cytokinin also dampened IAA transport, which was further decreased in the auf1-2 background (Fig. 4A). The inhibition of root growth by cytokinin could be mediated in part by the ability of cytokinin to stimulate ethylene production, which in turn blocks cell elongation (Chae et al., 2003). In support, we found that simultaneous treatment with kinetin and the ethylene biosynthesis inhibitor 2-aminoethoxyvinylglycine abolished the kinetin hypersensitivity of auf1-2 roots (Supplemental Fig. S6), further highlighting the important hormonal cross talk that regulates root growth.
One initial response to exogenous cytokinin is the rapid and transient increase in transcripts encoding the type A negative effectors ARR5 and ARR15 (D’Agostino et al., 2000; Kiba et al., 2002). The increases occur between 10 and 15 min after cytokinin treatment, with the mRNA levels quickly returning back to pretreatment levels after 30 min. This rise is abolished in cre1-12 ahk3-3 double mutants, thus implicating the two affected cytokinin receptors (Fig. 6, C and D; Higuchi et al., 2004). When ARR5/15 transcript accumulation was similarly tested in the auf backgrounds by semiquantitative RT-PCR and qRT-PCR, we found that the levels of both were substantially up-regulated by 0.5 μm kinetin in auf1-1 and auf1-2 plants as compared with wild-type and auf2-1 plants (Fig. 6, C and D). The rise and fall of mRNA levels were similar among the time courses, indicating that the magnitude but not the duration of the responses was accentuated by AUF1 inactivation. By comparison, the transcript levels for neither AUF1 nor AUF2 were increased by kinetin (Fig. 1C), consistent with a scenario in which the decrease of cytokinin responsiveness through AUF1 is mediated indirectly by its auxin up-regulation.
Cytokinin affects root development by increasing the rate of cell differentiation, which in turn reduces the size of the RM (Dello Ioio et al., 2008; Moubayidin et al., 2010). To determine if the cytokinin effect on auf1 plants was through an effect on RM size, we compared the number of cortical cells in the file that span the distance from the root QC to the first noticeably elongated cell marking the beginning of the EDZ. Similar to previous reports (Dello Ioio et al., 2008), we found that exogenous kinetin (0.5 μm) substantially reduced the number of cells in the wild-type RM. However, the RM of auf1-2 roots was similar in size to the wild type without kinetin and was reduced to an equivalent amount upon kinetin treatment (Fig. 6,E and F). Collectively, these data imply that AUF1 does not control RM size but instead may affect elongation/differentiation of the downstream EDZ in response to cytokinin.
AUF1 Is Required for the Cytokinin-Mediated Promotion of Root Cell Differentiation
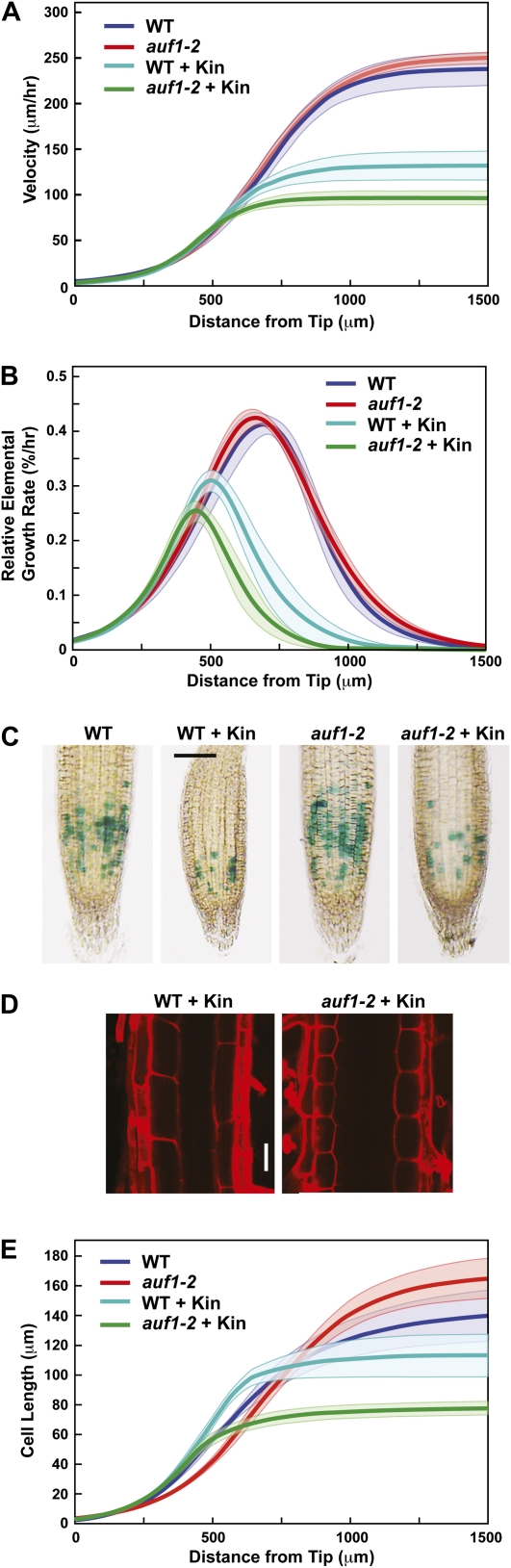
To help localize where AUF1 controls root growth in response to cytokinin, we used computer-assisted kinematic analyses of individual roots to describe their growth dynamics in detail (Beemster and Baskin, 1998, 2000; Miller et al., 2007). Growth velocity profiles along the length of wild-type roots revealed that cell growth begins to accelerate approximately 200 μm away from the tip near the base of the RM and then increases steeply over the next 500 μm in the region encompassing the proximal EDZ (Fig. 7A). Cytokinin suppresses growth velocity to produce shorter roots at any point in time. Whereas inactivation of AUF1 did little to affect growth velocity in the absence of kinetin (approximately 230 versus approximately 250 μm h−1), growth velocity of the mutant was significantly reduced in its presence (approximately 130 to approximately 90 μm h−1; Fig. 7A), thus explaining why auf1 mutant roots are shorter than wild-type roots after cytokinin treatment.
