Abstract
Pentatricopeptide repeat (PPR) proteins (PPRPs) are encoded by a large gene family in Arabidopsis (Arabidopsis thaliana), and their functions are largely unknown. The few studied PPRPs are implicated in different developmental processes through their function in RNA metabolism and posttranscriptional regulation in plant organelles. Here, we studied the functions of Arabidopsis PENTATRICOPEPTIDE REPEAT PROTEIN FOR GERMINATION ON NaCl (PGN) in plant defense and abiotic stress responses. Inactivation of PGN results in susceptibility to necrotrophic fungal pathogens as well as hypersensitivity to abscisic acid (ABA), glucose, and salinity. Interestingly, ectopic expression of PGN results in the same phenotypes as the pgn null allele, indicating that a tight regulation of the PGN transcript is required for normal function. Loss of PGN function dramatically enhanced reactive oxygen species accumulation in seedlings in response to salt stress. Inhibition of ABA synthesis and signaling partially alleviates the glucose sensitivity of pgn, suggesting that the mutant accumulates high endogenous ABA. Accordingly, induction of NCED3, encoding the rate-limiting enzyme in stress-induced ABA biosynthesis, is significantly higher in pgn, and the mutant has higher basal ABA levels, which may underlie its phenotypes. The pgn mutant has altered expression of other ABA-related genes as well as mitochondria-associated transcripts, most notably elevated levels of ABI4 and ALTERNATIVE OXIDASE1a, which are known for their roles in retrograde signaling induced by changes in or inhibition of mitochondrial function. These data, coupled with its mitochondrial localization, suggest that PGN functions in regulation of reactive oxygen species homeostasis in mitochondria during abiotic and biotic stress responses, likely through involvement in retrograde signaling.
Plants display diverse survival mechanisms against microbial infection and other environmental stresses. While specialized host responses do occur, many components of the molecular events underlying plant responses to abiotic and biotic stresses are common. Passive defenses, such as the cuticle, aid in drought tolerance and protection from UV damage while also acting as a deterrent of herbivory and barrier against pathogen infection (Reina-Pinto and Yephremov, 2009). Similarly, the cellular and biochemical processes associated with active responses to different abiotic and biotic stimuli also share functional overlaps (Fujita et al., 2006). These induced responses are largely mediated by plant hormones and their interactions, which range from simple synergism or antagonism to intricate networks of cross-regulation (Grant and Jones, 2009). Responses to pathogen infection are modulated by salicylate (SA), jasmonate (JA), and ethylene (ET) with a growing role for abscisic acid (ABA), auxin, and GAs. ABA, a major regulator of environmental stress responses, is generally regarded as a negative regulator of plant defense, with exogenous application or increased endogenous levels typically correlating with plant susceptibility to pathogens (Mauch-Mani and Mauch, 2005; Fujita et al., 2006). However, there are instances of ABA positively contributing to disease resistance through modulation of callose deposition, stomatal closure, defense gene expression, and accumulation of reactive oxygen species (ROS; Mauch-Mani and Mauch, 2005).
Overall, plant responses to different stresses share significant overlap and points of convergence defined by regulatory factors that integrate signaling from various pathways (Fujita et al., 2006; Robert-Seilaniantz et al., 2010). Among these, the Arabidopsis (Arabidopsis thaliana) R2R3MYB transcription factor BOS1 is a mediator of abiotic and biotic stress responses, its loss of function resulting in susceptibility to necrotrophic infection as well as hypersensitivity to salt, osmotic, and oxidative stress (Mengiste et al., 2003). Similarly, overexpression of the ATAF1 transcription factor results in susceptibility to Botrytis cinerea and Blumeria graminis f. sp. hordei as well as decreased tolerance to ABA, salt, and oxidative stress (Jensen et al., 2007; Wu et al., 2009). PHYTOCHROME AND FLOWERING TIME1 (PFT1) regulates plant resistance to Alternaria brassicicola, B. cinerea, and Fusarium oxysporum through its function in the biosynthesis of anthocyanin, a flavonoid linked to numerous abiotic and biotic stress responses (Kidd et al., 2009). PFT1 encodes a subunit of the evolutionarily conserved Mediator complex, which was recently shown to promote transcription of microRNA (miRNA) genes by recruiting RNA polymerase II to their promoters (Kim et al., 2011). miRNAs and small interfering RNAs have emerged as important regulators of plant defense and stress tolerance known to affect gene expression, ROS accumulation, and plant cell death (Borsani et al., 2005; Katiyar-Agarwal et al., 2007; Sunkar et al., 2007; Xie and Qi, 2008). Natural cis-antisense small interfering RNAs have been associated with Arabidopsis salt tolerance as well as resistance to pathogens (Borsani et al., 2005; Katiyar-Agarwal et al., 2007; Xie and Qi, 2008). Natural small interfering RNA ATGB2 contributes to RPS2-mediated resistance to Pseudomonas syringae by repressing PENTATRICOPEPTIDE REPEAT PROTEIN-LIKE (PPRL) gene expression (Katiyar-Agarwal et al., 2007).
Members of the eukaryotic pentatricopeptide repeat (PPR) protein (PPRP) family contain tandem arrays of a degenerate 35-amino acid repeat and function in RNA or DNA modification through sequence-specific binding (Saha et al., 2007). As such, PPRPs have been associated with all stages of RNA processing, maturation, and translation (Saha et al., 2007; Schmitz-Linneweber and Small, 2008). PPRPs are classified into subgroups based on C terminus domains as well as the nature and order of their repeats (Small and Peeters, 2000; Lurin et al., 2004). Three conserved motifs, E-, E+, and DYW, in the C terminus define four subclasses of the PPRP family. These domains always require the one prior to be present in the protein; therefore, the subclasses consist of PPRPs with no C-terminal motifs, E-, E+ (preceded by E-), and DYW (preceded by E+ and E-; Lurin et al., 2004). The repeat motifs are defined as P, L, or S based on size and variability: P-type is the characteristic repeat defining the protein family, L-type is a long variant of the P repeat, and S-type is a short variant (Lurin et al., 2004). The Arabidopsis genome contains more than 450 PPRPs, yet surprisingly few have been studied, and their functions remain largely unknown (Schmitz-Linneweber and Small, 2008). The few studied PPRPs play diverse and crucial roles in plant growth and development, including embryogenesis, circadian rhythm, chloroplast development, and retrograde nuclear signaling (Lurin et al., 2004; Oguchi et al., 2004; Tzafrir et al., 2004; Cushing et al., 2005; Ding et al., 2006; Koussevitzky et al., 2007; Chi et al., 2008). GUN1, a DNA-binding chloroplast PPRP, is involved in retrograde signaling, regulation of ABI4 expression, and photooxidative stress responses (Zhang et al., 2006; Koussevitzky et al., 2007). ABI4 functions in light and sugar-induced stress responses as well as callose-mediated defense, ROS signaling, and resistance to fungal infection (Ton et al., 2009). LOVASTATIN INSENSITIVE1 (LOI1) is a PPRP that regulates biosynthesis of isoprenoids, metabolites known to affect defense gene expression in response to wounding and pathogen infection (Kishimoto et al., 2005; Kobayashi et al., 2007). The loi1 mutant has decreased sensitivity to two inhibitors of isoprenoid synthesis, the fungal phytotoxin lovastatin and the herbicide clomazone, as evidenced by higher sterol and chlorophyll accumulation compared to that of treated wild-type plants (Kobayashi et al., 2007). PPR40 is a mitochondrial PPRP involved in oxidative respiration that also contributes to abiotic stress tolerance in Arabidopsis (Zsigmond et al., 2008). The ppr40 mutant exhibits enhanced sensitivity to ABA and salinity that correlates with increased ROS accumulation and altered stress-responsive gene expression. Thus far, of the 450 predicted PPRPs, only GUN1, LOI1, PPRL, and PPR40 have been associated with Arabidopsis defense and/or stress tolerance (Katiyar-Agarwal et al., 2006; Kobayashi et al., 2007; Koussevitzky et al., 2007; Zsigmond et al., 2008). However, the functional link of many PPRPs in chloroplast and mitochondrial development and/or regulation suggests they may play a role in managing perturbations in cellular redox elicited by different types of stress (Lurin et al., 2004; Andres et al., 2007; Saha et al., 2007). Here, we describe the function of the Arabidopsis PENTATRICOPEPTIDE REPEAT PROTEIN FOR GERMINATION ON NaCl (PGN) in plant resistance to necrotrophic fungi and tolerance to abiotic stress.
