Abstract
Receptor-like kinase-mediated cell signaling pathways play fundamental roles in many aspects of plant growth and development. A pair of Arabidopsis (Arabidopsis thaliana) leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA (HAE) and HAESA-LIKE2 (HSL2), have been shown to activate the cell separation process that leads to organ abscission. Another pair of LRR-RLKs, EVERSHED (EVR) and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1, act as inhibitors of abscission, potentially by modulating HAE/HSL2 activity. Cycling of these RLKs to and from the cell surface may be regulated by NEVERSHED (NEV), a membrane trafficking regulator that is essential for organ abscission. We report here the characterization of CAST AWAY (CST), a receptor-like cytoplasmic kinase that acts as a spatial inhibitor of cell separation. Disruption of CST suppresses the abscission defects of nev mutant flowers and restores the discrete identity of the trans-Golgi network in nev abscission zones. After organ shedding, enlarged abscission zones with obscured boundaries are found in nev cst flowers. We show that CST is a dual-specificity kinase in vitro and that myristoylation at its amino terminus promotes association with the plasma membrane. Using the bimolecular fluorescence complementation assay, we have detected interactions of CST with HAE and EVR at the plasma membrane of Arabidopsis protoplasts and hypothesize that CST negatively regulates cell separation signaling directly and indirectly. A model integrating the potential roles of receptor-like kinase signaling and membrane trafficking during organ separation is presented.
Signaling by transmembrane receptor-like kinases (RLKs) underlies diverse aspects of plant growth and development. Surprisingly, a substantial number of plant RLKs do not contain either extracellular or transmembrane domains. Although receptor-like cytoplasmic kinases (RLCKs) account for at least 125 of the 610 annotated RLKs in Arabidopsis (Arabidopsis thaliana), much remains to be learned about their functions within cell signaling complexes (Shiu and Bleecker, 2001; Goring and Walker, 2004; Shiu et al., 2004; Jurca et al., 2008). Several of the 46 RLCKs assigned to the class VII subfamily have been found to function in pathogen response and developmental signaling pathways (Swiderski and Innes, 2001; Shao et al., 2003; Murase et al., 2004; Muto et al., 2004; Veronese et al., 2006; Ade et al., 2007; Lu et al., 2010; Zhang et al., 2010; Liu et al., 2011).
Functional studies of class VII RLCKs have identified four different modes of action; RLCKs can act as coreceptors of RLKs, in signal relays, as repressors, and as activators of signaling. The M-Locus Protein Kinase (MLPK) RLCK functions as a coreceptor of a ligand-binding RLK to transduce signaling. During the self-incompatibility response in Brassica rapa flowers, MLPK has been found to interact with the ligand-activated S-locus receptor kinase and is essential for the cell signaling leading to rejection of self pollen (Murase et al., 2004; Kakita et al., 2007). The BOTRYTIS-INDUCED KINASE1 (BIK1) RLCK functions in a signaling relay with an activated ligand-binding RLK and its coreceptor. BIK1 was shown to interact with two Leu-rich repeat receptor-like kinases (LRR-RLKs), the ligand-binding FLAGELLIN-SENSITIVE2 (FLS2) and its coreceptor BRI1-ASSOCIATED KINASE1 (BAK1; Veronese et al., 2006; Lu et al., 2010). FLS2 binding of the bacterial flagellin-derived peptide, flg22, triggers the interaction of FLS2 and BAK1 and downstream signaling for pathogen-associated molecular patterns (PAMP)-triggered immunity (Chinchilla et al., 2007; Heese et al., 2007). BIK1, which independently associates with FLS2 and BAK1 in the absence of ligand, is rapidly phosphorylated by BAK1 upon flg22 treatment (Lu et al., 2010). According to the model of Lu et al. (2010), phosphorylated BIK1 subsequently transphosphorylates FLS2 and BAKI, leading to an activated FLS2-BAK1-BIK1 complex and the promotion of pathogen immune responses downstream of FLS2.
The AvrPphB SUSCEPTIBLE1 (PBS1) RLCK acts as a repressor. PBS1 is cleaved by an effector of the pathogen Pseudomonas syringae, activating the nucleotide-binding site (NB)-LRR protein, RPS5, which triggers programmed cell death (Warren et al., 1999; Swiderski and Innes, 2001; Shao et al., 2003; Ade et al., 2007). A recent study has revealed that, like PBS1, BIK1 and several PBS1-like (PBL) RLCKs are also substrates of the bacterial AvrPphB protease effector (Zhang et al., 2010). This discovery and other work suggests that a bacterial effector can suppress PAMP-triggered immunity in plants by cleaving RLCKs known (BIK1) or proposed (PBL1, PBL2) to positively interact with RLKs that bind PAMPs, including FLS2, CHITIN ELICITOR RECEPTOR KINASE1, and the EF-Tu RECEPTOR (Zipfel et al., 2006; Miya et al., 2007; Zhang et al., 2010). During effector-triggered immunity, another RLCK, RPM1-INDUCED PROTEIN KINASE (RIPK), is involved in activating the NB-LRR protein RPM1 (Liu et al., 2011). RIPK interacts with the P. syringae effector AvrB and either directly phosphorylates the immune regulator RPM1-INTERACTING PROTEIN4 (RIN4) or promotes AvrB-mediated RIN4 phosphorylation; phosphorylated RIN4 then activates RPM1 (Chung et al., 2011; Liu et al., 2011).
In Arabidopsis, organ abscission is controlled by the competing activities of several LRR-RLKs. The HAESA (HAE) and HAESA-LIKE2 (HSL2) LRR-RLKs redundantly activate a mitogen-activated protein kinase signaling cascade that leads to cell separation and release of the outer floral organs (Jinn et al., 2000; Cho et al., 2008). The predicted signaling ligand for HAE/HSL2 is INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), a small, secreted peptide (Butenko et al., 2003; Cho et al., 2008; Stenvik et al., 2008). Two inhibitors of organ separation that may directly regulate HAE/HSL2 are the EVERSHED (EVR) and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) LRR-RLKs (Leslie et al., 2010; Lewis et al., 2010). Mutations in EVR or SERK1 were found to restore abscission in plants defective for NEVERSHED (NEV), a gene that encodes an ADP-ribosylation factor GTPase-activating protein (ARF-GAP). NEV has been proposed to regulate the movement of proteins essential for activating cell separation (Liljegren et al., 2009).
Here, we show that CAST AWAY (CST), a membrane-associated class VII RLCK, acts as a spatial inhibitor of abscission zone (AZ) signaling. We have found that CST interacts at the plasma membrane with EVR and HAE, LRR-RLKs that inhibit and promote organ separation, respectively. Our studies of CST suggest a distinct mode of RLCK action in which an RLCK and RLK partner may act in a step-wise fashion to inhibit the activity or alter the location of a ligand-binding RLK.
RESULTS
Organ Separation Is Restored in nev cst Flowers
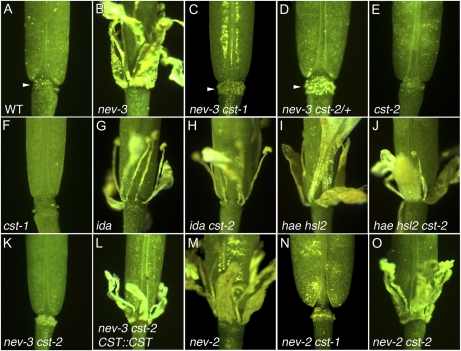
To identify novel regulators of organ abscission, a genetic screen was conducted for mutations that restored abscission in nev-3 mutant flowers (Lewis et al., 2010). A recessive mutation identified in this screen, cst-1, was found to rescue organ separation in nev flowers (Fig. 1, A–C). A second mutant allele of CST from the SAIL T-DNA collection (cst-2 [SAIL_296_A06]; Sessions et al., 2002) dominantly rescues organ abscission in nev-3 flowers (Fig. 1D). Flowers with mutations in CST alone have a wild-type appearance, and organ shedding occurs normally (Fig. 1, E and F).
Figure 1.