Figure 7.
AUF1 controls root cell elongation but not division in the presence of cytokinin. A and B, Growth velocity profile and REGR of wild-type (WT) and auf1-2 roots, respectively. Growth and REGR measurements were made along the length of the primary root beginning from the QC toward the shoot in the absence or presence of 0.5 μm kinetin (Kin). Thick lines represent smoothed averages, whereas thin lines represent se. Six to eight roots were analyzed for each condition. C, Mitotic activity of auf1-2 RMs. Wild-type and auf1-2 roots expressing the CYCLINB1;1pro::GUS-DBox reporter were grown for 7 d in the absence or presence of 0.5 μm kinetin and then stained for GUS activity. D, Representative images of cells in the root maturation zone from wild-type and auf1-2 plants treated with kinetin. Shown are the cell walls from cortical cells stained with propidium iodide and observed by fluorescence confocal microscopy. Bar = 50 μm. E, Average cell length of the cortical cell file in wild-type and auf1-2 roots. Thick lines represents smoothed averages, whereas thin lines represent se. In each experiment, the primary root from six to eight seedlings grown for 7 d on medium supplemented without or with 0.5 μm kinetin was analyzed.
The first derivative of the velocity profiles was used to produce axial relative elemental growth rate (REGR) profiles (Beemster and Baskin, 2000). As shown in Figure 7B, inactivation of AUF1 by itself affected neither how fast the root isodiametrically expanded nor where along the axis this expansion occurred. Both peaked at approximately 45% h−1 approximately 700 μm from the root tip. Kinetin compressed the REGR profile by shifting the basal boundary closer toward the tip by approximately 200 μm. This accelerated cessation of expansion by cytokinin could be explained by an apical shift in root cell maturation. Loss of AUF1 exacerbated this cytokinin effect by further compressing the elongation zone, especially in the region between 500 and 750 μm from the tip encompassing the proximal EDZ, thus leading to shorter roots (Fig. 7B).
Differences in the REGR profiles of auf1 versus wild-type roots exposed to cytokinin could reflect differences in the rates of cell production by the RM and/or differences in cell expansion. To observe an effect on cell division, we introgressed the mitotic reporter CYCLINB1;1pro::GUS-DBox into auf1-2 plants, which transiently accumulates GUS at the G2 stage of the cell cycle (Colon-Carmona et al., 1999). From counts of GUS-stained cells, we found that both wild-type and auf1-2 RMs had similar mitotic numbers without treatment and that the numbers decreased equally upon 0.5 μm kinetin treatment, indicating that AUF1 does not modify cell division rates in the RM. To test for changes in cell expansion rates, we measured the lengths of cortical cells along the entire root axis. Here, a significant effect on cytokinin-treated auf1-2 roots was observed. From initial microscopic analysis of the EDZ region, it was clear that the auf1-2 cortical cells were considerably smaller than similarly treated wild-type cells (Fig. 7, D and E). Whereas 0.5 μm kinetin decreased final cell size in the wild type by approximately 12% (140–114 μm), the hormone decreased the final cell size in auf1-2 by 53% (163–77 μm). Detailed analysis of the entire cortical file from the RM to the EDZ revealed that this reduced elongation begins closer to the tip, with kinetin-treated auf1-2 cells prematurely terminating elongation closer to the apex than wild-type cells (Fig. 7E).
