Abstract
Understanding the relationship of the size and shape of an organism to the size, shape, and number of its constituent cells is a basic problem in biology; however, numerous studies indicate that the relationship is complex and often nonintuitive. To investigate this problem, we used a system for the inducible expression of genes involved in the G1/S transition of the plant cell cycle and analyzed the outcome on leaf shape. By combining a careful developmental staging with a quantitative analysis of the temporal and spatial response of cell division pattern and leaf shape to these manipulations, we found that changes in cell division frequency occurred much later than the observed changes in leaf shape. These data indicate that altered cell division frequency cannot be causally involved in the observed change of shape. Rather, a shift to a smaller cell size as a result of the genetic manipulations performed correlated with the formation of a smoother leaf perimeter, i.e. appeared to be the primary cellular driver influencing form. These data are discussed in the context of the relationship of cell division, growth, and leaf size and shape.
Leaf initiation occurs by a process of organogenesis at the shoot apical meristem. As a result, a group of dividing cells is separated from the meristem and undergoes a series of programmed developmental events that leads to the eventual formation of a mature leaf (Fleming, 2005; Tsukaya, 2006; Barkoulas et al., 2007). Significant progress has been made in the identification of the conserved transcriptional and signaling modules involved in the control of leaf form, notably, the process of differential lateral lamina growth, which distinguishes leaves as being, for example, serrated, lobed, or compound (Grigg et al., 2005; Hay and Tsiantis, 2006; Koyama et al., 2007; Blein et al., 2008; Bayer et al., 2009; Larue et al., 2009). These different shapes are defined by differences in the degree and distribution of edge curvature, and an outstanding question in plant developmental biology is how the patterns of transcription factors and signaling modules that regulate margin form (Bilsborough et al., 2011) are actually translated into the differential growth around the leaf perimeter that leads to curvature. An intuitive expectation is that division patterns of the constituent cells of the leaf play a major role in this process.
At leaf initiation, all constituent cells are in a proliferative state. As development proceeds, a controlled termination of cell division occurs in different regions of the leaf at different times. Although there is clearly a stochastic element to this process, for any leaf type, there is a robust overall spatial-temporal pattern of termination/maintenance of cell proliferation (Poethig and Sussex, 1985; Donnelly et al., 1999). The maintenance of cell division provides the building blocks for future growth, whereas termination of cell division is generally associated with a transition to vacuolar-linked cell expansion, the main driver for actual growth. The balance between these two cellular states is thought to have a significant influence on the growth and form of plant organs, including leaves.
A number of lines of evidence do indeed indicate that altered cell division pattern does influence leaf shape. For example, mutants in which leaf morphology is altered often display an altered pattern of division termination (Nath et al., 2003), and an extended phase of cell proliferation has frequently been associated with alterations in leaf size and shape (Mizukami and Fischer, 2000; Autran et al., 2002; Clay and Nelson, 2005). Furthermore, experiments in which genes encoding components of the cell cycle have been misexpressed have often led to altered leaf morphology. For example, overexpression of cyclin-dependent kinase inhibitors led to deeper sinus formation (Wang et al., 2000; De Veylder et al., 2001), and overexpression of a D-type cyclin led to altered leaf size and shape (Dewitte et al., 2003). Such observations support the view that the temporal and spatial pattern of cell division during development is intrinsically linked to the control of leaf morphogenesis. However, this view is complicated by the observation that mutants have been created in which, although overall leaf morphology is not greatly changed from the wild type, there are clear differences in component cell size and shape (Hemerly et al., 1995; Smith et al., 1996; Tsukaya, 2006). Thus, the fundamental issue of the functional relationship between the size and shape of leaf and the size, shape, and number of its constituent cells remains a conundrum.
There are a number of problems in gaining a true appraisal of the situation from the published literature. First, in most of the reported experiments, an accurate, quantitative, spatial, and temporal analysis of the endogenous pattern of cell division during normal development, the changes induced by the manipulations performed, and the precise resultant morphological changes are not provided or are incomplete. This is especially true for the very early stages of organ development when a number of key events determining leaf form occur (Sinha, 1999). Second, many of the genetic manipulations that have been analyzed involve misexpression of gene products that almost certainly are not directly involved in cell division or at least probably influence the expression of a large number of genes, only a subset of which are involved in cell division (Autran et al., 2002; Nath et al., 2003; Palatnik et al., 2003; Dinneny et al., 2004). Thus, although altered cell division patterns occur in these mutants (and it is generally inferred that these changes in cell proliferation lead to the morphological alterations observed), it is possible to argue that these are indirect affects that are not causally related to morphogenesis. Third, much of this work has been performed on constitutive mutants, i.e. target gene expression has been absent or altered throughout the development of the organism. The fact that observations can be made on developing organs at all indicates that the organism has coped with the altered genetic factor during embryogenesis and germination, raising the question of to what extent the plant has accommodated to the loss (or gain) of gene function.
In a previous investigation, we manipulated the expression of cell cycle regulators during tobacco (Nicotiana tabacum) leaf development and showed that local promotion of cell division at an early stage of development led to the nonintuitive outcome of an eventual decrease in lamina growth in that area (Wyrzykowska et al., 2002). However, the technical difficulty of dissecting tobacco leaf primordia and the lack of a system to reliably quantify the outcome on cell division and leaf shape at time points after our manipulations meant that our investigation lacked sufficient resolution to quantify the relationship of cell division pattern and change in leaf shape. To provide a more detailed and quantitative analysis of this relationship, we report here on a system in Arabidopsis (Arabidopsis thaliana) to inducibly express genes whose products are involved in the regulation of the G1/S transition in the plant cell cycle. Our aim was to use these genes as tools to artificially manipulate the system and by observing how the system responded to perturbation, gain an insight into the potential rules relating division pattern and leaf form. Coupled with a careful developmental staging and leaf dissection, this system allowed us to perform a quantitative analysis of the temporal and spatial response of cell division pattern to the manipulation of these genes at different time points in development. Moreover, we coupled these manipulations with a quantitative temporal analysis of shape change during early leaf development. Surprisingly, measurable changes in cell division frequency occurred much later than the measurable change in leaf shape, indicating that altered cell division frequency is not causally involved in this aspect of morphogenesis. Rather, a shift to a smaller cell size as a result of these manipulations correlated with the formation of a smoother leaf perimeter. These data are discussed in relation to the interpretation of experiments in which cell cycle genes are misexpressed and the relationship of the cell cycle, growth, and leaf shape.