Loss of CST rescues floral organ shedding in nev plants. A to D, Sepals, petals, and stamens are shed from wild-type (WT) flowers by floral stage 17 (A) and remain attached in nev-3 flowers (B). Organ separation is recessively restored in nev-3 cst-1 plants (C), while the cst-2 allele acts dominantly to restore floral organ shedding in the nev-3 background (D). The AZ regions of nev cst flowers (C and D; arrowheads) are enlarged and visibly rougher than those of wild-type flowers (A; arrowhead). E and F, In the cst-2 (E) and cst-1 (F) single mutants, the organ AZs appear like those of wild-type flowers and shedding occurs normally. G to J, The abscission defects of the ida (G) and hae hsl2 (I) mutants are not rescued by the cst-2 allele (H and J). K and L, The nev-3 cst-2 mutant phenotype (K) can be complemented by a CST::CST transgene (L). Presence of the CST transgene blocks organ abscission, restoring the nev mutant phenotype. M to O, The abscission defects of nev-2 flowers (M) are rescued by cst-1 (N) but not by cst-2 (O) mutant alleles. [See online article for color version of this figure.]
Since NEV, IDA, HAE, and HSL2 each regulates the cell separation stage of organ separation (Butenko et al., 2003; Cho et al., 2008; Stenvik et al., 2008; Liljegren et al., 2009), we tested whether disruption of CST activity might also suppress the ida and hae hsl2 mutant phenotypes. We found that mutations in CST do not rescue the shedding defects of ida or hae hsl2 flowers (Fig. 1, G–J). These results suggest that CST acts upstream of IDA and HAE/HSL2 or in a parallel pathway that converges at HAE/HSL2 activity or farther downstream.
The Organ AZs of nev cst Flowers Are Enlarged and Disorganized
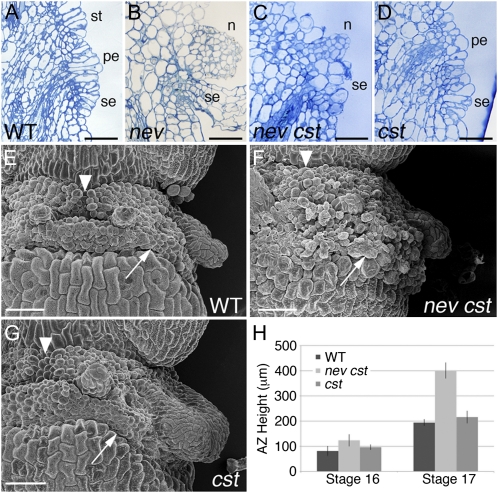
Although organ separation occurs in nev-3 cst-1, nev-3 cst-2/+, and nev-3 cst-2 flowers, the AZ regions have a visibly rough appearance compared with the smooth surfaces of the organ detachment sites in wild-type flowers (Fig. 1, A, C, and D). To further characterize this phenotype, we examined longitudinal sections and scanning electron micrographs of nev cst flowers at the time of shedding compared with wild-type and cst single mutant flowers (Fig. 2, A and C–G). While the remaining AZ cells of wild-type flowers expand to form discrete scars (Fig. 2, A and E), cells in the AZ regions of nev cst flowers have a disordered appearance and show increased, uneven cell expansion (Fig. 2, C and F). After organ shedding, nev cst AZ regions were found to be significantly enlarged compared with the wild type (Fig. 2H), and the boundaries between individual organ detachment sites and with the floral stem were notably obscured (Fig. 2, E–G). These results suggest that CST acts as a spatial inhibitor of signaling that modulates AZ cell adhesion and expansion.
Figure 2.
nev cst AZs are disorganized and enlarged. A to D, Longitudinal sections of flowers (stage 17) stained with toluidine blue. The remaining AZ cells of wild-type (WT; A) and cst-2 (D) flowers show coordinated cell expansion, while the floral organs remain attached in nev flowers (B). Although organ abscission is rescued in nev cst-2 flowers (C), the AZ cells have a disordered appearance. The petal (pe), sepal (se), and stamen (st) AZs and nectaries (n) are indicated. Bars = 50 μm. E to G, Scanning electron micrographs of flowers after organ separation (stage 17). Distinct AZs are apparent in wild-type (E) and cst (G) flowers, whereas the AZ regions of nev cst flowers have formed an enlarged, disorganized band of cells at the fruit base (F). In nev cst flowers, the junction between the medial stamen AZs is no longer visible (arrowheads), and the border between the sepal AZ and the floral stem is not clearly defined (arrows). Bars = 500 μm. H, Quantification of AZ size in wild-type and mutant flowers. The distance between the lower border of the sepal AZ and the upper border of the stamen AZ was measured in stage 16 and the first stage 17 flowers (n ≥ 4 per genotype). nev cst flowers contain significantly enlarged AZs after organ shedding compared with wild-type and cst flowers. [See online article for color version of this figure.]
Disruption of CST Activity Suppresses the Subcellular Defects of nev flowers
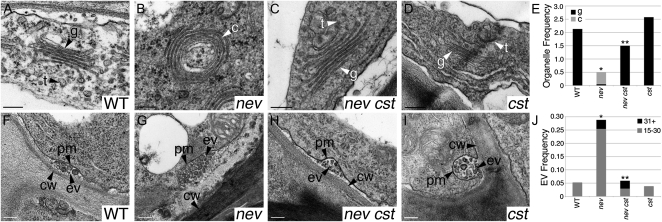
Our studies have previously shown that mutations in NEV are associated with a unique set of trafficking defects in flowers undergoing organ separation (Liljegren et al., 2009). To determine whether disruption of CST activity suppresses these subcellular changes as well as restores organ separation in nev cst mutant flowers, we carried out transmission electron microscopy of wild-type and mutant flowers shortly after organ abscission (stage 17; Fig. 2, A–D). Whereas the structure and organization of the Golgi cisternae and trans-Golgi network (TGN) are altered in nev-3 mutant cells (Fig. 3, B and E) compared with the wild type (Fig. 3, A and E), we found that these organelles are unaffected in nev-3 cst-2 (Fig. 3, C and E) and cst-2 (Fig. 3, D and E) cells. We also discovered that the hyperaccumulation of extracellular vesicles in nev-3 cells (Fig. 3, G and J) compared with wild-type (Fig. 3, F and J) and cst-2 (Fig. 3, I and J) cells is significantly reduced in nev-3 cst-2 cells (Fig. 3, H and J). These results suggest that specific mutations in CST are sufficient to alleviate the disruption of vesicular traffic in nev cells that blocks organ separation.
Figure 3.
Mutations in CST rescue the subcellular defects of nev mutant flowers. Transmission electron micrographs show cells in the sepal AZs of wild-type (WT) and mutant flowers (stage 17). A to D, The linear stacks of Golgi cisternae (g) and associated TGN (t) seen in wild-type (A) and cst-2 (D) flowers are replaced by circularized multilamellar structures (c) in nev-3 flowers (B). In nev-3 cst-2 flowers (C), the discrete structures of the Golgi and TGN are restored. E, Frequency of Golgi cisternae (black bars) and circularized structures (gray bars) per cell in sections of wild-type and mutant sepal AZs. Statistical differences between the type of multilamellar structure observed in nev and the wild type, and in nev cst and nev tissues are indicated by single and double asterisks, respectively (Fisher’s exact test; P = 0). A statistical difference was not detected between cst and wild-type tissues. n ≥ 26 cells per genotype. F to I, Extracellular vesicles are frequently observed in the apoplastic space of nev-3 AZ cells (G). In wild-type (F), nev-3 cst-2 (H), and cst-2 (I) AZs, the appearance of extracellular vesicles is significantly reduced. J, Frequency of extracellular vesicles (clusters of 15–30, gray bars; clusters of 31+, black bars) in wild-type and mutant sepal AZ cells. Statistical differences between nev and the wild type and between nev cst and nev tissues are indicated by single (Fisher’s exact test; P < 0.012) and double (P < 0.02) asterisks, respectively. A statistical difference was not detected between cst and wild-type tissues. n ≥ 26 cells per genotype. cw, Cell wall; ev/EV, extracellular vesicles; pm, plasma membrane; t, TGN. Bars = 200 nm.