RESULTS
Identification of the PGN Gene and Its Role in Resistance to Necrotrophic Pathogens
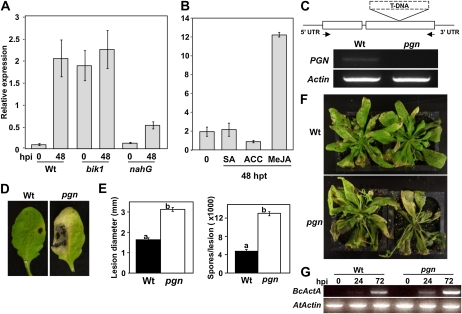
Previously, Arabidopsis BOTRYTIS-INDUCED KINASE1 (BIK1) was found to play a contrasting role in plant defense, positively contributing to resistance against B. cinerea but functioning as a negative regulator of resistance against virulent P. syringae (Veronese et al., 2006). In an effort to further define the role of BIK1 in defense and identify genes involved in resistance, we compared the genome-wide transcript profiles of wild-type and bik1 plants prior to and following B. cinerea inoculation (Dhawan et al., 2009). From this, AT1G56570 (designated PGN), encoding a PPRP, was identified as a potential BIK1 target due to its increased basal expression in the mutant and significant induction in wild-type plants following infection (Fig. 1A). Interestingly, Arabidopsis nahG lines have reduced B. cinerea-induced PGN expression (Fig. 1A). nahG plants are SA deficient, whereas the bik1 mutation leads to high basal and induced SA accumulation (Delaney et al., 1994; Veronese et al., 2006), suggesting that PGN expression in response to B. cinerea is at least partially dependent on SA levels. Yet, exogenous application of SA does not affect PGN expression, whereas treatment with methyl-jasmonate (MeJA) leads to significant induction and the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC) causes minor suppression (Fig. 1B). Remarkably, PGN expression significantly correlates with the expression of 13 other PPRP genes as well as several genes associated with RNA synthesis or processing (Supplemental Fig. S1A; Obayashi et al., 2009). PGN is also highly expressed in dry and imbibed seed tissue as well as the shoot apex throughout development (Supplemental Fig. S1B).
Figure 1.
Expression of the PGN gene and characterization of the pgn mutant for disease resistance. A and B, Expression of PGN in response to B. cinerea (A) and exogenous application of plant hormones (B). C, Genomic organization of the pgn T-DNA insertion and loss of PGN expression (arrows indicate primer locations used to assay expression). D and E, Disease symptoms (D), lesion diameter (E), and number of spores per lesion 4 d after inoculation with A. brassicicola. F and G, Disease symptoms (F) and B. cinerea ActinA (G; BcActA) accumulation as a measure of fungal growth following spray inoculation with B. cinerea. Images were taken 5 (D) and 7 d after inoculation (F). Inoculation and quantification of disease symptoms/fungal growth were performed as described in “Materials and Methods.” Data for lesion diameters and spores/lesion represent the mean ± se from a minimum of 20 inoculated leaves. hpi, Hours postinoculation; hpt, hours posttreatment; UTR, untranslated region; Wt, wild type. Experiments were repeated at least three times with similar results.
To determine the biological relevance of its B. cinerea-induced gene expression, we characterized plants harboring a null T-DNA insertion allele of the PGN gene (SALK_141937; (Fig. 1C). The pgn mutation resulted in enhanced susceptibility to A. brassicicola as evidenced by increased chlorosis and necrosis at the site of inoculation (Fig. 1D). Compared to the wild type, pgn leaves develop significantly larger disease lesions and support increased fungal proliferation (Fig. 1E). The pgn mutant is also susceptible to B. cinerea. Following spray inoculation, pgn plants display increased chlorosis and tissue maceration 4 d after inoculation that progresses into abundant leaf decay around 7 d after inoculation (Fig. 1F). Despite relatively slow symptom development, fungal growth in the pgn mutant is higher than the wild type just 24 h after inoculation based on transcript accumulation of the constitutive B. cinerea ActinA gene (Fig. 1G).
Additionally, we assayed pgn plants for altered resistance to the bacterial pathogen P. syringae to further clarify PGN function in BIK1-regulated defense responses and determine if the altered susceptibility of the mutant to necrotrophic infection is a result of hormone-mediated defense antagonism. Plant immune responses to necrotrophic infection are regulated by JA/ET-mediated signaling events known to antagonize SA-dependent defenses associated with biotrophic resistance (Koornneef and Pieterse, 2008). No difference was observed in bacterial growth between wild-type plants and the pgn mutant inoculated with virulent (DC3000) or avirulent (DC3000AvrRpm1) strains of P. syringae (Supplemental Fig. S2). Interestingly, B. cinerea-induced expression of PR-1 and PDF1.2, considered molecular markers of SA- and JA/ET-dependent defense responses, respectively, are not altered in the pgn mutant (Supplemental Fig. S3, A and B). Overall, these results suggest the function of PGN in pathogen resistance is specific to necrotrophs and not associated with SA-, JA-, or ET-mediated defenses.
PGN Encodes a PPRP Localized to Mitochondria
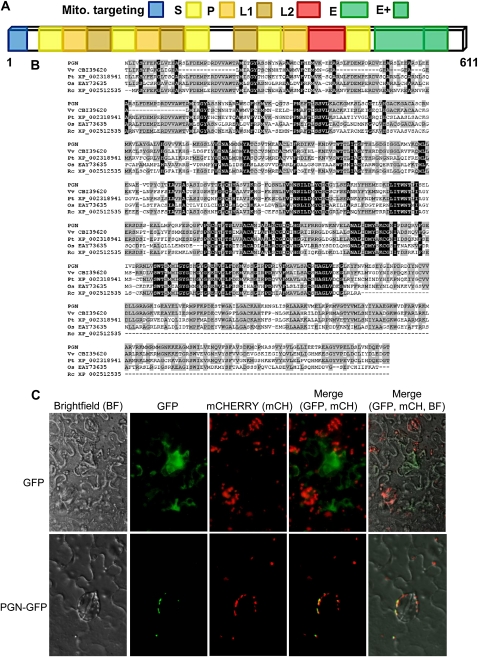
PGN is predicted to encode a PPRP (Fig. 2A) with significant homology to proteins from different plant species and Arabidopsis spanning full-length amino acid sequences (Fig. 2B; Supplemental Fig. S4). The PGN protein shows the characteristics of the E+ PPRP subclass with a 43-S-P-L-S-P-L-S-34-L-S-1-P-L2-S-4-E-E+-19 repeat and motif arrangement (numbers indicate the amount of residues in between motifs; Fig. 2A). The repeats are predicted to form a superhelix compromised of two antiparallel α-helices able to bind DNA or RNA with substrate specificity dependent on the residue properties within the groove (Delannoy et al., 2007). In Arabidopsis, there are more than 450 predicted PPRPs, with all described proteins localized to mitochondria, chloroplast, or nucleus (Small and Peeters, 2000; Lurin et al., 2004; Schmitz-Linneweber and Small, 2008). Of the E+ PPRP subfamily, more than 40% are predicted to localize to mitochondria (Lurin et al., 2004). Prediction programs indicate PGN has a cleavable targeting sequence (MSITKLARSNAFKPIPNFVRSSLRN) and a 96% likelihood of subcellular localization to mitochondria (Claros and Vincens, 1996). To experimentally determine the subcellular localization of PGN, 35S:PGN-GFP was transiently expressed in Nicotiana benthamiana leaves in conjunction with different cellular markers. PGN-GFP colocalized with a mitochondrial marker (mCHERRY) consistent with the in silico predictions (Fig. 2C).
Figure 2.