RESULTS
Induction of AtCYCLIND3;1 and Repression of AtRBR1 Leads to Leaves with Smaller Constituent Cells But Does Not Initially Lead to an Increase in Cell Division Frequency
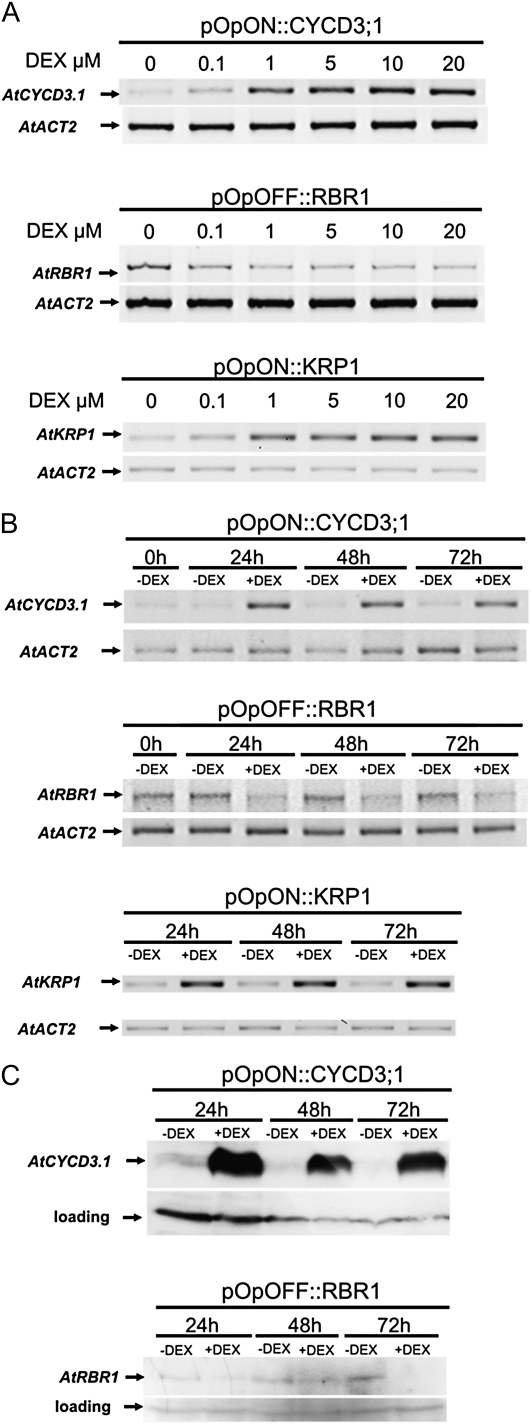
A significant body of data now describes the complex network of genes involved in controlling cell division in plants (De Veylder et al., 2007). As in other eukaryotes, the G1/S phase transition is key to the decision of whether progression through the plant cell cycle occurs, with the cyclin/cyclin-dependent kinase/retinoblastoma protein module being core to this regulatory step. Published data indicate that either overexpression of AtCYCLIND3;1 (AtCYCD3;1) or repression of RETINOBLASTOMA-RELATED1 (AtRBR1) can be used as a tool to promote cell division in Arabidopsis (Dewitte et al., 2003; Wildwater et al., 2005). Therefore, to investigate the relationship of cell division and leaf form in Arabidopsis, we created a series of transgenic plants in which either AtCYCD3;1 expression could be induced or AtRBR1 expression could be repressed via the supply of an exogenous chemical, dexamethasone (DEX; Supplemental Fig. S1). Molecular analysis of transgenic plants generated using these constructs confirmed that following treatment with DEX there was either an accumulation of AtCYCD3;1 mRNA and protein or a decrease in transcript and protein level for AtRBR1 (Fig. 1). These changes were detectable within 24 h of treatment with DEX at a concentration of 10 μm.
Figure 1.
Molecular characterization of pOpON::CYCD3;1, pOpOFF::RBR, and pOpON::KRP1 plants. A, RT-PCR analysis of transgenic plants (pOpON::CYCD3;1, pOpOFF::RBR, and pOpON::KRP1) treated for 48 h with various concentrations of DEX. Gene-specific primers were used to amplify AtCYCD3;1, AtRBR1, or AtKRP1 with amplification of an actin gene (AtACT2) being used as an internal control. B, RT-PCR analysis of transgenic plants (pOpON::CYCD3;1, pOpOFF::RBR, and pOpON::KRP1) treated with (+DEX) or without (−DEX) 10 μm DEX for different time periods prior to RNA extraction. Induction was started at day 12, and analysis was performed at 24, 48, or 72 h using gene-specific primers to amplify AtCYCD3;1, AtRBR1, AtKRP1, or AtACT2 as indicated. C, Western-blot analysis of transgenic plants (pOpON::CYCD3;1 or pOpOFF::RBR) treated with (+DEX) or without (−DEX) 10 μm DEX for different time periods (as in B) prior to protein extraction. Blots were incubated with an antibody raised against either AtCYCD3;1 or AtRBR (as indicated) prior to visualization of signal. A loading control to indicate total protein amount in each sample is shown.