ARR1 Is Epistatic to AUF1
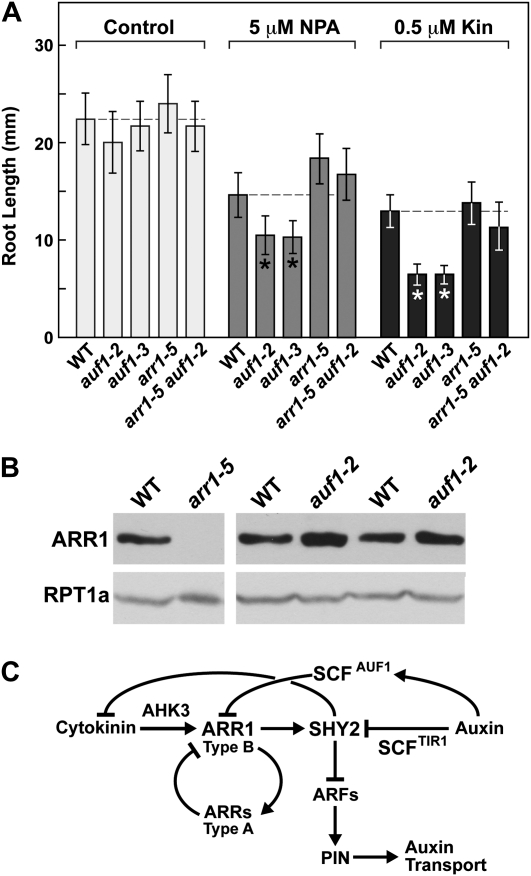
Prior studies connected cytokinin to auxin transport and root growth via the action of the type B response regulator ARR1 (and possibly ARR12), whose expression is concentrated in the transition zone encompassing the distal RM and the proximal EDZ (Dello Ioio et al., 2007). To examine whether AUF1 and ARR1 might interact genetically, we compared the response of auf1-2 and auf1-3 roots to cytokinin and NPA with that of the arr1-5 null mutant of ARR1 and an arr1-5 auf1-2 double mutant. As shown in Figure 8A, untreated roots from all the genetic backgrounds grew similar to the wild type. However, whereas the two auf1 mutants were significantly shorter than the wild type in the presence of 5 μm NPA or 0.5 μm kinetin, the arr1-5 and arr1-5 auf1-2 plants resembled the wild type. This rescue demonstrated that the hypersensitivity of auf1 roots to both cytokinin and NPA can be reversed by removing ARR1 (Fig. 8A).
Figure 8.
AUF1 potentially functions in a pathway involving ARR1. A, Removal of ARR1 reverses the hypersensitivity of auf1-2 roots to the auxin transport inhibitor NPA and to cytokinin. Primary root length was measured on 7-d-old wild-type (WT), auf1-2, auf1-3, arr1-5, and arr1-5 auf1-2 seedlings grown without or with 5 μm NPA or 0.5 μm kinetin (Kin). Error bars indicate sd (n > 20). Asterisks identify treatments with significant differences between the wild type and the mutants containing the auf1 alleles (Student’s t test; P ≤ 0.0012). B, Immunoblot detection of ARR1. Crude extracts were prepared from roots harvested from 10-d-old seedlings and subjected to SDS-PAGE and immunoblot analysis with anti-ARR1 antibodies. Duplicate loads are shown to account for lane-to-lane variation. Equal protein loading was confirmed by immunoblot analysis with antibodies against the 26S proteasome subunit RPT1a. C, Possible model describing the role of the SCFAUF1 E3 in the cross talk between cytokinin and auxin transport.
The apparent rescue of auf1 roots by the arr1-5 mutation suggested that ARR1 is in the same pathway and epistatic to AUF1. An obvious possibility is that the ARR1 protein is the direct target of SCFAUF1, with the corollary expectation that ARR1 is stabilized and thus more abundant in auf1 plants. However, immunoblot analyses of ARR1 in whole root extracts revealed that its abundance with or without kinetin or IAA pretreatment was not consistently increased in the auf1-2 background (Fig. 8B; data not shown). This lack of effect could imply a more complex relationship between ARR1 and AUF1 or the possibility that localized ARR1 turnover is masked when whole roots are examined.
DISCUSSION
Previous studies identified a set of transcriptional and proteolytic feedback loops that antagonistically connect auxin and cytokinin signaling during root growth and differentiation (Blilou et al., 2005; Dello Ioio et al., 2007, 2008). Here, we add another loop to this cross talk with the discovery that the Arabidopsis FBX protein AUF1 (and possibly AUF2) regulates root auxin transport. AUF1 expression is rapidly and transiently up-regulated by auxin, and its disruption generates a hypersensitivity to the auxin transport inhibitors NPA and TIBA as well as to cytokinin. Both hypersensitivities were confirmed with multiple auf1 alleles and by complementation of the auf1-2 phenotype with tagged AUF1 transgenes. Altered auxin transport in auf1 plants was demonstrated by (1) reduced [3H]IAA transport rates, (2) altered expression patterns of the auxin-sensitive DR5pro::GUS reporter after IAA or NPA application, and (3) altered expression of several PIN auxin efflux facilitators, which at least for PIN2 reduced its protein abundance. A role for AUF1 in cytokinin perception was supported by a hypersensitivity of auf1 root growth to exogenous cytokinin and by the transiently enhanced up-regulation of several type A ARRs that negatively regulate cytokinin signaling. Part of the cytokinin effect may involve an enhanced synthesis of ethylene, which can also alter auxin transport (Vandenbussche et al., 2003; Negi et al., 2007). While the collective data indicate that AUF1 plays a role in modulating auxin transport in roots, it is notable that AUF1 is not essential for either root or shoot growth under normal conditions. Consequently, AUF1 must play a more subtle role in fine-tuning the process(es) that dictate auxin movements, auxin maxima, and hormonal cross talk, which seem to remain somewhat robust in its absence.