Main features of the PGN protein and its subcellular localization. A, Conserved domains of the PGN protein as defined by Lurin et al. (2004). B, Multiple sequence alignment of PGN and closely related proteins in other species. Black shading indicates conserved residues, and gray indicates residues identical to PGN. Proteins were aligned using ClustalW with default gap penalties (Thompson et al., 1994). C, PGN is localized to mitochondria. GFP, PGN-GFP, and the mitochondrial marker mCHERRY were transiently expressed in N. benthamiana leaf cells. Localization of tagged proteins was examined using the appropriate fluorescence wavelengths with images merged to show colocalization. Localization experiments were repeated at least three times with similar results. Vv, Vitis vinifera; Pt, Populus trichocarpa; Os, Oryza sativa; Rc, Ricinus communis.
Loss or Gain of PGN Function Causes Hypersensitivity to ABA, NaCl, and Glc
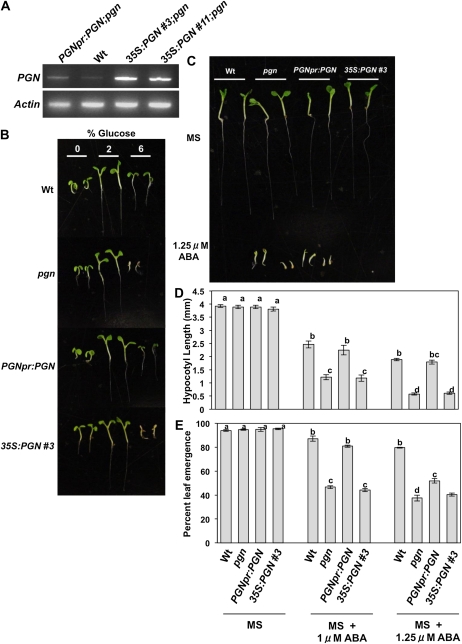
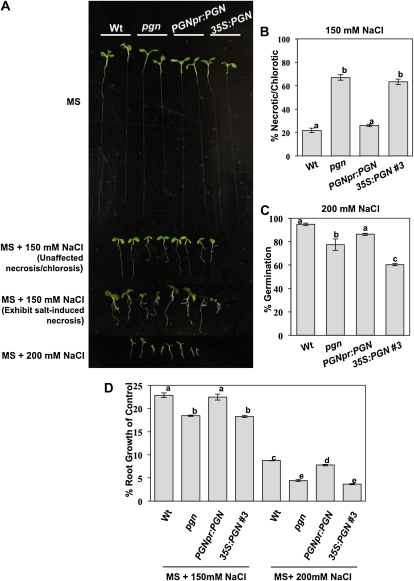
To further study the function of PGN, overexpression (35S:PGN;pgn) and complementation lines (PGNpr:PGN;pgn) were generated (Fig. 3A). The different PGN genotypes were assayed for altered responses to hormones and abiotic stress agents in an effort to clarify the mechanism of PGN function in Arabidopsis defense. The pgn mutant displays germination hypersensitivity to media supplemented with ABA and increased Glc (Fig. 3, B–E). Overexpression of PGN also results in hypersensitivity to ABA and Glc comparable to that of the mutant (Fig. 3, B–E). Similarly, the pgn mutant and overexpression lines have increased sensitivity to salt, displaying increased salt-induced necrosis and chlorosis as well as reduced germination and root growth compared to wild-type seedlings (Fig. 4). Alternatively, both pgn and 35S:PGN;pgn plants have enhanced growth on Murashige and Skoog (MS) media lacking Glc relative to the wild type (Fig. 3B). In general, transformation with genomic PGN driven by its native promoter (PGNpr:PGN;pgn) restores mutant sensitivity to wild-type levels (Figs. 3, B–E, and 4). However, at higher concentrations of ABA, salt, and Glc, PGNpr:PGN;pgn seedlings have significantly reduced leaf emergence, root growth, and smaller cotyledons, respectively, relative to the wild type (Figs. 3, C–E, and 4, A, C, and D). This disparity could be attributed to the slightly higher PGN expression of these lines (Fig. 3A) compared to the wild type and further supports the hypersensitivity exhibited by 35S:PGN;pgn plants. Overall, these data suggest that, in addition to defense against necrotrophic infection, PGN regulates plant responses to abiotic stress, and its expression level is an important determinant of function. No altered seedling growth was observed when pregerminated 5-d seedlings were transferred to media supplemented with Glc, ABA, or indole-3-acetic acid (Supplemental Fig. S5, A–C). Also, germination on media supplemented with ACC, MeJA, SA, GA, indole-3-acetic acid, or hydrogen peroxide (H2O2) is not affected by the pgn mutation, limiting the role of PGN to a specific subset of stress and hormone responses (Supplemental Fig. S5D).
Figure 3.
Loss or gain of PGN function causes hypersensitivity to Glc and ABA at germination. A, PGN expression in wild-type (Wt), complemented (PGNpr:PGN;pgn), and overexpression (35S:PGN;pgn) lines. B and C, Seedling growth of transgenic PGN lines on Glc (B) and ABA-supplemented media (C). D and E, Mean hypocotyl length (D) and percentage of leaf emergence (E) on ABA-supplemented media. Data represent the mean ± se from a minimum of 40 seedlings. Experiments were repeated at least three times with similar results.
Figure 4.
Altered PGN expression causes impaired germination and growth responses to increased salinity. A, Comparison of seedling growth on NaCl-supplemented media. The representative growth phenotypes of wild-type (Wt) and pgn seedlings on 200 mm NaCl (fourth row), exhibiting salt-induced necrosis and/or chlorosis on 150 mm NaCl (third row), and those that did not on 150 mm NaCl (second row) are presented. B, Percentage of seedlings exhibiting salt-induced chlorosis and/or necrosis on 150 mm NaCl. C, Percentage of germination on 200 mm NaCl. D, Percentage of reduction in root growth on NaCl relative to MS controls. Data represent the mean ± se from a minimum of 40 seedlings. Experiments were repeated at least three times with similar results.
The Glc Hypersensitivity of the pgn Mutant Is Partially Restored by ABA Antagonists
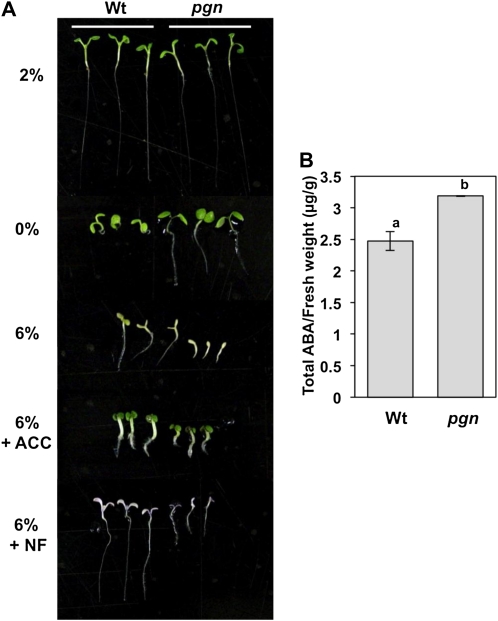
Increased concentrations of Glc are known to delay Arabidopsis germination, with ABA levels determining the severity of inhibition (Gazzarrini and McCourt, 2001; Dekkers et al., 2004). Many mutants exhibiting enhanced growth on media containing high exogenous sugars are also ABA insensitive, whereas even minimal ABA increases act additively to sugar-mediated seedling growth inhibition (Laby et al., 2000; Gazzarrini and McCourt, 2001; Gibson et al., 2001). To determine if pgn hypersensitivity to Glc and ABA is a result of increased endogenous ABA, we assayed growth responses to 6% Glc in the presence of two ABA inhibitors: the ET precursor ACC and norflurazon (NF). ET antagonizes ABA function in germination, likely promoting emergence through suppression of ABA signaling and synthesis initiated by Glc and ABA (Beaudoin et al., 2000; Ghassemian et al., 2000; León and Sheen, 2003; Matilla and Matilla-Vazquez, 2008). NF inhibits ABA accumulation through disruption of carotenoid biosynthesis upstream of ABA biosynthesis (Bartels and Watson, 1978; Zeevaart and Creelman, 1988). Addition of ACC or NF partially relieved pgn hypersensitivity to Glc relative to 6% Glc alone (Fig. 5A). Interestingly, whereas ACC restored pgn growth to a degree comparable to respective wild-type controls, the effect of NF resulted in seedlings nearly half the size of corresponding wild type. These data suggest that the increased Glc sensitivity exhibited by the pgn mutant is likely due to high levels of endogenous ABA. Subsequent analysis of total ABA content indicates that pgn seedlings do have significantly higher basal levels of ABA compared to the wild type (Fig. 5B). The growth variation between treatments may be explained by disparities in the mechanism by which they inhibit ABA responses. ACC suppresses ABA accumulation and signaling, whereas NF only disrupts ABA biosynthesis. Thus, based on its phenotypes, the pgn mutant may be altered in ABA levels as well as signaling. Alternatively, variation may be a result of secondary effects of treatment, such as NF inhibition of GA synthesis. However, based on the normal responses of the mutant to GA and other plant hormones (Supplemental Fig. S5D), we limited our conclusions to the effects of these chemicals on ABA responses for which pgn is clearly altered. The independent restoration of growth responses with either ACC or NF also serves to further strengthen an ABA-dependent mechanism. Interestingly, low levels of Glc also have an inhibitory effect on germination, yet the effect is lessened in ABA-sensitive and -insensitive mutants (Garciarrubio et al., 1997; Price et al., 2003). Thus, the enhanced growth of pgn on media lacking Glc lends further support to its increased sensitivity to ABA (Fig. 5A).