CST Encodes a RLCK with Dual Specificity
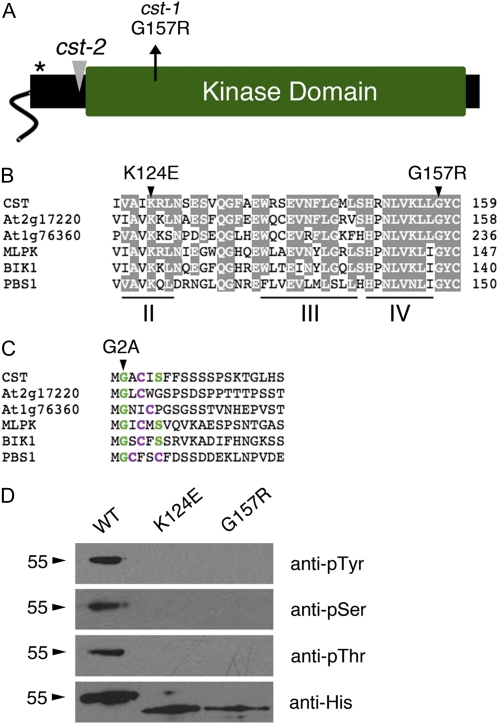
The cst-1 mutation was mapped to chromosome 4 and found to affect At4g35600, a gene encoding a predicted RLCK of the class VII subfamily (Fig. 4A). Although an early study suggested that the At4g35600 gene product shared sequence homology with animal connexins (Meiners et al., 1991), subsequent analysis has shown that the first open reading frame predicted was incorrect (Mushegian and Koonin, 1993; Arabidopsis Genome Initiative, 2000; Yamada et al., 2003). No evidence of connexin homology in the accepted sequence of the At4g35600 gene product has been found.
Figure 4.
CST encodes a dual-specificity RLCK. A, CST encodes a RLCK with a predicted myristoylation site at its N terminus (asterisk). The cst-1 point mutation affects a conserved residue within the kinase domain, and the cst-2 allele contains a T-DNA insertion upstream of the kinase domain. B, Sequence alignment of kinase subdomains II to IV from CST and other class VII RLCKs from Arabidopsis and B. rapa. The cst-1 mutation (G157R) alters an amino acid that is invariant among class VII RLCKs. Conserved amino acids between CST and other proteins are shaded. C, Sequence alignment of the N terminus of CST and other related RLCKs. Residues predicted to undergo myristoylation are highlighted in green, and expected palmitoylation sites are highlighted in purple. D, Recombinant CST autophosphorylates on Ser, Thr, and Tyr residues. Mutations in subdomain II (K124E) and next to subdomain IV (G157R) interfere with the kinase activity of CST. Antiserum that recognizes the 6xHis tag labels approximately 55- and 49-kD phosphorylated and unphosphorylated CST fusion proteins, respectively. WT, Wild type. [See online article for color version of this figure.]
The cst-1 mutation introduces an amino acid substitution immediately after subdomain IV of the kinase domain, which is involved in binding ATP (Fig. 4B; Hanks, 2003). Although residues in this region are not highly conserved among eukaryotic protein kinases (Hanks and Hunter, 1995), the affected Gly is invariant in the kinase domains of all 46 predicted class VII RLCKs in Arabidopsis (Fig. 4B; data not shown). The cst-2 mutant allele contains a T-DNA insertion upstream of the kinase domain and is predicted to cause production of a truncated protein (Fig. 4A).
To test CST kinase activity, full-length proteins of wild type (WT), a traditional kinase-dead mutant (K124E; Horn and Walker, 1994), and the cst-1 mutant (G157R) were expressed as N-terminal 6xHis-tagged fusion proteins in Escherichia coli. Whereas a His antibody recognizes purified, presumably phosphorylated CSTWT protein migrating as a single band of approximately 55 kD, it recognizes purified CSTK124E and CSTG157R proteins migrating as single bands of approximately 49 kD, in agreement with the predicted size of the tagged protein (49 kD; Fig. 4D). Phospho-Ser, phospho-Thr, and phospho-Tyr antisera were used to detect phosphorylated residues on the recombinant proteins. Each of these antisera recognized the CSTWT protein and neither of the mutant proteins (Fig. 4D). These results suggest that CST is a dual-specificity kinase that autophosphorylates Ser, Thr, and Tyr residues in vitro and that its kinase activity is abolished by the cst-1 mutation.
Analysis of Allele-Specific Interactions between NEV and CST
We have observed allele-specific differences in the number of copies of cst-1 and cst-2 required to restore organ shedding in nev-3 flowers. While a single cst-2 allele is sufficient to dominantly rescue abscission in nev-3 flowers (Fig. 1, B and D), both copies of the cst-1 mutant allele must be present to restore organ shedding in this background (Fig. 1, B and C).
To determine whether we could uncover additional allele-specific interactions between CST and NEV, the cst-1 and cst-2 alleles were crossed to the nev-2 and nev-6 mutant alleles. While we have not detected significant phenotypic differences between the nev-2, nev-3, and nev-6 mutants in a Landsberg erecta (Ler) background, the molecular nature of the respective mutations is quite different. The nev-3 mutation introduces an amino acid substitution in the ARF-GAP domain at an invariant Arg (R59K), which is known to be essential for ARF-GAP enzymatic activity (Luo et al., 2007; Liljegren et al., 2009). The nev-2 mutation introduces a stop codon downstream of the ARF-GAP domain (Q198*) and is predicted to cause the production of a truncated protein with an ARF-GAP domain. The nev-6 allele contains a T-DNA insertion in the first intron and is expected to produce a truncated protein without an ARF-GAP domain.
While the cst-1 mutation recessively rescues organ shedding in nev-2 and nev-6 flowers (Fig. 1, M and N; data not shown) and cst-2 dominantly rescues shedding in nev-6 flowers (data not shown), the cst-2 allele was unable to restore abscission in nev-2 flowers even if both mutant copies of CST were present (Fig. 1O). These results are partially consistent with an allele-specific compensatory mutation, in which the suppressor mutation restores a physical interaction between the affected components (Michels, 2002). However, if the truncated cst-2 mutant protein were to interact with the nev-3 mutant protein in such a way that its ARF-GAP activity was restored, one would expect that the cst-2 mutant allele should also not be able to rescue abscission in a nev mutant protein missing the ARF-GAP domain.
To address whether the dominant rescue of organ shedding in nev flowers by the cst-2 allele is due to either a dominant-negative interaction or haploinsufficiency, a wild-type copy of the CST cDNA driven by its predicted 1.4-kb promoter was introduced into nev-3 cst-2 homozygous mutant plants. We predicted that one copy of CST would not suppress organ shedding in either a dominant-negative or haploinsufficient situation, whereas a block of organ shedding by two copies of CST would be consistent with haploinsufficiency but not a dominant-negative interaction. We observed that for two independent T1 lines with T2 kanamycin-resistance segregation ratios characteristic of a single insertional locus, the presence of the CST::CST transgene was sufficient to block organ abscission in nev cst flowers, restoring the nev mutant phenotype in plants hemizygous and homozygous for the transgene (Fig. 1, K and L; Supplemental Table S1). Since multiple T-DNA insertions can be present at a single locus (Jorgensen et al., 1987), these results do not rule out a dominant-negative interaction involving truncated CST protein produced from the cst-2 allele. Multiple copies of wild-type CST could efficiently dilute the dominant-negative effect of a single locus producing truncated CST protein.
Localization of CST to the Plasma Membrane Is Supported by N-Terminal Myristoylation
CST is predicted to associate with membranes in part via myristoylation of its N terminus (Fig. 4C), as previously shown for the PBS1, MLPK, and BIK1 class VII RLCKs (Warren et al., 1999; Boisson et al., 2003; Murase et al., 2004; Veronese et al., 2006; Kakita et al., 2007). Palmitoylation of N-terminal Cys residues is also predicted to allow reversible membrane association for CST and many other class VII RLCKs (Fig. 4C; Sorek et al., 2009; Zhang et al., 2010). To visualize CST protein within Arabidopsis cells, we generated a CST-GFP fusion construct driven by the constitutive viral 35S promoter that could be transfected into mesophyll protoplasts. Attempts to visualize CST-YFP (for yellow fluorescent protein) under the control of its native promoter in vivo were unsuccessful, likely due to the limited expression of CST in roots and leaves (Fig. 6, F, G, and J; data not shown). CST-GFP transformed protoplasts exhibit fluorescent localization of the protein to the plasma membrane and some internal, punctate structures (Fig. 5, A and A′).
