Abstract
Despite the accumulation of data on the genetic and molecular understanding of heterosis, there is little information on the regulation of heterosis at the physiological level. In this study, we performed a quantitative analysis of endogenous gibberellin (GA) content and expression profiling of the GA metabolism and signaling genes to investigate the possible relationship between GA signaling and heterosis for seedling development in rice (Oryza sativa). The materials used were an incomplete diallele set of 3 × 3 crosses and the six parents. In the growing shoots of the seedlings at 20 d after sowing, significant positive correlations between the contents of some GA species and performance and heterosis based on shoot dry mass were detected. Expression analyses of GA-related genes by real-time reverse transcription-polymerase chain reaction revealed that 13 out of the 16 GA-related genes examined exhibited significant differential expression among the F1 hybrid and its parents, acting predominantly in the modes of overdominance and positive dominance. Expression levels of nine genes in the hybrids displayed significant positive correlations with the heterosis of shoot dry mass. These results imply that GAs play a positive role in the regulation of heterosis for rice seedling development. In shoots plus root axes of 4-d-old germinating seeds that had undergone the deetiolation, mimicking normal germination in soil, the axis dry mass was positively correlated with the content of GA29 but negatively correlated with that of GA19. Our findings provide supporting evidence for GAs playing an important regulatory role in heterosis for rice seedling development.
Heterosis or hybrid vigor refers to the phenomenon in which hybrids outperform their inbred parents in yield, biomass, biotic and abiotic stress tolerance, or other traits. Heterosis has been widely exploited to increase the productivity of crop plants for many decades (Stuber, 1994; Yuan, 1998). Generally, the life cycle of rice (Oryza sativa) plants can be divided into vegetative and reproductive stages. In previous studies, heterosis was demonstrated in yield and yield component traits of rice (Zhang et al., 1994; Xiao et al., 1995; Yu et al., 1997; Li et al., 2001; Luo et al., 2001; Hua et al., 2003), but very few studies have been conducted to systematically examine heterosis in seedling traits, which is the basis for canopy development and yield.
The biological basis of heterosis has been of primary interest for biology researchers for many years due to its scientific and practical significance. Complete elucidation of the mechanism for this phenomenon requires knowledge at three levels: genetic, molecular, and physiological. Genetic models of heterosis currently rely on information from quantitative trait locus analyses of yield-related traits in various plant species, which show that dominance, overdominance, and epistasis are all involved in the manifestation of heterosis, with various degrees of significance according to experimental design and different species (Stuber et al., 1992; Xiao et al., 1995; Yu et al., 1997; Hua et al., 2002, 2003; Kusterer et al., 2007; Melchinger et al., 2007, Garcia et al., 2008; Li et al., 2008).
To unravel the molecular basis of heterosis, high-throughput expression profiling of heterotic crosses in various plant species was carried out, which identified large numbers of differentially regulated genes with diverse functions (Guo et al., 2006; Huang et al., 2006; Swanson-Wagner et al., 2006; Zhang et al., 2008; Wei et al., 2009; He et al., 2010). Although no consensus results have emerged, it is thought that allelic variants at a large number of loci acting through partial to complete dominance would provide favorable combinations resulting in superior hybrid phenotypes (Springer and Stupar, 2007). In addition, allelic variations in genic sequences, genome structure, DNA methylation patterns, and chromatin structure also contribute to the molecular basis of heterosis (Springer and Stupar, 2007).
In contrast, knowledge regarding the physiological basis of heterosis is sporadic and is mainly focused on some specific traits, such as freezing tolerance in Arabidopsis (Arabidopsis thaliana; Korn et al., 2010). With the development of the technologies for high-throughput metabolic profiling, it is now possible to predict heterosis using both DNA and physiologically metabolic markers (Gärtner et al., 2009; Andorf et al., 2010), which will be of considerable value for finding physiological clues for heterosis.
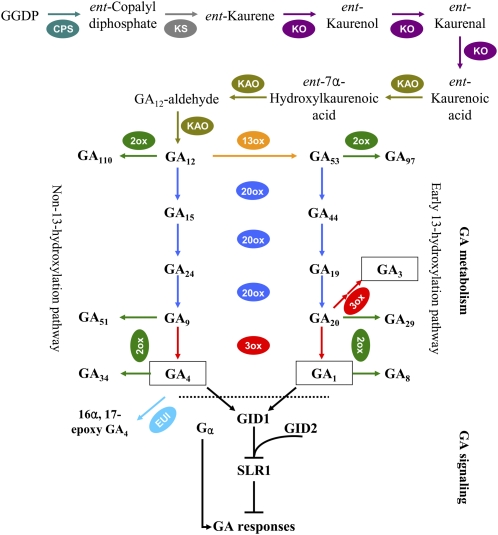
GAs are a group of tetracyclic diterpene phytohormones that control diverse aspects of plant growth and development, from seed germination, leaf expansion, and stem elongation to flower initiation and the development of flowers and fruits (Davies, 2004; Fleet and Sun, 2005). Bioactive GAs are synthesized from the common diterpene precursor geranylgeranyl diphosphate (Fig. 1), which is first converted to ent-kaurene by two kinds of diterpene cyclases, ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS), followed by sequential oxidations to produce GA12, catalyzed by two cytochrome P450 monooxygenases, ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO). At the final stage of bioactive GA synthesis, GA53/GA12 is converted to GA1/GA4 by two 2-oxoglutarate-dependent dioxygenases, GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox). Deactivation of bioactive GAs and their precursors is catalyzed by another dioxygenase, GA 2-oxidase (GA2ox), and a P450 monooxygenase, Elongated Uppermost Internode (EUI; Yamaguchi, 2008). GA signaling proceeds by derepression of the DELLA protein, a repressor of GA signaling, with the aid of a GA receptor, GIBBERELLIN INSENSITIVE DWARF1 (GID1), and an F-box protein, GID2 (Sasaki et al., 2003; Ueguchi-Tanaka et al., 2005). In addition, the α-subunit of the heterotrimeric G protein is thought to be a positive regulator of GA signaling (Ueguchi-Tanaka et al., 2000). In recent years, the aforementioned GA metabolism and signaling genes have been cloned in rice (Sakamoto et al., 2004; Zhu et al., 2006; Lo et al., 2008), and these genes are regulated by development, hormones, light, temperature, and stress (Yamaguchi, 2008).
Figure 1.
Schematic diagram of GA metabolism and signaling pathways in rice. Bioactive GAs are shown in black rectangles. For each step in the metabolic pathway, the reaction is highlighted in color. GA7 (13-nonhydroxy GA3) is biosynthesized from GA9 in a similar pathway to the synthesis of GA3 from GA20, but it is not shown here. EUI is a P450 designated as CYP714D1 that preferentially epoxidizes the 16,17-double bond of non-13-hydroxylated GAs, including GA4, GA9, and GA12, to deactivate them, and only the reaction on GA4 is shown here. Bioactive GAs are perceived by the soluble GA receptor GID1 (Ueguchi-Tanaka et al., 2005). SLR1, a DELLA protein, acts as a negative regulator of the GA responses (Ikeda et al., 2001). SLR1 within the GA-GID1-SLR1 complex is degraded through the SCFGID2 complex, resulting in GA responses (Sasaki et al., 2003; Hirano et al., 2008). Apart from these signaling factors, the α-subunit of the heterotrimeric G protein (Gα) is suggested to function as a positive regulator of GA signaling (Ueguchi-Tanaka et al., 2000). GGDP, Geranylgeranyl diphosphate.
There have been studies on the role of GAs in the regulation of heterosis. For example, Rood et al. (1988) analyzed the differences in responsiveness to the exogenous application of GA3 and endogenous levels of GAs between F1 hybrids and their inbred parents of diallele combinations in maize (Zea mays). They found that inbreds were more responsive than the hybrids to the exogenous GA3, while the hybrids had higher concentrations of endogenous GAs than their parental inbreds. They concluded that the increased endogenous concentration of GA in the hybrids could provide a phytohormone basis for heterosis for shoot growth.
Plants maintain the levels of bioactive GAs via feedback and feed-forward regulation of GA metabolism (Hedden and Phillips, 2000; Olszewski et al., 2002), and the dioxygenases, namely GA20oxs, GA3oxs, and GA2oxs, are the main targets of regulation by GA signaling to establish homeostasis. For example, an elevated level of bioactive GA usually suppresses the expression of GA20ox and GA3ox while stimulating the expression of GA2ox; conversely, a drop in bioactive GA level usually up-regulates the expression of GA20ox and GA3ox and down-regulates the expression of GA2ox. This kind of delicate homeostatic balance of GA metabolism makes it difficult to accurately interpret the results of individual experiments with respect to the physiological implications of changes in GA biosynthetic gene expression, which reinforces the necessity of measuring GA contents not only for the bioactive hormones but also for their precursors and catabolites.
In this study, we investigated the possible role of GAs in the regulation of heterosis in rice. Using an incomplete diallele set consisting of six elite inbreds and corresponding hybrids with varying heterotic responses, we measured the amounts of both bioactive GAs and their precursors and catabolites and analyzed the expression levels of GA metabolism and signaling genes. The results showed positive correlations between not only several GA species but also some GA-related genes and rice seedling heterosis.