Figure 5.
The Glc sensitivity of the pgn mutant is partially reversed by ACC and NF. A, Germination responses of wild-type (Wt) and pgn seedlings on varying concentrations of Glc and 6% Glc supplemented with ACC or NF. B, ABA content of 10-d-old seedlings determined by liquid chromatography-tandem mass spectrometry. Experiments were performed as stated in “Materials and Methods” and repeated at least two times with similar results.
Altered PGN Expression Enhances the Triple Response
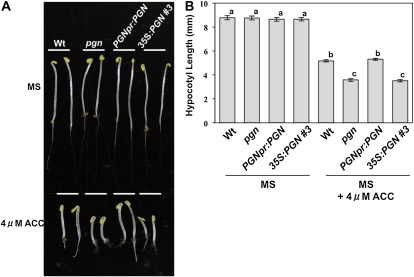
Generally, ABA and ET function antagonistically, with most ET-insensitive mutants showing sensitivity to ABA and vice versa (Gazzarrini and McCourt, 2001). Our data suggest that pgn mutation leads to disrupted ABA responses that can be partially restored through ET-mediated antagonism (Fig. 5). This implies that loss of PGN function is likely to cause insensitivity to ET. However, instead of increased tolerance, the pgn mutant exhibits enhanced ET sensitivity as observed by an altered triple response (Fig. 6). The triple response is induced when plants are grown in the dark on media containing ET or its precursor ACC, producing seedlings with exaggerated apical hooks, swollen hypocotyls, and inhibited root/hypocotyl elongation (Bleecker et al., 1988). In response to ACC, pgn seedlings produce shorter hypocotyls with limited root growth compared to the wild type (Fig. 6). Ectopic PGN expression also results in an enhanced triple response comparable to that of the mutant, consistent with either loss or gain of PGN function resulting in the same phenotypes.
Figure 6.
Loss or gain of PGN function enhances the triple response. Triple response phenotypes (A) and mean hypocotyl lengths (B) of the PGN genotypes following growth in the dark on ACC-supplemented media. Data represent the mean ± se from a minimum of 40 seedlings. Experiments were repeated at least three times with similar results. Wt, Wild type. [See online article for color version of this figure.]
The pgn Mutant Has Altered ABA-Related Gene Expression
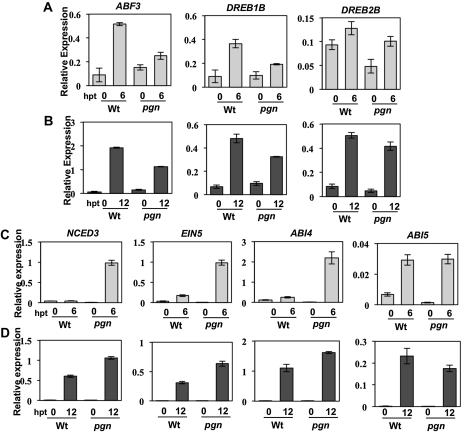
Loss of PGN function results in hypersensitivity to ABA as well as salt and Glc. To determine if this decreased tolerance is due to altered ABA signaling, we assayed the mutant for altered expression of ABA-responsive marker genes associated with activation of abiotic stress responses. Salt-induced expression of RD29A, RAB18, and COR6.6 is not altered by the pgn mutation (Supplemental Fig. S3C). RD29A, RAB18, and COR6.6 are ABA-responsive genes with varying roles in salt, sugar, and osmotic stress tolerance (Mahajan and Tuteja, 2005; Chinnusamy et al., 2006). By contrast, ABF3, DREB1B, and DREB2B are all induced at lower levels in the pgn mutant in response to salt or ABA (Fig. 7, A and B). Salt-induce.d ABI5 expression is also slightly reduced in pgn, but induction by ABA was unaffected by mutation (Fig. 7, C and D). Interestingly, pgn has higher NCED3, EIN5, and ABI4 expression in response to both ABA and NaCl compared to the wild type (Fig. 7, C and D). ABI4 is the most significantly affected by pgn mutation, showing nearly 5 times greater expression in the mutant compared to wild-type ABI4 induction in response to ABA (Fig. 7C). ABI4 encodes a transcription factor involved in ABA signaling and functions downstream of the PPRP GUN1 in photooxidative stress responses (Berrocal-Lobo et al., 2002; Koussevitzky et al., 2007).
Figure 7.
Expression of genes involved in ABA-related functions in the pgn mutant. Expression of ABA-related genes in ABA- (A and C) or NaCl-treated (B and D) seedlings. hpt, Hours posttreatment; Wt, wild type. Treatment and quantification of expression was performed as described in “Materials and Methods.”
B. cinerea-Induced PGN Expression Is Enhanced in the ET and ABA Response Mutants etr1 and abi5
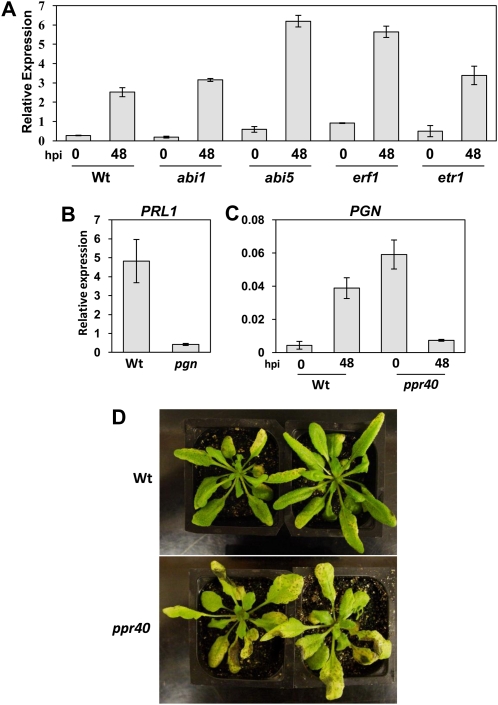
To further define PGN function in resistance to necrotrophic infection with respect to ABA and ET signaling, we examined its expression in mutants with altered ABA and ET responses. In response to B. cinerea, PGN expression was not altered in etr1 or abi1 (Fig. 8A). Mutation in the ET receptor ETR1 results in significantly decreased ABI1 expression, which is consistent with our results and suggests neither of these genes contribute to regulation of PGN during infection. By contrast, inoculated abi5 and erf1 mutants exhibit markedly higher PGN induction. ERF1 is a positive regulator of JA/ET-mediated defenses that functions partially through antagonism of ABA responses (Hildmann et al., 1992; Lorenzo et al., 2004). Thus, PGN expression may increase as ABA responses are relieved from ERF1-mediated repression. However, mutation in ABI5, a positive regulator of ABA signaling, also causes increased induction, suggesting that altered expression in these mutants may be linked to a secondary effect or that PGN regulation is not solely dependent on ABA responses (Berrocal-Lobo et al., 2002). In addition, basal expression of PLEIOTROPIC REGULATORY LOCUS1 (PRL1) is significantly repressed in the pgn mutant (Fig. 8B). PRL1 encodes a WD-40 protein that functions in global regulation of sugar, stress, and hormone responses as well as basal defense in Arabidopsis (Németh et al., 1998; Palma et al., 2007). Based on literature searches, prl1 was the only mutant we could identify with increased sensitivity to Glc, ABA, and ET similar to that observed for pgn.