Our results with AUF1 provide further support for a role of cytokinin in controlling auxin transport; in particular, we identify a role for AUF1 in affecting PIN gene expression and ultimately PIN protein accumulation in roots. Presumably, reduced PIN levels dampen auxin distributions sufficiently to make auf1 seedlings more sensitive to auxin transport inhibitors such as NPA and TIBA. One striking feature of auf1-2 plants treated with NPA is the dramatic accumulation of auxin (as observed by DR5pro::GUS expression) in the region encompassing the distal RM and proximal EDZ. Together with kinematic analysis of root growth, the data imply that AUF1 has its greatest effect on auxin concentrations in this small region, which importantly coincides with the region where auxin transport is predicted to be most affected by cytokinin (Dello Ioio et al., 2008). One interesting point from the study of Ruzicka et al. (2009) and observed here is that cytokinin or the elimination of AUF1 does not dampen the expression of all PIN genes but have contrasting roles in decreasing PIN2, PIN3, and possibly PIN4 transcript levels while simultaneously increasing PIN7 transcript levels. These differential effects could trap auxin in particular cells/tissues within the RM and EDZ by encouraging the acropetal transport responsible for delivering shoot-derived auxin into this region while simultaneously discouraging the basipetal transport responsible for moving auxin away from the apical maximum.
Interestingly, auf1 mutants share similar phenotypes with several described mdr/abcb mutants, thus further supporting a connection between AUF1 and auxin transport. For instance, the mdr1-1 mutant responds like the wild type to exogenous auxin and has normal gravitropism, yet it exhibits an 80% reduction of acropetal auxin transport in the root (Lewis et al., 2007). Mutants in PIS1, which encodes the ABCG37 transporter, were shown previously to be hypersensitive to TIBA and NPA, as measured by an increased inhibition of root growth, but they respond normally to exogenous IAA (Fujita and Syono, 1997). Recently, it was demonstrated that ABCG37 primarily aids in transporting the auxinic compound indole-3-butyric acid instead of IAA (Ruzicka et al., 2010), thus raising the possibility that AUF1 also controls the movement of auxins besides IAA. Dual effects on ABCB and PIN auxin transport facilitators may explain why auf1 plants are defective in both acropetal and basipetal auxin transport. However, at least for the major auxin transporters in roots AUX1, ABCB4, and ABCB19 (Yang et al., 2006; Lewis et al., 2007; Wu et al., 2007), AUF1 appears to have no role in controlling their expression.
The exact role(s) of Arabidopsis AUF1 (and possibly AUF2) in the cross talk between auxin transport and cytokinin signaling remains unclear. Based on the strong homology within the predicted FBX domains, we expect that both assemble into SCF E3 complexes that recognize the same or similar target(s). Unfortunately, attempts to confirm this assembly have been unsuccessful, primarily due to (1) our failure to express sufficient quantities of tagged AUF1 variants (Flag or GFP) in planta that could be used to isolate the entire SCF complex, and (2) the propensity of AUF1 to autoactivate yeast two-hybrid assays, thus precluding its use in paired interaction studies with Arabidopsis SKP1 proteins. The poor expression of AUF1 is not without precedent, as a number of other FBX proteins have been shown to express poorly and/or be inherently unstable as a result of an intrinsic autoubiquitylation activity of SCF E3s, which triggers turnover of the FBX subunit by the 26S proteasome (Bosu and Kipreos, 2008; An et al., 2010). In our case, even pretreating AUF1-Flag or AUF1-GFP plants with MG132 failed to permit transgenic AUF1 protein detection with anti-Flag or anti-GFP antibodies. In the absence of direct data, we note that AUF1 and AUF2 phylogenetically cluster based on their FBX domains close to several well-characterized Arabidopsis FBX proteins (Gagne et al., 2002; Hua et al., 2011), including SLEEPY1, which has been shown previously to incorporate into an SCF E3 complex (McGinnis et al., 2003).
While we presume based on sequence homology that AUF2 functions like AUF1, we failed to associate these potential paralogs either genetically or by expression studies. Unlike AUF1, expression of AUF2 is not up-regulated by auxin. Furthermore, auf2 mutants were indistinguishable from the wild type under all conditions tested, and the auf1-2 auf2-1 double mutant did not display any exaggerated phenotypes compared with the auf1-2 single mutant. Consistent with the low expression of AUF2 relative to AUF1, we propose that AUF2, if active, plays a minor, more constitutive role in whatever process(es) these two FBX proteins control.