RESULTS
Heterosis of Rice Seedlings and Germinating Seeds
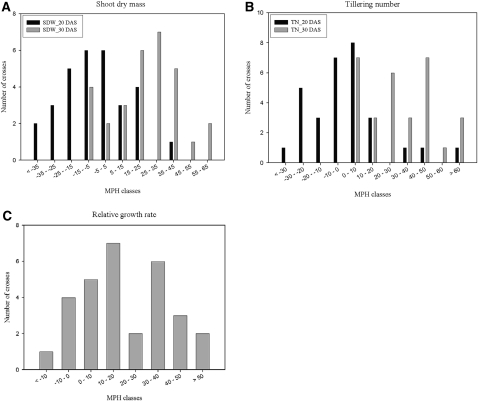
A survey of the occurrence and the degree of heterosis related to rice seedling growth was conducted with the incomplete diallele set of 30 F1 hybrids and their 11 parents, including five maintainers and six restorers. A number of seedling growth traits were assayed at 20 and 30 d after sowing (DAS; five to 10 individuals per genotype) for the 41 entries (Supplemental Table S2). Shoot dry weight and tiller number per seedling exhibited high levels of heterosis, especially at 30 DAS. A derived trait, relative growth rate, based on shoot dry weight at two stages, also showed high heterosis. The distribution of midparent heterosis (MPH) of the three traits for rice seedling growth is shown in Figure 2. At 20 DAS, very little or even moderate negative heterosis was observed for shoot dry weight and tiller number in some of the crosses. In contrast, high levels of heterosis were observed at 30 DAS for shoot dry weight in the majority of the 30 crosses and for tiller number in all of the crosses. MPH ranged from −9% to 61.7% for shoot dry weight and from 1.4% to 73.3% for tiller number. Two crosses, Maxie × Minghui 63 and II-32 × 6078, expressed 50% or greater MPH for both traits. There was a highly significant correlation between shoot dry weight and tiller number in both F1 performance (r = 0.577, P < 0.01) and heterosis (r = 0.724, P < 0.01), indicating a contribution of heterosis to the F1 performance at this stage.
Figure 2.
Distribution of MPH for the three traits of rice seedling growth in the 5 × 6 incomplete diallele set. The horizontal axis of each chart is the interval of MPH in percentage, and the vertical axis is the number of crosses. Shown are the distribution of MPH of shoot dry mass per seedling (A), tiller number per seedling (B), and relative growth rate per seedling (C). Averages of three replicates were used for calculation of MPH. SDW_20 DAS and SDW_30 DAS in A indicate shoot dry mass of rice seedling at 20 DAS and 30 DAS, respectively; TN_20 DAS and TN_30 DAS in B indicate tiller number of rice seedling at 20 DAS and 30 DAS, respectively.
From the original incomplete diallele set, three restorer lines (Ce 64, Minghui 63, and Mianhui 725) and three maintainer lines (Jin 23, Maxie, and Zhenshan 97) were selected and intermated with each other (3 × 3) to form a subset of incomplete diallele of nine crosses to represent low, intermediate, and high levels of seedling heterosis for GA quantification and gene expression analyses. The measurements of shoot dry weight, tiller number, and heterosis of these genotypes are presented in Tables I and II.
Table I. Measurements and MPH of shoot dry weight of rice seedling at 20 and 30 DAS and shoot plus root axis dry weight of 4-d-old germinating seeds in the 3 × 3 incomplete diallele set.
Data shown are means ± se from three biological replicates. For shoot dry weight, the values were measured on a per seedling basis.
| Sample Name | Genotype | Shoot Dry Weight of 20-DAS Seedlings |
Shoot Dry Weight of 30-DAS Seedlings |
Shoot Plus Root Axis Dry Weight of 4-d-Old Germinating Seeds |
|||
| Average | MPH | Average | MPH | Average | MPH | ||
| g | % | g | % | g | % | ||
| InDi_01 | Jin 23 | 0.25 ± 0.038 | 1.02 ± 0.15 | 2.30 ± 0.11 | |||
| InDi_02 | Maxie | 0.25 ± 0.067 | 0.84 ± 0.09 | 2.73 ± 0.21 | |||
| InDi_03 | Zhenshan 97 | 0.23 ± 0.065 | 0.96 ± 0.004 | 1.98 ± 0.23 | |||
| InDi_04 | Ce 64 | 0.22 ± 0.035 | 1.02 ± 0.18 | 2.39 ± 0.34 | |||
| InDi_05 | Minghui 63 | 0.28 ± 0.072 | 0.90 ± 0.16 | 2.42 ± 0.12 | |||
| InDi_06 | Mianhui 725 | 0.19 ± 0.028 | 0.70 ± 0.14 | 2.42 ± 0.03 | |||
| InDi_07 | Jin 23 × Ce 64 | 0.20 ± 0.009 | −17.6 | 1.01 ± 0.05 | −5.3 | 3.06 ± 0.15 | 30.6 |
| InDi_08 | Maxie × Ce 64 | 0.17 ± 0.037 | −29.8 | 1.25 ± 0.39 | 34.0 | 3.09 ± 0.10 | 20.5 |
| InDi_09 | Zhenshan 97 × Ce 64 | 0.19 ± 0.011 | −14.5 | 0.93 ± 0.07 | 3.1 | 3.52 ± 0.37 | 61.1 |
| InDi_10 | Jin 23 × Minghui 63 | 0.28 ± 0.078 | 11.6 | 1.19 ± 0.42 | 18.8 | 2.44 ± 0.21 | 3.7 |
| InDi_11 | Maxie × Minghui 63 | 0.25 ± 0.031 | 4.9 | 1.41 ± 0.36 | 61.7 | 3.05 ± 0.08 | 18.4 |
| InDi_12 | Zhenshan 97 × Minghui 63 | 0.30 ± 0.030 | 16.4 | 1.13 ± 0.13 | 22.0 | 3.61 ± 0.19 | 64.4 |
| InDi_13 | Jin 23 × Mianhui 725 | 0.27 ± 0.093 | 16.4 | 1.40 ± 0.42 | 43.1 | 3.32 ± 0.14 | 41.0 |
| InDi_14 | Maxie × Mianhui 725 | 0.20 ± 0.039 | −10.1 | 1.19 ± 0.11 | 41.0 | 3.55 ± 0.08 | 38.0 |
| InDi_15 | Zhenshan 97 × Mianhui 725 | 0.22 ± 0.052 | 0.6 | 1.28 ± 0.10 | 53.1 | 3.37 ± 0.09 | 53.5 |
Table II. Measurements and MPH of tiller number of rice seedlings at 20 and 30 DAS and relative growth rate in the 3 × 3 incomplete diallele set.
| Sample Name | Genotype | Tiller No. of 20-DAS Seedlings |
Tiller No. of 30-DAS Seedlings |
Relative Growth Rate |
|||
| Average | MPH | Average | MPH | Average | MPH | ||
| % | % | d−1 | % | ||||
| InDi_01 | Jin 23 | 2.6 ± 0.1 | 4.0 ± 0.4 | 0.14 | |||
| InDi_02 | Maxie | 2.0 ± 0.3 | 3.8 ± 0.2 | 0.12 | |||
| InDi_03 | Zhenshan 97 | 1.1 ± 0.07 | 3.8 ± 0.1 | 0.14 | |||
| InDi_04 | Ce 64 | 2.6 ± 0.3 | 7.5 ± 0.8 | 0.14 | |||
| InDi_05 | Minghui 63 | 1.9 ± 0.4 | 5.4 ± 0.8 | 0.12 | |||
| InDi_06 | Mianhui 725 | 1.4 ± 0.2 | 3.4 ± 0.4 | 0.15 | |||
| InDi_07 | Jin 23 × Ce 64 | 2.4 ± 0.06 | −7.7 | 6.1 ± 0.8 | 6.2 | 0.16 | 9.1 |
| InDi_08 | Maxie × Ce 64 | 1.8 ± 0.1 | −20.9 | 7.0 ± 1.0 | 23.9 | 0.20 | 47.4 |
| InDi_09 | Zhenshan 97 × Ce 64 | 2.0 ± 0.3 | 9.2 | 5.7 ± 0.6 | 1.4 | 0.16 | 13.5 |
| InDi_10 | Jin 23 × Minghui 63 | 2.1 ± 0.4 | −6.0 | 6.0 ± 1.3 | 26.6 | 0.14 | 4.6 |
| InDi_11 | Maxie × Minghui 63 | 2.2 ± 0.2 | 14.5 | 6.9 ± 0.9 | 50.4 | 0.17 | 33.5 |
| InDi_12 | Zhenshan 97 × Minghui 63 | 2.2 ± 0.1 | 49.3 | 6.0 ± 0.1 | 31.7 | 0.13 | 3.2 |
| InDi_13 | Jin 23 × Mianhui 725 | 2.1 ± 0.5 | 4.1 | 5.4 ± 1.0 | 47.1 | 0.16 | 13.5 |
| InDi_14 | Maxie × Mianhui 725 | 1.5 ± 0.2 | −11.5 | 5.3 ± 0.2 | 48.8 | 0.18 | 32.2 |
| InDi_15 | Zhenshan 97 × Mianhui 725 | 1.7 ± 0.3 | 32.9 | 5.0 ± 0.5 | 40.8 | 0.18 | 32.2 |
Since seed germination is also an important trait, we analyzed seed germination in F1 hybrids and parents of the 3 × 3 incomplete diallele set in moisturized petri dishes incubated in a growth chamber. At 4 d after imbibition, shoot plus root axis was removed from germinating seeds and their dry weights were determined for the 15 entries (100–200 grains per entry with three independent replicates). A highly significant difference (P < 0.001) in shoot plus root axis dry weight from 4-d-old germinating seeds was found among the nine crosses and the six parents using an F test. The F1s producing the highest dry mass had Zhenshan 97 and Mianhui 725 as one or both of the parents (Table I). It is interesting that the cross Zhenshan 97 × Minghui 63, producing an F1 hybrid named Shanyou 63, which is one of the most widely cultivated rice hybrids in China during the last two decades, demonstrated the highest axis dry mass.
Substantial heterosis was detected for axis dry weight, with MPH varying from 3.7% for the cross Jin 23 × Minghui 63 to 64.4% for the cross Zhenshan 97 × Minghui 63 (Table I). Thus, the crosses showing the lowest heterosis and the one with the highest heterosis both involved Minghui 63. Moreover, in five out of the nine crosses (InDi_09 and InDi_12–InDi_15; for genotypes, see Table I), there were significant differences between parents and F1 hybrids via one-way ANOVA (P < 0.01), and all of the five crosses exhibited significant high-parent heterosis (F1 shows better performance than the higher parent) for the character, with the level of high-parent heterosis ranging from 30.1% to 49.5%. There was no significant correlation between performance or heterosis for shoot plus root axis dry mass and seedling growth traits.