Figure 8.
Impact of the ABA response pathway and the PPR40 gene on PGN expression and B. cinerea resistance. A and C, PGN expression in ABA/ET response mutants (A) and the ppr40 mutant (C) in response to B. cinerea. B, Basal PRL1 expression in pgn and wild-type (Wt) plants. D, The ppr40 mutant has increased susceptibility to B. cinerea. Images were taken 5 d after inoculation. Inoculation and quantification of gene expression were performed as described in “Materials and Methods.” hpi, Hours postinoculation.
The pgn Mutation Decreases Chlorophyll Content in Seedlings But Does Not Impair Oxidative Stress Tolerance
Recently, the PPR40 gene was isolated from a screen for ABA-sensitive Arabidopsis mutants (Zsigmond et al., 2008). In addition to ABA, the ppr40 mutant is also sensitive to salt and oxidative stress. PPR40 encodes a mitochondrial PPRP important for the function of Complex III in electron transport (Zsigmond et al., 2008). Due to the remarkable similarity of stress response phenotypes and shared subcellular localization of PGN and PPR40, we studied their functional relationship. The ppr40 mutant has significantly higher basal PGN expression that becomes repressed after B. cinerea infection (Fig. 8C). This pattern of expression is in direct contrast to that in wild-type plants, suggesting that PPR40 may function in the regulation of PGN transcription. Interestingly, ppr40 mutation results in a loss of resistance to B. cinerea comparable to that resulting from loss of PGN function (Fig. 8D).
We also assayed the pgn mutant for tolerance to oxidative stresses. Detached leaf treatment with methyl viologen (MV), an herbicide that generates ROS, did not indicate any impaired oxidative stress tolerance in the pgn mutant (Supplemental Fig. S6A). Additionally, there was no significant difference in chlorophyll content between pgn and wild-type seedlings treated with H2O2 or MV as observed for the ppr40 mutant (Zsigmond et al., 2008; Supplemental Fig. S6B). However, untreated pgn seedlings had significantly lower total chlorophyll content compared to the wild type. This difference was not observed in 4-week-old plants (Supplemental Fig. S6C). Thus, it appears that the pgn mutation does not affect oxidative stress tolerance in response to MV or H2O2, though it does lead to reduced chlorophyll levels in seedlings. Thus, PPR40 and PGN appear to have overlapping as well as distinct functions in Arabidopsis stress and defense responses.
The pgn Mutation Affects Mitochondrial Retrograde and Electron Transport Gene Expression
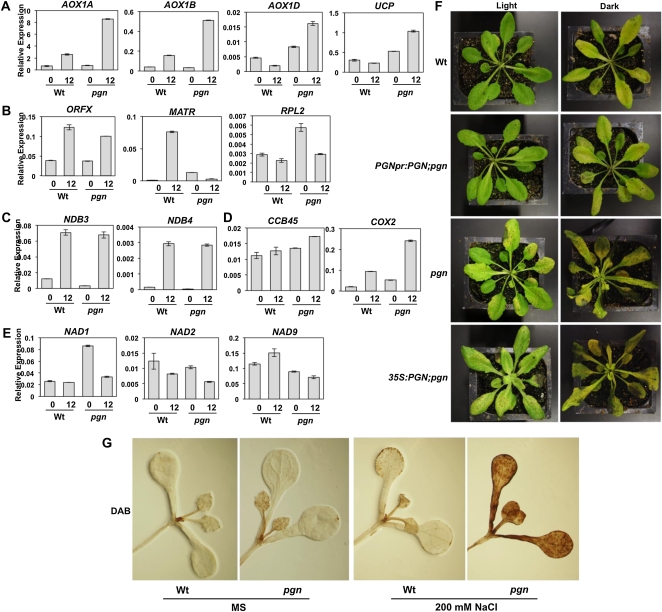
PGN’s localization to mitochondria and putative function in RNA metabolism prompted us to assay expression of mitochondria-associated transcripts in the mutant. pgn has significantly higher salt-induced ALTERNATIVE OXIDASE 1a (AOX1a), AOX1b, AOX1d, UCP, CCB45, and COX2 expression, with AOX1d, UCP, and COX2 also showing increased basal expression (Fig. 9, A and D). The AOX proteins and UCP serve as nonphosphorylating bypasses of mitochondrial electron transport, whereas CCB45 and COX2 are associated with cytochrome c biogenesis and oxidation, respectively (van Dongen et al., 2011). AOX induction also serves as a marker of mitochondrial retrograde responses in Arabidopsis. No significant differences in expression were observed for the extensively edited mitochondrial ORFX transcript, NAD2, NDB3, or NDB4 encoding a protein of unknown function, an NAD(P)H dehydrogenase subunit, and NAD(P)H dehydrogenases B3 and B4, respectively (Fig. 9, B–E). Yet, the pgn mutant has significantly higher basal expression of the NAD1 transcript, which is trans-spliced by the PPRP OTP43 and encodes an NAD(P)H dehydrogenase subunit associated with mitochondrial respiratory chain complex I (de Longevialle et al., 2007). RPL2, encoding a constituent of the large subunit of the mitochondrial ribosome, is also elevated in the mutant but gets repressed following salt treatment (Fig. 9B). NAD9 and MATR are expressed at lower levels in pgn both before and after treatment, with the RNA maturase gene MATR showing the highest degree of suppression caused by pgn mutation (Fig. 9, B and E).
Figure 9.
Expression profile of mitochondria-associated genes in the pgn mutant and light-dependent variations in disease resistance. A to E, Expression of genes associated with mitochondrial electron transport (A), transcription (B), oxidation/reduction (C), cytochrome c regulation (D), and NAD(P)H dehydrogenase subunits (E) in wild-type (Wt) and pgn seedlings after NaCl treatment. F, Disease responses of B. cinerea-inoculated plants kept in continuous dark or light conditions. Image was taken 4 d after inoculation. G, Accumulation of H2O2 in seedlings as revealed by DAB staining. DAB polymerizes in the presence of H2O2, producing a visible brown stain. Seedling treatment, plant inoculation, and quantification of gene expression were performed as described in “Materials and Methods.”
Altered ROS Metabolism in the pgn Mutant Contributes to Its Susceptibility to B. cinerea and Hypersensitivity to Salt
Consistent with germination assays in which both high and low PGN levels led to increased sensitivity to ABA, salt, and Glc, PGN overexpression resulted in enhanced susceptibility to B. cinerea similar to that observed for the mutant (Fig. 9F). Infected wild-type and complemented (PGNpr:PGN;pgn) plants shared similar levels of resistance without any observable differences in disease symptoms. Thus, the level of PGN expression appears to influence the extent of plant susceptibility to B. cinerea, consistent with results obtained for germination responses to abiotic stress agents. However, the threshold for transcript levels that alter responses appears to be lower for germination as the marginal increase in PGNpr:PGN;pgn plants results in weak sensitivity to abiotic stresses but has no effect on resistance to B. cinerea.
To determine if altered mitochondrial function in management of ROS is a contributing factor in pgn susceptibility, the PGN genotypes were inoculated with B. cinerea and incubated under constant light or dark. Under dark conditions, pgn and the overexpression line (35S:PGN;pgn) had significantly enhanced susceptibility relative to infected wild-type and complemented (PGNpr:PGN;pgn) plants as well as all plants incubated in light (Fig. 9F). Although light-grown plants maintained susceptibility, continuous darkness significantly enhanced disease symptoms. Thus, pgn susceptibility appears to be linked to impaired B. cinerea-induced ROS detoxification as, under dark conditions, this process becomes largely dependent on mitochondrial functions. The effect of pgn mutation on ROS metabolism was further confirmed by histochemical staining for the presence of H2O2 using 3,3′-diaminobenzidine (DAB) in salt-treated seedlings. Whereas minimal increases in H2O2 were observed in the leaf margins of wild-type seedlings, leaves of pgn seedlings incubated in 200 mm NaCl were intensely and wholly stained (Fig. 9G).