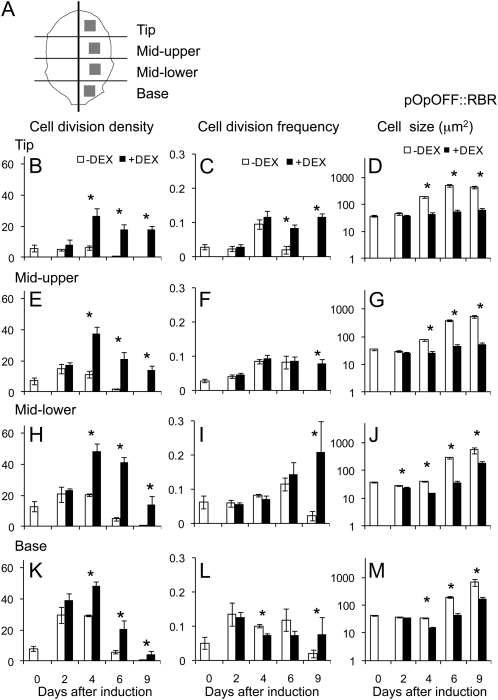
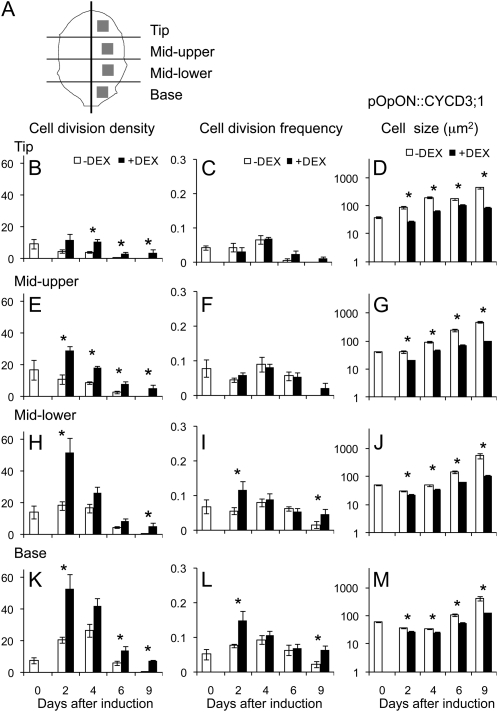
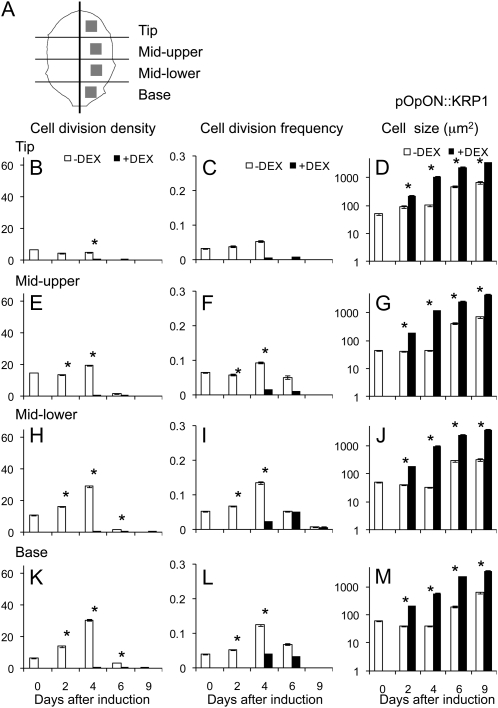
We set out to use these lines as tools to manipulate cell division pattern during specific phases of leaf growth. During early development, Arabidopsis leaves undergo a controlled change in form as well as increasing in size (Supplemental Fig. S2). The precise dynamics of these changes depend on the developmental stage of the plant, and in the experiments reported here, we focused on leaf number 6, thus allowing a comparison of leaves of similar developmental stage in different experiments. Preliminary investigation indicated that at 10 d after sowing (DAS), leaf 6, which was approximately 200 μm long, displayed a simple form and that cell divisions were present in all regions of the leaf. When pOpON::CYCD3;1 or pOpOFF::RBR plants were induced with DEX from 10 DAS and observed 9 d later, altered cell division patterns were observed in leaf 6 (Supplemental Fig. S3, A–D), with smaller epidermal cells resulting from either induced overexpression of AtCYCD3;1 or repression of AtRBR (as expected based on previous publications; Dewitte et al., 2003; Desvoyes et al., 2006). To quantify these changes, we used a fluorescent staining technique (Supplemental Fig. S4; Kuwabara and Nagata, 2006) to visualize and count the distribution of new cell plates in leaves at different times after DEX induction. Cell divisions were recorded in four regions of the leaves along the proximal-distal axis (tip, mid-upper, mid-lower, and base) and at different times after induction at 10 DAS for both pOpOFF::RBR (Fig. 2) and pOpON::CYCD3;1 plants (Fig. 3). The data were initially calculated either on an area basis (number of cell divisions/leaf area; termed cell division density) or a per cell basis (number of cell divisions/number of cells; termed cell division frequency).
Figure 2.
Spatial-temporal changes in cell division density, frequency, and size in the epidermis after induction of leaf 6 of pOpOFF::RBR1 plants. A, Schematic showing the regions of leaf 6 from which cell division data were collected. Cell division density (divisions/area; B, E, H, and K), cell division frequency (divisions/number of cells; C, F, I, and L), and cell size (D, G, J, and M) were calculated in the epidermis of pOpOFF::RBR1 leaves either induced (+DEX, black bars) or not induced (−DEX, white bars) from day 10. Data were analyzed in regions of the tip (B–D), mid-upper (E–G), mid-lower (H–J), and base (K–M) of the leaf at different time points (days after induction). Mean values with se are shown (n > 30 from at least five independent leaves for each data point). Pairwise comparisons showing significant difference at a confidence limit of at least 0.05 are indicated with an asterisk.
Figure 3.
Spatial-temporal changes in cell division density, frequency, and size in the epidermis after induction of leaf 6 of pOpON::CYCD3;1 plants. A, Schematic showing the regions of leaf 6 from which cell division data were collected. Cell division density (divisions/area; B, E, H, and K), cell division frequency (divisions/number of cells; C, F, I, and L), and cell size (D, G, J, and M) were calculated in the epidermis of pOpON::CYCD3;1 leaves either induced (+DEX, black bars) or not induced (−DEX, white bars) from day 10. Data were analyzed in regions of the tip (B–D), mid-upper (E–G), mid-lower (H–J), and base (K–M) of the leaf at different time points (days after induction). Mean values with se are shown (n > 30 from at least five independent leaves for each data point). Pairwise comparisons showing significant difference at a confidence limit of at least 0.05 are indicated with an asterisk.
Considering the cell division density data for the pOpOFF::RBR lines, a significant (P < 0.05) increase occurred within 4 d of induction in all parts of the leaf (Fig. 2, B, E, H, and K). This increase was maintained at all time points so that even 9 d after induction an increase in cell division density was detectable in the induced leaves. However, when these data were expressed as cell division frequency, no significant increase was calculated for any region of the pOpOFF::RBR leaves until 6 d after induction, with a consistent increase in all regions only being observed 9 d after induction (Fig. 2, C, F, I, and L). An analysis of the data for the pOpON::CYCD3;1 lines indicated a similar result. When expressed as cell division density, there was a significant (P < 0.05) increase in most regions of the leaf within 2 d of induction and cell division was maintained at time points when cell division was no longer detectable in noninduced leaves, 9 d after induction (Fig. 3, B, E, H, and K). However, when expressed as cell division frequency, although a transient increase was observed in the lower part of the leaf 2 d after induction, a consistent and significant increase in all regions of the leaf was calculated only at 9 d after induction (Fig. 3, C, F, I, and L).