Levels of Endogenous GAs in Rice Seedlings and Germinating Seeds
GAs have a well-characterized involvement in promoting vegetative growth in rice (Kaneko et al., 2003; Sakamoto et al., 2004). According to our survey and previous studies (Akita et al., 1990; de Leon et al., 2001), heterosis in rice seedling growth could be detected as early as 16 to 23 DAS to varying degrees. To assay the accumulation levels of GAs, including active forms as well as their metabolites and precursors (Fig. 1), at early stages of rice seedling growth, which may be part of the hormonal basis of seedling heterosis, the growing shoots from 20-DAS seedlings and the shoot plus root axis from 4-d-old germinating seeds of the incomplete diallele set of nine crosses were harvested and the endogenous GA levels were analyzed by combined gas chromatography-mass spectrometry.
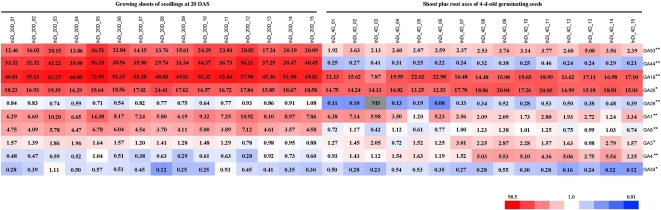
The concentrations and distribution patterns of 10 quantified GA species, all of which, except GA4 and GA34, belong to the early-13-hydroxylation pathway (Fig. 1) that is the dominant pathway for GAs in vegetative growth of rice, are shown in the heat map (Fig. 3). The concentrations of GA19 and GA20 were highest in both tissues, although they varied among the 15 entries. The level of GA53 was high in shoots at 20 DAS but moderate in 4-d-old germinating seeds, compared with GA44, which was present at very high levels in shoots at 20 DAS but very low levels in 4-d-old germinating seeds. Low to moderate levels of GA1, GA8, GA3, and GA4 were detected in both tissues, also with considerable variation among the 15 entries. Finally, GA29 and GA34 were detected at very low levels in both tissues in all the entries.
Figure 3.
Heat map of the distribution of GA species in the 3 × 3 incomplete diallele set. Growing shoots of rice seedlings at 20 DAS and shoot plus root axes of 4-d-old germinating seeds were harvested, and the endogenous concentrations of the GA species were analyzed by gas chromatography-mass spectrometry. The relative accumulation patterns are shown in the heat map based on the average value for each GA species. Red and blue colors indicate higher and lower concentrations, respectively. The color scale is shown at the bottom. The value in each block is the concentration (average value; n = 3) as ng g−1 dry weight. InDi_20D_01 to InDi_20D_15, Growing shoots of 20-DAS seedlings of the parental lines and crosses from the 3 × 3 incomplete diallele set; InDi_4D_01 to InDi_4D_15, shoot plus root axes of 4-d-old germinating seeds of the parental lines and crosses from the 3 × 3 incomplete diallele set; ND, not detected under the quantification limit. The correspondence between the line number and the line identity is the same as in Table I. Asterisks indicate that the GA contents were significantly different between the two tissues by one-sided pairwise t test (* P < 0.05, ** P < 0.01).
The levels for eight of the 10 GA molecules (except for GA4 and GA3) were significantly (P < 0.01) higher in samples from 20-DAS seedlings than 4-d-old germinating seeds (Fig. 3), which is consistent with the presence of GAs mainly in actively growing and elongating tissues. It is noteworthy that the accumulation of GA44, an intermediate of the reaction in the GA biosynthesis pathway catalyzed by GA20ox (Fig. 1), increased 80 to 236 times in the growing shoots of 20-DAS seedlings compared with that in the axes of 4-d-old germinating seeds. By contrast, the contents of GA20, the product of GA20ox and the immediate precursor for bioactive GA1 (Fig. 1), did not change much in these two stages, although the overall levels were high. In addition, in 20-DAS seedlings, the levels of GA1 were much higher than GA4, which is consistent with previous reports that GA1 acts dominantly during the vegetative stages of rice while GA4 is the predominant bioactive form in the reproductive organs.
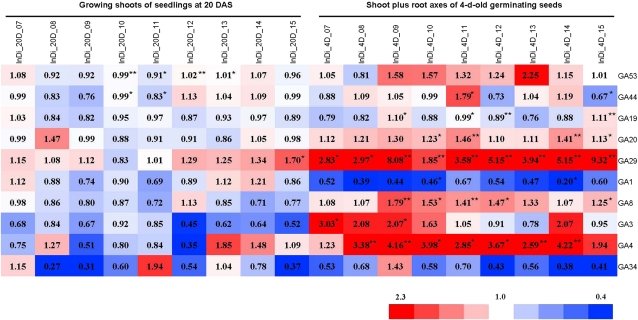
We also displayed the GA contents in the hybrids relative to their parents (Fig. 4) to illustrate heterosis in GA contents. As shown in Figure 4, except for GA4 and GA29 in the axes of germinating seeds, heterotic responses of GA contents in F1 hybrids were low, with some negative heterosis observed for the contents of GA1 and GA34 in the axes of germinating seeds.
Figure 4.
Heat map of the relative levels of GA species in the F1 hybrids versus those in the parents of the 3 × 3 incomplete diallele set. The heterotic responses of GA contents in the F1 hybrids are shown in the heat map for each GA species. Red and blue colors indicate higher and lower heterotic responses, respectively. The color scale is shown at the bottom. The value in each block is the ratio of the GA content in the F1 hybrid relative to the average of its two parents (H-MP). Other details are the same as in Figure 3. Asterisks indicate that the difference among the F1 hybrid and its parents was significant (* P < 0.05, ** P < 0.01).
In the shoots of 20-DAS seedlings, the contents of GA53, GA44, GA19, and GA1 were significantly (P < 0.05) different among the nine crosses and the six parents. However, in most cases (each of the 10 GA species in the nine crossing combinations), the differences in GA contents between the hybrids and the parents were not significant (Fig. 4), and the values of the hybrids were close to the means of the parents. Even in some rare cases where GA contents between the hybrids and the parents were significantly different (Fig. 4), for example, GA53 in the cross of Jin 23 × Minghui 63, with a hybrid-parent (H-MP) ratio of 0.99 (Fig. 4), the contents in the hybrids were still close to the means of the parents, reflected by the H-MP ratios being about 1.0, which implies the additive effect of the parental genotypes. One exception appeared for the contents of GA29 in the cross of Zhenshan 97 × Mianhui 725, where GA29 in the F1 hybrid (1.08 ± 0.22 ng g−1 dry weight) was significantly (P < 0.05) higher than that in both parents (Figs. 3 and 4). The concentrations of GA53, GA44, GA1, and GA8 in this tissue displayed significant positive correlations (P < 0.05) with the shoot dry mass, with correlation coefficients of 0.581, 0.661, 0.567, and 0.523, respectively (Supplemental Table S3). This is consistent with recent reports that bioactive GAs are responsible for elongating shoots and biomass production (Eriksson et al., 2000; Biemelt et al., 2004). There is a positive relationship between the contents of bioactive GA1 and GA8, its physiologically inactive catabolic product (r = 0.773, P < 0.01), reflecting the normal metabolism from GA1 to GA8 (Fig. 1).
In the shoot plus root axis of 4-d-old germinating seeds, except for GA53 and GA44, highly significant differences (P < 0.05) in GA contents were observed among the nine crosses and the six parents. In 36 (40%) of 90 cases (10 GA species in nine crossing combinations), GA levels in the hybrids showed significant differences (P < 0.05) compared with their parents, and in 20 (22%) cases, the hybrids had significantly (P < 0.05) higher GA levels relative to the high-value parents (Fig. 4). There was only one case where GA1 content in an F1 hybrid, Maxie × Mianhui 725 (1.24 ± 0.53 ng g−1 dry weight), was significantly (P < 0.05) lower than both parents (Figs. 3 and 4). We also found that while the levels of bioactive GA1 in the F1 hybrids were similar to the low-value parents (Fig. 3), the levels of GA8, the deactivated form of GA1 (Fig. 1), in the hybrids were close to the high-value parents (Fig. 3) and the levels of the bioactive GA4 in the hybrids were much higher than both parents (Fig. 4). As observed for the 20-DAS seedlings, correlations between GA contents and axis dry mass were also found (Supplemental Table S3). The level of GA29, a deactivated product (catalyzed by GA2ox) of GA20 (Fig. 1), which is the immediate precursor for bioactive GA1, exhibited a significant positive correlation (r = 0.9, P < 0.01) with the axis dry mass, whereas in contrast, the content of bioactive GA1 showed a significant negative correlation (r = −0.554, P < 0.05), implying that bioactive GA1 plays a negative role in the regulation of dry mass production in the germinating seeds. It is reported that GA action suppresses many facets of photomorphogenesis (Alabadí et al., 2004); therefore, suppression of GA accumulation in specific tissues would be necessary to allow the photomorphogenic development of young seedlings. This may explain the negative correlation between GA1 level and axis dry mass in the tissues of the 4-d-old germinating seeds examined here, likely achieved by phytochrome-mediated induction of GA2ox expression in response to light (Zhao et al., 2007).