The pgn Mutation Does Not Affect Retrograde Signaling between the Chloroplast and Nucleus in Response to Photooxidative Stress
Recent studies have shown that GUN1 is a DNA-binding PPRP involved in retrograde signaling from the chloroplast (Koussevitzky et al., 2007). The gun1 mutant exhibits higher LIGHT HARVESTING CHLOROPHYLL BINDING1 (LHCB1) expression than the wild type after treatment with NF or lincomycin (Koussevitzky et al., 2007). Both chemicals disrupt chloroplast protein biosynthesis leading to Mg-ProtoIX accumulation (Koussevitzky et al., 2007). Mg-ProtoIX acts as a signal repressing nuclear expression of photosynthetic genes, specifically the LHCB family (Strand et al., 2003). Many of the genes affected by gun1 mutation are involved in ABA signaling, with ABI4 acting as a downstream component of the GUN1-regulated retrograde response pathway to photooxidative stress (Koussevitzky et al., 2007). ABI4 is a transcriptional regulator of sugar signaling required for plant responses to ABA, Glc, and salt (Finkelstein et al., 1998; Huijser et al., 2000; Quesada et al., 2000; Bossi et al., 2009). Based on the hypersensitive phenotypes and altered ABI4 expression in the pgn mutant, we assayed the level of LHCB1 transcripts in NF-treated pgn to determine if it plays a role in plastid-to-nucleus signaling. In response to NF, LHCB1 was repressed in both wild-type and pgn seedlings without any observable difference in the level of suppression, suggesting that PGN does not function in photooxidative retrograde signaling (Supplemental Fig. S7).
DISCUSSION
The Arabidopsis PPRP family consists of hundreds of proteins whose function is largely unexplored (Lurin et al., 2004). In this study, we describe the biological function of PGN, encoding a mitochondrial PPRP, in plant stress responses. Our study provides molecular and genetic evidence for the critical function of PGN in plant responses to a subset of abiotic and biotic stresses. The PGN gene shows increased expression in response to necrotrophic infection, and its loss of function results in susceptibility to the fungal pathogens A. brassicicola and B. cinerea. The susceptibility of pgn is independent of SA-, ET-, and JA-mediated responses, and the mutant shows no altered resistance to virulent or avirulent strains of the bacterial pathogen P. syringae, suggesting the defense function of PGN is restricted to necrotrophic infection. However, the altered germination responses of the pgn mutant to ABA, ACC, Glc, and salt also suggest a critical role for PGN in abiotic stress tolerance (Gazzarrini and McCourt, 2001; Dekkers et al., 2004; Tuteja, 2007). Importantly, the pgn mutant accumulates increased levels of ABA and stress-induced ROS, both of which may underpin its various phenotypes. Furthermore, the expression profiles of many nuclear and mitochondrial genes, particularly those involved in retrograde and ABA signaling are altered by pgn mutation. These data, together with its mitochondrial localization, uniquely link PGN function to the regulation of stress-induced ABA responses and mitochondrial ROS homeostasis in Arabidopsis.
In contrast to SA, JA, and ET, which have long been known for their functions in plant defense, ABA is primarily known for its role in regulating abiotic stress responses (Fujita et al., 2006). Many mutants altered in ABA responses exhibit hypersensitivity to salt or Glc similar to that observed for pgn (Xiong et al., 2002; León and Sheen, 2003). More recently, ABA synthesis and signaling have been implicated in plant defense against pathogens (Mauch-Mani and Mauch, 2005). Although the mechanism is currently not known, PGN regulates ABA levels, which, in turn, is likely responsible for the observed mutant phenotypes including impaired disease resistance. The high levels of endogenous ABA caused by the pgn mutation may be due to an indirect effect of cellular perturbations, including loss of ROS homeostasis in the mitochondria or a function of the direct regulatory effect of PGN on genes involved in ABA biosynthesis. Consistent with this, in response to salt and ABA, pgn seedlings have markedly increased expression of the NCED3 gene, encoding a key enzyme responsible for stress-induced ABA biosynthesis. The expression of NCED3 directly correlates with increased ABA accumulation (Thompson et al., 2000; Iuchi et al., 2001). Thus, the germination responses of pgn may be a result of increased NCED3-mediated ABA synthesis leading to a misregulation of ABA-induced ROS generation. This notion is further supported by the ABA, salt, and sugar insensitivity conferred by NCED3 loss of function as well as the partial alleviation of pgn Glc hypersensitivity by addition of ABA antagonists (Ruggiero et al., 2004; Huang et al., 2008). Alternatively, impaired ROS metabolism in the mutant may underlie its altered ABA accumulation.
ET and ABA are known to function antagonistically in a number of signaling events occurring at germination and throughout development, including the regulation of abiotic and biotic stress responses (Beaudoin et al., 2000; Ghassemian et al., 2000; Mauch-Mani and Mauch, 2005; Fujita et al., 2006). Many ABA-hypersensitive mutants have been independently isolated in genetic screens for ET insensitivity (León and Sheen, 2003). The partial restoration of pgn growth sensitivity to Glc by ACC is likely due to a suppression of ABA signaling and synthesis. The same can be said for the relieved growth inhibition caused by NF, an inhibitor of ABA biosynthesis (Bartels and Watson, 1978; Zeevaart and Creelman, 1988). However, pgn also has heightened sensitivity to ET, exhibiting an enhanced triple response characterized by decreased hypocotyl length and root elongation relative to wild-type seedlings, which is in direct contrast with the majority of published data. To our knowledge, prl1 is the only other Arabidopsis mutation resulting in concurrent hypersensitivity to ET, ABA, and Glc (Németh et al., 1998). PRL1 is part of a spliceosome-associated complex that may contribute to regulation of alternative RNA splicing or miRNA/small interfering RNA generation (Palma et al., 2007). Interestingly, expression of PRL1 is significantly repressed in the pgn mutant, suggesting that these two may have functional interaction in RNA metabolism in response to different abiotic and biotic stresses. In addition, prl1 plants are resistant to the methylerythritol phosphate pathway inhibitor clomazone, a phenotype observed with loss of function of LOI1, which encodes a mitochondrial PPRP (Kobayashi et al., 2007; Flores-Pérez et al., 2010).
Though some are known to have DNA substrates, PPRPs are largely sequence-specific RNA-binding proteins associated with all stages of posttranscriptional regulation including translation (Delannoy et al., 2007). PPRPs function in RNA splicing, degradation, editing, and maturation by recruiting processing enzymes to bound transcripts (Delannoy et al., 2007; Schmitz-Linneweber and Small, 2008). Potentially, PGN may regulate stress-induced expression of transcripts involved in ABA responses or the regulation of ROS homeostasis. Alternatively, PGN may be a positive regulator of genes involved in abiotic and biotic resistance or suppress negative regulators through RNA degradation. The altered expression of both nuclear and mitochondrial genes in the mutant is consistent with a role for PGN in general stress tolerance rather than acting as a specific regulator of defense against pathogens. Particularly, the differential expression of several mitochondria-encoded transcripts, including NAD1, RPL2, NAD9, and MATR, in the mutant suggests that PGN has a critical regulatory role in mitochondrial gene expression. In addition, pgn shows significantly higher induction of AOX1 gene expression. AOX genes encode alternative oxidases that function in the maintenance of ROS levels in the mitochondria and are considered markers of mitochondrial retrograde signaling (Maxwell et al., 1999; Giraud et al., 2009).