An increase in cell divisions/area with limited or no increase in cell division frequency indicates that mean cell size is altered. Measurement of cell size in the different regions of the pOpOFF::RBR and pOpON::CYCD3;1 leaves either with or without induction supported this hypothesis. At all time points after induction, and in all regions, there was a tendency for mean cell size to be smaller in the induced tissue compared to the noninduced tissue. By 14 DAS (4 d after induction), this difference in cell size was significant (P < 0.05) in all regions of the leaf in both pOpOFF::RBR (Fig. 2, D, G, J, and M) and pOpON::CYCD3;1 lines (Fig. 3, D, G, J, and M), with the pOpON::CYCD3;1 leaves already showing a significant decrease in cell size 2 d after induction. The shifts in cell size distribution underpinning these differences in mean cell size values are shown in Supplemental Figure S5.
Change of Leaf Shape in Response to Increased AtCYCD3;1 Expression or Repression of AtRBR Occurs before an Increase in Cell Division Frequency But Coincides with a Decrease in Cell Size
The induction of AtCYCD3;1 and the repression of AtRBR1 led to a change in final leaf shape. To investigate this alteration in morphology, we analyzed leaf shape at various time points after induction, but it was very difficult to judge by simple visual inspection exactly when this divergence of shape occurred relative to controls (Fig. 4). To facilitate a comparison of the timing and extent of shape change with the timing and extent of the altered cell division patterns described above, we performed a quantitative analysis of leaf shape change during development. To do this, we calculated the bending energy (BE) of the leaf perimeter for leaves at different points after induction (Backhaus et al., 2010). This parameter is essentially an integrated value of the square of a contour’s curvature along the perimeter that allows scale-independent quantitative analysis and comparison of the contour curvature of any shape (Supplemental Fig. S6). In addition, since BE can be calculated for any portion of a perimeter, it allows the description and comparison of curvature in defined regions around the leaf shape.
Figure 4.
Spatial-temporal changes in leaf shape after induction of cell cycle genes. Silhouettes of leaf 6 from pOpON::CYCD3;1 (A), pOpOFF::RBR (B), and pOpON::KRP1 (C) plants during normal growth (−DEX) or at time points after induction (+DEX). Silhouettes have been normalized for size to facilitate shape comparison.
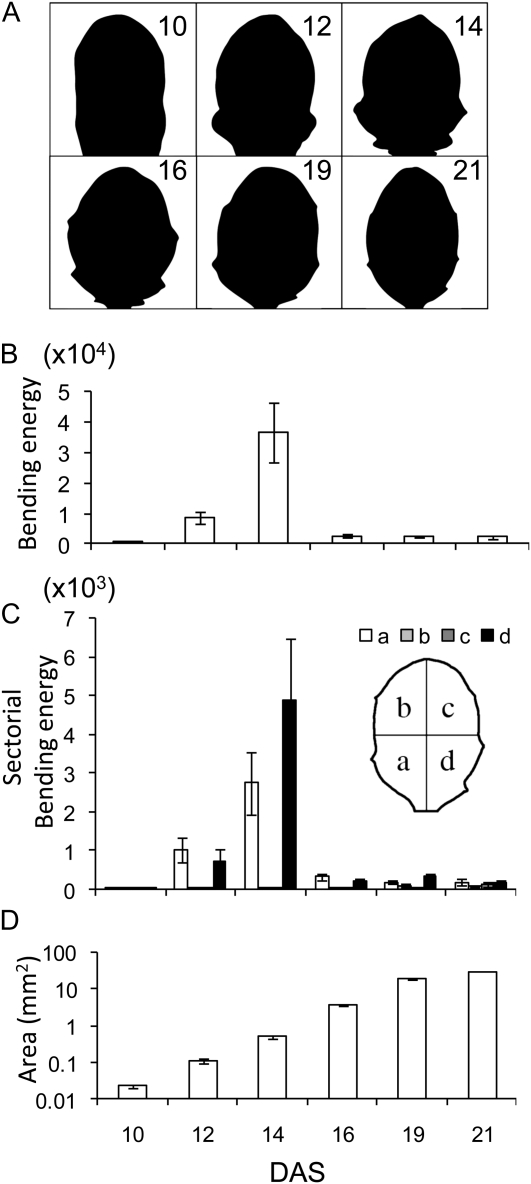
Considering first of all wild-type leaves, the earliest stages of noninduced leaves (10–12 DAS; Fig. 5A) have relatively low values of total BE (Fig. 5B). BE then increases to a maximum at 14 DAS, before gradually decreasing by 19 to 21 DAS. This progression of BE values indicates a leaf perimeter that is initially smooth, becomes less smooth, then becomes again relatively smooth, as validated by visual inspection of the images in Figure 5A and Supplemental Figure S2. A sectoral analysis of the distribution of BE around the leaf perimeter shows that the higher total BE value at 14 DAS is mainly due to a relative increase in BE in the more proximal regions of the leaf (Fig. 5C), fitting with the observed serration pattern of the wild-type leaves (Fig. 5A). This progression in shape change is accompanied by an increase in leaf area (Fig. 5D).
Figure 5.
Analysis of shape change during Arabidopsis leaf development. A, Outlines of representative Arabidopsis leaf number 6 at different stages (10–21 DAS). The images have been normalized for size to facilitate shape comparison. B, BE values for the entire leaf contours of the leaf stages shown in A. Mean values are shown with se (n = 5). BE values at 12 and 14 DAS are significantly different from each other and the values at other stages of leaf development at the 0.05 confidence limit. C, Sectoral BE values for the leaf stages shown in A. Each leaf was split into four sectors (a–d, as shown in the schematic inlay). Mean values with se (n = 5). D, Change of leaf area during the developmental progression shown in A. Mean values with se (n = 5).