Expression Levels of GA Metabolism and Signaling Genes
GA metabolism, including the biosynthesis of bioactive GAs from transgeranylgeranyl diphosphate and their deactivation, can be divided into three stages: (1) the formation of ent-kaurene; (2) the conversion of ent-kaurene to GA12; and (3) the formation and deactivation of bioactive GAs (Fig. 1; Hedden and Phillips, 2000; Yamaguchi, 2008). Eight types of enzymes are required for this process (Fig. 1), namely CPS, KS, KO, KAO, GA20ox, GA3ox, GA2ox, and CYP714D1 (EUI; Yamaguchi, 2008). Once bioactive GAs are synthesized, they are perceived by a soluble GA receptor, GID1 (Ueguchi-Tanaka et al., 2005), which, upon binding with GA, becomes capable of interacting with the DELLA protein SLENDER RICE1 (SLR1 in rice), a negative regulator of GA response (Ikeda et al., 2001). GID2, an F-box protein (Sasaki et al., 2003), forms an SCFGID2 complex that functions as an SCF E3 ubiquitin ligase targeting SLR1 within the GA-GID1-SLR1 complex for degradation, resulting in the derepression of GA responses (Hirano et al., 2008; Fig. 1). In addition, the rice Dwarf1 (D1) gene, encoding the α-subunit of heterotrimeric G protein, is thought to function as a positive regulator of GA signaling (Ueguchi-Tanaka et al., 2000; Fig. 1). In rice, the enzymes that catalyze the early steps of GA biosynthesis, CPS, KS, KO, and KAO, are encoded by single genes, while those that catalyze the late steps in the pathway, including GA20ox, GA3ox, and GA2ox, are encoded by multigene families, having four, two, and 10 gene members, respectively (Sakamoto et al., 2004; Lo et al., 2008). As far as the GA signaling factors GID1, GID2, SLR1, and D1 are concerned, they are encoded by single genes.
To reveal the expression profiles of GA metabolism and signaling genes in the F1 hybrids and their parents in the 3 × 3 incomplete diallele set, total RNAs from growing shoots of 20-DAS seedlings were extracted and reverse transcribed for real-time reverse transcription (RT)-PCR analysis. SEMIDWARF1 (SD1) encoding GA20ox2, also known as the “Green Revolution gene” in rice (Ashikari et al., 2002), has several naturally occurring mutants, one of which, designated as sd1-1, caused by a 383-bp deletion from the genome, has been widely used in rice breeding programs (Sasaki et al., 2002). Through PCR amplification using SD1-specific primers encompassing the 383-bp deletion area, it was confirmed that the sd1-1 allele was widespread in the parental lines (including Ce 64, Minghui 63, and Mianhui 725) of the incomplete diallele set used in this study. Furthermore, in the RT-PCR analyses using three pairs of specific primers located within the 5′, middle, and 3′ regions of SD1 transcript, transcription could be only detected by the primer pairs from the 3′ region in the samples tested (data not shown). Therefore, OsGA20ox2 was excluded from the analysis. Meanwhile, although 10 OsGA2oxs have been identified in the rice genome (OsGA2ox1–OsGA2ox10, with OsGA2ox10 being a pseudogene; Lo et al., 2008), only OsGA2ox1 to OsGA2ox6 are included in the analysis. In addition, because the transcripts of four genes, OsGA20ox3, OsGA3ox1, OsGA2ox2, and OsEUI, could not be detected in seedling samples (data not shown), they were also excluded from the real-time RT-PCR analysis. Thus, the expression of a total of 16 GA-related genes was eventually assayed by real-time quantitative RT-PCR (Supplemental Table S1).
For the comparative threshold cycle (CT) method of relative quantitation to be valid, the efficiency of the target amplification and that of the reference (endogenous control) must be approximately equal. To check if the assays for the 16 GA-related genes fulfilled this criterion, two genes, OsSLR1 and OsCPS1, were chosen randomly for a validation experiment as described in “Materials and Methods.” The absolute values of the slope for these two genes were 0.028 and 0.090, respectively, which passed this test because both were less than 0.1, indicating that the comparative CT method is valid for our assays.
Except for OsGA3ox2, significant differences in gene expression levels among the 15 genotypes from the 3 × 3 incomplete diallele set (Table III) were detected with P < 0.01 (P < 0.05 for OsGA2ox4). To determine differentially expressed genes between inbred parents or between parental lines and their F1 hybrids, one-way ANOVA of quantitative RT-PCR data within each triad of the nine crosses was carried out for each of the 16 genes. Thirteen out of the 16 (81%) genes were identified as differentially expressed at the level of P < 0.05 in at least one of the nine hybrid-parent triads (Table III). OsKO2, OsGA3ox2, and OsSLR1 did not display significant differential expression in any of the nine crosses investigated. The modes of gene action for the 13 genes identified as differentially expressed were further investigated in individual crossing combinations (Table III). Differential expression was identified (P < 0.05) in 46 (32%) of the 144 cases, in which only two cases showed an expression pattern that was not distinguishable from additivity, while 95% (44 of 46) exhibited nonadditive expression patterns (Table III). The nonadditive differential expression in 44 cases was further classified into four distinct modes based on multiple comparisons (P < 0.05): 28 exhibited overdominance, 14 exhibited positive dominance, and two exhibited negative dominance, while no underdominance was detected. Thus, the differential expression of these genes in one or more of the nine crosses is usually characterized by up-regulation in the hybrid relative to the mean of the two parents showing predominantly overdominant gene action, which is consistent with a recent study of microarray analysis of rice hybrids (Zhang et al., 2008) showing that genes involved in biosynthesis of the hormones exhibited a strong overdominance/underdominance mode of gene action. When we looked at the expression patterns of the 13 differentially regulated genes in the nine F1 hybrids, it was demonstrated that a variety of modes of gene action could be seen simultaneously for one given gene in different crosses (Table III). For example, four distinct modes of gene action, negative dominance, additivity, positive dominance, and overdominance, could be detected for OsGA2ox5 in eight of the nine crosses analyzed. Three kinds of modes, negative dominance, additivity, and positive dominance, appeared for OsGA2ox3, while two modes, positive dominance and overdominance, were identified at the same time for OsKAO, OsGA20ox1, and OsGA20ox4. These findings are consistent with a recent study in maize (Swanson-Wagner et al., 2006) that supports the involvement of multiple modes of gene action in association with heterosis, and they also imply that the mode of gene action is regulated by genetic background. On the other hand, the individual hybrid-parent triad having the largest number of differentially transcribed genes is from the cross Jin 23 × Mianhui 725, with 12 genes expressed predominantly in overdominance in the F1 hybrid, followed by Maxie × Mianhui 725 and Zhenshan 97 × Minghui 63, having nine and eight differentially expressed genes, respectively. For the cross Jin 23 × Ce 64, only one gene, OsGA2ox3, was differentially regulated with negative dominance in the F1 hybrid.
Table III. Heterotic effect of expression levels of GA-related genes in the F1 hybrids of the 3 × 3 incomplete diallele set in the shoots of 20-DAS rice seedlings.
Each value represents a ratio of fold change between the F1 and midparent value of gene expression level.
| Gene Name | InDi_07a | InDi_08 | InDi_09 | InDi_10 | InDi_11 | InDi_12 | InDi_13 | InDi_14 | InDi_15 |
| OsCPS1**b | 0.929c | 0.926 | 1.091 | 1.365 | 1.318 | 1.582 | 1.448 | 1.478 | 1.481 |
| F > M≈Pc | F > M≈P | ||||||||
| OsKS1** | 0.794 | 0.840 | 1.138 | 1.240 | 1.227 | 1.550 | 1.879 | 1.778 | 1.425 |
| F > M≈P | F > M≈P | F > M≈P | |||||||
| OsKO2** | 0.921 | 0.909 | 1.050 | 1.400 | 1.082 | 1.276 | 1.375 | 1.490 | 1.292 |
| OsKAO** | 1.037 | 0.925 | 1.152 | 1.241 | 1.426 | 1.506 | 2.211 | 1.631 | 1.434 |
| F > M≈P | F > M≈P | F > M≈P | F > M≈P | F≈M > P | |||||
| OsGA20ox1** | 0.869 | 1.035 | 1.096 | 1.183 | 1.105 | 1.076 | 1.276 | 1.468 | 1.229 |
| F≈M > P | F≈M > P | F≈M > P | F > M > P | F≈M > P | |||||
| OsGA20ox4** | 1.243 | 1.322 | 1.245 | 1.451 | 1.794 | 1.305 | 2.010 | 2.102 | 1.596 |
| F≈P > M | F > M≈P | F > M≈P | F > M≈P | F≈M > P | |||||
| OsGA3ox2 | 0.770 | 0.969 | 1.099 | 1.354 | 0.959 | 1.338 | 1.125 | 1.686 | 1.057 |
| OsGA2ox1** | 0.898 | 0.870 | 1.008 | 1.341 | 1.038 | 1.217 | 1.496 | 1.220 | 1.294 |
| F≈P > M | |||||||||
| OsGA2ox3** | 0.755 | 0.983 | 1.097 | 1.132 | 1.248 | 1.445 | 1.413 | 1.509 | 1.535 |
| F≈P < M | F≈MP(M > P) | F≈M > P | |||||||
| OsGA2ox4* | 0.953 | 1.061 | 1.228 | 1.109 | 1.297 | 2.153 | 1.906 | 1.928 | 2.142 |
| F > M≈P | F > M≈P | F > M≈P | |||||||
| OsGA2ox5** | 1.228 | 0.818 | 1.489 | 1.145 | 0.983 | 1.292 | 2.710 | 1.681 | 1.408 |
| F≈P < M | F≈M > P | F≈M > P | F≈MP(M > P) | F≈M > P | F > M > P | F≈M > P | F≈M > P | ||
| OsGA2ox6** | 0.825 | 0.787 | 1.286 | 1.697 | 1.134 | 2.232 | 2.672 | 2.247 | 2.007 |
| F > M≈P | F > M≈P | F > M≈P | F > M≈P | ||||||
| OsGID1** | 0.854 | 0.893 | 0.971 | 1.270 | 1.112 | 1.178 | 1.507 | 1.455 | 1.192 |
| F > M≈P | |||||||||
| OsGID2** | 0.822 | 0.915 | 1.059 | 1.292 | 1.188 | 1.315 | 1.690 | 1.706 | 1.198 |
| F > M≈P | F > M≈P | ||||||||
| OsSLR1** | 0.870 | 0.967 | 1.008 | 1.219 | 0.931 | 1.022 | 1.390 | 1.389 | 1.224 |
| OsD1** | 0.884 | 1.009 | 0.941 | 1.232 | 1.196 | 1.477 | 1.690 | 1.660 | 1.385 |
| F > M≈P | F > M≈P | F > M≈P | F > M≈P |
See Table I for the identity of each genotype.