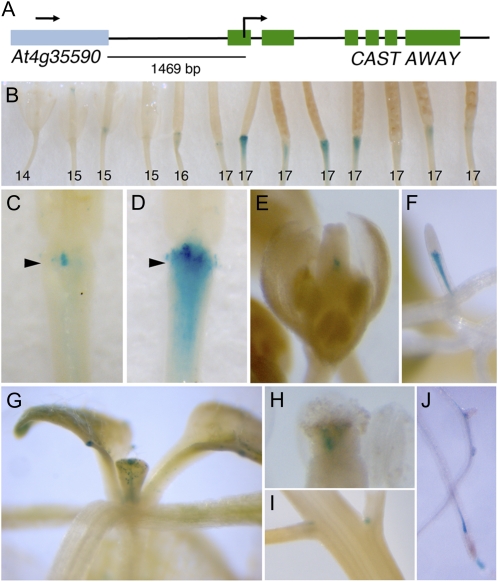
Figure 6.
CST is expressed in floral organ AZs and other specific tissues. A, A translational fusion of the CST regulatory region to the GUS reporter was created. B, Expression of GUS is first detected in stage 15 flowers prior to organ shedding (stage 16) and diminishes by mid stage 17. Flowers at consecutive positions along the inflorescence are shown, with the floral stages indicated below. C and D, During and after organ abscission, a dynamic pattern of GUS expression is seen in epidermal AZ cells. The early patches of GUS expression in subepidermal cells of the floral pedicel (C) expand into a broad, diffuse domain within the gynophore and pedicel of older flowers (D). Arrowheads indicate the positions of the sepal AZs. E to J, GUS expression was also detected in seedling roots (F and J) and leaves (G), the axils of floral stems (I), and developing (E) and mature (H) gynoecia.
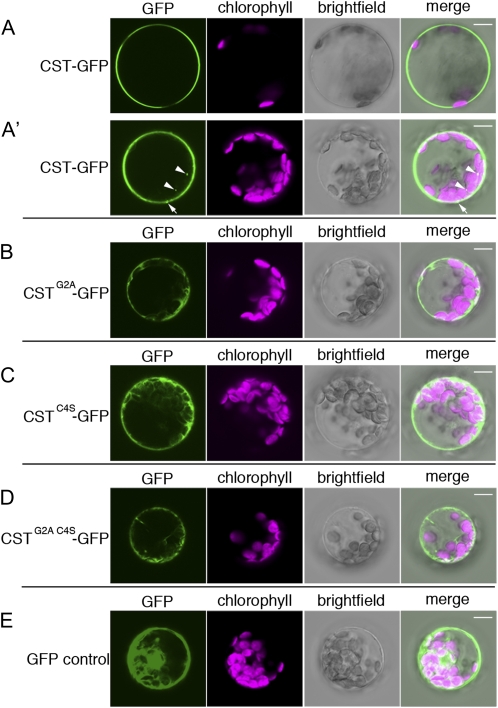
Figure 5.
CST localizes to the plasma membrane. Transfected Arabidopsis leaf protoplasts were imaged using confocal microscopy. GFP fluorescence (green), chlorophyll autofluorescence (magenta), bright-field, and merged images are shown for each protoplast. A and A′, CST (CST-GFP) localizes to the plasma membrane (A) and in internal speckles (A′; arrowheads). Spots of increased CST aggregation associated with the plasma membrane are observed (A′; arrows). Images of the same protoplast at different focal planes are shown. B to D, Mutation of either the predicted myristoylation site (CSTG2A-GFP) or the predicted palmitoylation site (CSTC4S-GFP) results in localization of the mutant proteins throughout the cytoplasm (B and C). Localization of the mutant proteins is still observed at or near the plasma membrane. Mutation of both sites (CSTG2A C4S-GFP) does not appear to result in additional disruption of plasma membrane localization. Protoplast focal planes are equivalent to that shown in A′. E, GFP is localized in a broad pattern throughout the cytoplasm and near the plasma membrane. Bars = 10 μm.
Mutation of the predicted myristoylation site (G2A; Fig. 4C) causes a partial redistribution of CSTG2A-GFP to the cytoplasm (Fig. 5B; Supplemental Fig. S1A), as independently observed in tobacco (Nicotiana tabacum) epidermal cells (Stael et al., 2011). Mutation of the predicted palmitoylation site (C4S; Fig. 4C) also results in partial cytoplasmic localization of CSTC4S-GFP (Fig. 5C; Supplemental Fig. S1B). Mutations in both sites do not appear to cause further redistribution of CSTG2A C4S-GFP to the cytoplasm (Fig. 5D; Supplemental Fig. S1C). Protoplasts transfected with a GFP tag alone (Fig. 5E; Supplemental Fig. S1D) show a more complete cytoplasmic localization pattern than any of the mutant CST proteins tested. These results suggest that myristoylation and palmitoylation of CST support but are not solely required for its localization at the plasma membrane.
The EVR, SERK1, and HAE LRR-RLKs, which contain transmembrane domains, have been previously shown to be associated with the plasma membrane or closely associated membrane structures (Jinn et al., 2000; Shah et al., 2001; Alexandersson et al., 2004; Leslie et al., 2010). To compare the localization profile of CST with HAE and EVR in protoplasts, we also generated 35S::HAE-GFP and 35S::EVR-GFP transfection constructs. HAE-GFP and EVR-GFP were observed at the plasma membrane and in structures with the appearance of the endoplasmic reticulum (ER) network (Supplemental Fig. S2, A, A′, C, and C′). SERK1-YFP was previously reported to localize to endosomal compartments and the ER network as well as the plasma membrane in Arabidopsis leaf protoplasts; this distribution was found to vary with respect to time after transfection (Aker et al., 2006). The distinct localization profile of CST compared with those of EVR, SERK1, and HAE may indicate that the mechanism of CST inhibition of abscission differs from that of EVR and SERK1.
CST Is Expressed in Organ AZs, Lateral Roots, and Developing Guard Cells
To determine the expression pattern directed by CST regulatory regions, a construct with a translational fusion of the predicted 1.4-kb promoter region to the GUS reporter was generated (Fig. 6A). Of 35 CST::CST-GUS transgenic lines analyzed, five showed GUS expression in floral organ AZs. Expression of GUS first appears prior to organ shedding (stage 15) in subepidermal cells within the floral pedicel underlying the organ attachment sites (Fig. 6, B and C). During and after organ shedding (stages 16 and 17), individual epidermal cells in the AZ regions show GUS expression in a dynamic fashion, and increased GUS expression is observed in subepidermal cells of the gynophore (fruit stem) and pedicel (Fig. 6, B and D). Expression of GUS in the AZ regions decreases as the fruit matures (mid stage 17). GUS expression is also observed within the gynoecium, in the style of the developing fruit, and in the axils of the floral stems (Fig. 6, E, H, and I).
Expression of GUS in vegetative tissues was observed in 24 of the 35 CST::CST-GUS transgenic lines analyzed. In seedlings, GUS expression is found within the lateral roots (Fig. 6, F and J) and in the guard cells and large pores (hydathodes) at leaf edges (Fig. 6G). These results suggest that expression of CST is restricted to specific cell types and tissues and that CST may function during other phases of plant development.
CST Interacts with the HAE and EVR RLKs at the Plasma Membrane
To test for interactions between CST and other RLKs that modulate abscission, we used the bimolecular fluorescence complementation (BiFC) assay in Arabidopsis mesophyll protoplasts (Walter et al., 2004; Yoo et al., 2007). This approach has been successfully used to detect interactions between membrane-bound LRR-RLKs, such as the SERK family members (SERK1, SERK2, BAK1, and BAK1-LIKE1), and BAK1-INTERACTING RECEPTOR-LIKE KINASE1 (Russinova et al., 2004; Albrecht et al., 2005; Karlova et al., 2006; Gao et al., 2009). To facilitate this analysis, constructs to express full-length versions of CST, HAE, and EVR with C-terminal translational fusions to either the N-terminal half of YFP (YFPn) or the C-terminal half of YFP (YFPc) were generated. When these plasmids are transfected into protoplasts, the gene fusions are expressed under the control of the constitutive 35S promoter. The presence of YFP fluorescence in protoplasts visualized about 16 h after plasmid transfection indicates a likely interaction between the two target proteins, as the N- and C-terminal halves of YFP were brought into close enough proximity to reconstitute the fluorescent protein.