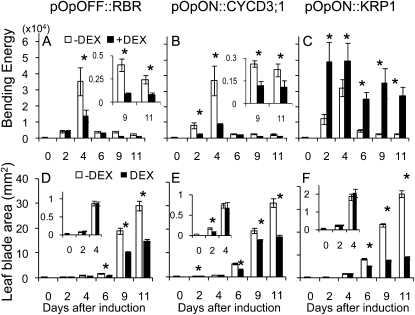
After overexpression of AtCYCD3;1 or suppression of AtRBR1 from 10 DAS, there was a severe damping of shape change, with the maximal value of BE achieved at 4 d after induction being significantly lower (P < 0.05) than in control leaves (Fig. 6, A and B). Thus, although some increase in BE occurred after induction (parallel to the pattern observed in control leaves), the magnitude of this change was limited and the final BE was lower than in control leaves (P < 0.05). This decrease in leaf BE was detectable during the first 2 to 4 d after induction for both the pOpON::CYCD3; 1 and pOpOFF::RBR transgenic lines, which correlated with the timing of decreased cell size after induction (Figs. 2 and 3), whereas altered cell division frequency for both lines was first detectable consistently at least 6 d after induction or later. The induction of AtCYCD3;1 and repression of AtRBR also led to the formation of smaller leaves (Fig. 6, D and E), although a significant decrease in leaf blade area was only consistently measured at 6 d after induction or later, i.e. after the change in BE.
Figure 6.
Induction of CYCD3;1 and repression of RBR leads to a damping of leaf shape change, whereas induction of KRP1 leads to an increase in BE. Total BE (A–C) and blade area (D–F) of leaf number 6 for pOpOFF::RBR (A and D), pOpON::CYCD3;1 (B and E), and pOpON::KRP1 (C and F) plants either induced (+DEX, black bars) or not induced (−DEX, white bars) with 20 μm DEX from 10 DAS. Leaves were harvested at time points after induction (days after induction) and the BE and area calculated. Mean values are shown with se (n = 5). Pairwise comparisons showing significant difference at a confidence limit of at least 0.05 are indicated with an asterisk.
The results from our analysis of pOpON::CYCD3;1 and pOpOFF::RBR plants indicated that altered expression of genes expected to promote the G1/S transition led to smoothing of the leaf perimeter (decreased BE), which correlated with a smaller cell size. This raised the question of whether the corollary is correct, i.e. does the expression of genes expected to inhibit the G1/S transition lead to increased BE? Previous work in which an inhibitor of cyclin-dependent kinase (and, thus, G1/S transition), AtKRP1, was constitutively expressed showed an increase in leaf serration and lobing (Wang et al., 2000; De Veylder et al., 2001), which would be consistent with this hypothesis. To quantify this response, we created a series of transgenic lines (pOpON::KRP1) in which expression of AtKRP1 could be induced in a similar manner to that described for AtCYCD3;1 and AtRBR1 (Fig. 1). Induction of pOpON::KRP1 plants led to smaller leaves containing larger cells (Supplemental Fig. S3), as shown in previous work (De Veylder et al., 2001). Quantitative analysis of cell division parameters in leaf 6 of these plants (Fig. 7) indicated a significant (P < 0.05) decrease in cell division density and frequency in all regions of the leaf by 2 d after induction, with a concomitant increase in cell size observed by 2 d in all regions. Analysis of leaf curvature indicated a significant increase (P < 0.05) of BE 2 d after induction, which was maintained at 4 d after induction (Fig. 6C). This was followed by a decline in BE in older leaves, although the BE level still remained significantly higher (P < 0.05) than in control noninduced leaves. As with the pOpON::CYCD3;1 and pOpOFF::RBR leaves, a significant decrease in leaf area was only measured at 6 d after induction (Fig. 6H), 2 d after the first recorded increase in BE.
Figure 7.
Spatial-temporal changes in cell division density, frequency, and size in the epidermis after induction of leaf 6 of pOpON::KRP1 plants. A, Schematic showing the regions of leaf 6 from which cell division data were collected. Cell division density (divisions/area; B, E, H, and K), cell division frequency (divisions/number of cells; C, F, I, and L), and cell size (D, G, J, and M) were calculated in the epidermis of pOpON::KRP1 leaves either induced (+DEX, black bars) or not induced (−DEX, white bars) from day 10. Data were analyzed in regions of the tip (B–D), mid-upper (E–G), mid-lower (H–J), and base (K–M) of the leaf at different time points (days after induction). Mean values with se are shown (n > 30 from at least five independent leaves for each data point). Pairwise comparisons showing significant difference at a confidence limit of at least 0.05 are indicated with an asterisk.
DISCUSSION
The Role of Cell Division in Leaf Morphogenesis
The idea that cell division plays a key role in defining plant form is deeply embedded in most plant biology textbooks. However, whether the observed patterns of cell division truly underpin the complex shapes of plants remains debatable (Fleming, 2005). Following the identification of the plethora of genes implicated in the plant cell cycle, investigators (ourselves included) have taken the approach of generating transgenic plants in which the expression of genes regulating the cell cycle can be altered, with the aim of observing the resultant altered pattern of cell division and the outcome this has on plant phenotype (De Veylder et al., 2001; Wyrzykowska et al., 2002, 2006; Dewitte et al., 2003). In this study, we refined this approach by combining an inducible gene expression system to alter cell cycle gene expression with methods to quantify the pattern of cell plate formation and the change in shape of a particular target organ, the leaf, in a carefully staged developmental analysis. A main conclusion from this analysis is that, although an increase in cell division frequency was indeed observed as a result of these manipulations, these changes occurred well after measurable changes in leaf shape, indicating that the frequency of cell division could not be causally involved in this aspect of the observed phenotype. For example, our data show that although after manipulation of CYCD3;1 or RBR expression a change of leaf BE was detectable within 2 to 4 d, an increase in cell division frequency was only detectable at day 6 or later. The only exception to this was an observed transient increase in cell division frequency at the base of the leaf after induction of pOpON::CYCD3;1 leaves (Fig. 3, I and L). However, this increase was not observed after induction of pOpOFF::RBR leaves (Fig. 2, I and L), yet manipulation of both pOpON::CYCD3;1 and pOpOFF::RBR led to a very similar change in leaf shape (BE; Fig. 6). This again suggests that an increase in cell division frequency is not required for change in leaf shape. Rather, the primary effect at the cellular level of all the manipulations described here was to decrease mean cell size, with the timing of this change correlating with the timing of observed leaf shape change. These changes in cell size and leaf shape occurred between days 2 and 4 after induction, before any measurable decrease in leaf growth (day 6). This is consistent with the idea that during this phase of the experiment, proliferating cells in the induced leaves were dividing at a smaller mother cell size (and more rapidly) but that the relative number of cells undergoing division was unaltered.