The central unresolved question pertains to the identity of the AUF1/2 substrate(s). The widespread distribution of AUF-type genes within the plant kingdom implies that these substrate(s) direct a key conserved step in root/rhizoid development that appeared early in land plant evolution. Based on the auf1 phenotypes, we propose that SCFAUF1/2 targets for ubiquitylation a positive regulator in the cross talk between cytokinin signaling and auxin transport. Overaccumulation of this regulator in auf1 roots accentuates the responsiveness to cytokinin, which through the SHY2 feedback loop (and possibly aminoethoxyvinylglycine-sensitive ethylene synthesis) dampens the expression of specific PIN genes (e.g. PIN2, PIN3, and PIN4; Dello Ioio et al., 2008; Ruzicka et al., 2009). Concomitant reductions in protein levels (e.g. PIN2) reduce auxin transport below a critical threshold, which, in turn, makes auf1 roots more sensitive to auxin transport inhibitors. Coincidently, coexpression correlations derived from nearly all available Arabidopsis microarray data sets found that the expression patterns of AUF1 most closely matched that of SHY2 (Pearson correlation coefficient = 0.53 [http//atted.jp]), proposed to be at the core of the cytokinin/auxin transport connection.
Previous studies showed that SHY2 expression is up-regulated by cytokinin via the type B ARR1 transcription factor and that its increased protein abundance then down-regulates the expression of multiple PIN genes, thus attenuating auxin transport and altering auxin maxima (Dello Ioio et al., 2007). The end result is shorter roots caused by increased differentiation rates in the RM and EDZ. Auxin, conversely, targets SHY2 for breakdown via the SCFTRI1/ABF1-3 E3s, thus relieving this repression. Taken together, we propose a model that explains the auf1 phenotypes and their restoration in the arr1-5 auf2-1 combination. The model states that SCFAUF1 targets ARR1 for ubiquitylation and subsequent turnover in the absence of cytokinin. Low ARR1 levels would dampen SHY2 expression, thus attenuating its repressive effects on the expression of some, but not all, PIN genes. Increased PIN levels driving robust auxin transport would then promote maintenance of the RM and delay elongation/differentiation, thus increasing root growth. The auxin-induced expression of AUF1 in the RM/EDZ could reinforce UPS-mediated ARR1 turnover and subsequent SHY2 down-regulation by increasing the concentration of the SCFAUF1/2 complex. Cytokinin, in contrast, could antagonize this promotional effect on growth by blocking ARR1 breakdown by SCFAUF1/2. ARR1 stabilization by cytokinin or AUF1 inactivation increases SHY2 transcription, with the increased SHY2 protein levels then repressing both cytokinin synthesis and PIN expression, resulting in reduced auxin transport. As observed here for auf1 mutants, increased ARR1 would also up-regulate the transcription of the ARR5 and ARR15 type A negative effectors of cytokinin signaling (Taniguchi et al., 2007).
While the data are consistent with ARR1 being the SCFAUF1/2 substrate, we failed to detect a marked increase of ARR1 protein in auf1 backgrounds from the immunoblot analysis of whole seedlings or even just roots with or without auxin and cytokinin pretreatment. Consequently, other modes of ARR1 down-regulation are possible, including AUF1 affecting the abundance and/or activity of other positive/negative regulators associated with cytokinin signaling, including a secondary effect on auxin transport by ethylene. However, given that the SCFAUF1/2 complex may have a highly restricted effect on auxin transport (i.e. distal RM and proximal EDZ), the ability to detect ARR1 stabilization in auf1 plants may require focused analysis of just the small zone of root tissue where AUF1 and ARR1 overlap (i.e. distal RM and proximal EDZ; Brady et al., 2007; Dello Ioio et al., 2007). Whatever the SCFAUF1/2 substrate(s), their identities will likely reveal an additional regulatory mechanism within the root tip that dynamically responds to auxin/cytokinin cross talk.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana Col-0) plants were used in all analyses. Prior to germination, seeds were vapor-phase sterilized and incubated in sterile water at 4°C for 2 d in the dark. For phenotypic assays, seedlings were grown vertically on half-strength Murashige and Skoog (MS) medium with 1% Suc and 1% agar at 21°C in continuous white light. For liquid-grown seedlings, seeds were germinated and cultured in half-strength MS liquid medium under continuous white light. For various hormone and inhibitor treatments, the compounds were added to the germination medium. Effects of the compounds on root elongation were quantified using the ImageJ software package (http://rsb.info.nih.gov/ij/).
AUF Protein Sequence Alignment and Phylogenetic Analysis
The full-length AUF1 protein sequence was used as a query to search the National Center for Biotechnology Information nonredundant protein database by the BLASTP algorithm (Altschul et al., 1990). Sequences with scores above 200 were chosen as potential AUF1 orthologs. Amino acid sequences were compared by ClustalW alignment followed by manual adjustment and displayed by MACBOXSHADE version 2.11 (U.K. Institute of Health). FBX domains were predicted by HMMER using the PFAM database (Finn et al., 2010; Hua et al., 2011). Phylogenetic trees were generated in MEGA3.1 (Kumar et al., 2004) by neighbor-joining analysis using the Poisson distance method, pairwise deletion of gaps, and the default assumptions that the substitution patterns among lineages and substitution rates among sites were homogeneous.