To allow for the detection of protoplasts successfully transfected with plasmid DNA, as well as for the calculation of transfection efficiency, we cotransfected all protoplasts with a previously described cyan fluorescent protein (CFP)-tagged mitochondrial marker (CD3-986; Nelson et al., 2007). Only those experiments with a protoplast transfection efficiency of greater than or equal to 50% were used for BiFC analysis.
Since multiple RLKs are known to form homodimers (Russinova et al., 2004; Hink et al., 2008; Zhu et al., 2010), we first tested for an interaction of CST with itself. CST was found to self-interact at the plasma membrane, as cotransfection of CST-YFPn/CST-YFPc resulted in a similar pattern of fluorescence as CST-GFP (Figs. 5A and 7A). When HAE-YFPn/HAE-YFPc and EVR-YFPn/EVR-YFPc were cotransfected, reconstituted YFP was also detected in a similar pattern to what we observed for HAE-GFP and EVR-GFP: localization to the plasma membrane and internal structures (Supplemental Fig. S2, A, A′, C, and C′). While the SERK1 and BRASSINOSTEROID INSENSITIVE1 (BRI1) LRR-RLKs were both found to homodimerize in cowpea (Vigna unguiculata) protoplasts, BAK1 did not, suggesting that not all LRR-RLKs can self-interact (Hink et al., 2008).
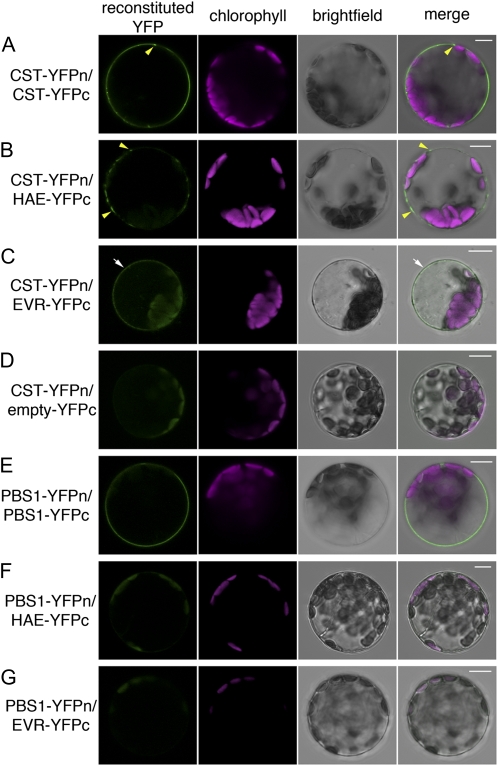
Figure 7.
CST interacts with HAE and EVR at the plasma membrane. BiFC assays of transfected Arabidopsis protoplasts are shown. Reconstituted YFP fluorescence (green), chlorophyll autofluorescence (magenta), bright-field, and merged images are shown for each protoplast. A, CST (CST-YFPn/CST-YFPc) homodimerizes at the plasma membrane. An area of increased aggregation is indicated (arrowheads). B, CST (CST-YFPn) interacts with HAE (HAE-YFPc) in discrete subdomains (arrowheads) at the plasma membrane. C, CST (CST-YFPn) interacts with EVR (EVR-YFPc) in a uniform pattern at the plasma membrane (arrows). D, CST (CST-YFPn) does not interact with YFP (YFPc). E, The PBS1 class VII RLCK (PBS1-YFPn/PBS1-YFPc) homodimerizes at the plasma membrane. F and G, PBS1 (PBS1-YFPn) does not interact with either HAE (HAE-YFPc) or EVR (EVR-YFPc). Bars = 10 μm.
We next tested the hypothesis that CST may inhibit abscission signaling by forming receptor heterocomplexes with HAE. Upon cotransfection of either CST-YFPn/HAE-YFPc or HAE-YFPn/CST-YFPc, reconstituted YFP was detected at the plasma membrane (Fig. 7B; Supplemental Fig. S3A). Interestingly, unlike the uniform localization of CST-GFP (Fig. 5A), the CST-HAE interaction appears to be restricted to subdomains of the plasma membrane (Fig. 7B; Supplemental Fig. S3A, arrowheads). Cotransfection of CST-YFPn/EVR-YFPc or EVR-YFPn/CST-YFPc also revealed interactions of CST and EVR at the plasma membrane in a uniform pattern (Fig. 7C; Supplemental Fig. S3B).
As a control for nonspecific interactions between RLCKs and YFP alone, cotransfection of CST-YFPn with YFPc (Walter et al., 2004) alone was performed (Fig. 7D). As depicted, no fluorescence was observed in the majority of protoplasts that were successfully transfected with the CFP-tagged mitochondrial expression control, indicating that CST and the C terminus of YFP do not interact.
As a control for nonspecific interactions between RLCKs and LRR-RLKs, we tested for interactions between the PBS1 class VII RLCK and the HAE and EVR LRR-RLKs. Cotransfection of PBS1-YFPn/PBS1-YFPc resulted in uniform plasma membrane fluorescence (Fig. 7E), indicating that PBS1 is also able to homodimerize. However, cotransfection with PBS1-YFPn/HAE-YFPc (Fig. 7F), PBS1-YFPn/EVR-YFPc (Fig. 7G), HAE-YFPn/PBS1-YFPc (Supplemental Fig. S3C), or EVR-YFPn/PBS1-YFPc (Supplemental Fig. S3D) did not show evidence of interactions between PBS1 and either HAE or EVR at the plasma membrane.
Taken together, these results suggest that CST inhibits signaling that promotes abscission both directly and indirectly by physically interacting with HAE and EVR at the cell surface.
DISCUSSION
We report here the identification of CST, a membrane-associated RLCK that functions as an inhibitor of organ abscission. Like the EVR and SERK1 LRR-RLKs (Leslie et al., 2010; Lewis et al., 2010), CST appears to restrict the extent of cell separation signaling, such that only cells within designated domains at the base of each outer organ undergo cell loosening, separation, and expansion. In nev cst flowers, the boundaries between the individual organ attachment sites and the border with the floral pedicel become notably obscured after organ abscission (Fig. 2F). Enlarged AZs that form visible collars of rough tissue are also found in nev evr, nev serk1, and 35S::IDA flowers instead of the smooth, discrete AZ scars of the wild type (Stenvik et al., 2006; Leslie et al., 2010; Lewis et al., 2010). The remarkable similarity of these phenotypes suggests that the rescue of organ abscission, AZ enlargement, and AZ boundary blurring in nev cst flowers may be due to ectopic, prolonged signaling of HAE and HSL2, the putative receptors of the IDA peptide.
We have found that the cst-2 mutation restores the structure of the Golgi and the location of the TGN in nev flowers and significantly reduces the hyperaccumulation of extracellular vesicles (Fig. 3). Golgi-derived structures like those in nev mutant flowers have also been seen in pollen mutants affecting the a1 subunit (VHA-a1) of the Arabidopsis vacuolar H+-ATPase proton transporter or when cells are treated with a specific V-ATPase inhibitor, concanamycin A (Dettmer et al., 2005; Liljegren et al., 2009). Recent studies have shown that the concanamycin A-induced structures are labeled with VHA-a1 and SYP61, markers of the TGN/early endosome (Viotti et al., 2010). These results suggest the possibility that the circularized structures in nev flowers are also chimeric fusions of the Golgi cisternae and the TGN and that traffic through the TGN/early endosome is affected by mutations in NEV. Disruption of CST, EVR, or SERK1 activity appears to restore the identity and independence of the TGN from the Golgi in nev flowers (Leslie et al., 2010; Lewis et al., 2010). Defects in membrane trafficking are thought to underlie the inappropriate fusion of the TGN and the Golgi, as V-ATPase-mediated endomembrane acidification is required to recruit ARF G-proteins, ARF-GEFs, and coat components involved in vesicle budding (Zeuzem et al., 1992; Aniento et al., 1996; Maranda et al., 2001; Hurtado-Lorenzo et al., 2006; Viotti et al., 2010). As an ARF-GAP, NEV may regulate the activity and recruitment of the same TGN-localized ARF G-protein(s) and adaptor molecules as the V-ATPase complex. The significant rescue of extracellular vesicle defects in nev cst flowers, which was not observed in nev serk1 or nev evr flowers (Leslie et al., 2010; Lewis et al., 2010), suggests the possibility that CST may interact more directly with NEV and in a different manner from EVR and SERK1 to inhibit abscission. Alternatively, if EVR and SERK1 act redundantly, suppression of extracellular vesicle accumulation may also be observed in nev serk1 evr flowers.