Both AtCYCD3;1 and AtRBR play a role in controlling the G1/S phase transition; thus, manipulating expression of these genes is predicted to lead to a change in progression through the cell cycle. At the same time, due to the central role of the CYC/RBR node at the nexus of division and differentiation (De Veylder et al., 2007), manipulation of this pathway will also lead to altered gene expression not directly linked to the cell cycle. We suspect that this is indeed the case in the experiments reported here. A primary outcome of our manipulations at the cellular level was that cell size decreased (and cell density increased), suggesting that the relationship of timing of division and cell size was altered. Interestingly, recent investigations into the role of retinoblastoma-related proteins in algal cells have revealed a role for these proteins in controlling cell size, suggesting that the cellular phenotype observed in Arabidopsis reflects an ancient role of this protein (Umen and Goodenough, 2001; Kianianmomeni et al., 2008; Borghi et al., 2010; Bosco, 2010). We conjecture that in multicellular plant organs, RBR plays a role in coordinating division and growth at the cellular level and that disruption of this pathway via suppression of RBR (or manipulation of associated proteins such as CYCD3;1) leads to cell division occurring at a smaller mother cell size. The molecular mechanism by which this occurs is still obscure, but it is likely to play a key role in the control of organ growth. We suggest that it is via its influence on cell size rather than division frequency (which occurs at a later time point) that the RBR cell cycle node impinges on leaf form. Further investigation of the transcriptional targets of RBR potentially involved in cell growth will help resolve this problem (Borghi et al., 2010).
Recent evidence indicates that the growth factor auxin and the CUC2 transcription factor network interact along the leaf perimeter in Arabidopsis to influence leaf shape (Bilsborough et al., 2011). Local regions of auxin accumulation are envisaged to promote outgrowth, whereas adjacent areas of CUC2 activity might lead to growth repression (Malinowski et al., 2011), thus acting to coordinate local morphogenesis. The outputs of this model are yet to be defined, i.e. how exactly the patterning elements influence growth. Our data indicate that cell division size could act as a modulator of the outputs of this system. Thus, there is a significant body of evidence linking auxin with cell cycle genes (e.g. Jurado et al., 2010), leading to the possibility that auxin might modulate growth locally in the leaf margin via cell cycle gene expression but that the eventual influence on growth might occur not via altered cell division frequency (as might be intuitively expected) but rather via an influence on cell division size. This raises the question of how altered cell size can influence form.
How Could Decrease in Cell Size Act to Dampen Shape Change?
In leaves in which cell size was decreased, the final lamina area was smaller than in noninduced leaves (Fig. 6). One possibility is that the observed decrease in perimeter deformation in leaves that have undergone induction of CYC/RBR genes reflects a developmental retardation so that leaves that are smaller automatically have a simpler morphology. A number of observations argue against this. First, analysis of normal Arabidopsis leaf development (Fig. 5) indicates that older (and, thus, larger) leaves actually have a lower BE than younger leaves. Second, the imposition of decreased growth via overexpression of a cyclin-dependent kinase inhibitor (AtKRP1) leads to leaves that are highly deformed and that have a higher BE (Fig. 6C; De Veylder et al., 2001). Third, in the manipulations reported here, altered BE was first recorded at between 2 and 4 d after gene induction, whereas a decrease in leaf area was generally observed later (6 d after induction). It seems highly unlikely that the loss of perimeter deformation observed in our experiments consequent to induction of CYCD3;1 or suppression of RBR is directly related to the smaller final size of the leaves.
An alternative possibility is that our manipulations led to the loss of a gradient of cell divisions/area required for shape change to occur. Although an increase in cell division density was calculated in all parts of the leaf after induction of CYCD3;1/suppression of RBR, an endogenous gradient of cell division density was still observed along the leaf proximal-distal gradient (Figs. 2 and 3). It thus seems unlikely that the measured loss of shape change was due to a loss of cell division gradient along the leaf proximal-distal axis. This still leaves the possibility that our manipulations destroyed local gradients of cell division within the region where shape change should have occurred (i.e. that our analysis lacked the resolution to detect the cell division gradients involved in morphogenesis). It is of course very difficult to totally discount the possibility of transient local gradients of cell division that were not picked up by our analysis. We attempted to calculate cell division frequencies in more localized regions of the leaf than those described in Figures 2 and 3; however, at these finer scales, the number of divisions counted per area was low and the variation between leaves was such that it is difficult to draw strong conclusions. Coupled with the limitations imposed by the use of static images to visualize a dynamic situation, this point must remain unresolved. The development of techniques that allow real-time capture of cell division patterns over the time spans required to assess changes in leaf morphology should allow the resolution of this issue.
A third alternative is that altered cell size has a knock-on effect on the movement of growth factors that influence morphogenesis. There is substantial evidence that auxin flux occurs around the leaf perimeter mediated by PIN/CUC2 patterning system and that this system is linked to leaf serration/lobbing (Bilsborough et al., 2011). Altered cell density resulting from a decrease in cell size would influence the number of cell boundaries across which PIN-mediated auxin transport would have to occur, and this could alter the dynamics and output of the system. Future investigation of the distribution of the components of the PIN/CUC2 module in the CYC/RBR manipulated system described here would help clarify this, in conjunction with analysis using established computational modeling tools.
A final alternative is as follows. Morphogenesis requires differences in growth rate between adjacent regions of an organ. The actual growth rate of a piece of tissue is liable to be affected by a number of factors, including, for example, cell wall extensibility, capacity to generate turgor pressure, and the ability to synthesize material required for growth (Fleming, 2005). We propose that the cell cycle phase of cells within a growing tissue is an additional factor that influences the growth rate of that tissue and that it does so in a nonlinear fashion. For example, data from the animal field indicate that small cells (in an early phase of cell cycle) have a lower growth rate than larger cells (late phase of cell cycle; Tzur et al., 2009). Equivalent data for plant cells are sparse, although the presence of smaller cells has been correlated with a lower growth rate in roots (Qi and John, 2007). In plants, there is the added complication that cells can switch from cell division-associated growth to cell division-independent growth (characterized by vacuole expansion). The tools used in this investigation to manipulate the cell cycle are liable to influence the switch between these two linked but alternate forms of cell growth (i.e. control the switch from maintenance of cell proliferation to differentiation). Such switching would tend to amplify any nonlinearity between cell cycle stage and cell growth rate.