ROS are produced not only as byproducts of cellular metabolism during electron transport but also as a result of plant exposure to abiotic and biotic stresses (Moller, 2001; Blokhina et al., 2003). Thus, the altered electron transport-associated gene expression coupled with the high level of salt-induced ROS accumulation in the mutant strongly suggests that loss of PGN function results in a stress-induced misregulation of ROS metabolism. Potentially, PGN and its network of coexpressed PPRPs may coordinately affect global mitochondrial gene expression through transcript editing, thereby contributing to plant defense and maintaining cellular redox. During germination, ROS alleviate dormancy by suppressing ABA responses; however, past a threshold, they lead to oxidative stress and elicit ABA-mediated responses. Therefore, ROS generated from altered mitochondrial function may be responsible for the increased levels of ABA in pgn seedlings. PPR40, encoding a PPRP important for Complex III function in mitochondrial electron transport, was suggested to function in a similar manner (Zsigmond et al., 2008). PPR40 was identified through a screen for ABA-hypersensitive mutants, with loss of function also resulting in decreased tolerance to salt and osmotic stress (Zsigmond et al., 2008). ppr40 mutants generate elevated levels of ROS coincident with increased AOX expression (Zsigmond et al., 2008). In addition, we found ppr40 plants are susceptible to B. cinerea and have significantly higher basal PGN expression that is repressed after infection. Based on the similarity of the phenotypes and the aberrant expression pattern of PGN in the ppr40 mutant, PPR40 may regulate PGN transcription.
The ACC sensitivity of the pgn mutant is likely also a reflection of disrupted cellular redox. Both the hypocotyls and roots of ACC-treated seedlings accumulate increased ROS in their elongation zones, thereby leading to the decreased growth characteristic of the triple response (De Cnodder et al., 2005). In plants, ROS accumulation and scavenging is largely mediated by redox events in chloroplasts and mitochondria (Noctor et al., 2007). As the triple response occurs under dark conditions, the majority of ROS production resulting from ACC treatment must occur in the mitochondria. Thus, the heighted ACC sensitivity of the mutant is likely a reflection of altered mitochondrial function rather than impaired ET signaling. This is further supported by the increased AOX expression, ROS accumulation, and enhanced susceptibility of the mutant to necrotrophic infection under dark conditions. Incubation in light allows inoculated plants to use chloroplasts and mitochondria in the removal of harmful ROS, thereby contributing to defense, whereas incubation in the dark necessitates relying largely only on mitochondria for ROS scavenging. This suggests the weak susceptibility of the mutant under light conditions is due to altered redox regulation in the mitochondria, with the more apparent susceptibility in the dark likely a result of loss of chloroplast contribution to ROS removal.
Its mitochondrial localization, involvement in the regulation of mitochondria-associated transcripts, and the phenotypes resulting from its loss of function all suggest a critical role for PGN in mitochondrial function during abiotic and biotic stress responses. Based on our data, the impaired ROS metabolism resulting from loss of PGN function likely leads to an up-regulation of ABA synthesis and signaling, or vice versa, accounting for the altered stress responses associated with PGN over- or underexpression. The relative specificity of the pgn mutant phenotypes, displaying altered responses only to certain pathogens and abiotic stresses, suggests the mutant is not impaired in general cell death responses due to constitutively high levels of ROS related downstream responses. However, we cannot exclude the possibility of impaired antioxidant activation in the mutant during responses to certain pathogens or stress responses. This is supported by the lack of runaway cell death in the mutant, its unaltered responses to MV, H2O2, and virulent P. syringae as well as an avirulent strain of this pathogen known to induce the hypersensitive response. Overall, the exact mechanism of function of PGN in plant defense and abiotic stress tolerance remains unclear. Future investigation into the molecular and biochemical basis of PGN regulation of ABA accumulation, ROS generation, target gene expression, and involvement in retrograde signaling is needed to further elucidate its functions in Arabidopsis stress responses.
MATERIALS AND METHODS
Plant Growth and Treatments
Plants were grown in soil under fluorescent light (150 μE m−2 s−1) at 23°C ± 4°C with 60% relative humidity and a 12-h-light/12-h-dark cycle. Seeds were vernalized in water for 2 d at 4°C prior to planting. For axenic plant growth, all seeds were surface sterilized as previously described (Mengiste et al., 2003). Seeds were plated on supplemented or nonsupplemented media consisting of MS salts (PhytoTechnology Laboratories) with 2% (w/v) Suc and 0.8% (w/v) agar, pH 5.7. Transgenic seedlings were identified by selecting on MS media supplemented with appropriate antibiotics. Seed germination and seedling growth assays were also performed using MS media supplemented with different concentrations of different chemicals. All plates were vernalized at 4°C for 2 d followed by incubation at room temperature. To assay for the triple response, seeds were surface sterilized and plated on MS media containing 0 and 4 μm ACC (Sigma-Aldrich). Plates were kept in the dark and incubated 2 d at 4°C followed by 3 d at 23°C ± 2°C.
For gene expression analysis, 4-week-old soil-grown plants were sprayed with 2.5 × 104 Botrytis cinerea spores/mL, 5 mm SA, 1 mm MeJA, 0.5 mm ACC, or 100 μm ABA (Sigma-Aldrich). Ten-day-old seedlings grown in vitro were treated with 200 mm NaCl or 100 μm ABA supplemented liquid MS media and shaken at room temperature for 0, 6, and 12 h or 0, 3, and 6 h, respectively. Seedlings were treated with NF (Chem Service) as previously described (Koussevitzky et al., 2007). Plants were treated with H2O2 and MV to assay chlorophyll degradation/content as previously described (Zsigmond et al., 2008). DAB staining for H2O2 was performed on 10-d-old seedlings following 3 h incubation in 200 mm NaCl supplemented or unsupplemented MS media according to the manufacture’s protocol (Sigmafast DAB; Sigma-Aldrich).
ABA Extraction and Quantification
Total ABA was extracted from 1 to 1.5 g of tissue from 10-d-old seedlings as previously described with the modifications of using an acetone/water/acetic acid (80%/19%/1%) solution for homogenization and (±)-3′,5′,5′,7′,7′,7′-d6 ABA as an internal standard (National Research Council Canada; Cheng et al., 2002). ABA content was then determined using liquid chromatography-tandem mass spectrometry.
RNA Extraction, Blots, and Expression Analyses
For RNA blots, total RNA was isolated as described (Lagrimini et al., 1987), separated on 1.2% agarose-formaldehyde gels, and blotted to Hybond N+ nylon membranes (Amersham Pharmacia Biotech). Probes were labeled with 32P using the random labeling system (Redi Prime II; GE Healthcare) and hybridized to blots as previously described (Church and Gilbert, 1984). Membranes were exposed to film for 24 h at −80°C (Biomax XAR Film; Kodak). Ethidium bromide staining of rRNA was used as a loading control. RNA extraction, cDNA synthesis, and quantitative reverse transcription (qRT)-PCR expression analyses were carried out as previously described (Dhawan et al., 2009). cDNA of all samples was synthesized using 3 μg DNAse-treated template RNA, AMV reverse transcriptase (Promega), and oligo(dT15) primers according to standard protocols. RT-PCR and qRT-PCR were performed using gene-specific primers with Arabidopsis (Arabidopsis thaliana) Actin2 as an endogenous reference for normalization. For qRT-PCR, a minimum of three technical replicates were used for each sample with a minimum of two biological replicates. Expression levels were calculated by the comparative cycle threshold method (Applied Biosystems). Normalization to the control was performed as previously described (Bluhm and Woloshuk, 2005). Primers used are listed in Supplemental Table S1.
Fungal Cultures and Disease Assays
Fungi were cultured and maintained as described (Mengiste et al., 2003). Disease assays using B. cinerea strain BO5-10 and Alternaria brassicicola strain MUCL20297 were performed as previously described (Mengiste et al., 2003). Disease assays for virulent/avirulent Pseudomonas syringae pv tomato were done as described (Mengiste et al., 2003).