Functional studies of class VII RLCKs have revealed that several act in plant defense cell signaling pathways; CST is one of the first RLCKs found to regulate a developmental process (Table I). The mlpk allele discovered in the Yellow Sarson variety of B. rapa was found to allow pollen self-compatibility in this species; MLPK and the ligand-activated S-locus receptor kinase are proposed to form a heteromeric complex in stigmatic cells that mediates signaling, leading to the rejection of self pollen (Murase et al., 2004; Kakita et al., 2007). As with the cst mutant, redundancy appears to mask the phenotype of the Arabidopsis constitutive differential growth1 (cdg1) single mutant (Muto et al., 2004). CDG1 is predicted to play a role in brassinosteroid signal transduction, based on the phenotype of a dominant, activation-tagged line with stunted growth, epinastic leaves, and twisted stems; cdg1 loss-of-function alleles were identified as intragenic suppressors of the gain-of-function phenotype (Muto et al., 2004). Since CST regulatory regions direct GUS expression in the lateral roots, gynoecium, and developing guard cells, CST may function in other aspects of plant development.
Table I. Mutations in class VII RLCKs.
Mutations previously identified in the PBS1, MLPK, CDG1, BIK1, PBL1, PBL2, and RIPK RLCKs are shown. EMS, Ethyl methanesulfonate.
| Mutant | Cause | Effect | Reference |
| pbs1-1 | Fast neutron | 3′ deletion of open reading frame | Warren et al. (1999); Swiderski and Innes (2001) |
| pbs1-2 | EMS | G252R (kinase subdomain VIII) | Warren et al. (1999); Swiderski and Innes (2001) |
| mlpk | Natural variation | G194R (kinase subdomain VIa) | Murase et al. (2004) |
| cdg1-1 | EMS | W184* (before kinase subdomain VIa) | Muto et al. (2004) |
| cdg1-2 | EMS | Splicing disruption (after kinase subdomain V) | Muto et al. (2004) |
| bik1 | T-DNA insertion | Truncation in kinase subdomain IX | Veronese et al. (2006) |
| pbl1 | T-DNA insertion | Truncation near kinase subdomain II | Zhang et al. (2010) |
| pbl2 | T-DNA insertion | Truncation near kinase subdomain II | Zhang et al. (2010) |
| ripk | T-DNA insertion | Truncation before kinase subdomain XI | Liu et al. (2011) |
Like CST, many RLCKs contain consensus sites for N-terminal myristoylation and palmitoylation (Zhang et al., 2010). For instance, MLPK shows cytoplasmic localization upon mutation of its myristoylation site (Murase et al., 2004), and the class II RLCK SHORT SUSPENSOR was found to lose its membrane association with the loss of either N-myristoylation or palmitoylation (Bayer et al., 2009). We have found that CST shows partial dissociation from the plasma membrane when mutations disrupt either or both of these modifications (Fig. 5, B–D; Supplemental Fig. S1, A–C). These results suggest that other unknown factors contribute to CST membrane association. In the future, it will be important to test the functional relevance of CST membrane association.
Although the genetic nature of the cst-2 allele has not been resolved, we speculate that it is likely acting as a dominant-negative mutation. If the truncated mutant protein is acting as a nonfunctional kinase and is capable of homodimerizing with wild-type CST protein and/or heterodimerizing with the HAE and EVR LRR-RLKs, it has the potential to interfere with the activity and regulation of its partners. Dominant-negative LRR-RLK mutants are associated with particular missense mutations in the extracellular LRR and kinase domains of CLAVATA1 (Diévart et al., 2003; Diévart and Clark, 2003) and with truncation of the entire kinase domain in some LRR-RLKs such as ERECTA (Shpak et al., 2003). While most class VII RLCK mutations either occur within the kinase domain or are expected to truncate part of the kinase domain, the cst-2 mutation affects a codon positioned upstream of the kinase domain (Fig. 4A; Table I). It is intriguing that cst-2 dominantly suppresses the nev-3 and nev-6 mutants but not the nev-2 mutant (Fig. 1, D and O; data not shown). Allele-specific interactions for genes with opposing functions in a biological process suggest that the CST and NEV proteins may function in a single complex and that the ARF-GAP domain may regulate this interaction. We plan to explore these possibilities in future genetic and biochemical studies.
Integrated Model for RLK Regulation of Organ Separation
Our studies suggest a model in which CST and EVR inhibit cell separation signaling by acting in a step-wise manner to mediate HAE/HSL2 receptor complex formation and internalization (Fig. 8; Leslie et al., 2010). First, CST may sequester the EVR RLK at the plasma membrane. While CST-GFP is primarily localized to the plasma membrane (Fig. 5, A and A′), EVR-GFP is distributed between the plasma membrane and unknown internal compartments having the appearance of the ER network (Supplemental Fig. S2, C and C′). CST-EVR complexes are uniformly localized to the plasma membrane (Fig. 7C; Supplemental Fig. S3B), suggesting that a direct interaction between CST and EVR could limit EVR movement from the plasma membrane and enable subsequent interactions of EVR with HAE. Second, CST-containing complexes may interact with the HAE RLK. In contrast to the uniform localization of CST-GFP at the plasma membrane (Fig. 5A), CST-HAE interactions are stabilized within subdomains of the plasma membrane or perhaps closely associated vesicles (Fig. 7B; Supplemental Fig. S3A). Receptor aggregation may be important for packaging into endocytic vesicles (Zappel and Panstruga, 2008). Third, we speculate that interactions of EVR and/or SERK1 with HAE may facilitate the internalization and trafficking of HAE-containing receptor complexes (Leslie et al., 2010; Lewis et al., 2010). Since CST, EVR, and SERK1 act as negative regulators of cell separation, the step-wise aggregation and internalization of HAE and HSL2 may function to attenuate signaling or target the receptors for degradation.
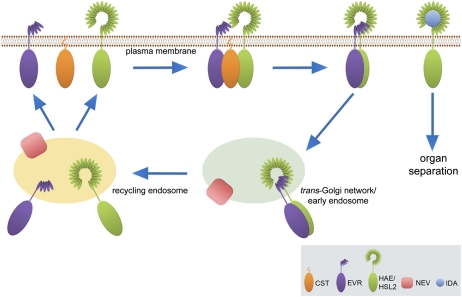
Figure 8.
An integrated model of RLK function and membrane trafficking during organ abscission. Interactions between a set of RLKs may modulate the timing and spatial extent of AZ cell loosening and separation. CST may sequester EVR at the plasma membrane and facilitate the formation of HAE/EVR and HSL2/EVR receptor complexes. Interactions between EVR and HAE/HSL2 may trigger internalization of the receptor complexes. NEV may regulate trafficking of the EVR and HAE/HSL2 receptors through the early endosome/TGN and recycling endosome, eventually restoring EVR and HAE/HSL2 to the plasma membrane. At the appropriate time in a discrete set of AZ cells, the secreted IDA peptide may bind to HAE/HSL2 at the cell surface, stabilizing the receptors and activating the downstream mitogen-activated protein kinase cascade that leads to cell separation and abscission.
NEV, as a global regulator of membrane trafficking, may be required for the trafficking of both positive (HAE, HSL2) and negative (EVR, SERK1) regulators of abscission (Fig. 8). From its locations at the TGN/early endosome and the recycling endosome (Liljegren et al., 2009), NEV may function to traffic receptors within the early endosomal system and ultimately recycle them back to the plasma membrane (Fig. 8). Loss of NEV could lead to the hyperaccumulation of inactivated receptors within endosomal compartments, while a secondary loss of CST, EVR, or SERK1 may stabilize the HAE/HSL2 RLKs at the plasma membrane. At the proper timing for abscission in wild-type flowers, our model predicts that IDA ligand binding stabilizes HAE/HSL2, leading to the activation of downstream signaling events required for the loss of cell adhesion (Fig. 8). As for activation of the FLS2 receptor by the bacterial elicitor flg22, which coincides with disassociation of the BIK1 RLCK (Lu et al., 2010), IDA peptide binding may trigger intracellular phosphorylation of HAE/HSL2 and disassociation of CST, EVR, and/or SERK1.