Identification of auf1 and auf2 Mutant Alleles and Complementation Analyses
The auf1-1, auf1-2, and auf1-3 T-DNA insertion lines were identified in the SIGNAL T-DNA insertion collection (Alonso et al., 2003) available from the Arabidopsis Biological Resource Center (Ohio State University). The auf2-1 plants were obtained from the GABI-KAT T-DNA insertion collection (http://www.gabi-kat.de). The catalog numbers of the mutants are as follows: auf1-1 (SALK_069429), auf1-2 (SALK_026385), auf1-3 (SALK_006851), and auf2-1 (GABI-KAT 134D01). The genotype of each line was determined by PCR of total genomic DNA using gene-specific primers in combination with T-DNA left border-specific primers. Primers used in this study are listed in Supplemental Table S1. Prior to generating the auf1-2 auf2-1 double mutant and the various phenotypic analyses, the auf2-1 line was backcrossed three times to the wild type. For analysis of the various reporters, homozygous Arabidopsis lines harboring the reporter genes were crossed with homozygous auf1-2 or auf1-2 auf2-1 plants; the heterozygous plants were identified by genomic PCR and selfed, and the homozygous progeny were then identified by antibiotic resistance and/or genomic PCR. Transgenic Arabidopsis lines containing the DR5pro::GUS (Ulmasov et al., 1997), CYCLINB1;1pro::GUS-DBox (Colon-Carmona et al., 1999), and PIN2pro::PIN2-GFP (Pan et al., 2009) reporter genes in the Col-0 background were as described. The cre1-12 ahk3-3 double mutant (Higuchi et al., 2004) and the arr1-5 mutant (Sakai et al., 2001) were as reported. Immunoblot detection of ARR1 in total root extracts was accomplished with anti-Arabidopsis ARR1 antibodies provided by Dr. Jan Smalle, using anti-RPT1a antibodies to confirm equal protein loading (Book et al., 2010).
Complementation was examined using an AUF1 fragment generated by genomic PCR, which contained 2,500 bp of 5′ sequence upstream of the ATG translation initiation codon and the region encompassing the full coding region. The product was cloned into the pDONR221 entry vector (Invitrogen), sequence confirmed, and then recombined into the destination vectors pEARLYGATE302 containing the Flag epitope (DYKDDDDK) or pMDC107 containing the full coding region of GFP, which appended the tags in-frame to the 3′ end of the AUF1 coding region. The constructions were transformed into homozygous auf1-2 plants by the Agrobacterium tumefaciens-mediated floral dip method. Homozygous plants for all loci were confirmed by Basta resistance and by genomic PCR of T3 plants.
RNA Gel-Blot and qRT-PCR Analyses
For RNA gel-blot analysis, total RNA from 7-d-old roots was isolated according to Smalle et al. (2002). 32P-labeled Riboprobes were synthesized with T7 or SP6 polymerases using the Riboprobe system (Promega) and the linearized pGEMT (Promega) cDNA constructs of AUF1 and 18S rRNA. Membranes were hybridized overnight at 65°C and washed as described (Smalle et al., 2002) prior to autoradiography.
RT-PCR analysis was conducted with total RNA isolated from liquid-grown plants using the Trizol reagent (Invitrogen). The RNA was treated with DNase RQI (Promega) and used for first-strand cDNA synthesis by SuperScript II reverse transcriptase (Invitrogen) in combination with an oligo(dT)18 primer (Fermentas). Equal amounts of cDNA were subjected to 35 cycles of PCR, and the products were examined by ethidium bromide staining after agarose gel electrophoresis. mRNA levels were quantified by qRT-PCR. For the IAA1 transcript, RNA was isolated from roots dissected from 7-d-old seedlings that were treated with or without 0.1 μm IAA for 5 h. For ARR5 and ARR15 transcripts, 7-d-old liquid-grown seedlings were exposed to 0.5 μm kinetin for various times before harvest of the entire seedling. Two micrograms of total RNA was isolated as above and reverse transcribed into cDNA using the SuperScript II reverse transcriptase kit (Invitrogen). qRT-PCR amplification was performed with the MyiQ5 two-color real-time PCR detection system using SYBR Premix ExTaq (Takara). Relative expression was calculated by the comparative threshold cycle method using reactions with the ACTIN2 (ACT2) or ACT4 transcript as the internal control.
Auxin Transport Assays
Acropetal or basipetal transport of auxin was measured by applying agar droplets containing [3H]IAA to the root/shoot junction zone or to the root apex of 5-d-old seedlings grown under constant light. Measurement of IAA movement was as described (Lewis and Muday, 2009). Each measurement represented the average of three independent assays, each of which was performed with 15 to 20 seedlings.