Asterisks indicate genes whose expression levels exhibited significant differences among the 15 genotypes (* P < 0.05, ** P < 0.01). For each gene, comparisons of relative gene expression levels among the three genotypes from each of the nine hybrid-parent triads were carried out by one-way ANOVA, and where significant differential gene expression (P < 0.05) was identified, the detail mode of gene action is shown.
< and > indicate significantly (P < 0.05) larger or smaller, while ≈ indicates no significant difference. F, F1 hybrid; M, female parent; P, male parent; F > M≈P, overdominance; F≈M > P, positive dominance; F≈M > P, additivity; F≈P < M, negative dominance.
Additionally, the ΔCT values from real-time RT-PCR analyses produced by normalizing the respective CT value of each gene to that of the endogenous reference, OsActin1, can be used to compare the expression levels between the genes analyzed, although this sort of comparison may not, in a strict sense, be completely accurate. We used it merely to reflect the relative differences in expression levels among the genes when necessary. In rice seedlings of 20 DAS, the GA-related genes analyzed in this study showed varying expression levels. Among the genes in the early steps of GA biosynthesis (Fig. 1), namely OsCPS1, OsKS1, OsKO2, and OsKAO, the expression level of OsKS1 was higher than other genes, which is in agreement with a study in Arabidopsis (Yamaguchi et al., 1998) showing that KS expression in Arabidopsis is at a much higher level than that of CPS. Between the two members of OsGA20ox examined in this study, OsGA20ox4 exhibited a higher expression level than OsGA20ox1. For the five OsGA2oxs, OsGA2ox1 and OsGA2ox6 were expressed at similar levels but higher than other gene members. Interestingly, the transcript abundance for three GA signaling genes (Fig. 1), OsGID1, OsGID2, and OsD1, was much higher than GA metabolism genes, with OsD1 showing the highest expression level among all the genes. It is thought that GAs are present in most vegetative and floral tissues at low concentrations (0.1–100.0 ng g−1 fresh weight) and that their biosynthetic enzymes are similarly low in abundance (Hedden and Phillips, 2000).
Relationship of GA Contents and Expression of GA-Related Genes with Hybrid Performance and Heterosis of Rice Seedlings
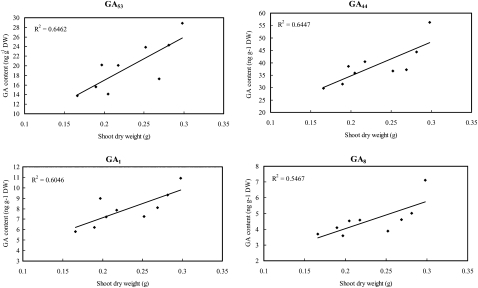
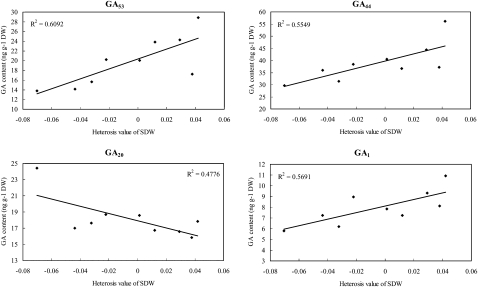
In the growing shoots of rice seedlings at 20 DAS, a significant positive correlation between the shoot dry weight and the content of each of four GA species, GA53, GA44, GA1, and GA8, was detected, with correlation coefficients of 0.804 (P < 0.01), 0.803 (P < 0.01), 0.778 (P < 0.05), and 0.739 (P < 0.05), respectively (Fig. 5; Supplemental Table S4). Similarly, significant positive correlations (P < 0.05) were also identified between heterosis of shoot dry mass and the contents of GAs, including GA53 (r = 0.781), GA44 (r = 0.745), and GA1 (r = 0.754), whereas a significant negative correlation was detected between heterosis and GA20 (r = −0.691), the immediate precursor of bioactive GA1 (Fig. 6; Supplemental Table S4). Besides shoot dry mass, positive correlations of heterosis for tiller number of 20-DAS seedlings with the contents of GAs (Supplemental Table S4) were significant at P < 0.05 for GA53 (r = 0.676), GA44 (r = 0.672), and GA8 (r = 0.684). We also calculated the correlation of GA contents in the seedlings at 20 DAS with seedling traits at 30 DAS observed in the paddy field. Only GA4 was found to be significantly correlated with both shoot dry weight (r = 0.809, P < 0.01) and heterosis of shoot dry weight (r = 0.716, P < 0.05). It should be noted that the above results and interpretation may be limited by the difference in planting conditions of the seedlings used for GA measurements in the growth chamber and those for morphological traits in the paddy field.
Figure 5.
Correlation between shoot dry weight (DW) and GA content of rice seedlings at 20 DAS. The GAs whose concentrations displayed significant correlations (P < 0.05) with shoot dry mass of 20-DAS seedlings are shown. The determination coefficient (R2) is shown for each plot.
Figure 6.
Correlation between heterosis of shoot dry weight (SDW) and GA content of rice seedlings at 20 DAS. The GAs whose concentrations exhibited significant correlations (P < 0.05) with the heterosis of shoot dry mass of 20-DAS seedlings are shown. Absolute MPH as described in “Materials and Methods” was used for correlation analyses. The determination coefficient (R2) is shown for each plot.
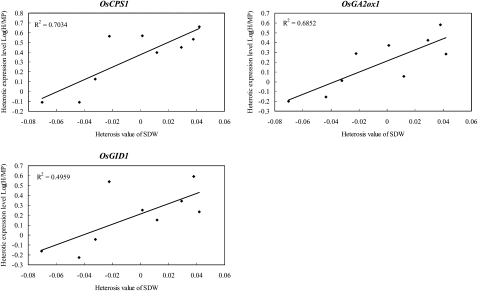
Similar results were obtained from gene expression analysis. Among the GA metabolic genes examined, the expression levels of OsCPS1 (r = 0.791) and OsGA2ox6 (r = 0.720) exhibited significant positive correlations (P < 0.05) with the heterosis of shoot dry mass, and those of OsCPS1 (r = 0.670) and OsKAO (r = 0.832) displayed significant positive correlations (P < 0.05) with the heterosis for tiller number (Supplemental Table S5). Intriguingly, significant positive correlations (P < 0.05) were frequently observed between the heterosis of shoot dry mass and the levels of heterotic expression indicated by the ratios of expression levels of hybrids relative to those for their parents (OsCPS1 [r = 0.839], OsKS1 [r = 0.718], OsKO2 [r = 0.707], OsKAO [r = 0.742], OsGA2ox1 [r = 0.828], and OsGA2ox6 [r = 0.757]; Fig. 7; Supplemental Table S6). Moreover, positive regulators in GA signaling, namely OsGID1, OsGID2, and OsD1, also exhibited significant positive correlations (P < 0.05) with the heterosis for shoot dry mass (Supplemental Tables S5 and S6).
Figure 7.
Correlations between heterosis of shoot dry mass and level of heterotic expression of GA-related genes. Three genes whose heterotic expression levels demonstrated significant correlations (P < 0.05) with the heterosis of shoot dry mass for 20-DAS seedlings are shown. They are OsCPS1, one of the genes involved in early GA biosynthesis, OsGA2ox1 from the GA2ox gene family, and OsGID1, one of the positive regulators of GA signaling. The ratio between the expression level of the F1 hybrid and the mean of the corresponding parental lines (H-MP) was log2 transformed and used in correlation analyses. The x axis is the absolute MPH, as described in “Materials and Methods,” for shoot dry weight (SDW) of 20-DAS seedlings, and the y axis is the level of heterotic gene expression in the F1 hybrid relative to the parents as log2 (H-MP). The determination coefficient (R2) is shown for each plot.
In the shoot plus root axis of 4-d-old germinating seeds, a positive correlation was observed between the axis dry mass and the contents of GA29 (r = 0.696, P < 0.05), while a significant negative correlation was found between the axis dry mass and GA19 (r = −0.666, P < 0.05; Supplemental Table S4). GA19 is an intermediate in the 20-oxidation steps (Fig. 1) that is often accumulated to a high level compared with other GA precursors during GA biosynthesis in rice (Kobayashi et al., 1988), while GA29 is the catabolic product via 2-oxidation of GA20 (Fig. 1), the immediate precursor for bioactive GA1. The same trends (Supplemental Table S4) were also found in terms of the GA19 content versus the heterosis of axis dry mass (r = −0.673, P < 0.05) and the heterotic content for GA29 versus the heterosis of axis dry mass (r = 0.779, P < 0.05). These results indicated that GAs may function as a negative regulator for heterosis regarding the development of germinated seeds of rice.
DISCUSSION
A comprehensive understanding of the biological mechanism for heterosis will clearly benefit from knowledge at three levels: genetic, molecular, and physiological. In rice, although many studies have been performed to contribute to a better understanding of the genetic and molecular bases of heterosis (Stuber et al., 1992; Xiao et al., 1995; Hua et al., 2003; Bao et al., 2005; Huang et al., 2006; Zhang et al., 2008; Wei et al., 2009; He et al., 2010; Song et al., 2010), little effort has been made toward understanding its physiological basis. Plant hormones play a vital role in the physiological regulation of plant growth and development, and it was proposed by Rood et al. (1988) that GAs provide a phytohormonal basis for heterosis in maize. The study presented here is designed to examine the hormonal basis for heterosis of rice through an in-depth investigation of the physiological role of GAs in the regulation of heterosis for seedling growth. For this purpose, we collected and analyzed data regarding endogenous GA levels together with expression profiles of the genes underlying GA metabolism and signaling in a 3 × 3 incomplete diallele set. It is worthy to note that all the molecules along the GA metabolic pathway (Fig. 1), including not only bioactive GAs but also their precursors and catabolites, are included in the quantification analysis. To our best knowledge, this study provides the most comprehensive coverage of GA molecular species for heterosis analysis in plants.