We have proposed that CST, EVR, and other RLKs act to sequester the HAE/HSL2 receptors at the cell surface prior to ligand binding and promote their internalization as a means to block signaling leading to organ abscission. This view of receptor internalization can be distinguished from the role of receptor endocytosis after ligand binding in facilitating downstream signaling. In the absence of brassinosteroids (BR), inactive BRI1 LRR-RLK receptors are thought to be constitutively internalized and recycled to the cell surface (Russinova et al., 2004; Geldner et al., 2007). Perception of BR leads to receptor activation; internalization of the activated receptor appears to enhance downstream signaling (Li and Chory, 1997; Kinoshita et al., 2005; Geldner et al., 2007). A recent study has revealed the role of receptor dephosphorylation in signaling for degradation of BRI1 (Wu et al., 2011). Signaling by BRI1 initiates a negative feedback loop that results in the methylation of a phosphatase that in turn dephosphorylates the internal pool of ligand-activated BRI1, marking it for degradation. How plant cells regulate the internalization of inactive receptors and target them for recycling to the cell surface or degradation remain open areas of investigation.
While a transient protoplast expression system is ideal for the rapid identification of interacting proteins, the interactions of CST with EVR and HAE must be confirmed by future experiments in floral tissues. By introducing epitope-tagged RLKs into the nev, nev cst, and nev evr mutants, the effect of NEV ARF-GAP activity upon RLK localization can also be tested. If NEV is required for the proper trafficking of RLKs that regulate AZ cell signaling, HAE and EVR may aggregate in aberrant locations in nev cells, either internally in endosomes or externally in the observed extracellular vesicles. Secondary loss of either the CST or EVR negative regulators may restore localization of the HAE/HSL2 RLKs to the plasma membrane. Since the primary function of CST may be to sequester RLKs at the plasma membrane and facilitate interactions between their intracellular kinase domains, future experiments will also investigate whether CST interacts with SERK1 and whether EVR and/or SERK1 interact with HAE.
There is a growing body of evidence that class VII RLCKs can mediate cell signaling by forming heteromeric complexes with RLKs. MLPK acts as a coreceptor for the S-locus receptor kinase during the self-incompatibility response (Murase et al., 2004; Kakita et al., 2007). BIK1 has been found to interact with the FLS2 and BAK1 LRR-RLKs to enable the PAMP-triggered immune response (Lu et al., 2010). Proposed interactions of the PBL1-like RLCKs with the EFR LRR-RLK and CERK1, an RLK with chitin oligosaccharide-recognizing LysM motifs in its extracellular domain, may promote PAMP-triggered immunity (Zhang et al., 2010). Our work highlights the potential significance of a sequential relay of receptor complex interactions and the regulated trafficking of receptor complexes to and from the cell surface as key mechanisms to modulate RLK-mediated cell signaling during plant growth and development.
MATERIALS AND METHODS
Plants and Growth Conditions
The cst-1 allele was identified through an ethyl methanesulfonate screen of nev-3 mutant Arabidopsis (Arabidopsis thaliana) plants (Ler ecotype) as described (Lewis et al., 2010). cst-1 is caused by a Gly-to-Arg substitution at amino acid 157. cst-2 (SAIL_296_A06; Columbia [Col-0] ecotype) contains a T-DNA insertion within codon 79 (Sessions et al., 2002). The primers and restriction enzyme for genotyping these alleles are described in Supplemental Table S2. Mutant alleles described previously include hae-1, hsl2-1, ida-2, nev-2, nev-3, and nev-6 (Cho et al., 2008; Stenvik et al., 2008; Liljegren et al., 2009).
For protoplast experiments, wild-type Arabidopsis (Col-0 ecotype) seeds were germinated on 1× Murashige and Skoog salts supplemented with 0.5% Suc and 0.8% agar. Seven-day-old seedlings were transferred to soil and grown in a 22°C chamber with a 12-h photoperiod for 3 to 4 additional weeks.
A 1.4-kb region of CST through the translational start site was PCR amplified from Col-0 DNA using 5′-CACCAAGAAGAGAGGAACTCTTGTA-3′ and 5′-CCACTCTGCAAATCCTTGAACA-3′, cloned into pENTR/D-TOPO (Life Technologies), and recombined into pGWB3 (Nakagawa et al., 2007) to create a translational fusion of CST to GUS. Of 35 CST::CST-GUS transgenic lines analyzed, 24 showed GUS expression in seedling leaf veins and developing guard cells; five of the 24 lines also showed GUS expression in floral AZs. GUS expression was not observed in any tissues for 11 of the 35 lines analyzed.
Mapping
The cst-1 mutation was mapped by crossing nev-3 cst-1 (Ler) and nev-6 (Col-0) flowers. Using DNA isolated from 518 nev cst F2 plants and PCR-based markers including several designed from Ler polymorphisms (http://www.Arabidopsis.org/Cereon/), cst-1 was located to the F8D20 bacterial artificial chromosome on chromosome 4. The coding regions of 11 of 15 genes in the 49-kb interval were sequenced from nev cst plants.
Kinase Activity
The CST open reading frame was amplified using 5′-CACCATGGGTGCTTGTATT-3′ and 5′-TTATTTTTCTACTGATCCAAACCGT-3′ from Col-0 DNA, to create CSTWT. This fragment was cloned into pENTR/D-TOPO, and site-directed mutagenesis (Stratagene) with 5′-GGTTCTGGTATGATCGTTGCCATCGAGAGATTGAATTCTGAGAGTGTTC-3′ and 5′-GAACACTCTCAGAATTCAATCTCTCGATGGCAACGATCATACCAGAACC-3′ or with 5′-CACCGAAATCTGGTGAAGTTATTGAGATACTGTCGTGAAGACAAAGAGC-3′ and 5′-GCTCTTTGTCTTCACGACAGTATCTCAATAACTTCACCAGATTTCGGTG-3′ was used to create CSTK124E and CSTG157R, respectively. Recombination with pDEST17 (Life Technologies) was used to generate 6x-His-tagged CST. Recombinant proteins were expressed in Escherichia coli and purified by Co2+ affinity chromatography (Clontech Laboratories). Antisera were used at the following dilutions: anti-polyHis-horseradish peroxidase (Sigma-Aldrich; 1:50,000), anti-phospho-Ser (Sigma-Aldrich; 1:2,000), anti-phospho-Tyr (Sigma-Aldrich; 1:2,000), anti-phospho-Thr (Zymed/Invitrogen; 1:800). Horseradish peroxidase-conjugated chicken anti-mouse (Santa Cruz Biotechnology) secondary antibodies were used at a 1:10,000 dilution.
Protein Interaction and Localization Assays
The coding regions for CST, HAE, and PBS1 were PCR amplified from the U18406 (Arabidopsis Biological Resource Center [ABRC]), pBS-HAE (kindly provided by John Walker, University of Missouri), and U12315 (ABRC) cDNA constructs and cloned into pENTR/D-TOPO (Life Technologies). The EVR coding region was amplified from genomic (Col-0) DNA and cloned into pENTR/D-TOPO. Using site-directed mutagenesis, 5′-GCCCCCTTCACCATGGCTGCTTGTATTTCGTTC-3′ and 5′-GAACGAAATACAAGCAGCCATGGTGAAGGGGGC-3′ were used to generate pENTR::CSTG2A, 5′-CCCCCTTCACCATGGGTGCTTCTATTTCGTTCTTTTCTTCTTCC-3′ and 5′-GGAAGAAGAAAAGAACGAAATAGAAGCACCCATGGTGAAGGGGG-3′ were used to generate pENTR::CSTC4S, and 5′-CCCCCTTCACCATGGCTGCTTCTATTTCGTTCTTTTCTTCTTCC-3′ and 5′-GGAAGAAGAAAAGAACGAAATAGAAGCAGCCATGGTGAAGGGGG-3′ were used to generate pENTR::CSTG2A C4S. Recombination with pHBT-gw-GFP, pUC-gw-SPYNE, and pUC-gw-SPYCE destination vectors (Punwani et al., 2010) using Gateway technology (Life Technologies) was used to generate C-terminal fusions of each RLK to GFP, YFPn, and YFPc, respectively. The pUC-SPYCE plasmid was used as a YFPc control (Walter et al., 2004).