Histochemical Analysis and MUG Assay
Histochemical staining for GUS activity was conducted as published (Malamy and Benfey, 1997) using the substrate 5-bromo-4-chloro-3-indolyl β-d-GlcA. For analysis of CYCLINB1;1pro::GUS-DBox expression, roots from 7-d-old seedlings were stained overnight prior to light microscopy. For quantitative MUG assays of DR5pro::GUS expression after IAA treatment, 40 seedlings were grown for 7 d on solid MS medium containing each concentration of IAA. Crude extracts were incubated with a reaction mix containing 0.3 mm MUG, 50 mm Na2HPO4 (pH 7.0), 10 mm 2-mercaptoethanol, 10 mm Na2EDTA, and 1 mm phenylmethylsulfonyl fluoride for 10 min. MUG product fluorescence was determined with a Wallac microtiter plate fluorometer. Values were normalized against total protein concentrations, which were determined by absorption at 280 nm with a Nanodrop spectrophotometer (Thermo Scientific). For quantitative MUG assays of DR5pro:GUS expression in the roots treated with NPA, the seedlings were grown for 4 d on solid MS medium with or without NPA. The 2- to 3-mm apical portion from 50 roots for each genotype was collected, homogenized, and then assayed in triplicate as above.
Laser Scanning Confocal Microscopy
GFP fluorescence was imaged with a Zeiss 510-Meta scanning laser confocal microscope using 488-nm light excitation and 500- to 530-nm light emission. For imaging of propidium iodide-stained roots, the 543-nm line of the helium/neon laser was used for excitation, and emission was detected at 590 to 620 nm. Laser, pinhole, and gain settings of the confocal microscope were kept identical among treatments for direct comparison. For analysis of PIN2 membrane cycling, 4-d-old liquid-grown PIN2pro::PIN2-GFP seedlings were pretreated with 50 μm cycloheximide for 30 min and then exposed for additional times to 10 μm BFA as described (Pan et al., 2009). To measure root meristem size, roots were stained with 10 μg mL−1 propidium iodide for 10 s prior to microscopy. Cells in the cortical cell file from the QC to the first noticeably more elongated cell were counted manually (Dello Ioio et al., 2007). Images were assembled using Photoshop version 4.0 (Adobe Systems).
Kinematic and Cell Length Measurements
For kinematic analyses of root growth, a thin layer of half-strength MS medium plus agar, supplemented with or without 0.5 μm kinetin, was poured over a glass cover slide and cooled. Seeds were sown on top of the agar, covered with a glass coverslip, and then allowed to germinate and grow vertically between the agar-glass interface. After 6.5 d, the sandwich containing the seedlings was mounted in a small growth chamber, placed in a vertical position, and then allowed to equilibrate for 1 h prior to data collection (Miller et al., 2007). Images of the apical 2 mm of root were acquired using a horizontal Nikon light microscope, a 10× objective, and an AVT Pike camera every 30 s for 20 min at a resolution of 1.77 μm per pixel. The resulting Nomarski image stacks were analyzed by customized software using an optical flow technique (Lucas and Kanade, 1981; Beemster and Baskin, 1998) to obtain tissue growth velocity along the axis.
To measure the REGR profiles, we selected approximately 30 points along the midline of the organ on the first frame of the image stack, and these points were tracked through the time series. Circular image patches surrounding the selected points were deformed and translated from the ith frame to best match corresponding image patches in the (i+1)th frame. The rate at which these image patches traversed the axis of the organ generated the tissue velocity. A flexible logistic function was fit to the velocity data, which resulted in a smooth representation of the velocity profile (Morris and Silk, 1992). The velocity profile was differentiated with respect to arc length to obtain the REGR profile. A detailed description of the technique and software will be published elsewhere (N.D. Miller and E.P. Spalding, unpublished data).
The average cell length in the cortical cell file was subsequently determined from the same roots used in the kinematic analysis. The roots were stained with propidium iodide and imaged with a confocal microscope (Zeiss LSM500) at 20× magnification. A series of overlapping root images were tiled to generate complete root cell pictures. The lengths of cells for a continuous single cortical cell file were measured along both sides of the root using the ImageJ software package (http://rsb.info.nih.gov/ij/). The position of each cell was calculated from the cumulative length of all cells between it and the root QC. Subsequently, a logistic function was fit to the data.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AAK96759 (AtAUF1), AAT06470 (AtAUF2), AAT69637 (OsAUF1), EDQ65221 (PpAUF1), EEE94383 (PtAUF1), EEF48483 (RcAUF1), EFJ08296 (SmAUF1), XP_00284195 (VvAUF1), and ACF78905 (ZmAUF1).
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure S1. Phylogenetic comparison of AUF proteins from land plants.
Supplemental Figure S2. Effect of AUF mutations on auxin response and distribution.
Supplemental Figure S3. Complementation of the auf1-2 growth defect in response to NPA and kinetin.
Supplemental Figure S4. AUF1 inactivation does not disturb PIN2 protein trafficking.