If there is a nonlinear relationship between cell size and growth rate, then a set decrease/increase in cell size will lead to a disproportionate decrease/increase in growth rate. In the context of the basal region of the Arabidopsis leaf, a tendency for cell division to occur at a smaller mother cell size (via altered CYCD3;1/RBR activity) would tend to smoothen the gradient of growth along the proximal-distal axis that must underpin any morphogenesis, thus leading to less shape change, i.e. a damping of morphogenesis. Conversely, premature cessation of cell division and consequent switching to a division-independent form of cell growth (via overexpression of KRP1) would tend to amplify differences in growth rate, leading to more pronounced shape change (higher BE). This interpretation clearly involves speculation on the relationship of plant cell size and growth rate, but the imaging methods to test such a hypothesis and to obtain quantitative in vivo data are being developed (Lee et al., 2006).
Irrespective of the mechanism, our data indicate that the relationship of cell division frequency and organ shape is not trivial. Simple inferences that, for example, altered cell cycle gene expression leads to increased cell division frequency, which then leads to increased growth rate, are not tenable. Rather, a fundamental relationship between cell size and growth rate could account for aspects of shape control, with cell cycle genes influencing growth rate indirectly by the setting of cell size.
MATERIALS AND METHODS
Plant Growth and Staging
Arabidopsis (Arabidopsis thaliana) seeds were kept at 4°C for 1 week before sowing on agar (0.8% [w/v]) medium containing half-strength Murashige and Skoog salt mix (Sigma-Aldrich) and 1% (w/v) Suc. Seedlings having approximately the same size of leaf number 5 (measured under a stereomicroscope) were selected after 10 d and taken forward for experimentation. Seedlings were transplanted onto medium supplemented with 0.1% (v/v) dimethyl sulfoxide with or without DEX for specific times before analysis. Growth conditions were 100 μmol m−2 s−1 light, a 16/8-h photoperiod, and temperature 20/18°C (light/dark).
Analysis of Leaf Shape
The sixth leaf formed in developmental series was used for analysis. Leaves were fixed in ethanol/acetic acid (7:1 [v/v]) and hydrated with 50% (v/v) aqueous ethanol. Images were taken with a CCD (DP71; Olympus) mounted on a light microscope (BX51; Olympus) or a CCD (Diagnostic Instruments) mounted on a stereomicroscope (MZFLIII; Leica Microsystems). To obtain leaf shape properties, we used in-house developed software to process digital images (LEAFPROCESSOR; Backhaus et al., 2010). Briefly, the software performs a contour detection, then uses active contours (coupled with manual correction) to fit a sequence of points around the leaf shape. A plane curve is then created through spline interpolation, allowing sampling of 500 points with equal curve length between each point per leaf. For BE, a curvature value was calculated at each point around the perimeter and then normalized to provide a scale-independent value. A brief description of how these values were obtained is provided in Supplemental Figure S6. For statistical comparison of BE, pairwise t tests or ANOVA were performed using the n values as indicated in the relevant figure legends, with differences being accepted as significant if they fell below the 0.05% confidence limit.
Analysis of Cell Division
Newly formed cell walls were detected according to Kuwabara and Nagata (2006) with a modification for Arabidopsis. Briefly, all trichomes were manually removed before staining in 0.02% (w/v) aniline blue in 100 mm phosphate buffer (pH 9.0) for 1 to 3 weeks at 4°C. Septum walls were observed using an epifluorescent microscope (BX51; Olympus) with a 4′,6-diamino-phenylindole filter setting. Images were taken with a CCD (DP71; Olympus). Frequency and orientation of cell division were analyzed using Scion Image (Scion). For leaves at days 10 and 12, each leaf was horizontally divided into four regions using cutoff lines at 25%, 50%, and 75% from the leaf base, and all cell divisions in a region were counted. For leaves at days 14 to 21, the number of cell divisions was counted in a square (100 × 100 μm2) placed at points 20%, 40%, 60%, and 80% from the leaf base on the midpoint between the leaf margin and the midrib. For each data point (dependent on leaf size and cell size), between five and 50 cells were counted from five individual staged leaves, so that the means displayed were calculated with n values ranging from 31 to 252 cells/data point (±se). For statistical comparison of cell divisions/area, cell division frequency, and cell size, pairwise t tests or ANOVA were performed using the n values as indicated in the relevant figure legends, with differences being accepted as significant if they fell below the 0.05% confidence limit.
Generation and Analysis of Transgenic Plants
The AtCYCD3;1 (AT4G34160) and AtKRP1 (AT2G23430) coding sequences were amplified using Pfu Turbo DNA Polymerase (Stratagene) and cloned into pENTR/D-TOPO (Invitrogen). LR Clonase II (Invitrogen) recombination reactions were carried out to insert AtCYCD3;1 or AtKRP1 into the pOpOn2.1 vector (Wielopolska et al., 2005). For the AtRBR1 (AT3G12280) silencing construct, a 169-bp region from the coding sequence in pDONR207 was obtained from the Nottingham Arabidopsis Stock Centre and verified using forward primer 5′-AATTGGCTGCTGTAAGAATCAATGG-3′ and reverse primer 5′-TTGGCCACTCCGTAGAAGCA-3′ (Hilson et al., 2004). The fragment was recombined into pOpOff2(hyg) vector (Wielopolska et al., 2005) as described above. Vectors were transformed into Agrobacterium tumefaciens and then plants (Arabidopsis Columbia-0) transformed by floral dipping (Clough and Bent, 1998). Several homozygous T3 generation plants were identified for each transgene and analysis performed on at least two separate lines. Similar phenotypes were observed irrespective of the line analyzed.