Protoplasts were isolated and transformed according to the method described previously (Yoo et al., 2007) with minor modifications. Mesophyll cells were exposed to digestive enzyme solution (1.5% Cellulase R-10 and 0.4% Macerozyme R-10; Yakult Pharmaceutical) for 1 h after removing the epidermal cell layer by the tape-Arabidopsis sandwich method (Wu et al., 2009). For each transfection reaction, 2 to 3 × 104 protoplasts were incubated with 10 to 20 μg of plasmid DNA in a 20% polyethylene glycol (Sigma) solution for 10 to 15 min. For BiFC assays, protoplasts were cotransfected with the experimental constructs and a mitochondrial CFP marker (ABRC CD3-986; Nelson et al., 2007) to track transfected cells and measure transfection efficiency. Cells were washed and incubated overnight in the dark to allow for gene expression. Protoplasts were imaged 16 to 20 h after transfection. Only those transfection experiments for which 50% or greater of the protoplasts showed expression of the cotransfected mitochondrial CFP marker were used for BiFC analysis.
Microscopy
Flowers were fixed and processed for scanning and transmission electron microscopy as described (Liljegren et al., 2009; Leslie et al., 2010). Confocal laser scanning microscopy of root epidermal cells was performed with an LSM-510 system (Carl Zeiss). Confocal laser scanning microscopy of leaf protoplasts was performed with a Zeiss LSM7 Duo system (Carl Zeiss). The following excitation (ex) lines and emission (em) ranges were used: GFP (ex 488, em 498–532), chlorophyll autofluorescence with GFP imaging (ex 560, em 572–716), YFP (ex 512, em 516–577), chlorophyll autofluorescence with YFP imaging (ex 512, em 603–732). Image brightness and contrast were adjusted with Photoshop CS4 (Adobe).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. CST localization is disrupted by mutations that affect N-terminal myristoylation or palmitoylation.
Supplemental Figure S2. EVR and HAE homodimerize at the plasma membrane.
Supplemental Figure S3. CST interacts with HAE and EVR at the plasma membrane.
Supplemental Table S1. Complementation of nev cst mutant phenotype.
Supplemental Table S2. Genotyping the cst mutant alleles.
Acknowledgments
We thank J. Walker, S. Patterson, M. Duncan, B. Goldstein, S. Ahmed, M. Peifer, J. Reed, and E.-H. Chung for helpful discussions; S. Hasty, P. Healy, and H. Kizer for technical assistance; J. Punwani and J. Kieber for protoplast transfection vectors and reagents; T. Perdue, S. Ray, and V. Madden for microscopy assistance; J. Walker and the ABRC for DNA and seed stocks; Monsanto for access to Ler polymorphisms; and Syngenta for access to the SAIL T-DNA collection.
References
- Ade J, DeYoung BJ, Golstein C, Innes RW. (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aker J, Borst JW, Karlova R, de Vries SC. (2006) The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the somatic embryogenesis receptor-like kinase 1 receptor at the plasma membrane. J Struct Biol 156: 62–71 [DOI] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersson E, Saalbach G, Larsson C, Kjellbom P. (2004) Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol 45: 1543–1556 [DOI] [PubMed] [Google Scholar]
- Aniento F, Gu F, Parton RG, Gruenberg J. (1996) An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol 133: 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Boisson B, Giglione C, Meinnel T. (2003) Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J Biol Chem 278: 43418–43429 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB. (2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC. (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E-H, da Cunha L, Wu A-J, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. (2011) Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Schubert D, Calvo-Weimar O, Stierhof YD, Schmidt R, Schumacher K. (2005) Essential role of the V-ATPase in male gametophyte development. Plant J 41: 117–124 [DOI] [PubMed] [Google Scholar]
- Diévart A, Clark SE. (2003) Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr Opin Plant Biol 6: 507–516 [DOI] [PubMed] [Google Scholar]
- Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, Clark SE. (2003) CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring DR, Walker JC. (2004) Self-rejection: a new kinase connection. Science 303: 1474–1475 [DOI] [PubMed] [Google Scholar]
- Hanks SK. (2003) Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol 4: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. (1995) Protein kinases 6: the eukaryotic protein kinase superfamily. Kinase (catalytic) domain structure and classification. FASEB J 9: 576–596 [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink MA, Shah K, Russinova E, de Vries SC, Visser AJ. (2008) Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys J 94: 1052–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn MA, Walker JC. (1994) Biochemical properties of the autophosphorylation of RLK5, a receptor-like protein kinase from Arabidopsis thaliana. Biochim Biophys Acta 1208: 65–74 [DOI] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, et al. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol 8: 124–136 [DOI] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC. (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R, Snyder C, Jones JG. (1987) T-DNA is organized predominantly in inverted repeat structures in plants transformed with Agrobacterium tumefaciens C58 derivatives. Mol Gen Genet 207: 471–477 [Google Scholar]
- Jurca ME, Bottka S, Fehér A. (2008) Characterization of a family of Arabidopsis receptor-like cytoplasmic kinases (RLCK class VI). Plant Cell Rep 27: 739–748 [DOI] [PubMed] [Google Scholar]
- Kakita M, Murase K, Iwano M, Matsumoto T, Watanabe M, Shiba H, Isogai A, Takayama S. (2007) Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries SC. (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Cano-Delgado A, Seto H, Hiranuma D, Fujioka S, Yoshida S, Chory J. (2005) Binding of brassinosteriods to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Leslie ME, Lewis MW, Youn JY, Daniels MJ, Liljegren SJ. (2010) The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development 137: 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Fulcher EH, Darnielle L, Healy PN, Youn JY, Liljegren SJ. (2010) The SERK1 receptor-like kinase regulates organ separation in Arabidopsis flowers. Plant J 62: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Leslie ME, Darnielle L, Lewis MW, Taylor SM, Luo R, Geldner N, Chory J, Randazzo PA, Yanofsky MF, et al. (2009) Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 136: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin Z-JD, Coaker G. (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Ahvazi B, Amariei D, Shroder D, Burrola B, Losert W, Randazzo PA. (2007) Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem J 402: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranda B, Brown D, Bourgoin S, Casanova JE, Vinay P, Ausiello DA, Marshansky V. (2001) Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J Biol Chem 276: 18540–18550 [DOI] [PubMed] [Google Scholar]
- Meiners S, Xu A, Schindler M. (1991) Gap junction protein homologue from Arabidopsis thaliana: evidence for connexins in plants. Proc Natl Acad Sci USA 88: 4119–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels CA. (2002) Suppression analysis. Michels CA, , Genetic Techniques for Biological Research: A Case Study Approach. John Wiley & Sons, Hoboken, NJ, pp 91–97 [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Shiba H, Iwano M, Che FS, Watanabe M, Isogai A, Takayama S. (2004) A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. (1993) The proposed plant connexin is a protein kinase-like protein. Plant Cell 5: 998–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Yabe N, Asami T, Hasunuma K, Yamamoto KT. (2004) Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol 136: 3124–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. (2010) The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J 62: 473–482 [DOI] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shah K, Gadella TW, Jr, van Erp H, Hecht V, de Vries SC. (2001) Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J Mol Biol 309: 641–655 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, Lakeman MB, Torii KU. (2003) Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Bloch D, Yalovsky S. (2009) Protein lipid modifications in signaling and subcellular targeting. Curr Opin Plant Biol 12: 714–720 [DOI] [PubMed] [Google Scholar]
- Stael S, Bayer RG, Mehlmer N, Teige M. (2011) Protein N-acylation overrides differing targeting signals. FEBS Lett 585: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB. (2006) Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW. (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J 26: 101–112 [DOI] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof Y-D, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, et al. (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Warren RF, Merritt PM, Holub E, Innes RW. (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J. (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zappel NF, Panstruga R. (2008) Heterogeneity and lateral compartmentalization of plant plasma membranes. Curr Opin Plant Biol 11: 632–640 [DOI] [PubMed] [Google Scholar]
- Zeuzem S, Feick P, Zimmermann P, Haase W, Kahn RA, Schulz I. (1992) Intravesicular acidification correlates with binding of ADP-ribosylation factor to microsomal membranes. Proc Natl Acad Sci USA 89: 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang Y, Li R, Song X, Wang Q, Huang S, Jin JB, Liu CM, Lin J. (2010) Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J 61: 223–233 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]