A systematic survey of heterosis for seedling growth in rice was performed using the 5 × 6 incomplete diallele set from 11 parents comprising elite germplasm for hybrid rice production. Our results (Supplemental Table S2) showed that heterosis for shoot biomass and tiller number could be detected as early as 20 DAS, although, at this stage, the extent of heterosis was not that large (highest MPH of 37%) and even moderate negative heterosis could be detected in a few crosses. With the development of rice seedlings, obvious and large heterosis for the same traits was observed at 30 DAS, with MPH up to 62% in shoot dry mass for Maxie × Minghui 63 and 73% in tiller number for Xieqingzao × Minghui 86. The extent of heterosis identified in our study is comparable to that from the research by Zhang et al. (2008), where the heterosis for shoot dry mass at the four-leaf stage (roughly equivalent to 20 DAS) of Liangyoupei 9, a super hybrid rice variety for commercial production in China, and Nipponbare × 93-11, an interspecific hybrid, was 20% and 68%, respectively, while the heterosis for tillering number at the six-leaf stage (roughly equivalent to 30 DAS) was 32% and 71%, respectively. Our results confirm the widespread occurrence of heterosis at the seedling stage in rice.
In the later steps of GA biosynthesis (Fig. 1), starting from GA12 or GA53, there are two parallel pathways leading to bioactive GAs: the early non-13-hydroxylation pathway and the early 13-hydroxylation pathway. It has been verified that the early 13-hydroxylation pathway prevails in vegetative shoots of rice, while the early non-13-hydroxylation pathway occurs predominantly in anther and floral organs (Choi et al., 1995; Hirano et al., 2008). Initially, we intended to quantify GAs in both pathways and, therefore, included deuterium-labeled GA internal standards for both 13-hydroxylated and non-13-hydroxylated GAs. However, with the exception of GA4 and GA34, the majority of non-13-hydroxylated GAs (even their internal standards) could not be detected. Comparison of GA profiles between 20-DAS seedlings and 4-d-old germinating seeds revealed that 13-hydroxylated GA levels were significantly more abundant in the former (P < 0.01, by t test), especially for two C20 GAs, GA53 and GA44 (Fig. 1), levels of which were very low in 4-d-old germinating seeds (Fig. 3). However, little change was observed on the level of GA20 (Fig. 3), the first C19 GA in the pathway (Fig. 1), between the two stages analyzed, indicating that the activity of GA20ox, which catalyzes the three steps from GA53 to GA20, is similar between the two stages. These results suggest that the much lower levels of GA53 and GA44 in 4-d-old seeds are more likely caused by the reduced activity of early GA biosynthetic steps, namely from transgeranylgeranyl diphosphate to GA53 (Fig. 1), or by elevated rates of catabolism conferred by the class C20 GA2oxs at the point of GA53 (Fig. 1) in 4-d-old germinating seeds.
On the other hand, the contents of GA4 in 4-d-old germinating seeds (Fig. 3) increased significantly in the hybrids relative to their parents, to levels comparable to or even exceeding those of GA1, implying higher flux through the early non-13-hydroxylation pathway in the tissue. In spite of a lack of data for non-13-hydroxylated precursors of GA4 due to technical reasons, it seems likely that the GA metabolism in this tissue is reallocated between the two parallel pathways. Considering the sample preparation process that, in order to mimic the real rice seed germination environment in soil, seeds were germinated under appropriate humidity and temperature in the dark for 3 d prior to harvesting the shoot and root axes under light within 1 h, it is similar to the well-known deetiolation process. It has been shown that the level of bioactive GA1 decreased to trace amounts within 4 h during deetiolation of pea seedlings (Ait-Ali et al., 1999; Gil and García-Martínez, 2000; Symons and Reid, 2003), and the suppression of GA biosynthesis during deetiolation may occur on the inhibition of CPS activity (Prisic and Peters, 2007), which is in agreement with our inference. In addition, we speculate, based on our observation, that the reduction of GA1 content, which is beneficial for photomorphogenesis of etiolated seedlings after irradiation, is accompanied by an increase in the level of GA4 that may act to ensure normal development of biological processes other than those related to the establishment of photomorphogenesis. The observation (Supplemental Table S3) that dry mass accumulation in 4-d-old germinating seeds prepared in this study correlated negatively with GA1 content (r = −0.555, P < 0.05) but positively with GA4 content (r = 0.662, P < 0.01) is in good agreement with this suggestion.
Differential gene expression (Table III) was detected for 13 out of the 16 GA-related genes analyzed in this study in at least one of the nine hybrid-parent triads from the 3 × 3 incomplete diallele set, and a surprisingly high level of nonadditive gene expression was observed, with over half of the cases demonstrating an overdominance mode of gene expression followed by high-parent dominance, indicating that most of the genes differentially expressed in hybrid-parent triads were up-regulated in the F1 hybrid. In previous studies of transcriptomic analyses of heterotic crosses in rice (Zhang et al., 2008; Wei et al., 2009; He et al., 2010), the enrichment of differentially expressed genes in some functional categories classified according to Gene Ontology was clearly observed and was considered to imply the involvement of such functional pathways underlain by the enriched genes in the growth vigor of the hybrid. Hence, the enrichment of differentially transcribed genes in GA metabolism and signaling revealed in our study implies that GAs may be involved in the heterosis of rice seedling growth.
Direct evidence supporting a role for endogenous GAs in the regulation of heterosis in rice comes from our correlation analyses of GA contents and GA-related gene expression with the performance and heterosis of rice seedling growth-related traits. As shown in Figure 6, significant positive correlations between endogenous amounts of several GA species, including bioactive GA1, the predominant effector for vegetative growth in rice, and the performance or heterosis of shoot dry mass were clearly identified. Interestingly, the content of GA8, the C-2 hydroxylated inactive GA acting oppositely to bioactive GA1, also showed positive correlations with the performance (r = 0.739, P < 0.05) and heterosis (r = 0.634, just below the critical value of 0.666) of shoot dry mass. This could be explained as follows: the deactivation reaction from GA1 to GA8 catalyzed by GA2ox (Fig. 1) may not be a rate-limiting step of the GA metabolic pathway in the shoots of 20-DAS seedlings, and GA1 could be converted to GA8 without limitation under the normal growing status of the plants in this study. This explanation is supported by the finding that there is a significant positive correlation between the contents of GA1 and GA8 (r = 0.773, P < 0.01). Conversely, in terms of GA20, the immediate precursor of GA1, its content was negatively correlated with the performance (r = −0.643, just beyond the critical value of −0.666) and heterosis (r = −0.691, P < 0.05) of shoot dry mass. This is because, as the very last step in bioactive GA1 synthesis, 3β-hydroxylation of GA20 to GA1 catalyzed by GA3ox (Fig. 1) may be a committed step for GA biosynthesis; the flow of GA precursors to bioactive GA1 might be strictly controlled at this point in the shoots of 20-DAS seedlings, so that the endogenous concentration of bioactive GA1 could be limited within the proper range necessary for normal seedling shoot growth. In rice, GA3ox is encoded by two genes, OsGA3ox1 and OsGA3ox2. Unlike OsGA3ox2, which is expressed universally in various organs throughout the rice life cycle, the expression of OsGA3ox1 is only detected in the tapetum of the anther and the embryo epithelium (Kaneko et al., 2003). Therefore, the existence of only one transcribed GA3ox gene, OsGA3ox2, in vegetative organs of rice pinpoints the importance of the control of its activity and supports our inference. Although no significant correlations were found between GA contents and tiller number of 20-DAS seedlings, significant positive correlations were observed between the contents of the same set of GA species as those for shoot dry mass and the heterosis of tiller number (data not shown). This difference in the correlations of GA contents with performance in terms of tiller number or the heterosis of the trait, respectively, reinforces our inference pertaining to the role of GAs in association with heterosis for seedling growth.
Additionally, the results from our gene expression analyses demonstrated that there were significant positive correlations between the levels of heterotic expression of GA biosynthetic genes and the heterosis of shoot dry mass (Fig. 7), suggesting the important role of heterotic expression frequently identified for GA-related genes in F1 hybrids caused by nonadditive gene expression in the regulation of heterosis. In agreement with the positive role of GA53 and GA44 in the regulation of heterosis, heterotic expression levels of all four genes involved in early GA biosynthesis, namely OsCPS1, OsKS1, OsKO2, and OsKAO (Fig. 1), exhibited significantly high correlations with the heterosis of shoot dry mass. Meanwhile, such positive relationships were also detected for OsGID1, OsGID2, and OsD1 (Fig. 1), three positive regulators for GA signaling, implying that not only GA metabolism but also GA signaling acts synergistically in the regulation of heterosis for rice seedling growth.
In this study, tiller numbers for young rice seedling were found to exhibit a substantially high degree of heterosis (Table II). Tillering is an agronomic trait of great importance for grain yield in rice. Rice tillers are formed by shoot branching, and this process appears to be regulated by complex interactions of phytohormones, which coordinate factors influenced by genetic, developmental, and environmental signals for axillary meristem activity (McSteen, 2009; Xing and Zhang, 2010). In a simplified model, auxin, strigolactone, and GA inhibit shoot branching, while cytokinin functions to promote it (Lo et al., 2008; McSteen, 2009), although the underlying molecular mechanisms, including the cross talk among these hormones, still remain elusive. Therefore, at least for this trait, investigation of the behavior of the hormones other than GA would be necessary for a better understanding of the hormonal basis for heterosis. In particular, besides its role in the regulation of the number of tillers in rice, cytokinin has also been shown to affect panicle size and spikelet number via its control of meristem activity (Ashikari et al., 2005; Kurakawa et al., 2007). Hence, physiological and molecular analyses of diverse endogenous cytokinins and the related genes are necessary in the future for evaluating their potential role in the regulation of heterosis.