For western-blot analysis of AtCYCD3;1, 40 μg of protein extract was isolated, separated on 10% SDS-PAGE gels, and transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories). Protein was detected using a 1:1,000 dilution of a polyclonal anti-CYCD3;1 antibody (a kind gift of J. Murray, University of Cardiff, UK), followed by incubation with a monoclonal alkaline phosphatase-conjugated anti-rabbit IgG antibody (Sigma-Aldrich). Signals were developed using the Lumiphos system (Thermo Fisher-Pierce). RBR1 protein was isolated using 7 m urea buffer and, after electrophoresis of 100 μg protein extract on an 8% SDS-PAGE gel, transferred and detected as described above with the difference that as primary antibody a 1:500 dilution of polyclonal anti-RBR1 (a kind gift of W. Gruissem, Eidgenössisch Technische Hochschule Zurich) was used. For semiquantitative reverse transcription (RT)-PCR, 4 μg of total RNA (isolated from seedlings according to Chomczynski and Sacchi, 1987) was used for cDNA synthesis using M-MLV reverse transcriptase (Promega). PCR amplification used BIO-Taq polymerase (BIOLINE). To determine the exponential phase of the amplification, 5-μL samples were taken at cycle numbers 22, 24, 26, 28, 30, and 32. Primers used were as follows: AtCYCD3;1, 5′-TACGATCTAATCCTCCAACTACC-3′ and 5′-TTATGGAGTGGCTACGATTG-3′; AtACT2 (AT1G49240), 5′-TCAGCACATTCCAGCAGATG-3′ and 5′-TTAACATTGCAAAGAGTTTCAAGGT-3′; AtRBR1, 5′-AATTGGCTGCTGTAAGAATCAATGG-3′ and 5′-TTGGCCACTCCGTAGAAGCA-3′.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Constructs pOpON::CYCD3;1 and pOpOFF::RBR.
Supplemental Figure S2. Developmental series of Arabidopsis leaves used in this investigation.
Supplemental Figure S3. Images of pOpON::CYCD3;1, pOpOFF::RBR, and pOpON::KRP1 leaf epidermis.
Supplemental Figure S4. Imaging of new cell plates.
Supplemental Figure S5. Cell size distribution for pOpON::CYCD3;1, pOpOFF::RBR1, and pOpON::KRP1 leaves.
Supplemental Figure S6. Calculation of BE.
Acknowledgments
We thank Ian Moore (University of Oxford) for providing the pOpON and pOpOFF vectors, Jim Murray (University Cardiff) for the AtCYCD3;1 antibody, and Wilhelm Gruissem (Eidgenössisch Technische Hochschule Zurich) for the AtRBR antibody. We thank members of the Fleming lab and Julie Gray (Sheffield) for critical reading of the manuscript.
References
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhaus A, Kuwabara A, Bauch M, Monk N, Sanguinetti G, Fleming A. (2010) LEAFPROCESSOR: a new leaf phenotyping tool using contour bending energy and shape cluster analysis. New Phytol 187: 251–261 [DOI] [PubMed] [Google Scholar]
- Barkoulas M, Galinha C, Grigg SP, Tsiantis M. (2007) From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol 10: 660–666 [DOI] [PubMed] [Google Scholar]
- Bayer EM, Smith RS, Mandel T, Nakayama N, Sauer M, Prusinkiewicz P, Kuhlemeier C. (2009) Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev 23: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. (2011) Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 108: 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. (2008) A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Futterer J, Laizet Y, Hennig L, Gruissem W. (2010) Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G. (2010) Cell cycle: retinoblastoma, a trip organizer. Nature 466: 1051–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clay NK, Nelson T. (2005) The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell 17: 1994–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua N-H, Gutierrez C. (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D. (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JMS, Jacqmard A, Kilby NJ, Murray JA. (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D. (2004) The role of JAGGED in shaping lateral organs. Development 131: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Fleming AJ. (2005) The control of leaf development. New Phytol 166: 9–20 [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. (2005) SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. (2006) The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hemerly A, Engler JA, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14(10B): 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S, Abraham Z, Manzano C, López-Torrejón G, Pacios LF, Del Pozo JC. (2010) The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianianmomeni A, Nematollahi G, Hallmann A. (2008) A gender-specific retinoblastoma-related protein in Volvox carteri implies a role for the retinoblastoma protein family in sexual development. Plant Cell 20: 2399–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara A, Nagata T. (2006) Cellular basis of developmental plasticity observed in heterophyllous leaf formation of Ludwigia arcuata (Onagraceae). Planta 224: 761–770 [DOI] [PubMed] [Google Scholar]
- Larue CT, Wen J, Walker JC. (2009) A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J 58: 450–463 [DOI] [PubMed] [Google Scholar]
- Lee K, Avondo J, Morrison H, Blot L, Stark M, Sharpe J, Bangham A, Coen E. (2006) Visualizing plant development and gene expression in three dimensions using optical projection tomography. Plant Cell 18: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowski R, Kasprzewska A, Fleming AJ. (2011) Targeted manipulation of leaf form via local growth repression. Plant J 66: 941–952 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E. (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Poethig RS, Sussex IM. (1985) The developmental morphology and growth dynamics of the tobacco leaf. Planta 165: 158–169 [DOI] [PubMed] [Google Scholar]
- Qi R, John PCL. (2007) Expression of genomic AtCYCD2;1 in Arabidopsis induces cell division at smaller cell sizes: implications for the control of plant growth. Plant Physiol 144: 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N. (1999) Leaf development in angiosperms. Annu Rev Plant Physiol Plant Mol Biol 50: 419–446 [DOI] [PubMed] [Google Scholar]
- Smith LG, Hake S, Sylvester AW. (1996) The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122: 481–489 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2006) Mechanism of leaf-shape determination. Annu Rev Plant Biol 57: 477–496 [DOI] [PubMed] [Google Scholar]
- Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. (2009) Cell growth and size homeostasis in proliferating animal cells. Science 325: 167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW. (2001) Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC. (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C. (2005) A high-throughput inducible RNAi vector for plants. Plant Biotechnol J 3: 583–590 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wyrzykowska J, Pien S, Shen WH, Fleming AJ. (2002) Manipulation of leaf shape by modulation of cell division. Development 129: 957–964 [DOI] [PubMed] [Google Scholar]
- Wyrzykowska J, Schorderet M, Pien S, Gruissem W, Fleming AJ. (2006) Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol 141: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]