Generation of PGN Transgenic Lines and Mutant Identification
PGN (AT1G56570) overexpression lines were generated by cloning the full-length PGN genomic sequence behind the cauliflower mosaic virus 35S promoter in pCAMBIA99-3XHA. PGN complementation lines were generated by cloning the full-length genomic sequence of PGN including 1.6 kb 5′ to the start codon into pCAMBIA1380. Sequences were amplified using the KOD DNA polymerase according to the manufacturer’s protocol with the addition of 1% dimethyl sulfoxide to each reaction (Novagen). Each binary vector was transferred into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis (Clough and Bent, 1998). Transgenic plants were selected on media containing hygromycin. The pgn (SALK_141937) mutant allele was identified from a segregating population obtained from the Arabidopsis Biological Resource Center using T-DNA and gene-specific primers (Sessions et al., 2002). Homozygous abi1, abi5, erf1, and etr1 mutant alleles were also obtained from the Arabidopsis Biological Resource Center (Chang et al., 1993; Gosti et al., 1999; Finkelstein and Lynch, 2000; Berrocal-Lobo et al., 2002). Primers used are listed in Supplemental Table S1.
Construction of PGN-GFP and Visualization
The full-length genomic sequence of PGN was inserted into pCAMBIA99-1-GFP to generate a 5′ PGN translational fusion. The construct was verified by sequencing and introduced into Agrobacterium (strain GV3101), which was subsequently used for transient expression in Nicotiana benthamiana via agroinfiltration. Agrobacteria carrying the empty pCAMBIA99-1-GFP vector was used as a control. The mCHERRY mitochondrial marker was previously described (Nelson et al., 2007). Localization was observed using a Nikon Eclipse E800 epifluorescence microscope. Primers used are listed in Supplemental Table S1.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At1g56570.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PGN expression at different developmental stages.
Supplemental Figure S2. PGN mutation does not alter plant resistance to P. syringae.
Supplemental Figure S3. The pgn mutant is not altered in expression of defense or abiotic stress response marker genes.
Supplemental Figure S4. Multiple sequence alignment of PGN and closely related Arabidopsis proteins.
Supplemental Figure S5. Germination and growth responses of the pgn mutant are not altered on ACC, SA, MeJA, GA, indole-3-acetic acid, or H2O2.
Supplemental Figure S6. The pgn mutant is not impaired in oxidative stress tolerance.
Supplemental Figure S7. The pgn mutant is not altered in retrograde signaling from the chloroplast.
Supplemental Table S1. List of primers used in PGN study.
Acknowledgments
We thank Dr. Zhibing Lai for comments on this manuscript.
References
- Andres C, Lurin C, Small ID. (2007) The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol Plant 129: 14–22 [Google Scholar]
- Bartels PG, Watson CW. (1978) Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Sci 26: 198–203 [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J. (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Blokhina O, Virolainen E, Fagerstedt KV. (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot (Lond) (Spec No) 91: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm BH, Woloshuk CP. (2005) Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol Plant Microbe Interact 18: 1333–1339 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu JH, Verslues PE, Sunkar R, Zhu JK. (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupré P, Mendoza MS, Román CS, León P. (2009) The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Ma JF, Zhang DY, Guo JK, Chen F, Lu CM, Zhang L. (2008) The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 147: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. (2006) Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng (N Y) 27: 141–177 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cushing DA, Forsthoefel NR, Gestaut DR, Vernon DM. (2005) Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221: 424–436 [DOI] [PubMed] [Google Scholar]
- De Cnodder T, Vissenberg K, Van Der Straeten D, Verbelen JP. (2005) Regulation of cell length in the Arabidopsis thaliana root by the ethylene precursor 1-aminocyclopropane- 1-carboxylic acid: a matter of apoplastic reactions. New Phytol 168: 541–550 [DOI] [PubMed] [Google Scholar]
- Dekkers BJW, Schuurmans JAMJ, Smeekens SCM. (2004) Glucose delays seed germination in Arabidopsis thaliana. Planta 218: 579–588 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID. (2007) Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans 35: 1643–1647 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID. (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YH, Liu NY, Tang ZS, Liu J, Yang WC. (2006) Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez U, Pérez-Gil J, Closa M, Wright LP, Botella-Pavía P, Phillips MA, Ferrer A, Gershenzon J, Rodríguez-Concepción M. (2010) PLEIOTROPIC REGULATORY LOCUS 1 (PRL1) integrates the regulation of sugar responses with isoprenoid metabolism in Arabidopsis. Mol Plant 3: 101–112 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA. (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203: 182–187 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. (2001) Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol 4: 387–391 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P. (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim DG. (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280: 196–203 [DOI] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LHM, Whelan J. (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Jones JDG. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Hildmann T, Ebneth M, Peña-Cortés H, Sánchez-Serrano JJ, Willmitzer L, Prat S. (1992) General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 4: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wu W, Abrams SR, Cutler AJ. (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot 59: 2991–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23: 577–585 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Hansen M, Joehnk N, Lyngkjaer MF, Collinge DB. (2007) The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol 65: 137–150 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21: 3123–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103: 18002–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K, Matsui K, Ozawa R, Takabayashi J. (2005) Volatile C6-aldehydes and Allo-ocimene activate defense genes and induce resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol 46: 1093–1102 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Suzuki M, Tang J, Nagata N, Seki H, Ohyama K, Kiuchi R, Kaneko Y, Matsumoto S, Yoshida S, Muranaka T. (2007) Lovastatin insensitive 1, encoding a novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in Arabidopsis. Plant Cell Physiol 48: 322–332 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ. (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim DG, Gibson SI. (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. (1987) Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA 84: 7542–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León P, Sheen J. (2003) Sugar and hormone connections. Trends Plant Sci 8: 110–116 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444: 139–158 [DOI] [PubMed] [Google Scholar]
- Matilla AJ, Matilla-Vazquez MA. (2008) Involvement of ethylene in seed physiology. Plant Sci 175: 87–97 [Google Scholar]
- Mauch-Mani B, Mauch F. (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8: 409–414 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L. (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R. (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM. (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Okrész L, Stabel S, et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, De Paepe R, Foyer CH. (2007) Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci 12: 125–134 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res (Database issue) 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi T, Sage-Ono K, Kamada H, Ono M. (2004) Genomic structure of a novel Arabidopsis clock-controlled gene, AtC401, which encodes a pentatricopeptide repeat protein. Gene 330: 29–37 [DOI] [PubMed] [Google Scholar]
- Palma K, Zhao QG, Cheng YT, Bi DL, Monaghan J, Cheng W, Zhang YL, Li X. (2007) Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev 21: 1484–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Li TC, Kang SG, Na JK, Jang JC. (2003) Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol 132: 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL. (2000) Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-Pinto JJ, Yephremov A. (2009) Surface lipids and plant defenses. Plant Physiol Biochem 47: 540–549 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Bari R, Jones JDG. (2010) A Biotic or Abiotic Stress? Springer, Dordrecht, The Netherlands [Google Scholar]
- Ruggiero B, Koiwa H, Manabe Y, Quist TM, Inan G, Saccardo F, Joly RJ, Hasegawa PM, Bressan RA, Maggio A. (2004) Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol 136: 3134–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Prasad AM, Srinivasan R. (2007) Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol Biochem 45: 521–534 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N. (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu JH, Zhu JK. (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12: 301–309 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Symonds RC, Mulholland BJ, Dadswell AR, Blake PS, Burbidge A, Taylor IB. (2000) Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J 23: 363–374 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2: 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Gupta KJ, Ramirez-Aguilar SJ, Araujo WL, Nunes-Nesi A, Fernie AR. (2011) Regulation of respiration in plants: a role for alternative metabolic pathways. J Plant Physiol 168: 1434–1443 [DOI] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, AbuQamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. (2006) Distinct roles of the membrane anchored Botrytis Induced Kinase 1 in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, et al. (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19: 1279–1290 [DOI] [PubMed] [Google Scholar]
- Xie ZX, Qi XP. (2008) Diverse small RNA-directed silencing pathways in plants. Biochim Biophys Acta 1779: 720–724 [DOI] [PubMed] [Google Scholar]
- Xiong LM, Schumaker KS, Zhu JK. (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA. (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39: 439–473 [Google Scholar]
- Zhang JH, Jia WS, Yang JC, Ismail AM. (2006) Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res 97: 111–119 [Google Scholar]
- Zsigmond L, Rigó G, Szarka A, Székely G, Otvös K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L. (2008) Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol 146: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]