Similar to the temporospatial feature of gene expression pattern, the concentration and distribution of a certain hormone, such as GA, would be specific to the tissue and stage of development under investigation. However, in most studies concerning the measurement of phytohormones, including the one presented here, samples analyzed are actually mixtures of tissues containing different cell types and organelles. Moreover, this situation is exacerbated by the extremely low concentrations of plant hormones, which are usually at the nanomolar level. Therefore, more specialized targeted technologies for phytohormone quantification are needed to precisely reflect the hormonal status within a plant. On the other hand, considering that plant hormones act in concert rather than independently to form a regulatory network governing a process of development, the simultaneous determination of multiple hormone concentrations according to the “omics”-based methodology, integrated with other molecular evidence, like transcriptome analysis of hormone-related genes, would improve our understanding of the phytohormonal basis for heterosis.
In summary, the data presented in this research confirm the widespread occurrence of heterosis at the seedling stage in rice. The combined analyses of fluctuations of endogenous GA contents and expression levels of GA-related genes in the incomplete diallele set reveal that GAs play a regulatory role in heterosis for rice seedling growth at the physiological level. Larger scale investigations of multiple plant hormones in more developmental stages of rice seedling growth are anticipated to confirm and extend these findings.
MATERIALS AND METHODS
Plant Materials
Eleven common indica parental lines widely adopted for commercial hybrid rice (Oryza sativa) production during different time periods in central China were used in this study, including six restorer lines (Ce 64, Mianhui 725, Minghui 63, Minghui 86, Shuhui 881, and 6078) and maintainers of five male-sterile lines (Jin 23, Maxie, Xieqingzao, Zhenshan 97, and II-32). By intermating between these male-sterile and restorer lines (5 × 6), an incomplete diallele set of 30 crosses was obtained, which were previously examined for yield and a number of agronomic traits in replicated field trials (Q. Ma, Y. Zhao, and Q. Zhang, unpublished data). Note that it was the maintainer lines, rather than the male-sterile lines, that were only used here for hybrid production with high efficiency and that were grown for all the analyses in this study.
Survey of Rice Seedling Growth
Dry seeds were soaked in water for 48 h at room temperature (25°C) and then pregerminated in moisture for 24 h at 37°C before sowing to the field. The field experiment was conducted under normal agricultural conditions in June and July at the experimental farm of Huazhong Agricultural University in Wuhan, China. Forty-one genotypes, including 11 parents and 30 F1 hybrids of the incomplete diallele set, were arranged in a randomized complete block design with three replications. Fifteen seedlings per genotype were grown in one row, with four rows in each plot. At 20 and 30 DAS, 10 seedlings in the middle of each row were sampled from each plot and measured for the following traits: seedling height, leaf age, tiller number, leaf length/width, shoot fresh weight, and shoot dry weight. For the measurement of dry weight, shoots of fresh seedlings were first quickly dried for 2 h at 120°C and then kept for 48 h at 80°C in an oven prior to weighing. Relative growth rate (RGR) was calculated as follows: RGR = [ln(M2) − ln(M1)]/(t2 − t1) (Wareing and Phillips, 1981), where M1 and M2 are the shoot dry mass at time points t1 and t2, respectively.
On the basis of the analysis of the performance and heterosis of rice seedling growth using the 5 × 6 incomplete diallele set, three restorer lines and three maintainer lines were chosen from the original 11 parental lines to intermate with each other (3 × 3) to form an incomplete diallele subset of nine crosses, which were employed for all of the following analyses: rice seed germination, GA quantification, and gene expression.
Survey of Rice Seed Germination
Rice seeds were surface sterilized with 2% NaOCl in water for 30 min and then rinsed with sterilized distilled water. The sterilized seeds were soaked in distilled water for 1 d at 37°C. The resulting germinated seeds were plated out (6 × 6 pattern) in sealed petri dishes (90 mm diameter) containing two layers of Whatman No. 1 filter paper moistened with 4.5 mL of distilled water. Dishes were incubated at 30°C for 3 d in darkness. Three independent replicates were set up for the 15 rice lines (nine crosses plus six parents) with 100 to 200 grains for each genotype in each replicate. Shoot plus root axes were dissected from grains, and the separated tissues were plunged directly into liquid N2 and stored at −80°C. After freeze drying, the dry masses of the shoot plus root axes were determined.
GA Quantification
Independent triplicate samples (approximately 0.5 g dry weight) of growing shoots from 20-DAS seedlings and shoot plus root axes from 4-d-old germinating seeds were analyzed for GA content. The samples from 4-d-old germinating seeds were described above. For the samples from 20-DAS seedlings, rice plants were grown in trays in the form of a randomized block design with three replications. Twenty seedlings of each genotype were taken as one sample for each replicate, with four seedlings of each line in each of five trays per block with 60 cells per tray. The experiment was conducted under a controlled environment at Rothamsted Research Institute in the United Kingdom, where plants were grown in a Gallenkamp 228 cabinet (Sanyo) under a photosynthetic photon flux density of approximately 300 μmol photons m−2 s−1 with a 14-h-light (30°C)/8-h-dark (22°C) photoperiod and 70% relative humidity and watered regularly. At 20 DAS, growing shoots were harvested from plants directly into liquid N2 and stored at −80°C prior to freeze drying.
Quantification of GAs was performed as described by Coles et al. (1999) with modifications. Briefly, freeze-dried samples were ground to a fine powder using a ball mill and then extracted by stirring overnight at 4°C in 80% (v/v) methanol-water (100 mL) containing appropriate amounts of [17-2H2]GAs as internal standards and 833 Bq each of the following tritiated GA standards: [1,2-3H2]GA1, [1,2-3H2]GA4, 16,17-dihydro[15,16,17-3H4]GA19, and [1,2,3-3H3]GA20. After filtration, the residue was reextracted with methanol (100 mL) for 2 h and refiltered. The combined methanol extracts were evaporated almost to dryness under reduced pressure. The residue was resuspended in water, adjusted to pH 8.0 (1 m KOH), and purified by QAE Sephadex A-25 (Pharmacia) anion-exchange column and C18 Solid Phase Extraction cartridge (500 mg; Thermo Fisher). The dried GAs were methylated twice with excess diazomethane, dissolved in ethyl acetate (1 mL), and partitioned against water (1 mL). The ethyl acetate phase was passed through a Bond-Elut NH2 cartridge (100 mg; Varian) that had been preconditioned with ethyl acetate (1 mL). The remaining water phase was partitioned twice more against ethyl acetate, with the organic phases being passed through the NH2 cartridge. The pooled ethyl acetate phases were evaporated to dryness in vacuo, and then the GA methyl esters were resolved by reverse-phase HPLC using conditions described previously (Croker et al., 1990). Collected fractions were separately pooled based on the locations of tritiated GAs, and the pooled fractions were analyzed as methyl ester trimethylsilyl ethers on a Thermo Finnigan GCQ mass spectrometer. Samples in N-trimethylsilylfluoroacetamide (10 μL) were diluted with dry ethyl acetate (20 μL) and injected (1 μL) into a TR-1 capillary column (30-m × 0.25-mm i.d. × 0.25-μm film thickness; Thermo Electron) at 50°C. The split valve (50:1) was opened after 2 min, and the temperature was increased at 20°C min−1 to 200°C and then at 4°C min−1 to 300°C. The instrument was operated in selective ion monitoring mode, with the selected ions for each GA and its 2H2-labeled internal standard as described by Croker et al. (1990). Amounts of endogenous GAs in the original extracts were determined from previously established calibration curves of the peak area ratios for unlabeled and deuterated GAs plotted against varying molar ratios of the two compounds. The same stock solutions of labeled GAs were used for production of the calibration curves.
Real-Time Quantitative RT-PCR Analysis
For expression analysis of GA metabolism and signaling genes, the incomplete diallele set of nine crosses plus their six parental lines was arranged in a paddy field in a randomized complete block design with three replications. Growing shoots of eight seedlings of 20 DAS from each line of each replicate were mixed and harvested directly into liquid N2 and stored at −80°C prior to RNA isolation.
Total RNA was extracted from 100 mg of tissue using TRIzol reagent (Invitrogen). First-strand cDNAs were synthesized from DNaseI-treated total RNA using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed on an optical 96-well plate on an ABI PRISM 7500 real-time PCR system (Applied Biosystems). Each reaction contained the cDNA equivalent of 50 ng of total RNA, 12.5 μL of 2× SYBR Green Master Mix reagent (Applied Biosystems), and 200 nm each of the gene-specific primers in a final volume of 25 μL. The thermal cycle was set as follows: 50°C for 2 min and 95°C for 10 min; 45 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 45 s; and finally, 72°C for 8 min. The specific primers for real-time PCR are listed in Supplemental Table S1, which were designed using Primer Express version 2.0 (Applied Biosystems) software on the basis of ClustalW analysis to ensure the specificity of primers among gene families. Reactions were carried out with three technical replicates, and the absence of genomic DNA and primer dimers was confirmed by analysis of RT-minus and water control samples and by examination of dissociation curves. The rice Actin1 gene was used as an endogenous reference to normalize all samples. A mixture of the reverse-transcribed cDNA templates from all of the 15 rice samples was prepared to serve as the common calibrator for all samples, and the same pool of calibrator cDNA was used throughout the study to provide for consistency. The comparative CT method (also known as the 2–ΔΔCT method; Livak and Schmittgen, 2001) was applied to calculate the relative amount of target gene expression, which is presented as the quantity normalized to the endogenous reference OsActin1 and relative to the calibrator.
A validation experiment was carried out according to the user manual from Applied Biosystems to demonstrate that the efficiencies of amplifications of target and reference are approximately equal, which is the prerequisite for a valid ΔΔCT calculation. Briefly, seven serial dilutions of cDNA of the calibrator by 5-fold were amplified by real-time PCR using primers for the target gene and OsActin1. The ΔCT value (CT,target – CT,OsActin1) was calculated for each cDNA dilution. A plot of the log cDNA dilution versus ΔCT was made. A general criterion for passing a validation experiment is that the absolute value of the slope is less than 0.1.