Supplemental Figure S5. Effect of the auf1-2 mutation on the expression of genes involved in auxin transport using qRT-PCR
Supplemental Figure S6. Inhibition of ethylene synthesis can overcome the hypersensitivity of auf1-2 plants to kinetin.
Supplemental Table S1. Oligonucleotide primers used during the genetic and expression analysis of AUF1 and AUF2.
Acknowledgments
We thank Joseph Walker, Dr. Zhihua Hua, Sarah Swanson, and Dr. Guosheng Wu for technical help and Dr. Jan Smalle for providing the anti-ARR1 antibodies. We also appreciate the helpful advice from Dr. Patrick Masson.
References
- Abel S, Oeller PW, Theologis A. (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA 91: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang Y-H, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Baskin TI. (1998) Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Baskin TI. (2000) STUNTED PLANT 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol 124: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Benkov E, Helariutta Y. (2011) Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr Opin Plant Biol 14: 10–16 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Book AJ, Gladman NP, Lee SS, Scalf M, Smith LM, Vierstra RD. (2010) Affinity purification of the Arabidopsis 26S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem 285: 25554–25569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. (2008) Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, et al. (2006) Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis p-glycoproteins. J Biol Chem 281: 30603–30612 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. (2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA 95: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner T. (1999) Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruere J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. (2005b) Plant development is regulated by a family of auxin receptor F-box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benkov E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, et al. (2002) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Science 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Fujita H, Syono K. (1997) PIS1, a negative regulator of the action of auxin transport inhibitors in Arabidopsis thaliana. Plant J 12: 583–595 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu S-H, Durski AM, Vierstra RD. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Heyl A, Schmuling T. (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6: 480–488 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Zou C, Shiu S-H, Vierstra RD. (2011) Phylogenetic comparison of F-box (FBX) superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 6: e16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Mizuno T. (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Kuderova A, Urbankova I, Valkova M, Malbeck J, Brzobohaty B, Nemethova D, Hejatko J. (2008) Effects of conditional IPT-dependent cytokinin overproduction on root architecture of Arabidopsis seedlings. Plant Cell Physiol 49: 570–582 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Miller ND, Splitt BL, Wu GS, Spalding EP. (2007) Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell 19: 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Muday GK. (2009) Measurement of auxin transport in Arabidopsis thaliana. Nat Protoc 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Wu GS, Ljung K, Spalding EP. (2009) Auxin transport into cotyledons and cotyledon growth depend similarly on the ABCB19 multidrug resistance-like transporter. Plant J 60: 91–101 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2006) Dynamic integration of auxin transport and signalling. Curr Biol 16: R424–R433 [DOI] [PubMed] [Google Scholar]
- Lomax TL, Muday GK, Rubery PH. (1995) Auxin transport. Davies PJ, , Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Kluwer Academic Press, Dordrecht, The Netherlands, pp 509–530 [Google Scholar]
- Lucas BD, Kanade T. (1981) An iterative image registration technique with an application to stereo vision. Hayes PJ, , Proceedings of the International Joint Conference on Artificial Intelligence. William Kaufmann Inc. Publishers, Los Altos, CA: pp 674–679 [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM. (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medford JI, Horgan R, El-Sawi Z, Klee HJ. (1989) Alterations of endogenous cytokinins in transgenic plants using a chimeric isopentenyl transferase gene. Plant Cell 1: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CO, Skoog F, Okumura FS, Von Saltza MH, Strong FM. (1956) Isolation, structure and synthesis of kinetin, a substance promoting cell division. J Am Chem Soc 78: 1375–1380 [Google Scholar]
- Miller ND, Parks BM, Spalding EP. (2007) Computer-vision analysis of seedling responses to light and gravity. Plant J 52: 374–381 [DOI] [PubMed] [Google Scholar]
- Morris AK, Silk WK. (1992) Use of a flexible logistic function to describe axial growth of plants. Bull Math Biol 54: 1069–1081 [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Muller B, Sheen J. (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. (2007) Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J 55: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Fujioka S, Peng J, Chen J, Li G, Chen R. (2009) The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell 21: 568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Friml J. (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmulling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Simeskova M, Duclercq J, Petrasek J, Zamimalova E, Simon S, Friml J, Van Montagu MCE, Benkova E. (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Strader LC, Bailly A, Yang H, Blakeslee J, Langowski L, Nejedla E, Fujita H, Itoh H, Syono K, et al. (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Salome PA, To JPC, Kieber JJ, McClung CR. (2006) Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18: 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Yanai O, Ori N. (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD. (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- To JPC, Deruere J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. (2007) Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19: 3901–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, Kieber JJ. (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Smalle J, Le J, Saibo NJ, De Paepe A, Chaerle L, Tietz O, Smets R, Laarhoven LJ, Harren FJ, et al. (2003) The Arabidopsis mutant alh1 illustrates a cross talk between ethylene and auxin. Plant Physiol 131: 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J. (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Anal Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GS, Lewis DR, Spalding EP. (2007) Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Yamashino T, Amano Y-I, Tajima Y, Imamura A, Sakakibara H, Mizuno T. (2007) Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol 48: 84–96 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]