Data Processing and Statistical Analysis
MPH was calculated as MPH = (F1 – MP)/MP in percentage, in which F1 is the performance of the hybrid and MP is the average performance of the two parents. The absolute MPH value of each cross, calculated as H = F1 − (P1 + P2)/2 (where H is the amount of heterosis, F1 is the trait measurement of the hybrid, and P1 and P2 are the measurements of the parents), was used for statistical analyses. For comparisons among the genotypes from the incomplete diallele set and among the F1 hybrid and its parents within a crossing combination, one-way ANOVA and appropriate multiple comparison were used; for pairwise comparisons between two groups of data, two-sided t tests were used. Pearson correlation coefficients were calculated between pairs of data in question.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Oligonucleotide primers for the GA metabolism and signaling genes subjected to real-time RT-PCR expression analysis.
Supplemental Table S2. Performance and MPH of rice seedling traits at 20 and 30 DAS for the 5 × 6 incomplete diallele set.
Supplemental Table S3. Correlation coefficients between endogenous GA levels and rice seedling traits.
Supplemental Table S4. Correlation coefficients between endogenous GA levels and performance or heterosis of rice seedling traits of F1 hybrids.
Supplemental Table S5. Correlation coefficients between expression levels of GA-related genes and heterosis of rice seedling traits.
Supplemental Table S6. Correlation coefficients between extent of heterotic expression of GA-related genes and heterosis of rice seedling traits.
Acknowledgments
We thank Caiguo Xu for producing and providing the rice seeds and for rice field management. We also thank Drs. John Lenton, Steve Croker, Paul Hopkins, Fan Gong, and Xianghua Li for excellent technical assistance.
References
- Ait-Ali T, Frances S, Weller JL, Reid JB, Kendrick RE, Kamiya Y. (1999) Regulation of gibberellin 20-oxidase and gibberellin 3β-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol 121: 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita S, Blanco L, Katayama K. (1990) Physiological mechanism of heterosis in seedling growth of indica F1 rice hybrids. Jpn J Crop Sci 59: 548–556 [Google Scholar]
- Alabadí D, Gil J, Blázquez MA, García-Martínez JL. (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorf S, Selbig J, Altmann T, Poos K, Witucka-Wall H, Repsilber D. (2010) Enriched partial correlations in genome-wide gene expression profiles of hybrids (A. thaliana): a systems biological approach towards the molecular basis of heterosis. Theor Appl Genet 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sasaki A, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice “Green Revolution.” Breed Sci 52: 143–150 [Google Scholar]
- Bao JY, Lee S, Chen C, Zhang XQ, Zhang Y, Liu SQ, Clark T, Wang J, Cao ML, Yang HM, et al. (2005) Serial analysis of gene expression study of a hybrid rice strain (LYP9) and its parental cultivars. Plant Physiol 138: 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemelt S, Tschiersch H, Sonnewald U. (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol 135: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, Kobayashi M, Fujioka S, Matsuno T, Hirosawa T, Sakurai A. (1995) Fluctuation of endogenous gibberellin levels in the early development of rice. Biosci Biotechnol Biochem 59: 285–288 [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, García-Lepe R, Lewis MJ, Hedden P. (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Croker SJ, Hedden P, Lenton JR, Stoddart JL. (1990) Comparison of gibberellins in normal and slender barley seedlings. Plant Physiol 94: 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. (2004) Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- de Leon JC, Abe T, Sasahara T. (2001) Variations in morpho-physiological traits relating to seedling vigor and heterosis in reciprocal crosses of rice. Breed Sci 51: 57–61 [Google Scholar]
- Eriksson ME, Israelsson M, Olsson O, Moritz T. (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol 18: 784–788 [DOI] [PubMed] [Google Scholar]
- Fleet CM, Sun TP. (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8: 77–85 [DOI] [PubMed] [Google Scholar]
- Garcia AAF, Wang SC, Melchinger AE, Zeng ZB. (2008) Quantitative trait loci mapping and the genetic basis of heterosis in maize and rice. Genetics 180: 1707–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner T, Steinfath M, Andorf S, Lisec J, Meyer RC, Altmann T, Willmitzer L, Selbig J. (2009) Improved heterosis prediction by combining information on DNA- and metabolic markers. PLoS ONE 4: e5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil JGarcía-Martínez JL (2000) Light regulation of gibberellin A(1) content and expression of genes coding for GA 20-oxidase and GA 3-hydroxylase in etiolated pea seedlings. Physiol Plant 180: 223–229 [Google Scholar]
- Guo M, Rupe MA, Yang XF, Crasta O, Zinselmeier C, Smith OS, Bowen B. (2006) Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet 113: 831–845 [DOI] [PubMed] [Google Scholar]
- He GM, Zhu XP, Elling AA, Chen LB, Wang XF, Guo L, Liang MZ, He H, Zhang HY, Chen FF, et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hirano K, Ueguchi-Tanaka M, Matsuoka M. (2008) GID1-mediated gibberellin signaling in plants. Trends Plant Sci 13: 192–199 [DOI] [PubMed] [Google Scholar]
- Hua J, Xing Y, Wu W, Xu C, Sun X, Yu S, Zhang Q. (2003) Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 100: 2574–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua JP, Xing YZ, Xu CG, Sun XL, Yu SB, Zhang Q. (2002) Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162: 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhang L, Zhang J, Yuan D, Xu C, Li X, Zhou D, Wang S, Zhang Q. (2006) Heterosis and polymorphisms of gene expression in an elite rice hybrid as revealed by a microarray analysis of 9198 unique ESTs. Plant Mol Biol 62: 579–591 [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M. (2003) Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J 35: 104–115 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Yamaguchi I, Murofushi N, Ota Y, Takahashi N. (1988) Fluctuation and localization of endogenous gibberellins in rice. Agric Biol Chem 52: 1189–1194 [Google Scholar]
- Korn M, Gärtner T, Erban A, Kopka J, Selbig J, Hincha DK. (2010) Predicting Arabidopsis freezing tolerance and heterosis in freezing tolerance from metabolite composition. Mol Plant 3: 224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kusterer B, Muminovic J, Utz HF, Piepho HP, Barth S, Heckenberger M, Meyer RC, Altmann T, Melchinger AE. (2007) Analysis of a triple testcross design with recombinant inbred lines reveals a significant role of epistasis in heterosis for biomass-related traits in Arabidopsis. Genetics 175: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LZ, Lu KY, Chen ZM, Mu TM, Hu ZL, Li XQ. (2008) Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics 180: 1725–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Luo LJ, Mei HW, Wang DL, Shu QY, Tabien R, Zhong DB, Ying CS, Stansel JW, Khush GS, et al. (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158: 1737–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JAD, Chen LJ, Yu SM. (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LJ, Li ZK, Mei HW, Shu QY, Tabien R, Zhong DB, Ying CS, Stansel JW, Khush GS, Paterson AH. (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158: 1755–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P. (2009) Hormonal regulation of branching in grasses. Plant Physiol 149: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchinger AE, Piepho HP, Utz HF, Muminovic J, Wegenast T, Törjék O, Altmann T, Kusterer B. (2007) Genetic basis of heterosis for growth-related traits in Arabidopsis investigated by testcross progenies of near-isogenic lines reveals a significant role of epistasis. Genetics 177: 1827–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Peters RJ. (2007) Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol 144: 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood SB, Buzzell RI, Mander LN, Pearce D, Pharis RP. (1988) Gibberellins: a phytohormonal basis for heterosis in maize. Science 241: 1216–1218 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Song GS, Zhai HL, Peng YG, Zhang L, Wei G, Chen XY, Xiao YG, Wang LL, Chen YJ, Wu B, et al. (2010) Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol Plant 3: 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM. (2007) Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res 17: 264–275 [DOI] [PubMed] [Google Scholar]
- Stuber CW. (1994) Heterosis in plant breeding. Plant Breed Rev 12: 227–251 [Google Scholar]
- Stuber CW, Lincoln SE, Wolff DW, Helentjaris T, Lander ES. (1992) Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132: 823–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103: 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Reid JB. (2003) Hormone levels and response during de-etiolation in pea. Planta 216: 422–431 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M. (2000) Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97: 11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing PF, Phillips IDJ. (1981) Growth and Differentiation in Plants, Ed 3 Pergamon Press, Oxford [Google Scholar]
- Wei G, Tao Y, Liu GZ, Chen C, Luo RY, Xia HA, Gan Q, Zeng HP, Lu ZK, Han YN, et al. (2009) A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc Natl Acad Sci USA 106: 7695–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Li J, Yuan L, Tanksley SD. (1995) Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YZ, Zhang QF. (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61: 11.1–11.22 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Sun T, Kawaide H, Kamiya Y. (1998) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SB, Li JX, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q, Saghai Maroof MA. (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 94: 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan LP. (1998) Hybrid rice breeding in china. Virmani SS, Siddiq EA, Muralidharan K, , Advances in Hybrid Rice Technology. International Rice Research Institute, Los Banos, Philippines, pp 27–33 [Google Scholar]
- Zhang HY, He H, Chen LB, Li L, Liang MZ, Wang XF, Liu XG, He GM, Chen RS, Ma LG, et al. (2008) A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol Plant 1: 720–731 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Gao YJ, Yang S, Ragab R, Saghai Maroof MA, Li ZB. (1994) A diallele analysis of heterosis in elite hybrid rice based on RFLPs and microsatellites. Theor Appl Genet 89: 185–192 [DOI] [PubMed] [Google Scholar]
- Zhao XY, Yu XH, Liu XM, Lin CT. (2007) Light regulation of gibberellin metabolism in seedling development. J Integr Plant Biol 49: 21–27 [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, et al. (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]